

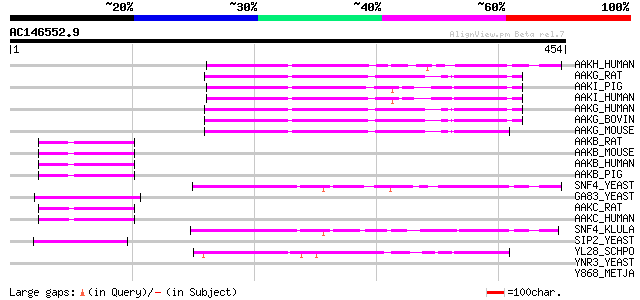
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146552.9 - phase: 0 /pseudo
(454 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
AAKH_HUMAN (Q9UGJ0) 5'-AMP-activated protein kinase, gamma-2 sub... 99 3e-20
AAKG_RAT (P80385) 5'-AMP-activated protein kinase, gamma-1 subun... 97 9e-20
AAKI_PIG (Q9MYP4) 5'-AMP-activated protein kinase, gamma-3 subun... 97 1e-19
AAKI_HUMAN (Q9UGI9) 5'-AMP-activated protein kinase, gamma-3 sub... 96 1e-19
AAKG_HUMAN (P54619) 5'-AMP-activated protein kinase, gamma-1 sub... 96 2e-19
AAKG_BOVIN (P58108) 5'-AMP-activated protein kinase, gamma-1 sub... 95 3e-19
AAKG_MOUSE (O54950) 5'-AMP-activated protein kinase, gamma-1 sub... 93 2e-18
AAKB_RAT (P80386) 5'-AMP-activated protein kinase, beta-1 subuni... 76 2e-13
AAKB_MOUSE (Q9R078) 5'-AMP-activated protein kinase, beta-1 subu... 76 2e-13
AAKB_HUMAN (Q9Y478) 5'-AMP-activated protein kinase, beta-1 subu... 76 2e-13
AAKB_PIG (P80387) 5'-AMP-activated protein kinase, beta-1 subuni... 74 1e-12
SNF4_YEAST (P12904) Nuclear protein SNF4 (Regulatory protein CAT3) 72 3e-12
GA83_YEAST (Q04739) Glucose repression protein GAL83 (SPM1 protein) 67 7e-11
AAKC_RAT (Q9QZH4) 5'-AMP-activated protein kinase, beta-2 subuni... 67 9e-11
AAKC_HUMAN (O43741) 5'-AMP-activated protein kinase, beta-2 subu... 67 9e-11
SNF4_KLULA (Q9P869) Nuclear protein SNF4 67 1e-10
SIP2_YEAST (P34164) SIP2 protein (SPM2 protein) 60 2e-08
YL28_SCHPO (Q10343) Hypothetical protein C1556.08c in chromosome I 58 4e-08
YNR3_YEAST (P53885) Hypothetical 40.3 kDa protein in RPS3-PSD1 i... 36 0.23
Y868_METJA (Q58278) Hypothetical protein MJ0868 35 0.40
>AAKH_HUMAN (Q9UGJ0) 5'-AMP-activated protein kinase, gamma-2
subunit (AMPK gamma-2 chain) (AMPK gamma2) (H91620p)
Length = 569
Score = 98.6 bits (244), Expect = 3e-20
Identities = 81/295 (27%), Positives = 139/295 (46%), Gaps = 39/295 (13%)
Query: 162 YLSMRTAYELLPESGKVVTLDVDLPVKQAFHILHEQGIPMAPLWDFCKGQFVGVLSVLDF 221
++ Y+++P S K+V D L VK+AF L G+ APLW+ K FVG+L++ DF
Sbjct: 264 FMRSHKCYDIVPTSSKLVVFDTTLQVKKAFFALVANGVRAAPLWESKKQSFVGMLTITDF 323
Query: 222 ILILRELGNHGSNLTE-EELETHTISAWKEGKWTLFSRRFIHAGPSDNLKDVALKILQNG 280
I IL + S + + ELE H I W+E + ++ P +L D +++N
Sbjct: 324 INILHRY--YKSPMVQIYELEEHKIETWRELYLQETFKPLVNISPDASLFDAVYSLIKNK 381
