

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146553.5 - phase: 1 /pseudo
(469 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
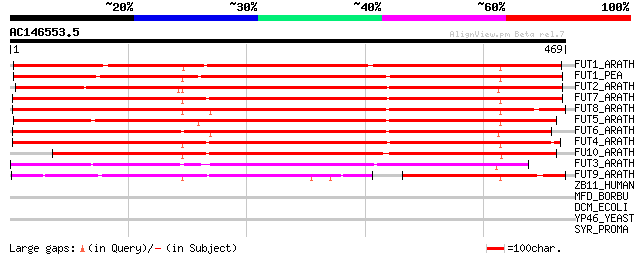
FUT1_ARATH (Q9SWH5) Galactoside 2-alpha-L-fucosyltransferase (EC... 422 e-118
FUT1_PEA (Q9M5Q1) Galactoside 2-alpha-L-fucosyltransferase (EC 2... 415 e-115
FUT2_ARATH (O81053) Probable fucosyltransferase 2 (EC 2.4.1.-) (... 414 e-115
FUT7_ARATH (Q9XI81) Probable fucosyltransferase 7 (EC 2.4.1.-) (... 392 e-108
FUT8_ARATH (Q9XI78) Probable fucosyltransferase 8 (EC 2.4.1.-) (... 389 e-108
FUT5_ARATH (Q9SJP4) Probable fucosyltransferase 5 (EC 2.4.1.-) (... 385 e-106
FUT6_ARATH (Q9XI80) Probable fucosyltransferase 6 (EC 2.4.1.-) (... 377 e-104
FUT4_ARATH (Q9SJP2) Probable fucosyltransferase 4 (EC 2.4.1.-) (... 376 e-104
FU10_ARATH (Q9SJP6) Putative fucosyltransferase 10 (EC 2.4.1.-) ... 355 1e-97
FUT3_ARATH (Q9CA71) Probable fucosyltransferase 3 (EC 2.4.1.-) (... 310 5e-84
FUT9_ARATH (Q9XI77) Probable fucosyltransferase 9 (EC 2.4.1.-) (... 224 4e-58
ZB11_HUMAN (O95625) Zinc finger and BTB domain containing protei... 33 1.2
MFD_BORBU (O51568) Transcription-repair coupling factor (TRCF) 32 4.6
DCM_ECOLI (P11876) DNA-cytosine methyltransferase (EC 2.1.1.37) ... 32 4.6
YP46_YEAST (Q12080) Hypothetical protein YPL146C 31 7.8
SYR_PROMA (Q7VE03) Arginyl-tRNA synthetase (EC 6.1.1.19) (Argini... 31 7.8
>FUT1_ARATH (Q9SWH5) Galactoside 2-alpha-L-fucosyltransferase (EC
2.4.1.69) (Xyloglucan alpha-(1,2)-fucosyltransferase)
(AtFUT1)
Length = 558
Score = 422 bits (1086), Expect = e-118
Identities = 221/472 (46%), Positives = 300/472 (62%), Gaps = 17/472 (3%)
Query: 4 EKLLDGLLVSGFDEASCVSRLQSHFYHKPSPHKPSPYLISKLRKYEELHRRCGPNTRAYN 63
+KLL GLL SGFDE SC+SR QS Y KPSP+KPS YLISKLR YE+LH+RCGP T +Y
Sbjct: 95 DKLLGGLLASGFDEDSCLSRYQSVHYRKPSPYKPSSYLISKLRNYEKLHKRCGPGTESYK 154
Query: 64 EDMKKITKSKKNGTSAATTCKYIIWLPANGLGNQIISMASSFLYALLTGRVMLVQFGKDK 123
+ +K++ + +G CKY++W+ +GLGN+I+S+AS FLYALLT RV+LV GKD
Sbjct: 155 KALKQLDQEHIDGDGE---CKYVVWISFSGLGNRILSLASVFLYALLTDRVLLVDRGKDM 211
Query: 124 EGLFCEPFLNSTWLLPEKSPFW------NAEKVQTYQSTIKMGGANTLNEDFPSALHVNL 177
+ LFCEPFL +WLLP P N E + Y +K +T E S L+++L
Sbjct: 212 DDLFCEPFLGMSWLLPLDFPMTDQFDGLNQESSRCYGYMVKNQVIDT--EGTLSHLYLHL 269
Query: 178 GYSPTSEERFFHCDHNQFLLSNIPLLFLEAGQYFVPSFFMTPIFKKELNKMFPEITSTFH 237
+ ++ F C+ +Q + +P L ++ YFVPS ++ P F ELNK+FP+ + FH
Sbjct: 270 VHDYGDHDKMFFCEGDQTFIGKVPWLIVKTDNYFVPSLWLIPGFDDELNKLFPQKATVFH 329
Query: 238 HLGRYLFHPSNEAWELITSFYQQNLAKANERIGLQIRVVDPKLTPHQVVMNQILNCTLGN 297
HLGRYLFHP+N+ W L+T +Y+ L+ A+E+IG+Q+RV D P Q VM+QI +CT
Sbjct: 330 HLGRYLFHPTNQVWGLVTRYYEAYLSHADEKIGIQVRVFDEDPGPFQHVMDQISSCTQKE 389
Query: 298 KLLPKVLGTKNIYLSSSDKNKKIMKAVLVSSLYRHYGENLKMTYMNKSTVSGEVIEVYQP 357
KLLP+V + S N KAVLV+SL Y ENLK Y T +GE+I V+QP
Sbjct: 390 KLLPEV---DTLVERSRHVNTPKHKAVLVTSLNAGYAENLKSMYWEYPTSTGEIIGVHQP 446
Query: 358 SGEEQQKFNDNKHNMKAWVDIYLLSLSDVLVTTYQSTFGYVAKALGNSRPWILYNP---V 414
S E Q+ HN KA ++YLLSL+D LVT+ STFGYVA+ LG +PWILY P
