

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146548.3 + phase: 0
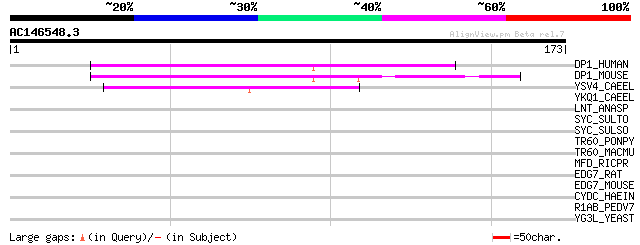
(173 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
DP1_HUMAN (Q00765) Polyposis locus protein 1 (TB2 protein) 70 2e-12
DP1_MOUSE (Q60870) Polyposis locus protein 1 homolog (TB2 protei... 68 1e-11
YSV4_CAEEL (Q10010) Hypothetical protein T19C3.4 in chromosome III 57 2e-08
YKQ1_CAEEL (P34296) Hypothetical protein C06E1.1 in chromosome III 38 0.010
LNT_ANASP (Q8YYI9) Apolipoprotein N-acyltransferase (EC 2.3.1.-)... 32 0.72
SYC_SULTO (Q96YC3) Cysteinyl-tRNA synthetase (EC 6.1.1.16) (Cyst... 31 1.6
SYC_SULSO (Q97WE6) Cysteinyl-tRNA synthetase (EC 6.1.1.16) (Cyst... 30 2.1
TR60_PONPY (Q645U7) Taste receptor type 2 member 60 (T2R60) 30 2.7
TR60_MACMU (Q645S2) Taste receptor type 2 member 60 (T2R60) 30 2.7
MFD_RICPR (O05955) Transcription-repair coupling factor (TRCF) 30 2.7
EDG7_RAT (Q8K5E0) Lysophosphatidic acid receptor Edg-7 (LPA rece... 30 2.7
EDG7_MOUSE (Q9EQ31) Lysophosphatidic acid receptor Edg-7 (LPA re... 29 4.7
CYDC_HAEIN (P45081) Transport ATP-binding protein cydC 29 4.7
R1AB_PEDV7 (Q91AV2) Replicase polyprotein 1ab (pp1ab) (ORF1ab po... 29 6.1
YG3L_YEAST (P48236) Hypothetical 51.6 kDa protein in RPL24B-RSR1... 28 8.0
>DP1_HUMAN (Q00765) Polyposis locus protein 1 (TB2 protein)
Length = 185
Score = 70.1 bits (170), Expect = 2e-12
Identities = 39/115 (33%), Positives = 55/115 (46%), Gaps = 1/115 (0%)
Query: 26 VISLVYPLYASVRAIESKSPVDDQQWLTYWVLYSLITLFELTFAKILEWIPIWPYAKLIL 85
+I YP Y S++AIES + DD QWLTYWV+Y + ++ E L W P + K
Sbjct: 57 LIGFGYPAYISIKAIESPNKEDDTQWLTYWVVYGVFSIAEFFSDIFLSWFPFYYMLKCGF 116
Query: 86 TCWLVLPY-FTGAAYVYEHYVRPFLANPQTINIWYVPRKKDVFTKQDDIITAAEK 139
W + P GA +Y+ +RPF ++ V KD + D IT K
Sbjct: 117 LLWCMAPSPSNGAELLYKRIIRPFFLKHESQMDSVVKDLKDKAKETADAITKEAK 171
>DP1_MOUSE (Q60870) Polyposis locus protein 1 homolog (TB2 protein
homolog) (GP106)
Length = 185
Score = 67.8 bits (164), Expect = 1e-11
Identities = 45/136 (33%), Positives = 65/136 (47%), Gaps = 10/136 (7%)
Query: 26 VISLVYPLYASVRAIESKSPVDDQQWLTYWVLYSLITLFELTFAKILEWIPIWPYAKLIL 85
+I YP Y S++AIES + DD QWLTYWV+Y + ++ E L W+P + K
Sbjct: 57 LIGFGYPAYISMKAIESPNKDDDTQWLTYWVVYGVFSIAEFFSDLFLSWLPFYYMLKCGF 116
Query: 86 TCWLVLPY-FTGAAYVYEHYVRP-FLANPQTINIWYVPRKKDVFTKQDDIITAAEKYIKE 143
W + P GA +Y +RP FL + ++ KDV K + A K +K
Sbjct: 117 LLWCMAPSPANGAEMLYRRIIRPIFLRHESQVD----SVVKDVKDKAKETADAISKEVK- 171
Query: 144 NGTEAFENLIHRADKS 159
+A NL+ KS
