

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146527.6 - phase: 0 /pseudo
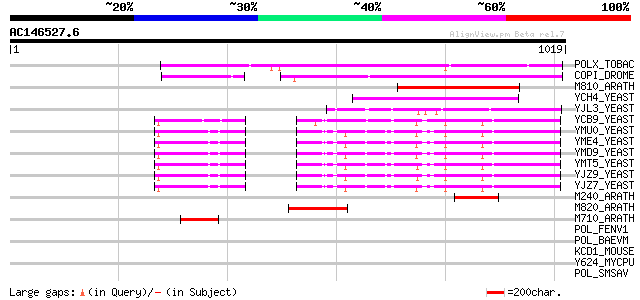
(1019 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
POLX_TOBAC (P10978) Retrovirus-related Pol polyprotein from tran... 474 e-133
COPI_DROME (P04146) Copia protein (Gag-int-pol protein) [Contain... 338 4e-92
M810_ARATH (P92519) Hypothetical mitochondrial protein AtMg00810... 197 2e-49
YCH4_YEAST (P25600) Transposon Ty5-1 34.5 kDa hypothetical protein 177 1e-43
YJL3_YEAST (P47024) Transposon Ty4 207.7 kDa hypothetical protein 120 2e-26
YCB9_YEAST (P25384) Transposon Ty2 protein B (Ty1-17 protein B) 106 4e-22
YMU0_YEAST (Q04670) Transposon Ty1 protein B 104 1e-21
YME4_YEAST (Q04711) Transposon Ty1 protein B 102 4e-21
YMD9_YEAST (Q03434) Transposon Ty1 protein B 100 2e-20
YMT5_YEAST (Q04214) Transposon Ty1 protein B 99 4e-20
YJZ9_YEAST (P47100) Transposon Ty1 protein B 99 6e-20
YJZ7_YEAST (P47098) Transposon Ty1 protein B 98 1e-19
M240_ARATH (P93290) Hypothetical mitochondrial protein AtMg00240... 92 5e-18
M820_ARATH (P92520) Hypothetical mitochondrial protein AtMg00820... 89 6e-17
M710_ARATH (P92512) Hypothetical mitochondrial protein AtMg00710... 63 5e-09
POL_FENV1 (P31792) Pol polyprotein [Contains: Reverse transcript... 39 0.054
POL_BAEVM (P10272) Pol polyprotein [Contains: Protease (EC 3.4.2... 38 0.12
KCD1_MOUSE (Q03719) Potassium voltage-gated channel subfamily D ... 33 3.0
Y624_MYCPU (Q98PU5) Hypothetical UPF0082 protein MYPU_6240 33 3.9
POL_SMSAV (P03359) Pol polyprotein [Contains: Reverse transcript... 33 5.1
>POLX_TOBAC (P10978) Retrovirus-related Pol polyprotein from
transposon TNT 1-94 [Contains: Protease (EC 3.4.23.-);
Reverse transcriptase (EC 2.7.7.49); Endonuclease]
Length = 1328
Score = 474 bits (1221), Expect = e-133
Identities = 291/770 (37%), Positives = 439/770 (56%), Gaps = 39/770 (5%)
Query: 278 GEYMSHSFQEFLQSNGIISQRSCPSTP*QNGVAERKNRHLLDVVRTLLLESHVPSRFWCE 337
GEY S F+E+ S+GI +++ P TP NGVAER NR +++ VR++L + +P FW E
Sbjct: 553 GEYTSREFEEYCSSHGIRHEKTVPGTPQHNGVAERMNRTIVEKVRSMLRMAKLPKSFWGE 612
Query: 338 ALSTAVHLINRMPSPSIGNESPFTRLYGHPPNYSTLRVFGCVCYVHLPPQERTKFTAQSV 397
A+ TA +LINR PS + E P +YS L+VFGC + H+P ++RTK +S+
Sbjct: 613 AVQTACYLINRSPSVPLAFEIPERVWTNKEVSYSHLKVFGCRAFAHVPKEQRTKLDDKSI 672
Query: 398 ECAFLGYSPHQKGFLCYDPNLRRIRVSRNVIFQENKYFFASHHDLVSSPI--SILPLFYD 455
C F+GY + G+ +DP +++ SR+V+F+E++ A+ +S + I+P F
Sbjct: 673 PCIFIGYGDEEFGYRLWDPVKKKVIRSRDVVFRESEVRTAAD---MSEKVKNGIIPNFVT 729
Query: 456 SHS-RQQPSKPLLTYKRRSTATHGP----PQDNSLVAG--PVEEPA-------PLRRSSR 501
S P+ T S P Q L G VE P PLRRS R
Sbjct: 730 IPSTSNNPTSAESTTDEVSEQGEQPGEVIEQGEQLDEGVEEVEHPTQGEEQHQPLRRSER 789
Query: 502 ESKPPERYINCMTATLSSIPIPSSYKQAM---ENDCWQKAIESELLALEENQTWDIVPCP 558
RY + +S P S K+ + E + KA++ E+ +L++N T+ +V P
Sbjct: 790 PRVESRRYPSTEYVLISDDREPESLKEVLSHPEKNQLMKAMQEEMESLQKNGTYKLVELP 849
Query: 559 SSVKPLGSKFVFSIKLRSDGSIDRYKAHLVVLGNKQEYGLDYDETFAPVAKMTTVRTILA 618
+PL K+VF +K D + RYKA LVV G +Q+ G+D+DE F+PV KMT++RTIL+
Sbjct: 850 KGKRPLKCKWVFKLKKDGDCKLVRYKARLVVKGFEQKKGIDFDEIFSPVVKMTSIRTILS 909
Query: 619 IAASQAWPLHQMDVKNAFLHGDLQEEVYIKLPNGMPTP-SPNTVCKLKRSLYGLKQAPRV 677