Sbjct: 447 SQEGYQQTEKKMHNGKALAEMYLLSLTDNLVTSAWSTFGYVAQGLGGLKPWILYRPENRT 506
Query: 415 YSNEICEREFTLEPCYHYPPLHYCNGKPIEDVGSSFPYIRHCKDYTFGVKLV 466
+ C R ++EPC+H PP + C K D G+ P++RHC+D ++G+KLV
Sbjct: 507 TPDPSCGRAMSMEPCFHSPPFYDCKAKTGIDTGTLVPHVRHCEDISWGLKLV 558
>FUT1_PEA (Q9M5Q1) Galactoside 2-alpha-L-fucosyltransferase (EC
2.4.1.69) (Xyloglucan alpha-(1,2)-fucosyltransferase)
(PsFT1)
Length = 565
Score = 415 bits (1067), Expect = e-115
Identities = 210/473 (44%), Positives = 294/473 (61%), Gaps = 17/473 (3%)
Query: 4 EKLLDGLLVSGFDEASCVSRLQSHFYHKPSPHKPSPYLISKLRKYEELHRRCGPNTRAYN 63
+KLL GLL GFDE SC+SR QS + K KPS YLIS+LRKYE H++CGP T +YN
Sbjct: 100 DKLLGGLLADGFDEKSCLSRYQSAIFGKGLSGKPSSYLISRLRKYEARHKQCGPYTESYN 159
Query: 64 EDMKKITKSKKNGTSAATTCKYIIWLPANGLGNQIISMASSFLYALLTGRVMLVQFGKDK 123
+ +K++ + S + CKY++W+ +GLGN+I+++ S+FLYALLT RV+LV G D
Sbjct: 160 KTVKELGSGQ---FSESVDCKYVVWISFSGLGNRILTLVSAFLYALLTDRVLLVDPGVDM 216
Query: 124 EGLFCEPFLNSTWLLPEKSPF------WNAEKVQTYQSTIKMGGANTLNEDFPSALHVNL 177
LFCEPF +++W +P P +N E Q + +K + N PS ++++L
Sbjct: 217 TDLFCEPFPDASWFVPPDFPLNSHLNNFNQESNQCHGKILKT--KSITNSTVPSFVYLHL 274
Query: 178 GYSPTSEERFFHCDHNQFLLSNIPLLFLEAGQYFVPSFFMTPIFKKELNKMFPEITSTFH 237
+ ++ F CD Q L N+PLL ++ YF+PS F+ P F++ELN +FP+ FH
Sbjct: 275 AHDYDDHDKLFFCDEEQLFLQNVPLLIMKTDNYFIPSLFLMPSFEQELNDLFPKKEKVFH 334
Query: 238 HLGRYLFHPSNEAWELITSFYQQNLAKANERIGLQIRVVDPKLTPHQVVMNQILNCTLGN 297
LGRYL HP+N W L+ +Y LAK +ERIG+QIRV D P Q V++Q+L CTL
Sbjct: 335 FLGRYLLHPTNNVWGLVVRYYDAYLAKVDERIGIQIRVFDTDPGPFQHVLDQVLACTLKE 394
Query: 298 KLLPKVLGTKNIYLSSSDKNKKIMKAVLVSSLYRHYGENLKMTYMNKSTVSGEVIEVYQP 357
+LP V +NI SS KAVL++SL Y E ++ Y T +GEV+ +YQP
Sbjct: 395 SILPDVNREQNINSSSGTPKS---KAVLITSLSSGYFEKVRDMYWEFPTETGEVVGIYQP 451
Query: 358 SGEEQQKFNDNKHNMKAWVDIYLLSLSDVLVTTYQSTFGYVAKALGNSRPWILYNP---V 414
S E Q+ HN KAW ++YLLSL+DVLVT+ STFGYVA+ LG +PWILY P
Sbjct: 452 SHEGYQQTQKQFHNQKAWAEMYLLSLTDVLVTSSWSTFGYVAQGLGGLKPWILYKPENRT 511
Query: 415 YSNEICEREFTLEPCYHYPPLHYCNGKPIEDVGSSFPYIRHCKDYTFGVKLVN 467
N C+R ++EPC+H PP + C K D G+ P++RHC+D ++G+KLV+
Sbjct: 512 APNPPCQRAMSMEPCFHAPPFYDCKAKRGTDTGALVPHVRHCEDMSWGLKLVD 564
>FUT2_ARATH (O81053) Probable fucosyltransferase 2 (EC 2.4.1.-)
(AtFUT2)
Length = 539
Score = 414 bits (1063), Expect = e-115
Identities = 217/471 (46%), Positives = 297/471 (62%), Gaps = 11/471 (2%)
Query: 6 LLDGLLVSGFDEASCVSRLQSHFYHKPSPHKPSPYLISKLRKYEELHRRCGPNTRAYNED 65
LL GLLVSGF + SC+SR QS+ Y K SP+KPS +L+SKLR YEELH+RCGP TR Y +
Sbjct: 69 LLGGLLVSGFKKESCLSRYQSYLYRKASPYKPSLHLLSKLRAYEELHKRCGPGTRQYT-N 127
Query: 66 MKKITKSKKNGTSAATTCKYIIWLPANGLGNQIISMASSFLYALLTGRVMLVQFGKDKEG 125
+++ K K+ G + CKY++W+ +GLGN+IIS+AS FLYA+LT RV+LV+ G+
Sbjct: 128 AERLLKQKQTGEMESQGCKYVVWMSFSGLGNRIISIASVFLYAMLTDRVLLVEGGEQFAD 187
Query: 126 LFCEPFLNSTWLLPEK----SPF--WNAEKVQTYQSTIKMGGANTLNEDFPSALHVNLGY 179
LFCEPFL++TWLLP+ S F + + +K N + S L+++L +
Sbjct: 188 LFCEPFLDTTWLLPKDFTLASQFSGFGQNSAHCHGDMLKRKLINESSVSSLSHLYLHLAH 247
Query: 180 SPTSEERFFHCDHNQFLLSNIPLLFLEAGQYFVPSFFMTPIFKKELNKMFPEITSTFHHL 239
++ F C+ +Q LL N+P L + +F PS F+ F++EL MFPE + FHHL