Sbjct: 172 ---KATVNLLGDVKKS 184
>YSV4_CAEEL (Q10010) Hypothetical protein T19C3.4 in chromosome III
Length = 229
Score = 57.0 bits (136), Expect = 2e-08
Identities = 23/81 (28%), Positives = 45/81 (55%), Gaps = 1/81 (1%)
Query: 30 VYPLYASVRAIESKSPVDDQQWLTYWVLYSLITLFELTFAKILE-WIPIWPYAKLILTCW 88
+YP Y S +A+ +K + +W+ YW+++++ + E +L W P + K++ W
Sbjct: 16 LYPAYRSYKAVRTKDTREYVKWMMYWIVFAIYSFLENLLDLVLAFWFPFYFQLKIVFIFW 75
Query: 89 LVLPYFTGAAYVYEHYVRPFL 109
L+ P+ GA+ +Y +V P L
Sbjct: 76 LLSPWTKGASILYRKWVHPTL 96
>YKQ1_CAEEL (P34296) Hypothetical protein C06E1.1 in chromosome III
Length = 161
Score = 38.1 bits (87), Expect = 0.010
Identities = 23/72 (31%), Positives = 42/72 (57%), Gaps = 6/72 (8%)
Query: 12 LMVLLRNFDVLAGPVISLVYPLYASVRAI-ESKSPVDDQQWLTYWVLYSLITLFELTFAK 70
+ VLL NF L +I + +P+ S + + K+P D+ L YW+LY++++LF+ +
Sbjct: 53 IYVLLGNFLPLFSHIICIFWPVKESFMILRQQKNPSDNI--LLYWILYAMVSLFDYS--- 107
Query: 71 ILEWIPIWPYAK 82
L +P + +AK
Sbjct: 108 ALPGVPFYYFAK 119
>LNT_ANASP (Q8YYI9) Apolipoprotein N-acyltransferase (EC 2.3.1.-)
(ALP N-acyltransferase)
Length = 500
Score = 32.0 bits (71), Expect = 0.72
Identities = 13/47 (27%), Positives = 25/47 (52%), Gaps = 2/47 (4%)
Query: 52 LTYWVLYSLITLFELTFAKILEW--IPIWPYAKLILTCWLVLPYFTG 96
LT+ + Y + LF +T ++W +P WP + + CW + ++ G
Sbjct: 43 LTWGIGYHGVALFWITGIHPMDWLGVPWWPSLAITIFCWSFISFYGG 89
>SYC_SULTO (Q96YC3) Cysteinyl-tRNA synthetase (EC 6.1.1.16)
(Cysteine--tRNA ligase) (CysRS)
Length = 471
Score = 30.8 bits (68), Expect = 1.6
Identities = 35/128 (27%), Positives = 56/128 (43%), Gaps = 19/128 (14%)
Query: 19 FDVLAGPVISLVYPLYASVRAIESKSPVDDQQWLTYWVLYSLITLFELTFAKIL-EWIPI 77
FD+ G I L++P + + RA + +QW+ YWV S +T+ + +K L IP+
Sbjct: 222 FDIHGGG-IDLIFPHHENERA--QSEALLGKQWVKYWVHVSYLTIRKEKMSKSLGNIIPL 278
Query: 78 WPYAKLILTCW--LVLPYFTGAAYVYEHYVRPFLANPQTINIWYVPRKKDVFTKQDDIIT 135
L W VL Y+ Y+ HY N ++ + K+ T+ D I+
Sbjct: 279 ----NEALKKWGPSVLRYW----YLSSHYRSSLDFNEDSLE-----QAKNALTRLKDAIS 325
Query: 136 AAEKYIKE 143
IKE
Sbjct: 326 IIRDVIKE 333
>SYC_SULSO (Q97WE6) Cysteinyl-tRNA synthetase (EC 6.1.1.16)
(Cysteine--tRNA ligase) (CysRS)
Length = 470
Score = 30.4 bits (67), Expect = 2.1
Identities = 31/144 (21%), Positives = 62/144 (42%), Gaps = 21/144 (14%)
Query: 19 FDVLAGPVISLVYPLYASVRAIESKSPVDDQQWLTYWVLYSLITLFELTFAKILEWI--- 75
FD+ G L++P + + RA + ++W+TYWV + +T+ + +K L I
Sbjct: 221 FDIHGGGA-DLIFPHHENERA--QTEALIGEKWVTYWVHSAFVTIRKEKMSKSLGNIIPL 277
Query: 76 --PIWPYAKLILTCWLVLPYFTGAAYVYEHYVRPFLANPQTIN--IWYVPRKKDVFTKQD 131
I + +L W Y+ HY P + + + + R KD