+AAS + Q+DVK AFLHGDL+EE+Y++ P G + VCKL +SLYGLKQAPR
Sbjct: 910 LAASLDLEVEQLDVKTAFLHGDLEEEIYMEQPEGFEVAGKKHMVCKLNKSLYGLKQAPRQ 969
Query: 678 WFEKFRSTLLGFEFSQSRYDPSLFLQR-TPKGMVVLLVYVDDIVVTGSDQDAISRIKNLL 736
W+ KF S + + ++ DP ++ +R + ++LL+YVDD+++ G D+ I+++K L
Sbjct: 970 WYMKFDSFMKSQTYLKTYSDPCVYFKRFSENNFIILLLYVDDMLIVGKDKGLIAKLKGDL 1029
Query: 737 HSTFHMKELGRLTYFLGLEVHYHHEG--VFLNQQKYIQDLVQLAGLTNATLVDTPMEVNV 794
+F MK+LG LG+++ ++L+Q+KYI+ +++ + NA V TP+ ++
Sbjct: 1030 SKSFDMKDLGPAQQILGMKIVRERTSRKLWLSQEKYIERVLERFNMKNAKPVSTPLAGHL 1089
Query: 795 KYRR--------DEGDHLDDPTQYRKLVGSLIYVTI-TRPDISFAVHTVSKFMQAPRHFH 845
K + ++G+ P Y VGSL+Y + TRPDI+ AV VS+F++ P H
Sbjct: 1090 KLSKKMCPTTVEEKGNMAKVP--YSSAVGSLMYAMVCTRPDIAHAVGVVSRFLENPGKEH 1147
Query: 846 LSAVQQIIRYLLGTLKRGLFFPVGSSIKLQAYSDADWAGCPDTRKSTTGWCMFLGNAPIS 905
AV+ I+RYL GT L F GS L+ Y+DAD AG D RKS+TG+ IS
Sbjct: 1148 WEAVKWILRYLRGTTGDCLCFG-GSDPILKGYTDADMAGDIDNRKSSTGYLFTFSGGAIS 1206
Query: 906 WKCKKQDSVSKSSTEAEYRAMSAACSEIIWLRGLLTELGFSQDQPTPLHADNTSAIQIAA 965
W+ K Q V+ S+TEAEY A + E+IWL+ L ELG Q + ++ D+ SAI ++
Sbjct: 1207 WQSKLQKCVALSTTEAEYIAATETGKEMIWLKRFLQELGLHQKEYV-VYCDSQSAIDLSK 1265
Query: 966 NPVYHERTKHIEVDCHSIREAYDRRIINLPHVSTSVQTADIFTKSLTRQR 1015
N +YH RTKHI+V H IRE D + + +ST+ AD+ TK + R +
Sbjct: 1266 NSMYHARTKHIDVRYHWIREMVDDESLKVLKISTNENPADMLTKVVPRNK 1315
>COPI_DROME (P04146) Copia protein (Gag-int-pol protein) [Contains:
Copia VLP protein; Copia protease (EC 3.4.23.-)]
Length = 1409
Score = 338 bits (867), Expect = 4e-92
Identities = 196/538 (36%), Positives = 297/538 (54%), Gaps = 22/538 (4%)
Query: 497 RRSSRESKPPERYINCMTATLSSI---------PIPSSYKQAMEND---CWQKAIESELL 544
RRS R P+ N +L+ + +P+S+ + D W++AI +EL
Sbjct: 856 RRSERLKTKPQISYNEEDNSLNKVVLNAHTIFNDVPNSFDEIQYRDDKSSWEEAINTELN 915
Query: 545 ALEENQTWDIVPCPSSVKPLGSKFVFSIKLRSDGSIDRYKAHLVVLGNKQEYGLDYDETF 604
A + N TW I P + + S++VFS+K G+ RYKA LV G Q+Y +DY+ETF
Sbjct: 916 AHKINNTWTITKRPENKNIVDSRWVFSVKYNELGNPIRYKARLVARGFTQKYQIDYEETF 975
Query: 605 APVAKMTTVRTILAIAASQAWPLHQMDVKNAFLHGDLQEEVYIKLPNGMPTPSPNTVCKL 664
APVA++++ R IL++ +HQMDVK AFL+G L+EE+Y++LP G+ S N VCKL
Sbjct: 976 APVARISSFRFILSLVIQYNLKVHQMDVKTAFLNGTLKEEIYMRLPQGISCNSDN-VCKL 1034
Query: 665 KRSLYGLKQAPRVWFEKFRSTLLGFEFSQSRYDPSLFL--QRTPKGMVVLLVYVDDIVVT 722
+++YGLKQA R WFE F L EF S D +++ + + +L+YVDD+V+
Sbjct: 1035 NKAIYGLKQAARCWFEVFEQALKECEFVNSSVDRCIYILDKGNINENIYVLLYVDDVVIA 1094
Query: 723 GSDQDAISRIKNLLHSTFHMKELGRLTYFLGLEVHYHHEGVFLNQQKYIQDLVQLAGLTN 782
D ++ K L F M +L + +F+G+ + + ++L+Q Y++ ++ + N
Sbjct: 1095 TGDMTRMNNFKRYLMEKFRMTDLNEIKHFIGIRIEMQEDKIYLSQSAYVKKILSKFNMEN 1154
Query: 783 ATLVDTPMEVNVKYRRDEGDHLDDPTQYRKLVGSLIYVTI-TRPDISFAVHTVSKFMQAP 841
V TP+ + Y D D T R L+G L+Y+ + TRPD++ AV+ +S++
Sbjct: 1155 CNAVSTPLPSKINYELLNSDE-DCNTPCRSLIGCLMYIMLCTRPDLTTAVNILSRYSSKN 1213
Query: 842 RHFHLSAVQQIIRYLLGTLKRGLFFPVGSSI--KLQAYSDADWAGCPDTRKSTTGWC--M 897
+++++RYL GT+ L F + K+ Y D+DWAG RKSTTG+ M
Sbjct: 1214 NSELWQNLKRVLRYLKGTIDMKLIFKKNLAFENKIIGYVDSDWAGSEIDRKSTTGYLFKM 1273
Query: 898 FLGNAPISWKCKKQDSVSKSSTEAEYRAMSAACSEIIWLRGLLTELGFSQDQPTPLHADN 957
F N I W K+Q+SV+ SSTEAEY A+ A E +WL+ LLT + + P ++ DN