Sbjct: 248 DYNEHDKMFFCEEDQNLLKNVPWLIMRTNNFFAPSLFLISSFEEELGMMFPEKGTVFHHL 307
Query: 240 GRYLFHPSNEAWELITSFYQQNLAKANERIGLQIRVVDPKLTPHQVVMNQILNCTLGNKL 299
GRYLFHPSN+ W LIT +YQ LAKA+ERIGLQIRV D K V QI++C L
Sbjct: 308 GRYLFHPSNQVWGLITRYYQAYLAKADERIGLQIRVFDEKSGVSPRVTKQIISCVQNENL 367
Query: 300 LPKVLGTKNIYLSSSDKNKKIMKAVLVSSLYRHYGENLKMTYMNKSTVSGEVIEVYQPSG 359
LP++ + Y S++ K +K+VLV+SL Y E LK Y TV+ +VI ++QPS
Sbjct: 368 LPRLSKGEEQYKQPSEEELK-LKSVLVTSLTTGYFEILKTMYWENPTVTRDVIGIHQPSH 426
Query: 360 EEQQKFNDNKHNMKAWVDIYLLSLSDVLVTTYQSTFGYVAKALGNSRPWILY---NPVYS 416
E Q+ HN KAW ++YLLSL+D LV + STFGYVA+ LG R WILY N
Sbjct: 427 EGHQQTEKLMHNRKAWAEMYLLSLTDKLVISAWSTFGYVAQGLGGLRAWILYKQENQTNP 486
Query: 417 NEICEREFTLEPCYHYPPLHYCNGKPIEDVGSSFPYIRHCKDYTFGVKLVN 467
N C R + +PC+H PP + C K D G+ P++RHC+D ++G+KLV+
Sbjct: 487 NPPCGRAMSPDPCFHAPPYYDCKAKKGTDTGNVVPHVRHCEDISWGLKLVD 537
>FUT7_ARATH (Q9XI81) Probable fucosyltransferase 7 (EC 2.4.1.-)
(AtFUT7)
Length = 526
Score = 392 bits (1006), Expect = e-108
Identities = 204/476 (42%), Positives = 296/476 (61%), Gaps = 14/476 (2%)
Query: 3 KEKLLDGLLVSGFDEASCVSRLQSHFYHKPSPHKPSPYLISKLRKYEELHRRCGPNTRAY 62
+++LL GLL + FDE SC+SR QS Y KPSP++ S YLISKLR YE LH+RCGP T AY
Sbjct: 48 RDRLLGGLLTADFDEDSCLSRYQSSLYRKPSPYRTSEYLISKLRNYEMLHKRCGPGTDAY 107
Query: 63 NEDMKKITKSKKN-GTSAATTCKYIIWLPANGLGNQIISMASSFLYALLTGRVMLVQFGK 121
+K+ +N G S+ CKYI+W+ GLGN+I+++AS FLYALLT R++LV K
Sbjct: 108 KRATEKLGHDHENVGDSSDGECKYIVWVAVYGLGNRILTLASVFLYALLTERIILVDQRK 167
Query: 122 DKEGLFCEPFLNSTWLLPEKSPF------WNAEKVQTYQSTIKMGGANTLNEDFPSALHV 175
D LFCEPF ++WLLP P +N E Y + +K N+ PS L++
Sbjct: 168 DISDLFCEPFPGTSWLLPLDFPLMGQIDSFNREYSHCYGTMLKNHTINSTT--IPSHLYL 225
Query: 176 NLGYSPTSEERFFHCDHNQFLLSNIPLLFLEAGQYFVPSFFMTPIFKKELNKMFPEITST 235
+L + +++ F C +Q L+ +P L +++ YF+PS ++ P F+ EL K+FP+ +
Sbjct: 226 HLLHDYRDQDKMFFCQKDQSLVDKVPWLVVKSNLYFIPSLWLNPSFQTELIKLFPQKDTV 285
Query: 236 FHHLGRYLFHPSNEAWELITSFYQQNLAKANERIGLQIRVVDPKLTPHQVVMNQILNCTL 295
F+HL RYLFHP+N+ W ++T Y L++A+E +G+Q+RV + Q VM+QI+ CT
Sbjct: 286 FYHLARYLFHPTNQVWGMVTRSYNAYLSRADEILGIQVRVFSRQTKYFQHVMDQIVACTQ 345
Query: 296 GNKLLPKVLGTKNIYLSSSDKNKKIMKAVLVSSLYRHYGENLKMTYMNKSTVSGEVIEVY 355
KLLP+ + ++++ K +KAVLV+SL Y NLK Y T +G+++EVY
Sbjct: 346 REKLLPEFAAQEEAQVTNTSNPSK-LKAVLVTSLNPEYSNNLKKMYWEHPTTTGDIVEVY 404
Query: 356 QPSGEEQQKFNDNKHNMKAWVDIYLLSLSDVLVTTYQSTFGYVAKALGNSRPWILYNP-- 413
QPS E Q+ + H+ KA ++YLLSL+D LVT+ STFGYVA+ LG +PWILY P
Sbjct: 405 QPSRERFQQTDKKLHDQKALAEMYLLSLTDKLVTSALSTFGYVAQGLGGLKPWILYTPKK 464
Query: 414 -VYSNEICEREFTLEPCYHYPPLHYCNGKPIEDVGSSFPYIRHCKDYT-FGVKLVN 467
N C R ++EPC+ PP+H C K + P++RHC+D +G+KLV+
Sbjct: 465 FKSPNPPCGRVISMEPCFLTPPVHGCEAKKGINTAKIVPFVRHCEDLRHYGLKLVD 520
>FUT8_ARATH (Q9XI78) Probable fucosyltransferase 8 (EC 2.4.1.-)
(AtFUT8)
Length = 500
Score = 389 bits (1000), Expect = e-108
Identities = 204/479 (42%), Positives = 296/479 (61%), Gaps = 18/479 (3%)
Query: 3 KEKLLDGLLVSGFDEASCVSRLQSHFYHKPSPHKPSPYLISKLRKYEELHRRCGPNTRAY 62
K+KLL GLL + FDE SC+SR +S Y KPSP+KPS YL+SKLR YE LH+RCGP T AY
Sbjct: 28 KDKLLGGLLTADFDEDSCLSRYESSLYRKPSPYKPSRYLVSKLRSYEMLHKRCGPGTEAY 87