Query: 281 ISTVPIIHSSSADGSFPQLLHLASLSGILRCILGVVLVHCPYFNFQFVQFLWAHGCPKLG 340
I +P+I S + L++ + IL+ L + + P F +LG
Sbjct: 382 IHRLPVIDPISGNA-----LYILTHKRILK-FLQLFMSDMPKPAFM------KQNLDELG 429
Query: 341 ----RQIAGL*QH*DQMLLLLQP*IY*FKFNIFFAAQVSSIPIVDESDSLLDIYCRSDIT 396
IA + H D ++ NIF ++S++P+VDES ++DIY + D+
Sbjct: 430 IGTYHNIAFI--HPDTPIIK--------ALNIFVERRISALPVVDESGKVVDIYSKFDVI 479
Query: 397 ALAKDRAYTHINLDEMTVHQALQLSQDAFNPNESRSQRCQMCLRTDSLHKVMERL 451
LA ++ Y ++ ++TV QALQ F + C + + L +++R+
Sbjct: 480 NLAAEKTYNNL---DITVTQALQHRSQYF-------EGVVKCNKLEILETIVDRI 524
>AAKG_RAT (P80385) 5'-AMP-activated protein kinase, gamma-1 subunit
(AMPK gamma-1 chain) (AMPKg)
Length = 330
Score = 97.1 bits (240), Expect = 9e-20
Identities = 76/262 (29%), Positives = 128/262 (48%), Gaps = 26/262 (9%)
Query: 160 STYLSMRTAYELLPESGKVVTLDVDLPVKQAFHILHEQGIPMAPLWDFCKGQFVGVLSVL 219
+T++ Y+L+P S K+V D L VK+AF L G+ APLWD K FVG+L++
Sbjct: 29 TTFMKSHRCYDLIPTSSKLVVFDTSLQVKKAFFALVTNGVRAAPLWDSKKQSFVGMLTIT 88
Query: 220 DFILILRELGNHGSNLTE-EELETHTISAWKEGKWTLFSRRFIHAGPSDNLKDVALKILQ 278
DFI IL + S L + ELE H I W+E + + P+ +L D +++
Sbjct: 89 DFINILHRY--YKSALVQIYELEEHKIETWREVYLQDSFKPLVCISPNASLFDAVSSLIR 146
Query: 279 NGISTVPIIHSSSADGSFPQLLHLASLSGILRCI-LGVVLVHCPYFNFQFVQFLWAHGCP 337
N I +P+I S + L++ + IL+ + L + P F + ++ L
Sbjct: 147 NKIHRLPVIDPESGN-----TLYILTHKRILKFLKLFITEFPKPEFMSKSLEELQIGTYA 201
Query: 338 KLGRQIAGL*QH*DQMLLLLQP*IY*FKFNIFFAAQVSSIPIVDESDSLLDIYCRSDITA 397
+ M+ P +Y IF +VS++P+VDE ++DIY + D+
Sbjct: 202 NIA------------MVRTTTP-VY-VALGIFVQHRVSALPVVDEKGRVVDIYSKFDVIN 247
Query: 398 LAKDRAYTHINLDEMTVHQALQ 419
LA ++ Y ++ +++V +ALQ
Sbjct: 248 LAAEKTYNNL---DVSVTKALQ 266
>AAKI_PIG (Q9MYP4) 5'-AMP-activated protein kinase, gamma-3 subunit
(AMPK gamma-3 chain) (AMPK gamma3)
Length = 514
Score = 96.7 bits (239), Expect = 1e-19
Identities = 74/261 (28%), Positives = 129/261 (49%), Gaps = 28/261 (10%)
Query: 162 YLSMRTAYELLPESGKVVTLDVDLPVKQAFHILHEQGIPMAPLWDFCKGQFVGVLSVLDF 221
++ T Y+ + S K+V D L +K+AF L G+ APLWD K FVG+L++ DF
Sbjct: 212 FMQEHTCYDAMATSSKLVIFDTMLEIKKAFFALVANGVRAAPLWDSKKQSFVGMLTITDF 271
Query: 222 ILILRELGNHGSNLTE-EELETHTISAWKEGKWTLFSRRFIHAGPSDNLKDVALKILQNG 280
IL+L + S L + E+E H I W+E + + P+D+L + +++N
Sbjct: 272 ILVLHRY--YRSPLVQIYEIEEHKIETWREIYLQGCFKPLVSISPNDSLFEAVYALIKNR 329
Query: 281 ISTVPIIHSSSADGSFPQLLHLASLSGILRC--ILGVVLVHCPYFNFQFVQFLWAHGCPK 338
