

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146549.11 + phase: 0
(597 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
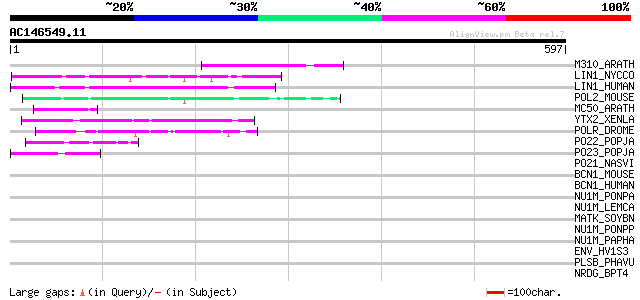
Score E
Sequences producing significant alignments: (bits) Value
M310_ARATH (P93295) Hypothetical mitochondrial protein AtMg00310... 87 9e-17
LIN1_NYCCO (P08548) LINE-1 reverse transcriptase homolog 78 8e-14
LIN1_HUMAN (P08547) LINE-1 reverse transcriptase homolog 74 8e-13
POL2_MOUSE (P11369) Retrovirus-related Pol polyprotein [Contains... 67 2e-10
MC50_ARATH (P92555) Hypothetical mitochondrial protein AtMg01250... 59 3e-08
YTX2_XENLA (P14381) Transposon TX1 hypothetical 149 kDa protein ... 52 4e-06
POLR_DROME (P16423) Retrovirus-related Pol polyprotein from type... 52 4e-06
PO22_POPJA (Q03274) Retrovirus-related Pol polyprotein from type... 49 5e-05
PO23_POPJA (Q05118) Retrovirus-related Pol polyprotein from type... 46 3e-04
PO21_NASVI (Q03278) Retrovirus-related Pol polyprotein from type... 35 0.73
BCN1_MOUSE (O88597) Beclin 1 (Coiled-coil myosin-like BCL2-inter... 35 0.73
BCN1_HUMAN (Q14457) Beclin 1 (Coiled-coil myosin-like BCL2-inter... 35 0.73
NU1M_PONPA (P92690) NADH-ubiquinone oxidoreductase chain 1 (EC 1... 33 2.1
NU1M_LEMCA (O78696) NADH-ubiquinone oxidoreductase chain 1 (EC 1... 33 2.1
MATK_SOYBN (Q9TKS6) Maturase K (Intron maturase) 33 2.8
NU1M_PONPP (P92718) NADH-ubiquinone oxidoreductase chain 1 (EC 1... 32 3.6
NU1M_PAPHA (Q9ZXY4) NADH-ubiquinone oxidoreductase chain 1 (EC 1... 32 3.6
ENV_HV1S3 (P19549) Envelope polyprotein GP160 precursor [Contain... 32 4.7
PLSB_PHAVU (Q43822) Glycerol-3-phosphate acyltransferase, chloro... 32 6.2
NRDG_BPT4 (P07075) Anaerobic ribonucleoside-triphosphate reducta... 32 6.2
>M310_ARATH (P93295) Hypothetical mitochondrial protein AtMg00310
(ORF154)
Length = 154
Score = 87.4 bits (215), Expect = 9e-17
Identities = 54/155 (34%), Positives = 77/155 (48%), Gaps = 10/155 (6%)
Query: 207 SLPVYALSFFRAPAGIISSIESILIKFFWGGSEDHRKIAWVDWNTICMNKE-AGGLGVRR 265
+LPVYA+S FR + + S + +F+W E+ RKI+WV W +C +KE GGLG R
Sbjct: 2 ALPVYAMSCFRLSKLLCKKLTSAMTEFWWSSCENKRKISWVAWQKLCKSKEDDGGLGFRD 61
Query: 266 LLEFNVALLGKWCWRCLVDREGLWFRVLSSRYGEQGGRLK-EGGMTGSAWWREIIKIQNG 324
L FN ALL K +R + L R+L SRY ++ G S WR II