Query: 63 NEDMKKITKSKKN-GTSAATTCKYIIWLPANGLGNQIISMASSFLYALLTGRVMLVQFGK 121
+ + + +N T + C+YI+W+ GLGN+I+++AS FLYALLT R+MLV
Sbjct: 88 KKATEILGHDDENHSTKSVGECRYIVWIAVYGLGNRILTLASLFLYALLTDRIMLVDQRT 147
Query: 122 DKEGLFCEPFLNSTWLLPEKSPF------WNAEKVQTYQSTIKMGGANTLNED--FPSAL 173
D LFCEPF ++WLLP P +N E + Y + +K N+ + PS L
Sbjct: 148 DISDLFCEPFPGTSWLLPLDFPLTDQLDSFNKESPRCYGTMLKNHAINSTTTESIIPSYL 207
Query: 174 HVNLGYSPTSEERFFHCDHNQFLLSNIPLLFLEAGQYFVPSFFMTPIFKKELNKMFPEIT 233
+ L + ++ F C+ +Q L+ +P L + YF+PS ++ P F+ EL+K+FP+
Sbjct: 208 CLYLIHDYDDYDKMFFCESDQILIRQVPWLVFNSNLYFIPSLWLIPSFQSELSKLFPQKE 267
Query: 234 STFHHLGRYLFHPSNEAWELITSFYQQNLAKANERIGLQIRVVDPKLTPHQVVMNQILNC 293
+ FHHL RYLFHP+N+ W +IT Y L++A+ER+G+Q+RV Q VM+QIL C
Sbjct: 268 TVFHHLARYLFHPTNQVWGMITRSYNGYLSRADERLGIQVRVFSKPAGYFQHVMDQILAC 327
Query: 294 TLGNKLLPKVLGTKNIYLSSSDKNKKIMKAVLVSSLYRHYGENLKMTYMNKSTVSGEVIE 353
T KLLP+V + ++S +K +KAVLV+SLY Y E L+ Y + +GE+I+
Sbjct: 328 TQREKLLPEVFVLETQVTNTSRSSK--LKAVLVTSLYPEYSEILRQMYWKGPSSTGEIIQ 385
Query: 354 VYQPSGEEQQKFNDNKHNMKAWVDIYLLSLSDVLVTTYQSTFGYVAKALGNSRPWILYNP 413
+YQPS E Q+ ++ H+ KA +IYLLSL+D +VT+ STFGYVA+ LG +PWILY P
Sbjct: 386 IYQPSQEIYQQTDNKLHDQKALAEIYLLSLTDYIVTSDSSTFGYVAQGLGGLKPWILYKP 445
Query: 414 ---VYSNEICEREFTLEPCYHYPPLHYCNGKPIEDVGSSFPYIRHCKDYTFGVKLVNAS 469
C R ++EPC+ PL+ C K + + P++ +C+D G+KLV+++
Sbjct: 446 KNHTAPEPPCVRAVSMEPCFLRAPLYGCQAKKV----NITPFVMYCEDRITGLKLVDSN 500
>FUT5_ARATH (Q9SJP4) Probable fucosyltransferase 5 (EC 2.4.1.-)
(AtFUT5)
Length = 533
Score = 385 bits (988), Expect = e-106
Identities = 194/469 (41%), Positives = 291/469 (61%), Gaps = 14/469 (2%)
Query: 4 EKLLDGLLVSGFDEASCVSRLQSHF-YHKPSPHKPSPYLISKLRKYEELHRRCGPNTRAY 62
+KL+ GLL + FDE SC+SR +F Y KPSP+KPS YL+SKLR YE LH+RCGP+T Y
Sbjct: 61 DKLIGGLLTADFDEGSCLSRYHKYFLYRKPSPYKPSEYLVSKLRSYEMLHKRCGPDTEYY 120
Query: 63 NEDMKKITKSKKNGTSAATTCKYIIWLPANGLGNQIISMASSFLYALLTGRVMLVQFGKD 122
E ++K+ S+ + + + C+YI+W+ GLGN+++++AS FLYALLT R++LV KD
Sbjct: 121 KEAIEKL--SRDDASESNGECRYIVWVAGYGLGNRLLTLASVFLYALLTERIILVDNRKD 178
Query: 123 KEGLFCEPFLNSTWLLPEKSPFWNAEKVQTYQSTIK-----MGGANTLNE-DFPSALHVN 176
L CEPF ++WLLP P N Y M +++N P L+++
Sbjct: 179 VSDLLCEPFPGTSWLLPLDFPMLNYTYAWGYNKEYPRCYGTMSEKHSINSTSIPPHLYMH 238
Query: 177 LGYSPTSEERFFHCDHNQFLLSNIPLLFLEAGQYFVPSFFMTPIFKKELNKMFPEITSTF 236
+ ++ F C +Q L+ +P L ++A YFVPS + P F+ EL K+FP+ + F
Sbjct: 239 NLHDSRDSDKLFVCQKDQSLIDKVPWLIVQANVYFVPSLWFNPTFQTELVKLFPQKETVF 298
Query: 237 HHLGRYLFHPSNEAWELITSFYQQNLAKANERIGLQIRVVDPKLTPHQVVMNQILNCTLG 296
HHL RYLFHP+NE W+++T +Y +L+KA+ER+G+QIRV + V++Q+++CT
Sbjct: 299 HHLARYLFHPTNEVWDMVTDYYHAHLSKADERLGIQIRVFGKPDGRFKHVIDQVISCTQR 358
Query: 297 NKLLPKVLGTKNIYLSSSDKNKKIMKAVLVSSLYRHYGENLKMTYMNKSTVSGEVIEVYQ 356
KLLP+ + ++ S K +K+VLV+SLY + NL + + + +GE++EVYQ
Sbjct: 359 EKLLPEFATPEESKVNISKTPK--LKSVLVASLYPEFSGNLTNMFSKRPSSTGEIVEVYQ 416
Query: 357 PSGEEQQKFNDNKHNMKAWVDIYLLSLSDVLVTTYQSTFGYVAKALGNSRPWILYNP--- 413
PSGE Q+ + H+ KA ++YLLSL+D +VT+ +STFGYV+ +LG +PW+LY P
Sbjct: 417 PSGERVQQTDKKSHDQKALAEMYLLSLTDNIVTSARSTFGYVSYSLGGLKPWLLYQPTNF 476