Sbjct: 278 NEAIKKWGPSVLRYW----------YLTSHYRSPIDFSEEALEQAKSALQRIKDSMAIIR 327
Query: 132 DIITAAEK-YIKENGTEAFENLIH 154
D+I+ K Y+K++ + + +++
Sbjct: 328 DVISEGPKFYVKDDDIKVYREILN 351
>TR60_PONPY (Q645U7) Taste receptor type 2 member 60 (T2R60)
Length = 318
Score = 30.0 bits (66), Expect = 2.7
Identities = 16/63 (25%), Positives = 30/63 (47%), Gaps = 4/63 (6%)
Query: 51 WLTYWVLYSLITLFELTFA----KILEWIPIWPYAKLILTCWLVLPYFTGAAYVYEHYVR 106
WL+ + + T F K+ EW+P ++ + L+ + + +F G VY+ Y+R
Sbjct: 113 WLSVFYCVKIATFTHPVFLWLKHKLSEWVPWMLFSSVGLSSFTTILFFIGNHRVYQSYLR 172
Query: 107 PFL 109
L
Sbjct: 173 NHL 175
>TR60_MACMU (Q645S2) Taste receptor type 2 member 60 (T2R60)
Length = 318
Score = 30.0 bits (66), Expect = 2.7
Identities = 15/63 (23%), Positives = 31/63 (48%), Gaps = 4/63 (6%)
Query: 51 WLTYWVLYSLITLFELTFA----KILEWIPIWPYAKLILTCWLVLPYFTGAAYVYEHYVR 106
WL+ + + T F K+ EW+P ++ + L+ + + +F G +Y++Y+R
Sbjct: 113 WLSVFYCVKIATFTHPVFLWLKHKLSEWVPWMFFSSVGLSSFTTILFFIGNHSIYQNYLR 172
Query: 107 PFL 109
L
Sbjct: 173 NHL 175
>MFD_RICPR (O05955) Transcription-repair coupling factor (TRCF)
Length = 1120
Score = 30.0 bits (66), Expect = 2.7
Identities = 28/106 (26%), Positives = 45/106 (42%), Gaps = 12/106 (11%)
Query: 68 FAKILEWIPIWPYAKLILTCWLVLPYFTGAAYVYEHYVRPFL-------ANPQTINIWYV 120
F K+ E I + K I+ C VL F + ++Y F A INI +
Sbjct: 355 FDKLFEIIKA-NFHKKIIICSSVLSSFERIKSIIQNYEYTFNEINKLDEAKASVINIGII 413
Query: 121 PRKKDVFTKQDDIITAA----EKYIKENGTEAFENLIHRADKSKWG 162
P + +TK+ IT++ EK + N + +N++ D K G
Sbjct: 414 PLNQSFYTKEYLFITSSELLEEKTLYTNTNKKLKNILLELDNLKEG 459
>EDG7_RAT (Q8K5E0) Lysophosphatidic acid receptor Edg-7 (LPA
receptor 3) (LPA-3) (snGPCR32)
Length = 354
Score = 30.0 bits (66), Expect = 2.7
Identities = 15/73 (20%), Positives = 38/73 (51%), Gaps = 4/73 (5%)
Query: 10 SFLMVLLRNFDVLAGPVISLVYPLYASVRAIESKSPVDDQQWLTYWVLYSLITLFELTFA 69
+ L++L+ + G V +L + ++ A S +P+ + +L +W + +L+ F +
Sbjct: 146 TLLILLVWAIAIFMGAVPTLGWNCLCNISACSSLAPIYSRSYLIFWTVSNLLAFFIM--- 202
Query: 70 KILEWIPIWPYAK 82
++ ++ I+ Y K
Sbjct: 203 -VVVYVRIYMYVK 214
>EDG7_MOUSE (Q9EQ31) Lysophosphatidic acid receptor Edg-7 (LPA
receptor 3) (LPA-3)
Length = 354
Score = 29.3 bits (64), Expect = 4.7
Identities = 15/73 (20%), Positives = 37/73 (50%), Gaps = 4/73 (5%)
Query: 10 SFLMVLLRNFDVLAGPVISLVYPLYASVRAIESKSPVDDQQWLTYWVLYSLITLFELTFA 69
+ L++L+ + G V +L + ++ A S +P+ + +L +W + +L+ F +
Sbjct: 146 TLLILLVWAIAIFMGAVPTLGWNCLCNISACSSLAPIYSRSYLIFWTVSNLLAFFIM--- 202
Query: 70 KILEWIPIWPYAK 82
+ ++ I+ Y K