Sbjct: 1274 FDFNL-ICWNTKRQNSVAASSTEAEYMALFEAVREALWLKFLLTSINIKLENPIKIYEDN 1332
Query: 958 TSAIQIAANPVYHERTKHIEVDCHSIREAYDRRIINLPHVSTSVQTADIFTKSLTRQR 1015
I IA NP H+R KHI++ H RE +I L ++ T Q ADIFTK L R
Sbjct: 1333 QGCISIANNPSCHKRAKHIDIKYHFAREQVQNNVICLEYIPTENQLADIFTKPLPAAR 1390
Score = 92.4 bits (228), Expect = 5e-18
Identities = 53/155 (34%), Positives = 87/155 (55%), Gaps = 5/155 (3%)
Query: 279 EYMSHSFQEFLQSNGIISQRSCPSTP*QNGVAERKNRHLLDVVRTLLLESHVPSRFWCEA 338
EY+S+ ++F GI + P TP NGV+ER R + + RT++ + + FW EA
Sbjct: 554 EYLSNEMRQFCVKKGISYHLTVPHTPQLNGVSERMIRTITEKARTMVSGAKLDKSFWGEA 613
Query: 339 LSTAVHLINRMPSPSI--GNESPFTRLYGHPPNYSTLRVFGCVCYVHLPPQERTKFTAQS 396
+ TA +LINR+PS ++ +++P+ + P LRVFG YVH+ ++ KF +S
Sbjct: 614 VLTATYLINRIPSRALVDSSKTPYEMWHNKKPYLKHLRVFGATVYVHI-KNKQGKFDDKS 672
Query: 397 VECAFLGYSPHQKGFLCYDPNLRRIRVSRNVIFQE 431
+ F+GY P+ GF +D + V+R+V+ E
Sbjct: 673 FKSIFVGYEPN--GFKLWDAVNEKFIVARDVVVDE 705
>M810_ARATH (P92519) Hypothetical mitochondrial protein AtMg00810
(ORF240b)
Length = 240
Score = 197 bits (500), Expect = 2e-49
Identities = 94/224 (41%), Positives = 147/224 (64%), Gaps = 1/224 (0%)
Query: 712 LLVYVDDIVVTGSDQDAISRIKNLLHSTFHMKELGRLTYFLGLEVHYHHEGVFLNQQKYI 771
LL+YVDDI++TGS ++ + L STF MK+LG + YFLG+++ H G+FL+Q KY
Sbjct: 3 LLLYVDDILLTGSSNTLLNMLIFQLSSTFSMKDLGPVHYFLGIQIKTHPSGLFLSQTKYA 62
Query: 772 QDLVQLAGLTNATLVDTPMEVNVKYRRDEGDHLDDPTQYRKLVGSLIYVTITRPDISFAV 831
+ ++ AG+ + + TP+ + + + DP+ +R +VG+L Y+T+TRPDIS+AV
Sbjct: 63 EQILNNAGMLDCKPMSTPLPLKLNSSVSTAKY-PDPSDFRSIVGALQYLTLTRPDISYAV 121
Query: 832 HTVSKFMQAPRHFHLSAVQQIIRYLLGTLKRGLFFPVGSSIKLQAYSDADWAGCPDTRKS 891
+ V + M P +++++RY+ GT+ GL+ S + +QA+ D+DWAGC TR+S
Sbjct: 122 NIVCQRMHEPTLADFDLLKRVLRYVKGTIFHGLYIHKNSKLNVQAFCDSDWAGCTSTRRS 181
Query: 892 TTGWCMFLGNAPISWKCKKQDSVSKSSTEAEYRAMSAACSEIIW 935
TTG+C FLG ISW K+Q +VS+SSTE EYRA++ +E+ W
Sbjct: 182 TTGFCTFLGCNIISWSAKRQPTVSRSSTETEYRALALTAAELTW 225
>YCH4_YEAST (P25600) Transposon Ty5-1 34.5 kDa hypothetical protein
Length = 308
Score = 177 bits (449), Expect = 1e-43
Identities = 106/308 (34%), Positives = 157/308 (50%), Gaps = 4/308 (1%)
Query: 630 MDVKNAFLHGDLQEEVYIKLPNGMPTP-SPNTVCKLKRSLYGLKQAPRVWFEKFRSTLLG 688
MDV AFL+ + E +Y+K P G +P+ V +L +YGLKQAP +W E +TL
Sbjct: 1 MDVDTAFLNSTMDEPIYVKQPPGFVNERNPDYVWELYGGMYGLKQAPLLWNEHINNTLKK 60
Query: 689 FEFSQSRYDPSLFLQRTPKGMVVLLVYVDDIVVTGSDQDAISRIKNLLHSTFHMKELGRL 748
F + + L+ + T G + + VYVDD++V R+K L + MK+LG++
Sbjct: 61 IGFCRHEGEHGLYFRSTSDGPIYIGVYVDDLLVAAPSPKIYDRVKQELTKLYSMKDLGKV 120
Query: 749 TYFLGLEVHYHHEG-VFLNQQKYIQDLVQLAGLTNATLVDTPMEVNVKYRRDEGDHLDDP 807
FLGL +H G + L+ Q YI + + L TP+ + HL D
Sbjct: 121 DKFLGLNIHQSTNGDITLSLQDYIAKAASESEINTFKLTQTPLCNSKPLFETTSPHLKDI 180
Query: 808 TQYRKLVGSLIYVTIT-RPDISFAVHTVSKFMQAPRHFHLSAVQQIIRYLLGTLKRGLFF 866
T Y+ +VG L++ T RPDIS+ V +S+F++ PR HL + ++++RYL T L +
Sbjct: 181 TPYQSIVGQLLFCANTGRPDISYPVSLLSRFLREPRAIHLESARRVLRYLYTTRSMCLKY 240
Query: 867 PVGSSIKLQAYSDADWAGCPDTRKSTTGWCMFLGNAPISWKCKK-QDSVSKSSTEAEYRA 925
GS + L Y DA D ST G+ L AP++W KK + + STEAEY
Sbjct: 241 RSGSQVALTVYCDASHGAIHDLPHSTGGYVTLLAGAPVTWSSKKLKGVIPVPSTEAEYIT 300
Query: 926 MSAACSEI 933
S EI
Sbjct: 301 ASETVMEI 308