Query: 414 VYSNEICEREFTLEPCYHYPPLHYCNGKPIEDVGSSFPYIRHCKDYTFG 462
N C R ++EPCY PP H C + G P++RHC+D +G
Sbjct: 477 TTPNPPCVRSKSMEPCYLTPPSHGCEADWGTNSGKILPFVRHCEDLIYG 525
>FUT6_ARATH (Q9XI80) Probable fucosyltransferase 6 (EC 2.4.1.-)
(AtFUT6)
Length = 537
Score = 377 bits (967), Expect = e-104
Identities = 196/467 (41%), Positives = 287/467 (60%), Gaps = 15/467 (3%)
Query: 3 KEKLLDGLLVSGFDEASCVSRLQ-SHFYHKPSPHKPSPYLISKLRKYEELHRRCGPNTRA 61
+++L+ GLL + FDE SC+SR Q S Y K SP+KPS YL+SKLR YE+LH+RCGP T A
Sbjct: 63 RDRLIGGLLTADFDEGSCLSRYQQSLLYRKASPYKPSEYLVSKLRSYEKLHKRCGPGTDA 122
Query: 62 YNEDMKKITKSKKNGTSAAT-TCKYIIWLPANGLGNQIISMASSFLYALLTGRVMLVQFG 120
Y + + + +N S + C+YI+W+ GLGN+I+++AS FLYALLT RV+LV
Sbjct: 123 YKKATEILGHDDENYASKSVGECRYIVWVAVYGLGNRILTLASVFLYALLTERVVLVDQS 182
Query: 121 KDKEGLFCEPFLNSTWLLPEKSPFWNAEKVQTYQSTIKMGGANTLNED------FPSALH 174
KD LFCEPF ++WLLP + P +++ Y LN P L+
Sbjct: 183 KDISDLFCEPFPGTSWLLPLEFPLM--KQIDGYNKGYSRCYGTMLNNQAINSTLIPPHLY 240
Query: 175 VNLGYSPTSEERFFHCDHNQFLLSNIPLLFLEAGQYFVPSFFMTPIFKKELNKMFPEITS 234
+++ + ++ F C +Q L+ +P L ++A YFVPS ++ P F+ EL K+FP+ +
Sbjct: 241 LHILHDSRDNDKMFFCQKDQSLVDKVPWLIVKANVYFVPSLWLNPTFQTELMKLFPQKEA 300
Query: 235 TFHHLGRYLFHPSNEAWELITSFYQQNLAKANERIGLQIRVVDPKLTPHQVVMNQILNCT 294
FHHL RYLFHP+N+ W LIT Y L++A+E +G+QIRV + Q VM+Q++ CT
Sbjct: 301 VFHHLARYLFHPTNQVWGLITRSYNAYLSRADETLGIQIRVFSDRAGYFQHVMDQVVACT 360
Query: 295 LGNKLLPKVLGTKNIYLSSSDKNKKIMKAVLVSSLYRHYGENLKMTYMNKSTVSGEVIEV 354
LLP+ + ++ S K +KAVLV+SLY Y E LK Y + + +GE+IEV
Sbjct: 361 RRENLLPEPAAQEEPKVNISRSQK--LKAVLVTSLYPEYSETLKNMYWERPSSTGEIIEV 418
Query: 355 YQPSGEEQQKFNDNKHNMKAWVDIYLLSLSDVLVTTYQSTFGYVAKALGNSRPWILY--- 411
YQPSGE Q+ + H+ KA ++YLLSL+D +VT+ +STFGYVA +LG +PW+LY
Sbjct: 419 YQPSGERVQQTDKKLHDQKALAEMYLLSLTDKIVTSARSTFGYVAHSLGGLKPWLLYQPT 478
Query: 412 NPVYSNEICEREFTLEPCYHYPPLHYCNGKPIEDVGSSFPYIRHCKD 458
P + C + +++PC+ PP H C + + G P++RHC+D
Sbjct: 479 GPTAPDPPCIQSTSMDPCHLTPPSHGCEPEWGTNSGKVVPFVRHCED 525
>FUT4_ARATH (Q9SJP2) Probable fucosyltransferase 4 (EC 2.4.1.-)
(AtFUT4)
Length = 503
Score = 376 bits (965), Expect = e-104
Identities = 194/475 (40%), Positives = 298/475 (61%), Gaps = 17/475 (3%)
Query: 3 KEKLLDGLLVSGFDEASCVSRLQSHF-YHKPSPHKPSPYLISKLRKYEELHRRCGPNTRA 61
+++L+ GLL + FDE SC+SR F Y KPSP+KPS YL+SKLR YE LH+RCGP T+A
Sbjct: 30 RDRLIGGLLTADFDEGSCLSRYHKTFLYRKPSPYKPSEYLVSKLRSYEMLHKRCGPGTKA 89
Query: 62 YNEDMKKITKSKK-NGTSAATTCKYIIWLPANGLGNQIISMASSFLYALLTGRVMLVQFG 120
Y E K ++ + N + + C+Y++WL GLGN+++++AS FLYALLT R++LV
Sbjct: 90 YKEATKHLSHDENYNASKSDGECRYVVWLADYGLGNRLLTLASVFLYALLTDRIILVDNR 149
Query: 121 KDKEGLFCEPFLNSTWLLPEKSPF------WNAEKVQTYQSTIKMGGANTLNEDFPSALH 174
KD L CEPF ++WLLP P ++ + Y + ++ N+ + FP L+
Sbjct: 150 KDIGDLLCEPFPGTSWLLPLDFPLMKYADGYHKGYSRCYGTMLENHSINSTS--FPPHLY 207
Query: 175 VNLGYSPTSEERFFHCDHNQFLLSNIPLLFLEAGQYFVPSFFMTPIFKKELNKMFPEITS 234
++ + ++ F C +Q L+ +P L A YFVPS + P F+ EL K+FP+ +
Sbjct: 208 MHNLHDSRDSDKMFFCQKDQSLIDKVPWLIFRANVYFVPSLWFNPTFQTELTKLFPQKET 267
Query: 235 TFHHLGRYLFHPSNEAWELITSFYQQNLAKANERIGLQIRVVDPKLTPHQVVMNQILNCT 294