I +P++ S +LH+ + +L+ I G +L P F ++ +Q L
Sbjct: 330 IHRLPVLDPVSG-----AVLHILTHKRLLKFLHIFGTLLPR-PSFLYRTIQDL------- 376
Query: 339 LGRQIAGL*QH*DQMLLLLQP*IY*FKFNIFFAAQVSSIPIVDESDSLLDIYCRSDITAL 398
G+ D ++L I +IF +VS++P+V+E+ ++ +Y R D+ L
Sbjct: 377 ------GIGTFRDLAVVLETAPIL-TALDIFVDRRVSALPVVNETGQVVGLYSRFDVIHL 429
Query: 399 AKDRAYTHINLDEMTVHQALQ 419
A + Y H+ +M V +AL+
Sbjct: 430 AAQQTYNHL---DMNVGEALR 447
>AAKI_HUMAN (Q9UGI9) 5'-AMP-activated protein kinase, gamma-3
subunit (AMPK gamma-3 chain) (AMPK gamma3)
Length = 464
Score = 96.3 bits (238), Expect = 1e-19
Identities = 74/261 (28%), Positives = 130/261 (49%), Gaps = 28/261 (10%)
Query: 162 YLSMRTAYELLPESGKVVTLDVDLPVKQAFHILHEQGIPMAPLWDFCKGQFVGVLSVLDF 221
++ T Y+ + S K+V D L +K+AF L G+ APLWD K FVG+L++ DF
Sbjct: 162 FMQEHTCYDAMATSSKLVIFDTMLEIKKAFFALVANGVRAAPLWDSKKQSFVGMLTITDF 221
Query: 222 ILILRELGNHGSNLTE-EELETHTISAWKEGKWTLFSRRFIHAGPSDNLKDVALKILQNG 280
IL+L + S L + E+E H I W+E + + P+D+L + +++N
Sbjct: 222 ILVLHRY--YRSPLVQIYEIEQHKIETWREIYLQGCFKPLVSISPNDSLFEAVYTLIKNR 279
Query: 281 ISTVPIIHSSSADGSFPQLLHLASLSGILRC--ILGVVLVHCPYFNFQFVQFLWAHGCPK 338
I +P++ S + +LH+ + +L+ I G +L P F ++ +Q L
Sbjct: 280 IHRLPVLDPVSGN-----VLHILTHKRLLKFLHIFGSLLPR-PSFLYRTIQDL------- 326
Query: 339 LGRQIAGL*QH*DQMLLLLQP*IY*FKFNIFFAAQVSSIPIVDESDSLLDIYCRSDITAL 398
G+ D ++L I +IF +VS++P+V+E ++ +Y R D+ L
Sbjct: 327 ------GIGTFRDLAVVLETAPIL-TALDIFVDRRVSALPVVNECGQVVGLYSRFDVIHL 379
Query: 399 AKDRAYTHINLDEMTVHQALQ 419
A + Y H+ +M+V +AL+
Sbjct: 380 AAQQTYNHL---DMSVGEALR 397
>AAKG_HUMAN (P54619) 5'-AMP-activated protein kinase, gamma-1
subunit (AMPK gamma-1 chain) (AMPKg)
Length = 331
Score = 95.5 bits (236), Expect = 2e-19
Identities = 75/262 (28%), Positives = 128/262 (48%), Gaps = 26/262 (9%)
Query: 160 STYLSMRTAYELLPESGKVVTLDVDLPVKQAFHILHEQGIPMAPLWDFCKGQFVGVLSVL 219
++++ Y+L+P S K+V D L VK+AF L G+ APLWD K FVG+L++
Sbjct: 30 TSFMKSHRCYDLIPTSSKLVVFDTSLQVKKAFFALVTNGVRAAPLWDSKKQSFVGMLTIT 89
Query: 220 DFILILRELGNHGSNLTE-EELETHTISAWKEGKWTLFSRRFIHAGPSDNLKDVALKILQ 278
DFI IL + S L + ELE H I W+E + + P+ +L D +++
Sbjct: 90 DFINILHRY--YKSALVQIYELEEHKIETWREVYLQDSFKPLVCISPNASLFDAVSSLIR 147
Query: 279 NGISTVPIIHSSSADGSFPQLLHLASLSGILRCI-LGVVLVHCPYFNFQFVQFLWAHGCP 337
N I +P+I S + L++ + IL+ + L + P F + ++ L
Sbjct: 148 NKIHRLPVIDPESGN-----TLYILTHKRILKFLKLFITEFPKPEFMSKSLEELQIGTYA 202