>YJL3_YEAST (P47024) Transposon Ty4 207.7 kDa hypothetical protein
Length = 1803
Score = 120 bits (301), Expect = 2e-26
Identities = 116/453 (25%), Positives = 199/453 (43%), Gaps = 35/453 (7%)
Query: 583 YKAHLVVLGNKQEYGLDYDETFAPVAKMTTVRTILAIAASQAWPLHQMDVKNAFLHGDLQ 642
YKA +V G+ Q Y ++ L IA ++ + +D+ +AFL+ L+
Sbjct: 1337 YKARIVCRGDTQSPDT-YSVITTESLNHNHIKIFLMIANNRNMFMKTLDINHAFLYAKLE 1395
Query: 643 EEVYIKLPNGMPTPSPNTVCKLKRSLYGLKQAPRVWFEKFRSTLLGFEFSQSRYDPSLFL 702
EE+YI P+ V KL ++LYGLKQ+P+ W + R L G + Y P L+
Sbjct: 1396 EEIYIPHPHDR-----RCVVKLNKALYGLKQSPKEWNDHLRQYLNGIGLKDNSYTPGLY- 1449
Query: 703 QRTPKGMVVLLVYVDDIVVTGSDQDAISRIKNLLHSTFHMKELGRL------TYFLGLEV 756
+T +++ VYVDD V+ S++ + N L S F +K G L T LG+++
Sbjct: 1450 -QTEDKNLMIAVYVDDCVIAASNEQRLDEFINKLKSNFELKITGTLIDDVLDTDILGMDL 1508
Query: 757 HYHHE--------GVFLNQ--QKYIQDLVQLAGLT----NATLVDTPMEVNVKYRRDEGD 802
Y+ F+N+ +KY ++L ++ + + +D +V + E +
Sbjct: 1509 VYNKRLGTIDLTLKSFINRMDKKYNEELKKIRKSSIPHMSTYKIDPKKDV---LQMSEEE 1565
Query: 803 HLDDPTQYRKLVGSLIYVT-ITRPDISFAVHTVSKFMQAPRHFHLSAVQQIIRYLLGTLK 861
+ ++L+G L YV R DI FAV V++ + P + +II+YL+
Sbjct: 1566 FRQGVLKLQQLLGELNYVRHKCRYDIEFAVKKVARLVNYPHERVFYMIYKIIQYLVRYKD 1625
Query: 862 RGLFF--PVGSSIKLQAYSDADWAGCPDTRKSTTGWCMFLGNAPISWKCKKQDSVSKSST 919
G+ + K+ A +DA G +S G ++ G + K + SST
Sbjct: 1626 IGIHYDRDCNKDKKVIAITDAS-VGSEYDAQSRIGVILWYGMNIFNVYSNKSTNRCVSST 1684
Query: 920 EAEYRAMSAACSEIIWLRGLLTELGFSQDQPTPLHADNTSAIQIAANPVYHERTKHIEVD 979
EAE A+ ++ L+ L ELG + + D+ AIQ + K +
Sbjct: 1685 EAELHAIYEGYADSETLKVTLKELGEGDNNDIVMITDSKPAIQGLNRSYQQPKEKFTWIK 1744
Query: 980 CHSIREAYDRRIINLPHVSTSVQTADIFTKSLT 1012
I+E + I L ++ AD+ TK ++
Sbjct: 1745 TEIIKEKIKEKSIKLLKITGKGNIADLLTKPVS 1777
Score = 35.8 bits (81), Expect = 0.60
Identities = 22/77 (28%), Positives = 39/77 (50%)
Query: 279 EYMSHSFQEFLQSNGIISQRSCPSTP*QNGVAERKNRHLLDVVRTLLLESHVPSRFWCEA 338
E+ + +E+ S GI + NG AER R ++ TLL +S++ +FW A
Sbjct: 700 EFTNDQIEEYFISKGIHHILTSTQDHAANGRAERYIRTIITDATTLLRQSNLRVKFWEYA 759
Query: 339 LSTAVHLINRMPSPSIG 355
+++A ++ N + S G
Sbjct: 760 VTSATNIRNYLEHKSTG 776
>YCB9_YEAST (P25384) Transposon Ty2 protein B (Ty1-17 protein B)
Length = 1770
Score = 106 bits (264), Expect = 4e-22
Identities = 124/520 (23%), Positives = 221/520 (41%), Gaps = 55/520 (10%)
Query: 527 KQAMENDCWQKAIESELLALEENQTWDIVPCPSS-----VKPLGSKFVFSIKLRSDGSID 581
K E D + +A E+ L + TWD K + S F+F+ K DG+
Sbjct: 1257 KDNKEKDRYVEAYHKEISQLLKMNTWDTNKYYDRNDIDPKKVINSMFIFNKK--RDGT-- 1312
Query: 582 RYKAHLVVLGNKQEYGLDYDETFAPVAKMTTVRTILAIAASQAWPLHQMDVKNAFLHGDL 641
+KA V G+ Q + + + T L+IA + + Q+D+ +A+L+ D+
Sbjct: 1313 -HKARFVARGDIQHPDTYDSDMQSNTVHHYALMTSLSIALDNDYYITQLDISSAYLYADI 1371
Query: 642 QEEVYIKLPNGMPTPSPNTVCKLKRSLYGLKQAPRVWFEKFRSTLLG-FEFSQSRYDPSL 700
+EE+YI+ P + + + +L++SLYGLKQ+ W+E +S L+ + + R +
Sbjct: 1372 KEELYIRPPPHLGL--NDKLLRLRKSLYGLKQSGANWYETIKSYLINCCDMQEVRGWSCV 1429
Query: 701 FLQRTPKGMVVLLVYVDDIVVTGSDQDAISRIKNLLHSTFHMKEL------GRLTY-FLG 753
F V + ++VDD+++ D +A +I L + K + + Y LG
Sbjct: 1430 F----KNSQVTICLFVDDMILFSKDLNANKKIITTLKKQYDTKIINLGESDNEIQYDILG 1485
Query: 754 LEVHYHHEGVFLNQQKYIQDLVQLAGLTNATLVDTPMEVNVKYRR--------------- 798
LE+ Y + KY++ ++ + ++ P+ K R