FHHLGRYLFHP N+ W+++T +Y +L+KA+ER+G+QIRV + +Q VM+Q+++CT
Sbjct: 268 VFHHLGRYLFHPKNQVWDIVTKYYHDHLSKADERLGIQIRVFRDQGGYYQHVMDQVISCT 327
Query: 295 LGNKLLPKVLGTKNIYLSSSDKNKKIMKAVLVSSLYRHYGENLKMTYMNKSTVSGEVIEV 354
KLLP++ + ++ S+ K KAVLV+SL Y + L+ + ++ ++GE+I+V
Sbjct: 328 QREKLLPELATQEESKVNISNIPKS--KAVLVTSLSPEYSKKLENMFSERANMTGEIIKV 385
Query: 355 YQPSGEEQQKFNDNKHNMKAWVDIYLLSLSDVLVTTYQSTFGYVAKALGNSRPWILYNP- 413
YQPSGE Q+ + H+ KA ++YLLSL+D +V + +STFGYVA +LG +PW+LY P
Sbjct: 386 YQPSGERYQQTDKKVHDQKALAEMYLLSLTDNIVASSRSTFGYVAYSLGGLKPWLLYLPN 445
Query: 414 --VYSNEICEREFTLEPCYHYPPLHYCNGKPI-EDVGSSFPYIRHCKDYTFGVKL 465
+ C R ++EPC+ PP H C + G P++R+C+D +G+KL
Sbjct: 446 DNKAPDPPCVRSTSMEPCFLTPPTHGCEPDAWGTESGKVVPFVRYCED-IWGLKL 499
>FU10_ARATH (Q9SJP6) Putative fucosyltransferase 10 (EC 2.4.1.-)
(AtFUT10) (Fragment)
Length = 440
Score = 355 bits (911), Expect = 1e-97
Identities = 175/437 (40%), Positives = 268/437 (61%), Gaps = 17/437 (3%)
Query: 37 PSPYLISKLRKYEELHRRCGPNTRAYNEDMKKITKSKKNGTSAATTCKYIIWLPANGLGN 96
PS YL+S+LR YE LH+RCGP+T+AY E +K+++ + + + C+YI+WL +GLGN
Sbjct: 2 PSEYLVSELRSYEMLHKRCGPDTKAYKEATEKLSRDEYYASESNGECRYIVWLARDGLGN 61
Query: 97 QIISMASSFLYALLTGRVMLVQFGKDKEGLFCEPFLNSTWLLPEKSPF--------WNAE 148
++I++AS FLYA+LT R++LV KD L CEPF ++WLLP P +N E
Sbjct: 62 RLITLASVFLYAILTERIILVDNRKDVSDLLCEPFPGTSWLLPLDFPMLNYTYAYGYNKE 121
Query: 149 KVQTYQSTIKMGGANTLNEDFPSALHVNLGYSPTSEERFFHCDHNQFLLSNIPLLFLEAG 208
+ Y + ++ N+ + P L+++ + ++ F C +Q + +P L ++
Sbjct: 122 YPRCYGTMLENHAINSTS--IPPHLYLHNIHDSRDSDKLFFCQKDQSFIDKVPWLIIQTN 179
Query: 209 QYFVPSFFMTPIFKKELNKMFPEITSTFHHLGRYLFHPSNEAWELITSFYQQNLAKANER 268
YFVPS ++ P F+ +L K+FP+ + FHHL RYLFHP+NE W+++T +Y +L+ A+ER
Sbjct: 180 AYFVPSLWLNPTFQTKLVKLFPQKETVFHHLARYLFHPTNEVWDMVTKYYDAHLSNADER 239
Query: 269 IGLQIRVVDPKLTPHQVVMNQILNCTLGNKLLPKVLGTKNIYLSSSDKNKKIMKAVLVSS 328
+G+QIRV + VM+Q++ CT KLLP+ + +S K +K VLV+S
Sbjct: 240 LGIQIRVFGKPSGYFKHVMDQVVACTQREKLLPEFEEESKVNISKPPK----LKVVLVAS 295
Query: 329 LYRHYGENLKMTYMNKSTVSGEVIEVYQPSGEEQQKFNDNKHNMKAWVDIYLLSLSDVLV 388
LY Y NL ++ + + +GE+IEVYQPS E Q+ + H+ KA ++YLLSL+D +V
Sbjct: 296 LYPEYSVNLTNMFLARPSSTGEIIEVYQPSAERVQQTDKKSHDQKALAEMYLLSLTDNIV 355
Query: 389 TTYQSTFGYVAKALGNSRPWILYNPV---YSNEICEREFTLEPCYHYPPLHYCNGKPIED 445
T+ STFGYV+ +LG +PW+LY PV N C R ++EPCYH PP H C +
Sbjct: 356 TSGWSTFGYVSYSLGGLKPWLLYQPVNFTTPNPPCVRSKSMEPCYHTPPSHGCEADWGTN 415
Query: 446 VGSSFPYIRHCKDYTFG 462
G P++RHC+D +G
Sbjct: 416 SGKILPFVRHCEDMMYG 432
>FUT3_ARATH (Q9CA71) Probable fucosyltransferase 3 (EC 2.4.1.-)
(AtFUT3)
Length = 493
Score = 310 bits (794), Expect = 5e-84
Identities = 178/443 (40%), Positives = 256/443 (57%), Gaps = 18/443 (4%)
Query: 1 DEKEKLLDGLLVSGFDEASCVSRLQSHFYHKPSPHKPSPYLISKLRKYEELHRRCGPNTR 60
++K +L + LV FD+ SC+SR ++ Y K SP K S YL +L++YE+LHRRCGP TR
Sbjct: 62 EKKAQLGNINLVPSFDKESCLSRYEASLYRKESPFKQSSYLDYRLQRYEDLHRRCGPFTR 121
Query: 61 AYNEDMKKITKSKKNGTSAATTCKYIIWLPANG-LGNQIISMASSFLYALLTGRVMLVQF 119
+YN + K+ KS + C+Y+IWL +NG LGN+++S+AS+FLYALLT R +LV+
Sbjct: 122 SYNLTLDKL-KSGDRSDGEVSGCRYVIWLNSNGDLGNRMLSLASAFLYALLTNRFLLVEL 180