Sbjct: 62 LGWFNQALLAKQSFRIIHQPHTLLSRLLRSRYFPHSSMMECSVGTRPSYAWRSII----- 116
Query: 325 VGVEGGSWFEESISKRLGNGFNTFFWSDCWLGTES 359
G + + +G+G +T W D W+ E+
Sbjct: 117 ---HGRELLSRGLLRTIGDGIHTKVWLDRWIMDET 148
>LIN1_NYCCO (P08548) LINE-1 reverse transcriptase homolog
Length = 1260
Score = 77.8 bits (190), Expect = 8e-14
Identities = 79/303 (26%), Positives = 140/303 (46%), Gaps = 32/303 (10%)
Query: 3 KMGFPTLWRKWMKECVSTATVSVLVNGSPTDEFYMQRGLRQGDPLSPFLFLLAAEGLNVL 62
K+G + K ++ S T ++++NG F ++ G RQG PLSP LF + E VL
Sbjct: 618 KIGIEGTFLKLIEAIYSKPTANIILNGVKLKSFPLRSGTRQGCPLSPLLFNIVME---VL 674
Query: 63 MTSAVNSNLFTGYSIGMHSPTVLSHLQFADDTLLLGVKSWANVRALRSILVIFENMSGLK 122
+ G IG + FADD ++ + + L ++ + N+SG K
Sbjct: 675 AIAIREEKAIKGIHIGSEE---IKLSLFADDMIVYLENTRDSTTKLLEVIKEYSNVSGYK 731
Query: 123 VNYNKS---LLVGINIAESWLQEAASILSCKLGKTPFMYLGLPIGGDARRLCFWDPVLER 179
+N +KS + N AE ++++ + + YLG+ + D + L + E
Sbjct: 732 INTHKSVAFIYTNNNQAEKTVKDS---IPFTVVPKKMKYLGVYLTKDVKDL--YKENYET 786
Query: 180 LRSRLSE----WKSRNLSYGGRLILLKSVLSSLPVYALSF----FRAPAGIISSIESILI 231
LR ++E WK+ S+ GR+ ++K +S LP +F +AP +E I++
Sbjct: 787 LRKEIAEDVNKWKNIPCSWLGRINIVK--MSILPKAIYNFNAIPIKAPLSYFKDLEKIIL 844
Query: 232 KFFWGGSEDHRKIAWVDWNTICMNK-EAGGLGVRRLLEFNVALLGKWCWRCLVDRE-GLW 289
F W ++ +IA T+ NK +AGG+ + L + +++ K W +RE +W
Sbjct: 845 HFIW--NQKKPQIA----KTLLSNKNKAGGITLPDLRLYYKSIVIKTAWYWHKNREVDVW 898
Query: 290 FRV 292
R+
Sbjct: 899 NRI 901
>LIN1_HUMAN (P08547) LINE-1 reverse transcriptase homolog
Length = 1259
Score = 74.3 bits (181), Expect = 8e-13
Identities = 64/290 (22%), Positives = 122/290 (42%), Gaps = 15/290 (5%)
Query: 1 MGKMGFPTLWRKWMKECVSTATVSVLVNGSPTDEFYMQRGLRQGDPLSPFLFLLAAEGLN 60
+ K+G + K ++ T ++++NG + ++ G RQG PLSP LL L
Sbjct: 616 LNKLGIDGTYLKIIRAIYDKPTANIILNGQKLEAPPLKTGTRQGCPLSP---LLPNIVLE 672
Query: 61 VLMTSAVNSNLFTGYSIGMHSPTVLSHLQFADDTLLLGVKSWANVRALRSILVIFENMSG 120
VL + G +G + FADD ++ + + L ++ F +SG
Sbjct: 673 VLARAIRQEKEIKGIQLGKEE---VKLSLFADDMIVYLENPIVSAQNLLKLISNFSKVSG 729
Query: 121 LKVNYNKSLLVGINIAESWLQEAASILSCKLGKTPFMYLGLPIGGDARRLC--FWDPVLE 178
K+N KS + S L + YLG+ + D + L + P+L
Sbjct: 730 YKINVQKSQAFLYTNNRQTESQIMSELPFTIASKRIKYLGIQLTRDVKDLFKENYKPLLN 789
Query: 179 RLRSRLSEWKSRNLSYGGRLILLKSVLSSLPVYALSF--FRAPAGIISSIESILIKFFWG 236
++ ++WK+ S+ GR+ ++K + +Y + + P + +E +KF W