Sbjct: 1486 LEIKY-------QRSKYMKLGMEKSLTEKLPKLNVPLNPKGKKLRAPGQPGHYIDQDELE 1538
Query: 799 -DEGDHLDDPTQYRKLVGSLIYVTIT-RPDISFAVHTVSKFMQAPRHFHLSAVQQIIRYL 856
DE ++ + + +KL+G YV R D+ + ++T+++ + P L ++I+++
Sbjct: 1539 IDEDEYKEKVHEMQKLIGLASYVGYKFRFDLLYYINTLAQHILFPSRQVLDMTYELIQFM 1598
Query: 857 LGTLKRGLFF----PVGSSIKLQAYSDADWAGCPDTRKSTTGWCMFLGNAPISWKCKKQD 912
T + L + P KL A SDA + P KS G L I K K
Sbjct: 1599 WDTRDKQLIWHKNKPTKPDNKLVAISDASYGNQP-YYKSQIGNIFLLNGKVIGGKSTKAS 1657
Query: 913 SVSKSSTEAEYRAMSAACSEIIWLRGLLTELGFSQDQPTPLHADNTSAIQIAANPVYHE- 971
S+TEAE A+S A + L L+ EL + L D+ S I I + +
Sbjct: 1658 LTCTSTTEAEIHAVSEAIPLLNNLSHLVQELN-KKPIIKGLLTDSRSTISIIKSTNEEKF 1716
Query: 972 RTKHIEVDCHSIREAYDRRIINLPHVSTSVQTADIFTKSL 1011
R + +R+ + + ++ T AD+ TK L
Sbjct: 1717 RNRFFGTKAMRLRDEVSGNNLYVYYIETKKNIADVMTKPL 1756
Score = 48.9 bits (115), Expect = 7e-05
Identities = 48/177 (27%), Positives = 83/177 (46%), Gaps = 14/177 (7%)
Query: 266 FIKNQN------ISL*QWGEYMSHSFQEFLQSNGIISQRSCPSTP*QNGVAERKNRHLLD 319
FIKNQ I + + EY + + +F + GI + + + +GVAER NR LL+
Sbjct: 717 FIKNQFNARVLVIQMDRGSEYTNKTLHKFFTNRGITACYTTTADSRAHGVAERLNRTLLN 776
Query: 320 VVRTLLLESHVPSRFWCEALSTAVHLINRMPSPSIGNESPFTRLYGHPP-NYSTLRVFGC 378
RTLL S +P+ W A+ + + N + SP N+ + G + +T+ FG
Sbjct: 777 DCRTLLHCSGLPNHLWFSAVEFSTIIRNSLVSPK--NDKSARQHAGLAGLDITTILPFGQ 834
Query: 379 VCYV--HLPPQERTKFTAQSVECAFLGYSPHQKGFLCYDPNLRRIRVSRNVIFQENK 433
V H P +K + + L S + G++ Y P+L++ + N + ++K
Sbjct: 835 PVIVNNHNPD---SKIHPRGIPGYALHPSRNSYGYIIYLPSLKKTVDTTNYVILQDK 888
>YMU0_YEAST (Q04670) Transposon Ty1 protein B
Length = 1328
Score = 104 bits (259), Expect = 1e-21
Identities = 132/532 (24%), Positives = 232/532 (42%), Gaps = 79/532 (14%)
Query: 527 KQAMENDCWQKAIESELLALEENQTWDIVPC--PSSVKP---LGSKFVFSIKLRSDGSID 581
K E + + +A E+ L + +TWD + P + S F+F+ K DG+
Sbjct: 815 KDIKEKEKYIQAYHKEVNQLLKMKTWDTDRYYDRKEIDPKRVINSMFIFNRK--RDGT-- 870
Query: 582 RYKAHLVVLGNKQEYGLDYDETFAPVAKMTTVR-----TILAIAASQAWPLHQMDVKNAF 636
+KA V G+ Q + +T+ P + TV T L++A + + Q+D+ +A+
Sbjct: 871 -HKARFVARGDIQ-----HPDTYDPGMQSNTVHHYALMTSLSLALDNNYYITQLDISSAY 924
Query: 637 LHGDLQEEVYIKLPNGMPTPSPNTVCKLKRSLYGLKQAPRVWFEKFRSTLL---GFEFSQ 693
L+ D++EE+YI+ P + + + +LK+SLYGLKQ+ W+E +S L+ G E +
Sbjct: 925 LYADIKEELYIRPPPHL--GMNDKLIRLKKSLYGLKQSGANWYETIKSYLIKQCGME--E 980
Query: 694 SRYDPSLFLQRTPKGMVVLLVYVDDIVVTGSDQDAISRIKNLLHSTFHMKEL------GR 747
R +F V + ++VDD+++ D +A +I L + K +
Sbjct: 981 VRGWSCVF----KNSQVTICLFVDDMILFSKDLNANKKIITTLKKQYDTKIINLGESDNE 1036
Query: 748 LTY-FLGLEVHYHHEGVFLNQQKYIQDLVQLAGLTNATLVDTP---MEVNVKYRR----- 798
+ Y LGLE+ Y + KY++ G+ N+ P + +N K R+
Sbjct: 1037 IQYDILGLEIKY-------QRGKYMK-----LGMENSLTEKIPKLNVPLNPKGRKLSAPG 1084
Query: 799 -------------DEGDHLDDPTQYRKLVGSLIYVTIT-RPDISFAVHTVSKFMQAPRHF 844
+E D+ + +KL+G YV R D+ + ++T+++ + P
Sbjct: 1085 QPGLYIDQQELELEEDDYKMKVHEMQKLIGLASYVGYKFRFDLLYYINTLAQHILFPSKQ 1144
Query: 845 HLSAVQQIIRYLLGTLKRGLFF----PVGSSIKLQAYSDADWAGCPDTRKSTTGWCMFLG 900
L ++I+++ T + L + PV + KL SDA + P KS G L
Sbjct: 1145 VLDMTYELIQFIWNTRDKQLIWHKSKPVKPTNKLVVISDASYGNQP-YYKSQIGNIYLLN 1203
Query: 901 NAPISWKCKKQDSVSKSSTEAEYRAMSAACSEIIWLRGLLTELGFSQDQPTPLHADNTSA 960
I K K S+TEAE A+S + + L L+ EL + L D+ S