Query: 338 KLGRQIAGL*QH*DQMLLLLQP*IY*FKFNIFFAAQVSSIPIVDESDSLLDIYCRSDITA 397
+ M+ P +Y IF +VS++P+VDE ++DIY + D+
Sbjct: 203 NIA------------MVRTTTP-VY-VALGIFVQHRVSALPVVDEKGRVVDIYSKFDVIN 248
Query: 398 LAKDRAYTHINLDEMTVHQALQ 419
LA ++ Y ++ +++V +ALQ
Sbjct: 249 LAAEKTYNNL---DVSVTKALQ 267
>AAKG_BOVIN (P58108) 5'-AMP-activated protein kinase, gamma-1
subunit (AMPK gamma-1 chain) (AMPKg)
Length = 330
Score = 95.1 bits (235), Expect = 3e-19
Identities = 75/262 (28%), Positives = 128/262 (48%), Gaps = 26/262 (9%)
Query: 160 STYLSMRTAYELLPESGKVVTLDVDLPVKQAFHILHEQGIPMAPLWDFCKGQFVGVLSVL 219
++++ Y+L+P S K+V D L VK+AF L G+ APLWD K FVG+L++
Sbjct: 30 TSFMKSHRCYDLIPTSSKLVVFDTSLQVKKAFFALVTNGVRAAPLWDSKKQSFVGMLTIT 89
Query: 220 DFILILRELGNHGSNLTE-EELETHTISAWKEGKWTLFSRRFIHAGPSDNLKDVALKILQ 278
DFI IL + S L + ELE H I W+E + + P+ +L D +++
Sbjct: 90 DFINILHRY--YKSALVQIYELEEHKIETWREVFLQDSFKPLVCISPNASLFDAVSSLIR 147
Query: 279 NGISTVPIIHSSSADGSFPQLLHLASLSGILRCI-LGVVLVHCPYFNFQFVQFLWAHGCP 337
N I +P+I S + L++ + IL+ + L + P F + ++ L
Sbjct: 148 NKIHRLPVIDPESGN-----TLYILTHKRILKFLKLFITEFPKPEFMSKSLEELQIGTYA 202
Query: 338 KLGRQIAGL*QH*DQMLLLLQP*IY*FKFNIFFAAQVSSIPIVDESDSLLDIYCRSDITA 397
+ M+ P +Y IF +VS++P+VDE ++DIY + D+
Sbjct: 203 NIA------------MVRTTTP-VY-VALGIFVQHRVSALPVVDEKGRVVDIYSKFDVIN 248
Query: 398 LAKDRAYTHINLDEMTVHQALQ 419
LA ++ Y ++ +++V +ALQ
Sbjct: 249 LAAEKTYNNL---DVSVTKALQ 267
>AAKG_MOUSE (O54950) 5'-AMP-activated protein kinase, gamma-1
subunit (AMPK gamma-1 chain) (AMPKg)
Length = 330
Score = 92.8 bits (229), Expect = 2e-18
Identities = 71/252 (28%), Positives = 122/252 (48%), Gaps = 23/252 (9%)
Query: 160 STYLSMRTAYELLPESGKVVTLDVDLPVKQAFHILHEQGIPMAPLWDFCKGQFVGVLSVL 219
++++ Y+L+P S K+V D L VK+AF L G+ APLWD K FVG+L++
Sbjct: 29 TSFMKSHRCYDLIPTSSKLVVFDTSLQVKKAFFALVTNGVRAAPLWDSKKQCFVGMLTIT 88
Query: 220 DFILILRELGNHGSNLTE-EELETHTISAWKEGKWTLFSRRFIHAGPSDNLKDVALKILQ 278
DFI IL + S L + ELE H I W+E + + P+ + D +++
Sbjct: 89 DFINILHRY--YKSALVQIYELEEHKIETWREVYLQDSFKPLVCISPNASSFDAVSSLIR 146
Query: 279 NGISTVPIIHSSSADGSFPQLLHLASLSGILRCI-LGVVLVHCPYFNFQFVQFLWAHGCP 337
N I +P+I S + L++ + IL+ + L ++ P F + +Q L
Sbjct: 147 NKIHRLPVIDPESGN-----TLYILTHKRILKFLKLFIIEFPKPEFMSKSLQELQIGTYA 201
Query: 338 KLGRQIAGL*QH*DQMLLLLQP*IY*FKFNIFFAAQVSSIPIVDESDSLLDIYCRSDITA 397
+ M+ P +Y IF +VS++P+VDE ++DIY + D+
Sbjct: 202 NIA------------MVRTTTP-VY-VALGIFVQHRVSALPVVDEKGRVVDIYSKFDVIN 247