Sbjct: 790 EIKEDTNKWKNIPCSWVGRINIVKMAILPKVIYRFNAIPIKLPMTFFTELEKTTLKFIW- 848
Query: 237 GSEDHRKIAWVDWNTICMNKEAGGLGVRRLLEFNVALLGKWCWRCLVDRE 286
++K A + +T+ +AGG+ + + A + K W +R+
Sbjct: 849 ----NQKRAHIAKSTLSQKNKAGGITLPDFKLYYKATVTKTAWYWYQNRD 894
>POL2_MOUSE (P11369) Retrovirus-related Pol polyprotein [Contains:
Reverse transcriptase (EC 2.7.7.49); Endonuclease]
Length = 1300
Score = 66.6 bits (161), Expect = 2e-10
Identities = 75/348 (21%), Positives = 138/348 (39%), Gaps = 42/348 (12%)
Query: 14 MKECVSTATVSVLVNGSPTDEFYMQRGLRQGDPLSPFLFLLAAEGLNVLMTSAVNSNLFT 73
+K S ++ VNG + ++ G RQG PLSP+LF + L VL +
Sbjct: 656 IKAIYSKPVANIKVNGEKLEAIPLKSGTRQGCPLSPYLFNIV---LEVLARAIRQQKEIK 712
Query: 74 GYSIGMHSPTVLSHLQFADDTLLLGVKSWANVRALRSILVIFENMSGLKVNYNKSLLVGI 133
G IG + ADD ++ + R L +++ F + G K+N NKS+
Sbjct: 713 GIQIGKEEVKI---SLLADDMIVYISDPKNSTRELLNLINSFGEVVGYKINSNKSMAFLY 769
Query: 134 NIAESWLQEAASILSCKLGKTPFMYLGLPIGGDARRLCFWDPVLERLRSRLSE----WKS 189
+ +E + YLG+ + + + L +D + L+ + E WK
Sbjct: 770 TKNKQAEKEIRETTPFSIVTNNIKYLGVTLTKEVKDL--YDKNFKSLKKEIKEDLRRWKD 827
Query: 190 RNLSYGGRLILLKSVLSSLPVYALSF--FRAPAGIISSIESILIKFFWGGSEDHRKIAWV 247
S+ GR+ ++K + +Y + + P + +E + KF W + +
Sbjct: 828 LPCSWIGRINIVKMAILPKAIYRFNAIPIKIPTQFFNELEGAICKFVWNNKKPR-----I 882
Query: 248 DWNTICMNKEAGGLGVRRLLEFNVALLGKWCWRCLVDREGLWFRVLSSRYGEQGGRLKEG 307
+ + + +GG+ + L + A++ K W W+R R +Q R+++
Sbjct: 883 AKSLLKDKRTSGGITMPDLKLYYRAIVIKTAW--------YWYR---DRQVDQWNRIEDP 931
Query: 308 GMTGSAWWREIIKIQNGVGVEGGSWFEESISKRLGNGFNTFFWSDCWL 355
M + I G + W ++SI FN + W + WL
Sbjct: 932 EMNPHTYGHLIF----DKGAKTIQWKKDSI-------FNNWCWHN-WL 967
>MC50_ARATH (P92555) Hypothetical mitochondrial protein AtMg01250
(ORF102)
Length = 122
Score = 59.3 bits (142), Expect = 3e-08
Identities = 31/69 (44%), Positives = 41/69 (58%), Gaps = 1/69 (1%)
Query: 26 LVNGSPTDEFYMQRGLRQGDPLSPFLFLLAAEGLNVLMTSAVNSNLFTGYSIGMHSPTVL 85
++NG+P RGLRQGDPLSP+LF+L E L+ L A G + +SP +
Sbjct: 13 IINGAPQGLVTPSRGLRQGDPLSPYLFILCTEVLSGLCRRAQEQGRLPGIRVSNNSPRI- 71
Query: 86 SHLQFADDT 94
+HL FADDT
Sbjct: 72 NHLLFADDT 80
>YTX2_XENLA (P14381) Transposon TX1 hypothetical 149 kDa protein
(ORF 2)
Length = 1308
Score = 52.0 bits (123), Expect = 4e-06
Identities = 60/256 (23%), Positives = 111/256 (42%), Gaps = 21/256 (8%)