Query: 120 GKDKEGLFCEPFLNSTWLLPEKSPFWNAEKVQTYQSTIKMGGANTLNEDFPSALHVNLGY 179
G D LFCEPF N+TW LP + P QS ++ G P + ++
Sbjct: 181 GVDMADLFCEPFPNTTWFLPPEFPL---NSHFNEQSLLRNSGN-------PMVAYRHVVR 230
Query: 180 SPTSEERFFHCDHNQFLLSNIPLLFLEAGQYFVPSFFMTPIFKKELNKMFPEITSTFHHL 239
+ +++ F C+ +Q LL P L L+A +F+PS F FK+EL +FPE + FH L
Sbjct: 231 DSSDQQKLFFCEDSQVLLEETPWLILKADSFFLPSLFSVSSFKQELQMLFPEKDTAFHFL 290
Query: 240 GRYLFHPSNEAWELITSFYQQNLAKANERIGLQIRVVDPKLTPHQVVMNQILNCTLGNKL 299
+YLFHP+N W LIT +Y LAKA++RIG+ I V + Q +++QIL C +KL
Sbjct: 291 SQYLFHPTNVVWGLITRYYNAYLAKADQRIGIYIGVSESGNEQFQHLIDQILACGTRHKL 350
Query: 300 LPKVLGTKNIYLSSSDKNKKIMKAVLVSSLYRHYGENLKMTYMNKSTVSGEVIEVYQPSG 359
LP+V +N L SS + KAV +SS Y ++++ Y TV GE+I V++PS
Sbjct: 351 LPEVDKQRN--LPSSQVLNRKSKAVFISSSSPGYFKSIRDVYWENPTVMGEIISVHKPSY 408
Query: 360 EEQQKFNDNKHNMKAWVDIYLLSLSDVLVTT-YQSTFGYVAKALGNSRPWIL---YNPVY 415
++ QK N + +AW +IYLLS SD LV T S+ VA LG +PW+L N
Sbjct: 409 KDYQKTPRNMESKRAWAEIYLLSCSDALVVTGLWSSLVEVAHGLGGLKPWVLNKAENGTA 468
Query: 416 SNEICEREFTLEPCYHYPPLHYC 438
C + ++EPC H C
Sbjct: 469 HEPYCVKARSIEPCSQATLFHGC 491
>FUT9_ARATH (Q9XI77) Probable fucosyltransferase 9 (EC 2.4.1.-)
(AtFUT9)
Length = 435
Score = 224 bits (571), Expect = 4e-58
Identities = 123/320 (38%), Positives = 189/320 (58%), Gaps = 20/320 (6%)
Query: 2 EKEKLLDGLLVSGFDEASCVSRLQSHFYHKPSPHKPSPYLISKLRKYEELHRRCGPNTRA 61
+ EKLL GLL +GF+E SC+SR KPSP+KPS +++SKLR YE LH+RCGP T+A
Sbjct: 44 QSEKLLGGLLATGFEEKSCLSRYDQSM-SKPSPYKPSRHIVSKLRSYEMLHKRCGPGTKA 102
Query: 62 YNEDMKKITKSKKNGTSAATTCKYIIWLPANGLGNQIISMASSFLYALLTGRVMLVQFGK 121
Y K++ ++ + S+ C+Y++W+P GLGN+++S+ S FLYALLT RVMLV
Sbjct: 103 YKRATKQLGHNELS--SSGDECRYVVWMPMFGLGNRMLSLVSVFLYALLTDRVMLVDQRN 160
Query: 122 DKEGLFCEPFLNSTWLLPEKSPF------WNAEKVQTYQSTIKMGGANTLNEDFPSALHV 175
D LFCEPF ++WLLP P +N E + Y + +K G N+ + PS L++
Sbjct: 161 DITDLFCEPFPETSWLLPLDFPLNDQLDSFNREHSRCYGTMLKNHGINSTSI-IPSHLYL 219
Query: 176 NLGYSPTSEERFFHCDHNQFLLSNIPLLFLEAGQYFVPSFFMTPIFKKELNKMFPEITST 235
++ + ++ F C+ +Q L + L +++ YFVPS +M P F+ +L K+FP+ +
Sbjct: 220 DIFHDSRDHDKKFFCEEDQAFLDKVTWLVVKSNLYFVPSLWMIPSFQTKLIKLFPQKETV 279
Query: 236 FHHLGRYLFHPSNEAWEL---ITSFYQQNLAKANERI------GLQIRVVDPKLTPHQVV 286
FHHL RYLFHP+N+ WE + + + + + E I G +I+ D KL Q
Sbjct: 280 FHHLARYLFHPTNQVWEYSDHLKNMFLEQASSTGETIEVYQPSGEKIQQTDKKL-HDQKA 338
Query: 287 MNQILNCTLGNKLLPKVLGT 306
+ +I +L ++L+ T
Sbjct: 339 LAEIYLLSLTDELVTSTRST 358
Score = 125 bits (315), Expect = 2e-28
Identities = 64/140 (45%), Positives = 92/140 (65%), Gaps = 7/140 (5%)
Query: 333 YGENLKMTYMNKSTVSGEVIEVYQPSGEEQQKFNDNKHNMKAWVDIYLLSLSDVLVTTYQ 392
Y ++LK ++ +++ +GE IEVYQPSGE+ Q+ + H+ KA +IYLLSL+D LVT+ +
Sbjct: 297 YSDHLKNMFLEQASSTGETIEVYQPSGEKIQQTDKKLHDQKALAEIYLLSLTDELVTSTR 356
Query: 393 STFGYVAKALGNSRPWILYNP---VYSNEICEREFTLEPCYHYPPLHYCNGKPIEDVGSS 449
STFGYVA+ LG +PWILY P N C R ++EPC+ PLH C K I+
Sbjct: 357 STFGYVAQGLGGLKPWILYEPRDKKTPNPPCVRAMSMEPCFLRAPLHGCQAKTIKIP--- 413
Query: 450 FPYIRHCKDYTFGVKLVNAS 469
P++R C+D+ G+KLV+ S
Sbjct: 414 -PFVRICEDWKTGLKLVDVS 432
>ZB11_HUMAN (O95625) Zinc finger and BTB domain containing protein