Query: 398 LAKDRAYTHINL 409
LA ++ Y ++++
Sbjct: 248 LAAEKTYNNLDV 259
>AAKB_RAT (P80386) 5'-AMP-activated protein kinase, beta-1 subunit
(AMPK beta-1 chain) (AMPKb) (40 kDa subunit)
Length = 269
Score = 76.3 bits (186), Expect = 2e-13
Identities = 37/79 (46%), Positives = 45/79 (56%), Gaps = 4/79 (5%)
Query: 24 PVRFVWPYGGRTVYLSGSFTRWSELLQMSPVEGCPTVFQVIHNLAPGYHQYKFFVDGEWR 83
P F W GG+ VYLSGSF WS+L P+ F I +L G HQYKFFVDG+W
Sbjct: 78 PTVFRWTGGGKEVYLSGSFNNWSKL----PLTRSQNNFVAILDLPEGEHQYKFFVDGQWT 133
Query: 84 HDEHTPHITGDYGIVNTVL 102
HD P +T G VN ++
Sbjct: 134 HDPSEPIVTSQLGTVNNII 152
>AAKB_MOUSE (Q9R078) 5'-AMP-activated protein kinase, beta-1 subunit
(AMPK beta-1 chain) (AMPKb)
Length = 269
Score = 76.3 bits (186), Expect = 2e-13
Identities = 37/79 (46%), Positives = 45/79 (56%), Gaps = 4/79 (5%)
Query: 24 PVRFVWPYGGRTVYLSGSFTRWSELLQMSPVEGCPTVFQVIHNLAPGYHQYKFFVDGEWR 83
P F W GG+ VYLSGSF WS+L P+ F I +L G HQYKFFVDG+W
Sbjct: 78 PTVFRWTGGGKEVYLSGSFNNWSKL----PLTRSQNNFVAILDLPEGEHQYKFFVDGQWT 133
Query: 84 HDEHTPHITGDYGIVNTVL 102
HD P +T G VN ++
Sbjct: 134 HDPSEPIVTSQLGTVNNII 152
>AAKB_HUMAN (Q9Y478) 5'-AMP-activated protein kinase, beta-1 subunit
(AMPK beta-1 chain) (AMPKb)
Length = 269
Score = 75.9 bits (185), Expect = 2e-13
Identities = 37/79 (46%), Positives = 45/79 (56%), Gaps = 4/79 (5%)
Query: 24 PVRFVWPYGGRTVYLSGSFTRWSELLQMSPVEGCPTVFQVIHNLAPGYHQYKFFVDGEWR 83
P F W GG+ VYLSGSF WS+L P+ F I +L G HQYKFFVDG+W
Sbjct: 78 PTVFRWTGGGKEVYLSGSFNNWSKL----PLTRSHNNFVAILDLPEGEHQYKFFVDGQWT 133
Query: 84 HDEHTPHITGDYGIVNTVL 102
HD P +T G VN ++
Sbjct: 134 HDPSEPIVTSQLGTVNNII 152
>AAKB_PIG (P80387) 5'-AMP-activated protein kinase, beta-1 subunit
(AMPK beta-1 chain) (AMPKb) (40 kDa subunit) (Fragment)
Length = 122
Score = 73.6 bits (179), Expect = 1e-12
Identities = 36/79 (45%), Positives = 44/79 (55%), Gaps = 4/79 (5%)
Query: 24 PVRFVWPYGGRTVYLSGSFTRWSELLQMSPVEGCPTVFQVIHNLAPGYHQYKFFVDGEWR 83
P F W GG+ VYLSGSF WS+L P+ F I +L G HQYKF VDG+W
Sbjct: 44 PTVFRWTGGGKEVYLSGSFNNWSKL----PLTRSHNNFVAILDLPEGEHQYKFLVDGQWT 99
Query: 84 HDEHTPHITGDYGIVNTVL 102
HD P +T G VN ++
Sbjct: 100 HDPSEPVVTSQLGTVNNII 118
>SNF4_YEAST (P12904) Nuclear protein SNF4 (Regulatory protein CAT3)
Length = 322
Score = 72.0 bits (175), Expect = 3e-12
Identities = 73/308 (23%), Positives = 136/308 (43%), Gaps = 39/308 (12%)