Sbjct: 1204 GKVIGGKSTKASLTCTSTTEAEIHAISESVPLLNNLSHLVQELN-KKPITKGLLTDSKST 1262
Query: 961 IQ-IAANPVYHERTKHIEVDCHSIREAYDRRIINLPHVSTSVQTADIFTKSL 1011
I I +N R + +R+ +++ ++ T AD+ TK L
Sbjct: 1263 ISIIISNNEEKFRNRFFGTKAMRLRDEVSGNHLHVCYIETKKNIADVMTKPL 1314
Score = 55.1 bits (131), Expect = 1e-06
Identities = 51/175 (29%), Positives = 81/175 (46%), Gaps = 10/175 (5%)
Query: 266 FIKNQN------ISL*QWGEYMSHSFQEFLQSNGIISQRSCPSTP*QNGVAERKNRHLLD 319
FIKNQ I + + EY + + +FL+ NGI + + +GVAER NR LLD
Sbjct: 294 FIKNQFQASVLVIQMDRGSEYTNRTLHKFLEKNGITPCYTTTADSRAHGVAERLNRTLLD 353
Query: 320 VVRTLLLESHVPSRFWCEALSTAVHLINRMPSP-SIGNESPFTRLYGHPPNYSTLRVFGC 378
RT L S +P+ W A+ + + N + SP S + L G + STL FG
Sbjct: 354 DCRTQLQCSGLPNHLWFSAIEFSTIVRNSLASPKSKKSARQHAGLAG--LDISTLLPFGQ 411
Query: 379 VCYVHLPPQERTKFTAQSVECAFLGYSPHQKGFLCYDPNLRRIRVSRNVIFQENK 433
V+ +K + + L S + G++ Y P+L++ + N + + K
Sbjct: 412 PVIVN-DHNPNSKIHPRGIPGYALHPSRNSYGYIIYLPSLKKTVDTTNYVILQGK 465
>YME4_YEAST (Q04711) Transposon Ty1 protein B
Length = 1328
Score = 102 bits (255), Expect = 4e-21
Identities = 131/532 (24%), Positives = 230/532 (42%), Gaps = 79/532 (14%)
Query: 527 KQAMENDCWQKAIESELLALEENQTWDIVPC--PSSVKP---LGSKFVFSIKLRSDGSID 581
K E + + +A E+ L + TWD + P + S F+F+ K DG+
Sbjct: 815 KDIKEKEKYIEAYHKEVNQLLKMNTWDTDKYYDRKEIDPKRVINSMFIFNRK--RDGT-- 870
Query: 582 RYKAHLVVLGNKQEYGLDYDETFAPVAKMTTVR-----TILAIAASQAWPLHQMDVKNAF 636
+KA V G+ Q + +T+ + TV T L++A + + Q+D+ +A+
Sbjct: 871 -HKARFVARGDIQ-----HPDTYDSGMQSNTVHHYALMTSLSLALDNNYYITQLDISSAY 924
Query: 637 LHGDLQEEVYIKLPNGMPTPSPNTVCKLKRSLYGLKQAPRVWFEKFRSTLL---GFEFSQ 693
L+ D++EE+YI+ P + + + +LK+SLYGLKQ+ W+E +S L+ G E +
Sbjct: 925 LYADIKEELYIRPPPHL--GMNDKLIRLKKSLYGLKQSGANWYETIKSYLIKQCGME--E 980
Query: 694 SRYDPSLFLQRTPKGMVVLLVYVDDIVVTGSDQDAISRIKNLLHSTFHMKEL------GR 747
R +F V + ++VDD+++ D +A +I L + K +
Sbjct: 981 VRGWSCVF----KNSQVTICLFVDDMILFSKDLNANKKIITTLKKQYDTKIINLGESDNE 1036
Query: 748 LTY-FLGLEVHYHHEGVFLNQQKYIQDLVQLAGLTNATLVDTP---MEVNVKYRR----- 798
+ Y LGLE+ Y + KY++ G+ N+ P + +N K R+
Sbjct: 1037 IQYDILGLEIKY-------QRGKYMK-----LGMENSLTEKIPKLNVPLNPKGRKLSAPG 1084
Query: 799 -------------DEGDHLDDPTQYRKLVGSLIYVTIT-RPDISFAVHTVSKFMQAPRHF 844
DE ++ + + +KL+G YV R D+ + ++T+++ + P
Sbjct: 1085 QPGLYIDQDELEIDEDEYKEKVHEMQKLIGLASYVGYKFRFDLLYYINTLAQHILFPSRQ 1144
Query: 845 HLSAVQQIIRYLLGTLKRGLFF----PVGSSIKLQAYSDADWAGCPDTRKSTTGWCMFLG 900
L ++I+++ T + L + P KL A SDA + P KS G L
Sbjct: 1145 VLDMTYELIQFMWDTRDKQLIWHKNKPTEPDNKLVAISDASYGNQP-YYKSQIGNIYLLN 1203
Query: 901 NAPISWKCKKQDSVSKSSTEAEYRAMSAACSEIIWLRGLLTELGFSQDQPTPLHADNTSA 960
I K K S+TEAE A+S + + L L+ EL + L D+ S
Sbjct: 1204 GKVIGGKSTKASLTCTSTTEAEIHAISESVPLLNNLSHLVQELN-KKPITKGLLTDSKST 1262
Query: 961 IQ-IAANPVYHERTKHIEVDCHSIREAYDRRIINLPHVSTSVQTADIFTKSL 1011
I I +N R + +R+ +++ ++ T AD+ TK L
Sbjct: 1263 ISIIISNNEEKFRNRFFGTKAMRLRDEVSGNHLHVCYIETKKNIADVMTKPL 1314
Score = 55.1 bits (131), Expect = 1e-06
Identities = 51/175 (29%), Positives = 81/175 (46%), Gaps = 10/175 (5%)
Query: 266 FIKNQN------ISL*QWGEYMSHSFQEFLQSNGIISQRSCPSTP*QNGVAERKNRHLLD 319
FIKNQ I + + EY + + +FL+ NGI + + +GVAER NR LLD
Sbjct: 294 FIKNQFQASVLVIQMDRGSEYTNRTLHKFLEKNGITPCYTTTADSRAHGVAERLNRTLLD 353