11
Length = 1053
Score = 33.5 bits (75), Expect = 1.2
Identities = 26/125 (20%), Positives = 57/125 (44%), Gaps = 5/125 (4%)
Query: 245 HPSNEAWELITSFYQQNLAKANERIGLQIRVVDPKLTPHQVVMNQILNCTLGNKLLPKVL 304
HP E+ +LIT Q L +N R + + PKL+ V+ + + +GN + + +
Sbjct: 406 HPEMESVDLITKNNQTELETSNNRENNTVSNIHPKLSKENVISSSPEDSGMGNDISAEDI 465
Query: 305 GTKNI--YLSSSDKNKKIMKAVLVSSLYRHYG---ENLKMTYMNKSTVSGEVIEVYQPSG 359
++I + D+ K + ++ S+ ++G + + +S G I +++
Sbjct: 466 CAEDIPKHRQKVDQPLKDQENLVASTAKTNFGPDDDTYRSRLRQRSVNEGAYIRLHKGME 525
Query: 360 EEQQK 364
++ QK
Sbjct: 526 KKLQK 530
>MFD_BORBU (O51568) Transcription-repair coupling factor (TRCF)
Length = 1125
Score = 31.6 bits (70), Expect = 4.6
Identities = 24/69 (34%), Positives = 38/69 (54%), Gaps = 4/69 (5%)
Query: 294 TLGNKLLPKVLGTKNIYLSSSDKNKKIMKAVLVSSLYR-HYGENLKMTYMNKSTVSGEVI 352
+L +K+ K KNIY +KN I A + +L Y + L++T + TV GE+I
Sbjct: 125 SLLSKIPDKNTLLKNIY--KIEKNTNINTADIEKTLITLGYEKTLRVTIPGEFTVKGEII 182
Query: 353 EVYQPSGEE 361
++Y P GE+
Sbjct: 183 DIY-PFGEQ 190
>DCM_ECOLI (P11876) DNA-cytosine methyltransferase (EC 2.1.1.37)
(M.EcoDcm)
Length = 472
Score = 31.6 bits (70), Expect = 4.6
Identities = 16/42 (38%), Positives = 23/42 (54%), Gaps = 1/42 (2%)
Query: 45 LRKYEELHRRCGPNTRAYNEDMKKITKSKKNGTSAATTCKYI 86
+R Y+ H C P T +NED++ IT S K G S ++I
Sbjct: 121 VRTYKANHY-CDPATHHFNEDIRDITLSHKEGVSDEAAAEHI 161
>YP46_YEAST (Q12080) Hypothetical protein YPL146C
Length = 455
Score = 30.8 bits (68), Expect = 7.8
Identities = 19/47 (40%), Positives = 29/47 (61%), Gaps = 6/47 (12%)
Query: 227 KMFPEITSTFHHLGRYLFHPSNEAW-ELITSFYQQNLAKANERIGLQ 272
K F EI H G+ ++P+N+AW ELI Y++ A+ +ERI L+
Sbjct: 215 KEFTEIP----HAGKS-YNPNNKAWSELINKEYKEEKAREDERIALE 256
>SYR_PROMA (Q7VE03) Arginyl-tRNA synthetase (EC 6.1.1.19)
(Arginine--tRNA ligase) (ArgRS)
Length = 603
Score = 30.8 bits (68), Expect = 7.8
Identities = 19/82 (23%), Positives = 37/82 (44%), Gaps = 4/82 (4%)
Query: 334 GENLKMTYMNKSTVSGEVIEVYQPSGEEQQK----FNDNKHNMKAWVDIYLLSLSDVLVT 389
GE +++ + +S +++ + EE +K F + N + LS +T
Sbjct: 410 GETVRLKDLLDEAISRAKLDIERRLNEENRKESQAFIEKVSNTIGIAAVKYADLSQNRIT 469
Query: 390 TYQSTFGYVAKALGNSRPWILY 411
YQ +F + GN+ P++LY
Sbjct: 470 NYQFSFDRMLALQGNTAPYLLY 491
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.319 0.136 0.414
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 58,244,179
Number of Sequences: 164201
Number of extensions: 2520411
Number of successful extensions: 5816
Number of sequences better than 10.0: 16
Number of HSP's better than 10.0 without gapping: 12
Number of HSP's successfully gapped in prelim test: 4
Number of HSP's that attempted gapping in prelim test: 5753
Number of HSP's gapped (non-prelim): 19
length of query: 469
length of database: 59,974,054
effective HSP length: 114
effective length of query: 355
effective length of database: 41,255,140
effective search space: 14645574700
effective search space used: 14645574700
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 68 (30.8 bits)
Medicago: description of AC146553.5