Query: 13 WMKECVSTATVSVLVNGSPTDEFYMQRGLRQGDPLSPFLFLLAAEGLNVLMTSAVNSNLF 72
++K ++A V +N S T RG+RQG PLS L+ LA E L+ +
Sbjct: 623 YLKTMYASAECLVKINWSLTAPLAFGRGVRQGCPLSGQLYSLAIEPFLCLLRKRLT---- 678
Query: 73 TGYSIGMHSPTVLSHLQFADDTLLLGVKSWANVRALRSILVIFENMSGLKVNYNKS--LL 130
+ + P + L D ++L + ++ + ++ S ++N++KS LL
Sbjct: 679 ---GLVLKEPDMRVVLSAYADDVILVAQDLVDLERAQECQEVYAAASSARINWSKSSGLL 735
Query: 131 VGINIAESWLQEAASILSCKLGKTPFMYLGLPIGGDARRLC-FWDPVLERLRSRLSEWK- 188
G ++ +L A +S + YLG+ + + + + + E + +RL +WK
Sbjct: 736 EG-SLKVDFLPPAFRDISWE--SKIIKYLGVYLSAEEYPVSQNFIELEECVLTRLGKWKG 792
Query: 189 -SRNLSYGGRLILLKSVLSSLPVYALSFFRAPAGIISSIESILIKFFWGGSEDHRKIAWV 247
++ LS GR +++ +++S Y L I+ I+ L+ F W G WV
Sbjct: 793 FAKVLSMRGRALVINQLVASQIWYRLICLSPTQEFIAKIQRRLLDFLWIGKH------WV 846
Query: 248 DWNTICMNKEAGGLGV 263
+ + GG GV
Sbjct: 847 SAGVSSLPLKEGGQGV 862
>POLR_DROME (P16423) Retrovirus-related Pol polyprotein from type II
retrotransposable element R2DM [Contains: Protease (EC
3.4.23.-); Reverse transcriptase (EC 2.7.7.49);
Endonuclease]
Length = 1057
Score = 52.0 bits (123), Expect = 4e-06
Identities = 65/252 (25%), Positives = 110/252 (42%), Gaps = 33/252 (13%)
Query: 28 NGSPTDEFYMQRGLRQGDPLSPFLFLLAAEGLNVLMTSAVNSNLFTGYSIGMHSPTVLSH 87
+G ++EF RG++QGDPLSP LF L + L + S + + + + +
Sbjct: 526 DGWSSEEFVPARGVKQGDPLSPILFNLVMDRLLRTLPSEIGAKV---------GNAITNA 576
Query: 88 LQFADDTLLLGVKSWANVRALRSILVIFENMSGLKVNYNKSLLVGIN----------IAE 137
FADD L+L ++ ++ L + F ++ GLK+N +K VGI A+
Sbjct: 577 AAFADD-LVLFAETRMGLQVLLDKTLDFLSIVGLKLNADKCFTVGIKGQPKQKCTVLEAQ 635
Query: 138 SWLQEAASILSCKLGKTPFMYLGLPIGGDARRLCFWDPVLERLRSRLSEWKSRNLSYGGR 197
S+ ++ I S K + YLG+ R C +P E + +L L R
Sbjct: 636 SFYVGSSEIPSLK-RTDEWKYLGINFTATGRVRC--NPA-EDIGPKLQRLTKAPLKPQQR 691
Query: 198 LILLKSVLSSLPVYALSFFRAPAGIISSIESILIKFF---WGGSEDHRKIAWVDWNTICM 254
L L++VL + L+ G++ + LI+++ W IA+V
Sbjct: 692 LFALRTVLIPQLYHKLALGSVAIGVLRKTDK-LIRYYVRRWLNLPLDVPIAFVH-----A 745
Query: 255 NKEAGGLGVRRL 266
++GGLG+ L
Sbjct: 746 PPKSGGLGIPSL 757
>PO22_POPJA (Q03274) Retrovirus-related Pol polyprotein from type I
retrotransposable element R2 [Contains: Reverse
transcriptase (EC 2.7.7.49); Endonuclease] (Fragment)
Length = 711
Score = 48.5 bits (114), Expect = 5e-05
Identities = 42/122 (34%), Positives = 63/122 (51%), Gaps = 11/122 (9%)