Query: 150 VDVQTSRQRISTYLSMRTAYELLPESGKVVTLDVDLPVKQAFHILHEQGIPMAPLWDFCK 209
++ Q + + I +L+ +T+Y++LP S +++ LD L VK++ ++L + I APLWD
Sbjct: 13 IEQQLAVESIRKFLNSKTSYDVLPVSYRLIVLDTSLLVKKSLNVLLQNSIVSAPLWDSKT 72
Query: 210 GQFVGVLSVLDFILILRELGNHGSNLTEEELETHTISAWKEGKWTL----FSRRFIHAGP 265
+F G+L+ DFI +++ ++ E ++ + K+ + L IH P
Sbjct: 73 SRFAGLLTTTDFINVIQYYFSNPDKF--ELVDKLQLDGLKDIERALGVDQLDTASIH--P 128
Query: 266 SDNLKDVALKILQNGISTVPIIHSSSADGSFPQLLHLASLSGILR--CILGVVLVHCPYF 323
S L + LK+L++ +P+I + H + +L IL V ++C
Sbjct: 129 SRPLFEACLKMLESRSGRIPLIDQD-------EETHREIVVSVLTQYRILKFVALNCRET 181
Query: 324 NFQFVQFLWAHGCPKLGRQIAGL*QH*DQMLLLLQP*IY*FKFNIFFAAQVSSIPIVDES 383
+F + G + Q D M + +VSS+PI+DE+
Sbjct: 182 HFLKIPI----GDLNIITQ--------DNMKSCQMTTPVIDVIQMLTQGRVSSVPIIDEN 229
Query: 384 DSLLDIYCRSDITALAKDRAYTHINLDEMTVHQALQLSQDAFNPNESRSQRCQMCLRTDS 443
L+++Y D+ L K Y ++L +V +AL D F + C + D
Sbjct: 230 GYLINVYEAYDVLGLIKGGIYNDLSL---SVGEALMRRSDDF-------EGVYTCTKNDK 279
Query: 444 LHKVMERL 451
L +M+ +
Sbjct: 280 LSTIMDNI 287
>GA83_YEAST (Q04739) Glucose repression protein GAL83 (SPM1 protein)
Length = 417
Score = 67.4 bits (163), Expect = 7e-11
Identities = 32/88 (36%), Positives = 49/88 (55%), Gaps = 1/88 (1%)
Query: 21 VLIPVRFVWPYGGRTVYLSGSFTRWSELLQMSPVEGCPTVFQVIHNLAPGYHQYKFFVDG 80
++ PV W GG VY++GSFT W +++ + PV G P + V L PG H+++F VD
Sbjct: 160 MMFPVDITWQQGGNKVYVTGSFTGWRKMIGLVPVPGQPGLMHVKLQLPPGTHRFRFIVDN 219
Query: 81 EWRHDEHTPHITGDYG-IVNTVLLATDP 107
E R ++ P T G VN + ++ P
Sbjct: 220 ELRFSDYLPTATDQMGNFVNYMEVSAPP 247
>AAKC_RAT (Q9QZH4) 5'-AMP-activated protein kinase, beta-2 subunit
(AMPK beta-2 chain)
Length = 271
Score = 67.0 bits (162), Expect = 9e-11
Identities = 32/79 (40%), Positives = 43/79 (53%), Gaps = 3/79 (3%)
Query: 24 PVRFVWPYGGRTVYLSGSFTRWSELLQMSPVEGCPTVFQVIHNLAPGYHQYKFFVDGEWR 83
P W GG+ V++SGSF WS + P+ F I +L G HQYKFFVDG+W
Sbjct: 77 PTVIRWSEGGKEVFISGSFNNWSTKI---PLIKSHNDFVAILDLPEGEHQYKFFVDGQWV 133
Query: 84 HDEHTPHITGDYGIVNTVL 102
HD P +T G +N ++
Sbjct: 134 HDPSEPVVTSQLGTINNLI 152
>AAKC_HUMAN (O43741) 5'-AMP-activated protein kinase, beta-2 subunit
(AMPK beta-2 chain)
Length = 272
Score = 67.0 bits (162), Expect = 9e-11
Identities = 32/79 (40%), Positives = 43/79 (53%), Gaps = 3/79 (3%)
Query: 24 PVRFVWPYGGRTVYLSGSFTRWSELLQMSPVEGCPTVFQVIHNLAPGYHQYKFFVDGEWR 83
P W GG+ V++SGSF WS + P+ F I +L G HQYKFFVDG+W