Query: 320 VVRTLLLESHVPSRFWCEALSTAVHLINRMPSP-SIGNESPFTRLYGHPPNYSTLRVFGC 378
RT L S +P+ W A+ + + N + SP S + L G + STL FG
Sbjct: 354 DCRTQLQCSGLPNHLWFSAIEFSTIVRNSLASPKSKKSARQHAGLAG--LDISTLLPFGQ 411
Query: 379 VCYVHLPPQERTKFTAQSVECAFLGYSPHQKGFLCYDPNLRRIRVSRNVIFQENK 433
V+ +K + + L S + G++ Y P+L++ + N + + K
Sbjct: 412 PVIVN-DHNPNSKIHPRGIPGYALHPSRNSYGYIIYLPSLKKTVDTTNYVILQGK 465
>YMD9_YEAST (Q03434) Transposon Ty1 protein B
Length = 1328
Score = 100 bits (249), Expect = 2e-20
Identities = 130/532 (24%), Positives = 230/532 (42%), Gaps = 79/532 (14%)
Query: 527 KQAMENDCWQKAIESELLALEENQTWDIVPC--PSSVKP---LGSKFVFSIKLRSDGSID 581
K E + + +A E+ L + +TWD + P + S F+F+ K DG+
Sbjct: 815 KDIKEKEKYIEAYHKEVNQLLKMKTWDTDEYYDRKEIDPKRVINSMFIFNKK--RDGT-- 870
Query: 582 RYKAHLVVLGNKQEYGLDYDETFAPVAKMTTVR-----TILAIAASQAWPLHQMDVKNAF 636
+KA V G+ Q + +T+ + TV T L++A + + Q+D+ +A+
Sbjct: 871 -HKARFVARGDIQ-----HPDTYDSGMQSNTVHHYALMTSLSLALDNNYYITQLDISSAY 924
Query: 637 LHGDLQEEVYIKLPNGMPTPSPNTVCKLKRSLYGLKQAPRVWFEKFRSTLL---GFEFSQ 693
L+ D++EE+YI+ P + + + +LK+SLYGLKQ+ W+E +S L+ G E +
Sbjct: 925 LYADIKEELYIRPPPHL--GMNDKLIRLKKSLYGLKQSGANWYETIKSYLIKQCGME--E 980
Query: 694 SRYDPSLFLQRTPKGMVVLLVYVDDIVVTGSDQDAISRIKNLLHSTFHMKEL------GR 747
R +F V + ++VDD+++ D +A +I L + K +
Sbjct: 981 VRGWSCVF----KNSQVTICLFVDDMILFSKDLNANKKIITTLKKQYDTKIINLGESDNE 1036
Query: 748 LTY-FLGLEVHYHHEGVFLNQQKYIQDLVQLAGLTNATLVDTP---MEVNVKYRR----- 798
+ Y LGLE+ Y + KY++ G+ N+ P + +N K R+
Sbjct: 1037 IQYDILGLEIKY-------QRGKYMK-----LGMENSLTEKIPKLNVPLNPKGRKLSAPG 1084
Query: 799 -------------DEGDHLDDPTQYRKLVGSLIYVTIT-RPDISFAVHTVSKFMQAPRHF 844
DE ++ + + +KL+G YV R D+ + ++T+++ + P
Sbjct: 1085 QPGLYIDQDELEIDEDEYKEKVHEMQKLIGLASYVGYKFRFDLLYYINTLAQHILFPSRQ 1144
Query: 845 HLSAVQQIIRYLLGTLKRGLFF----PVGSSIKLQAYSDADWAGCPDTRKSTTGWCMFLG 900
L ++I+++ T + L + P KL A SDA + P KS G L
Sbjct: 1145 VLDMTYELIQFMWDTRDKQLIWHKNKPTEPDNKLVAISDASYGNQP-YYKSQIGNIYLLN 1203
Query: 901 NAPISWKCKKQDSVSKSSTEAEYRAMSAACSEIIWLRGLLTELGFSQDQPTPLHADNTSA 960
I K K S+TEAE A+S + + L L+ EL + L D+ S
Sbjct: 1204 GKVIGGKSTKASLTCTSTTEAEIHAISESVPLLNNLSYLIQELN-KKPIIKGLLTDSRST 1262
Query: 961 IQIAANPVYHE-RTKHIEVDCHSIREAYDRRIINLPHVSTSVQTADIFTKSL 1011
I I + + R + +R+ + + ++ T AD+ TK L
Sbjct: 1263 ISIIKSTNEEKFRNRFFGTKAMRLRDEVSGNNLYVYYIETKKNIADVMTKPL 1314
Score = 55.1 bits (131), Expect = 1e-06
Identities = 51/175 (29%), Positives = 81/175 (46%), Gaps = 10/175 (5%)
Query: 266 FIKNQN------ISL*QWGEYMSHSFQEFLQSNGIISQRSCPSTP*QNGVAERKNRHLLD 319
FIKNQ I + + EY + + +FL+ NGI + + +GVAER NR LLD
Sbjct: 294 FIKNQFQASVLVIQMDRGSEYTNRTLHKFLEKNGITPCYTTTADSRAHGVAERLNRTLLD 353
Query: 320 VVRTLLLESHVPSRFWCEALSTAVHLINRMPSP-SIGNESPFTRLYGHPPNYSTLRVFGC 378
RT L S +P+ W A+ + + N + SP S + L G + STL FG
Sbjct: 354 DCRTQLQCSGLPNHLWFSAIEFSTIVRNSLASPKSKKSARQHAGLAG--LDISTLLPFGQ 411
Query: 379 VCYVHLPPQERTKFTAQSVECAFLGYSPHQKGFLCYDPNLRRIRVSRNVIFQENK 433
V+ +K + + L S + G++ Y P+L++ + N + + K
Sbjct: 412 PVIVN-DHNPNSKIHPRGIPGYALHPSRNSYGYIIYLPSLKKTVDTTNYVILQGK 465
>YMT5_YEAST (Q04214) Transposon Ty1 protein B
Length = 1328
Score = 99.4 bits (246), Expect = 4e-20