Sbjct: 203 -VAVYVRIYMYVK 214
>CYDC_HAEIN (P45081) Transport ATP-binding protein cydC
Length = 576
Score = 29.3 bits (64), Expect = 4.7
Identities = 13/35 (37%), Positives = 21/35 (59%)
Query: 1 MGGSGSGAGSFLMVLLRNFDVLAGPVISLVYPLYA 35
+G +GSG S L +L+RN+D G ++ P+ A
Sbjct: 371 LGKTGSGKSSLLQLLVRNYDANQGELLLAEKPISA 405
>R1AB_PEDV7 (Q91AV2) Replicase polyprotein 1ab (pp1ab) (ORF1ab
polyprotein) [Includes: Replicase polyprotein 1a (pp1a)
(ORF1a)] [Contains: p9; p87; p195 (EC 3.4.24.-)
(Papain-like proteinases 1/2) (PL1-PRO/PL2-PRO); Peptide
HD2; Unknown protein 1; 3C-like
Length = 6781
Score = 28.9 bits (63), Expect = 6.1
Identities = 33/127 (25%), Positives = 47/127 (36%), Gaps = 28/127 (22%)
Query: 23 AGPVISLVYPLYASVRAIESKSPVDDQQWLTYWVLYSLITLFELTFAKILEWIPIWPYAK 82
A ++S L+ASV W V Y + L F ++ I+ K
Sbjct: 3451 ASDILSCAMTLFASVTG----------NWFVGAVCYKVAVYMALRFPT---FVAIFGDIK 3497
Query: 83 LILTCWLVLPYFTGAAYVYEHYVRPFLANPQTINIWYVPRKKDVFTKQDDIITAAE-KYI 141
++ C+LVL YFT Y I W+ K D ++AAE KY+
Sbjct: 3498 SVMFCYLVLGYFTCCFY--------------GILYWFNRFFKVSVGVYDYTVSAAEFKYM 3543
Query: 142 KENGTEA 148
NG A
Sbjct: 3544 VANGLRA 3550
>YG3L_YEAST (P48236) Hypothetical 51.6 kDa protein in RPL24B-RSR1
intergenic region
Length = 432
Score = 28.5 bits (62), Expect = 8.0
Identities = 26/110 (23%), Positives = 45/110 (40%), Gaps = 6/110 (5%)
Query: 56 VLYSLITLFELTFAKIL--EWIPIWPYAKLILTCWLVLPYFTGAAYVYEHYVRPFLANPQ 113
++Y+L F F +L WI A L LT + GA Y +HY + F
Sbjct: 322 IIYTLSQYFYQLFTMLLCGIWIRYKLAAALFLTIVFLWASHNGATYYIDHYGKNFEKEVD 381
Query: 114 TINIWYVPRKKDVFTKQDDIITAA----EKYIKENGTEAFENLIHRADKS 159
+ + ++ + D +I+ A + Y+ N E F++ + KS
Sbjct: 382 RLRLEVENLQQKLQPDSDAVISDASVNDKDYLNVNRDEDFDDSSSVSSKS 431
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.323 0.139 0.446
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 21,860,027
Number of Sequences: 164201
Number of extensions: 896890
Number of successful extensions: 2221
Number of sequences better than 10.0: 15
Number of HSP's better than 10.0 without gapping: 6
Number of HSP's successfully gapped in prelim test: 9
Number of HSP's that attempted gapping in prelim test: 2212
Number of HSP's gapped (non-prelim): 17
length of query: 173
length of database: 59,974,054
effective HSP length: 103
effective length of query: 70
effective length of database: 43,061,351
effective search space: 3014294570
effective search space used: 3014294570
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 62 (28.5 bits)
Medicago: description of AC146548.3