Query: 18 VSTATVSVLVN-GSPTDEFYMQRGLRQGDPLSPFLFLLAAEGLNVLMTSAVNSNLFTGYS 76
+S +T ++ V GS T + ++RG++QGDPLSPFLF + L ++ S G +
Sbjct: 177 LSDSTTTIRVGPGSQTRKICIRRGVKQGDPLSPFLFNAVLDEL----LCSLQSTPGIGGT 232
Query: 77 IGMHSPTVLSHLQFADDTLLLGVKSWANVRALRSILVIFENMSGLKVNYNKSLLVGINIA 136
IG VL+ FADD LLL L ++ F + G+ +N KS V I++A
Sbjct: 233 IGEEKIPVLA---FADDLLLLEDNDVLLPTTLATVANFF-RLRGMSLNAKKS--VSISVA 286
Query: 137 ES 138
S
Sbjct: 287 AS 288
>PO23_POPJA (Q05118) Retrovirus-related Pol polyprotein from type I
retrotransposable element R2 [Contains: Reverse
transcriptase (EC 2.7.7.49); Endonuclease] (Fragment)
Length = 606
Score = 45.8 bits (107), Expect = 3e-04
Identities = 28/97 (28%), Positives = 47/97 (47%), Gaps = 7/97 (7%)
Query: 1 MGKMGFPTLWRKWMKECVSTATVSVLVNGSPTDEFYMQRGLRQGDPLSPFLFLLAAEGLN 60
M G + ++ ++ A +++V G T++ Y++ G++QGDPLSP LF N
Sbjct: 46 MRAFGIDDGMQDFIMSTITDAYTNIVVGGRTTNKIYIRNGVKQGDPLSPVLF-------N 98
Query: 61 VLMTSAVNSNLFTGYSIGMHSPTVLSHLQFADDTLLL 97
+++ V M ++ L FADD LLL
Sbjct: 99 IVLDELVTRLNDEQPGASMTPACKIASLAFADDLLLL 135
>PO21_NASVI (Q03278) Retrovirus-related Pol polyprotein from type I
retrotransposable element R2 [Contains: Reverse
transcriptase (EC 2.7.7.49); Endonuclease] (Fragment)
Length = 1025
Score = 34.7 bits (78), Expect = 0.73
Identities = 25/59 (42%), Positives = 31/59 (52%), Gaps = 9/59 (15%)
Query: 39 RGLRQGDPLSPFLFLLAAEGLNVLMTSAVNSNLFTGYSIGMHSPTVLSHLQFADDTLLL 97
RG+RQGDPLSP LF + VL N TG+ +G + L FADD +LL
Sbjct: 511 RGVRQGDPLSPLLFNCVMDA--VLRRLPEN----TGFLMGAEK---IGALVFADDLVLL 560
>BCN1_MOUSE (O88597) Beclin 1 (Coiled-coil myosin-like
BCL2-interacting protein)
Length = 448
Score = 34.7 bits (78), Expect = 0.73
Identities = 22/85 (25%), Positives = 39/85 (45%), Gaps = 13/85 (15%)
Query: 177 LERLRSRLSEWKSRNLSYGGRLILLKSVLSSLPVYALSFFRAPAGIISSIESIL------ 230
L RL S EW N ++G ++LL ++ + + + + P G S +ES+
Sbjct: 288 LGRLPSVPVEWNEINAAWGQTVLLLHALANKMGLKFQRYRLVPYGNHSYLESLTDKSKEL 347
Query: 231 -------IKFFWGGSEDHRKIAWVD 248
++FFW DH +A++D
Sbjct: 348 PLYCSGGLRFFWDNKFDHAMVAFLD 372
>BCN1_HUMAN (Q14457) Beclin 1 (Coiled-coil myosin-like
BCL2-interacting protein) (Protein GT197)
Length = 450
Score = 34.7 bits (78), Expect = 0.73
Identities = 22/85 (25%), Positives = 39/85 (45%), Gaps = 13/85 (15%)