Sbjct: 78 PTVIRWSEGGKEVFISGSFNNWSTKI---PLIKSHNDFVAILDLPEGEHQYKFFVDGQWV 134
Query: 84 HDEHTPHITGDYGIVNTVL 102
HD P +T G +N ++
Sbjct: 135 HDPSEPVVTSQLGTINNLI 153
>SNF4_KLULA (Q9P869) Nuclear protein SNF4
Length = 328
Score = 66.6 bits (161), Expect = 1e-10
Identities = 69/305 (22%), Positives = 140/305 (45%), Gaps = 34/305 (11%)
Query: 149 DVDVQTSRQRISTYLSMRTAYELLPESGKVVTLDVDLPVKQAFHILHEQGIPMAPLWDFC 208
+++ + + Q I +L +T+Y++LP S +++ LD L VK++ +IL + + APLWD
Sbjct: 17 ELEQKLAVQSIRVFLQSKTSYDVLPVSYRLIVLDTSLLVKKSLNILLQNNVVSAPLWDAQ 76
Query: 209 KGQFVGVLSVLDFILILRELGNHGSNLTEEELETHTISAWKEGKWTL----FSRRFIHAG 264
+F G+L+ DFI +++ ++ E ++ ++ K+ + + + R IH
Sbjct: 77 TSKFAGLLTSSDFINVIQYYFHNPDKF--ELVDKLQLNGLKDIERAIGIQPYDTRSIH-- 132
Query: 265 PSDNLKDVALKILQNGISTVPIIHSSSADGSFPQLLHLASLSGILRCILGVVLVHCPYFN 324
P L + +K++++ +P+I D + + ++ L+ IL V ++C
Sbjct: 133 PFRPLYEACVKMIESRSRRIPLI---DQDEETQREIVVSVLTQYR--ILKFVALNCKEIR 187
Query: 325 FQFVQFLWAHGCPKLGRQIAGL*QH*DQMLLLLQP*IY*FKFNIFFAAQVSSIPIVDESD 384
+ + R++ + + + P I + + A VSS+PIVDE
Sbjct: 188 Y----------LKRPLRELDIISTNNIMSCQMSTPVIDVIQL-LTLAGGVSSVPIVDEQG 236
Query: 385 SLLDIYCRSDITALAKDRAYTHINLDEMTVHQALQLSQDAFNPNESRSQRCQMCLRTDSL 444
L+++Y D+ L K Y ++L +V +AL D F + C D L
Sbjct: 237 KLVNVYEAVDVLGLIKGGIYNDLSL---SVGEALMRRSDDF-------EGVFTCTENDKL 286
Query: 445 HKVME 449
+++
Sbjct: 287 SSILD 291
>SIP2_YEAST (P34164) SIP2 protein (SPM2 protein)
Length = 414
Score = 59.7 bits (143), Expect = 2e-08
Identities = 27/77 (35%), Positives = 42/77 (54%)
Query: 20 TVLIPVRFVWPYGGRTVYLSGSFTRWSELLQMSPVEGCPTVFQVIHNLAPGYHQYKFFVD 79
++++PV W GG VY++GSFT+W +++ + P F V L PG H+++F VD
Sbjct: 160 SLMVPVEIRWQQGGSKVYVTGSFTKWRKMIGLIPDSDNNGSFHVKLRLLPGTHRFRFIVD 219
Query: 80 GEWRHDEHTPHITGDYG 96
E R + P T G
Sbjct: 220 NELRVSDFLPTATDQMG 236
>YL28_SCHPO (Q10343) Hypothetical protein C1556.08c in chromosome I
Length = 334
Score = 58.2 bits (139), Expect = 4e-08
Identities = 61/267 (22%), Positives = 117/267 (42%), Gaps = 26/267 (9%)
Query: 151 DVQTSRQ----RISTYLSMRTAYELLPESGKVVTLDVDLPVKQAFHILHEQGIPMAPLWD 206
DVQ +++ I ++ RT+Y++LP S +++ DV L VK + +L I APLWD
Sbjct: 3 DVQETQKGALKEIQAFIRSRTSYDVLPTSFRLIVFDVTLFVKTSLSLLTLNNIVSAPLWD 62
Query: 207 FCKGQFVGVLSVLDFILILRELGNHGSNLTE--EELETHTISAWK--EGKWTLFSRRFIH 262
+F G+L++ DF+ +++ S+ E E++ + + E K I+
Sbjct: 63 SEANKFAGLLTMADFVNVIKYY-YQSSSFPEAIAEIDKFRLLGLREVERKIGAIPPETIY 121
Query: 263 AGPSDNLKDVALKILQNGISTVPIIHSSSADGSFPQLLHLASLSGILRCILGVVLVHCPY 322
P +L D L + ++ +P+I GS + L IL + ++C
Sbjct: 122 VHPMHSLMDACLAMSKSRARRIPLIDVDGETGSEMIVSVLTQYR-----ILKFISMNCKE 176
Query: 323 FNFQFVQFLWAHGCPKLGRQIAGL*QH*DQMLLLLQP*IY*FKFNIFFAAQVSSIPIVDE 382
V L + G + ++ +Y + +S++PIV+
Sbjct: 177 TAMLRV---------PLNQMTIGTWS--NLATASMETKVY-DVIKMLAEKNISAVPIVNS 224
Query: 383 SDSLLDIYCRSDITALAKDRAYTHINL 409
+LL++Y D+ L +D Y++++L
Sbjct: 225 EGTLLNVYESVDVMHLIQDGDYSNLDL 251
>YNR3_YEAST (P53885) Hypothetical 40.3 kDa protein in RPS3-PSD1
intergenic region
Length = 366
Score = 35.8 bits (81), Expect = 0.23
Identities = 22/82 (26%), Positives = 32/82 (38%), Gaps = 3/82 (3%)
Query: 27 FVWPYGGRTVYLSGSFTRWSELLQM--SPVEGCPTVFQVIHNLAPGYHQYKFFVDGEWRH 84
F WP G + L+G+F W L M P V + +KF VDG+W
Sbjct: 10 FKWPKGPEAIILTGTFDDWKGTLPMVKDPSGAFEITLPVTFDSPSSKFYFKFIVDGQWLP 69
Query: 85 DEHTPHITGDYGIVNTVLLATD 106
+ + D G+ N + D
Sbjct: 70 SKDY-KVNIDEGVENNFITEED 90
>Y868_METJA (Q58278) Hypothetical protein MJ0868
Length = 127
Score = 35.0 bits (79), Expect = 0.40
Identities = 28/104 (26%), Positives = 50/104 (47%), Gaps = 21/104 (20%)
Query: 125 DNETFQRVVRLTDGTLSEVMPRISDVDVQTSRQRISTYLSMRTAYELLPESGKVVTLDVD 184
D ETF ++ TD V+ +D+D +TA E++ + +T+ +
Sbjct: 43 DGETFWGIITDTD-----VLKHYNDLD--------------KTAEEIMTTNP--ITVSPE 81
Query: 185 LPVKQAFHILHEQGIPMAPLWDFCKGQFVGVLSVLDFILILREL 228
P+++A I+ E+GI + C+ + VGVLS D I + +L
Sbjct: 82 APLEKAVEIMAEKGIHHLYVKSPCEDKIVGVLSSKDIIKLFSDL 125
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.328 0.142 0.446
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 53,270,600
Number of Sequences: 164201
Number of extensions: 2259526
Number of successful extensions: 5964
Number of sequences better than 10.0: 32
Number of HSP's better than 10.0 without gapping: 20
Number of HSP's successfully gapped in prelim test: 12
Number of HSP's that attempted gapping in prelim test: 5906
Number of HSP's gapped (non-prelim): 52
length of query: 454
length of database: 59,974,054
effective HSP length: 114
effective length of query: 340
effective length of database: 41,255,140
effective search space: 14026747600
effective search space used: 14026747600
T: 11
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.8 bits)
S2: 67 (30.4 bits)
Medicago: description of AC146552.9