Query: 177 LERLRSRLSEWKSRNLSYGGRLILLKSVLSSLPVYALSFFRAPAGIISSIESIL------ 230
L RL S EW N ++G ++LL ++ + + + + P G S +ES+
Sbjct: 290 LGRLPSVPVEWNEINAAWGQTVLLLHALANKMGLKFQRYRLVPYGNHSYLESLTDKSKEL 349
Query: 231 -------IKFFWGGSEDHRKIAWVD 248
++FFW DH +A++D
Sbjct: 350 PLYCSGGLRFFWDNKFDHAMVAFLD 374
>NU1M_PONPA (P92690) NADH-ubiquinone oxidoreductase chain 1 (EC
1.6.5.3)
Length = 318
Score = 33.1 bits (74), Expect = 2.1
Identities = 17/49 (34%), Positives = 28/49 (56%), Gaps = 1/49 (2%)
Query: 49 PFLFLLAAEGLNVLMTSAVNSNLFTGYSIGMHSPTVLSHLQFADDTLLL 97
PF AE N+++ +A+ + +F G + +HSP + + L F TLLL
Sbjct: 219 PFALFFMAEYTNIILMNALTTMIFLGTTFNIHSPELYTTL-FTIKTLLL 266
>NU1M_LEMCA (O78696) NADH-ubiquinone oxidoreductase chain 1 (EC
1.6.5.3)
Length = 318
Score = 33.1 bits (74), Expect = 2.1
Identities = 18/51 (35%), Positives = 26/51 (50%), Gaps = 1/51 (1%)
Query: 49 PFLFLLAAEGLNVLMTSAVNSNLFTGYSIGMHSPTVLSHLQFADDTLLLGV 99
PF AE N++M +A+ + LF G +H P + FA TLLL +
Sbjct: 219 PFALFFMAEYTNIIMMNALTTTLFLGALYNLHMPETYT-TSFAIKTLLLTI 268
>MATK_SOYBN (Q9TKS6) Maturase K (Intron maturase)
Length = 505
Score = 32.7 bits (73), Expect = 2.8
Identities = 26/103 (25%), Positives = 47/103 (45%), Gaps = 19/103 (18%)
Query: 61 VLMTSAVNSNLFTGYSIGMHSPTVLS----------HLQFADDTL--LLGVKSWANVRAL 108
++ T+ N N F GY+ +S +L LQF +L VKS+ N+R++
Sbjct: 68 IIFTNDSNKNPFMGYNNHFYSQIILEGFVVVVEILFSLQFCISSLREFEIVKSYNNLRSI 127
Query: 109 RSILVIFENM-------SGLKVNYNKSLLVGINIAESWLQEAA 144
SI FE+ S +++ Y L + + I W+++ +
Sbjct: 128 HSIFPFFEDKLIYLNHESDIRIPYPIHLEILVQILRYWIKDVS 170
>NU1M_PONPP (P92718) NADH-ubiquinone oxidoreductase chain 1 (EC
1.6.5.3)
Length = 318
Score = 32.3 bits (72), Expect = 3.6
Identities = 17/49 (34%), Positives = 28/49 (56%), Gaps = 1/49 (2%)
Query: 49 PFLFLLAAEGLNVLMTSAVNSNLFTGYSIGMHSPTVLSHLQFADDTLLL 97
PF AE N+++ +A+ + +F G + +HSP + + L F TLLL
Sbjct: 219 PFALFFMAEYTNIILMNALTAMIFLGTTFNIHSPELHTTL-FTIKTLLL 266
>NU1M_PAPHA (Q9ZXY4) NADH-ubiquinone oxidoreductase chain 1 (EC
1.6.5.3)
Length = 318
Score = 32.3 bits (72), Expect = 3.6
Identities = 17/49 (34%), Positives = 26/49 (52%), Gaps = 1/49 (2%)
Query: 49 PFLFLLAAEGLNVLMTSAVNSNLFTGYSIGMHSPTVLSHLQFADDTLLL 97
PF AE +N++M +A+ + +F G HSP + + F TLLL
Sbjct: 219 PFALFFMAEYMNIIMMNALTTTIFLGTFYPTHSPELFT-TSFTIKTLLL 266
>ENV_HV1S3 (P19549) Envelope polyprotein GP160 precursor [Contains:
Exterior membrane glycoprotein (GP120); Transmembrane
glycoprotein (GP41)]
Length = 852
Score = 32.0 bits (71), Expect = 4.7
Identities = 26/80 (32%), Positives = 33/80 (40%), Gaps = 8/80 (10%)
Query: 328 EGGSWFEESISKRLGNGFNTFFWSDCWLGTESFMVRFRRLYDLSIHKDLSVGEMNILGWG 387
EGG + S RL NGF FW D + + RL DL + V + GW
Sbjct: 731 EGGGERDRDRSTRLVNGFLALFWDDL---RSLCLFSYHRLTDLLLIVARIVELLGRRGW- 786
Query: 388 ENGEAWR-WRRRLLAWEEEL 406
E + W LL W +EL
Sbjct: 787 ---EVLKYWWNLLLYWSQEL 803
>PLSB_PHAVU (Q43822) Glycerol-3-phosphate acyltransferase,
chloroplast precursor (EC 2.3.1.15) (GPAT)
Length = 461
Score = 31.6 bits (70), Expect = 6.2
Identities = 17/47 (36%), Positives = 26/47 (55%)
Query: 75 YSIGMHSPTVLSHLQFADDTLLLGVKSWANVRALRSILVIFENMSGL 121
+SIG + V S DD L+ +K AN+RAL+ + ++ N S L
Sbjct: 272 FSIGRNLICVYSKKHMLDDPALVEMKRTANIRALKEMAMLLRNGSQL 318
>NRDG_BPT4 (P07075) Anaerobic ribonucleoside-triphosphate reductase
activating protein (EC 1.97.1.4) (Class III anaerobic
ribonucleotide reductase small component)
Length = 156
Score = 31.6 bits (70), Expect = 6.2
Identities = 22/80 (27%), Positives = 35/80 (43%), Gaps = 9/80 (11%)
Query: 409 EIRILLTNVTLQDTESDVWLWRPNIGDGYTVRGVYQMLMRQERHNYDVVSDAPWHKSAPL 468
EI L++ V + E D+WLW GY + Q+ M + DV+ D + K+ P
Sbjct: 82 EISNLVSWVKARFPEKDIWLW-----TGYKFEDIKQLEMLK---YVDVIIDGKYEKNLPT 133
Query: 469 KVSIWAWRLLRNRWPTKDNL 488
K +W + W D +
Sbjct: 134 K-KLWRGSDNQRLWSNTDGV 152
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.324 0.140 0.463
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 75,023,569
Number of Sequences: 164201
Number of extensions: 3259411
Number of successful extensions: 7902
Number of sequences better than 10.0: 23
Number of HSP's better than 10.0 without gapping: 12
Number of HSP's successfully gapped in prelim test: 11
Number of HSP's that attempted gapping in prelim test: 7862
Number of HSP's gapped (non-prelim): 36
length of query: 597
length of database: 59,974,054
effective HSP length: 116
effective length of query: 481
effective length of database: 40,926,738
effective search space: 19685760978
effective search space used: 19685760978
T: 11
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.5 bits)
S2: 69 (31.2 bits)
Medicago: description of AC146549.11