Identities = 131/532 (24%), Positives = 232/532 (42%), Gaps = 79/532 (14%)
Query: 527 KQAMENDCWQKAIESELLALEENQTWDIVPC--PSSVKP---LGSKFVFSIKLRSDGSID 581
K E + + +A E+ L + +TWD + P + S F+F+ K DG+
Sbjct: 815 KDIKEKEKYIEAYHKEVNQLLKMKTWDTDKYYDRKEIDPKRVINSMFIFNRK--RDGT-- 870
Query: 582 RYKAHLVVLGNKQEYGLDYDETFAPVAKMTTVR-----TILAIAASQAWPLHQMDVKNAF 636
+KA V G+ Q + +T+ + TV T L++A + + Q+D+ +A+
Sbjct: 871 -HKARFVARGDIQ-----HPDTYDSGMQSNTVHHYALMTSLSLALDNNYYITQLDISSAY 924
Query: 637 LHGDLQEEVYIKLPNGMPTPSPNTVCKLKRSLYGLKQAPRVWFEKFRSTLL---GFEFSQ 693
L+ D++EE+YI+ P + + + +LK+SLYGLKQ+ W+E +S L+ G E +
Sbjct: 925 LYADIKEELYIRPPPHL--GMNDKLIRLKKSLYGLKQSGANWYETIKSYLIKQCGME--E 980
Query: 694 SRYDPSLFLQRTPKGMVVLLVYVDDIVVTGSDQDAISRIKNLLHSTFHMKELG------R 747
R +F V + ++VDD+V+ + ++ RI + L + K +
Sbjct: 981 VRGWSCVF----ENSQVTICLFVDDMVLFSKNLNSNKRIIDKLKMQYDTKIINLGESDEE 1036
Query: 748 LTY-FLGLEVHYHHEGVFLNQQKYIQDLVQLAGLTNATLVDTP---MEVNVKYRR----- 798
+ Y LGLE+ Y + KY++ G+ N+ P + +N K R+
Sbjct: 1037 IQYDILGLEIKY-------QRGKYMK-----LGMENSLTEKIPKLNVPLNPKGRKLSAPG 1084
Query: 799 -------------DEGDHLDDPTQYRKLVGSLIYVTIT-RPDISFAVHTVSKFMQAPRHF 844
+E D+ + +KL+G YV R D+ + ++T+++ + P
Sbjct: 1085 QPGLYIDQQELELEEDDYKMKVHEMQKLIGLASYVGYKFRFDLLYYINTLAQHILFPSKQ 1144
Query: 845 HLSAVQQIIRYLLGTLKRGLFF----PVGSSIKLQAYSDADWAGCPDTRKSTTGWCMFLG 900
L ++I+++ T + L + PV + KL SDA + P KS G L
Sbjct: 1145 VLDMTYELIQFIWNTRDKQLIWHKSKPVKPTNKLVVISDASYGNQP-YYKSQIGNIYLLN 1203
Query: 901 NAPISWKCKKQDSVSKSSTEAEYRAMSAACSEIIWLRGLLTELGFSQDQPTPLHADNTSA 960
I K K S+TEAE A+S + + L L+ EL + L D+ S
Sbjct: 1204 GKVIGGKSTKASLTCTSTTEAEIHAISESVPLLNNLSYLIQELD-KKPITKGLLTDSKST 1262
Query: 961 IQ-IAANPVYHERTKHIEVDCHSIREAYDRRIINLPHVSTSVQTADIFTKSL 1011
I I +N R + +R+ +++ ++ T AD+ TK L
Sbjct: 1263 ISIIISNNEEKFRNRFFGTKAMRLRDEVSGNHLHVCYIETKKNIADVMTKPL 1314
Score = 55.1 bits (131), Expect = 1e-06
Identities = 51/175 (29%), Positives = 81/175 (46%), Gaps = 10/175 (5%)
Query: 266 FIKNQN------ISL*QWGEYMSHSFQEFLQSNGIISQRSCPSTP*QNGVAERKNRHLLD 319
FIKNQ I + + EY + + +FL+ NGI + + +GVAER NR LLD
Sbjct: 294 FIKNQFQASVLVIQMDRGSEYTNRTLHKFLEKNGITPCYTTTADSRAHGVAERLNRTLLD 353
Query: 320 VVRTLLLESHVPSRFWCEALSTAVHLINRMPSP-SIGNESPFTRLYGHPPNYSTLRVFGC 378
RT L S +P+ W A+ + + N + SP S + L G + STL FG
Sbjct: 354 DCRTQLQCSGLPNHLWFSAIEFSTIVRNSLASPKSKKSARQHAGLAG--LDISTLLPFGQ 411
Query: 379 VCYVHLPPQERTKFTAQSVECAFLGYSPHQKGFLCYDPNLRRIRVSRNVIFQENK 433
V+ +K + + L S + G++ Y P+L++ + N + + K
Sbjct: 412 PVIVN-DHNPNSKIHPRGIPGYALHPSRNSYGYIIYLPSLKKTVDTTNYVILQGK 465
>YJZ9_YEAST (P47100) Transposon Ty1 protein B
Length = 1755
Score = 99.0 bits (245), Expect = 6e-20
Identities = 131/532 (24%), Positives = 231/532 (42%), Gaps = 79/532 (14%)
Query: 527 KQAMENDCWQKAIESELLALEENQTWDIVPC--PSSVKP---LGSKFVFSIKLRSDGSID 581
K E + + +A E+ L + +TWD + P + S F+F+ K DG+
Sbjct: 1242 KDIKEKEKYIEAYHKEVNQLLKMKTWDTDEYYDRKEIDPKRVINSMFIFNKK--RDGT-- 1297
Query: 582 RYKAHLVVLGNKQEYGLDYDETFAPVAKMTTVR-----TILAIAASQAWPLHQMDVKNAF 636
+KA V G+ Q + +T+ + TV T L++A + + Q+D+ +A+
Sbjct: 1298 -HKARFVARGDIQ-----HPDTYDSGMQSNTVHHYALMTSLSLALDNNYYITQLDISSAY 1351
Query: 637 LHGDLQEEVYIKLPNGMPTPSPNTVCKLKRSLYGLKQAPRVWFEKFRSTLL---GFEFSQ 693
L+ D++EE+YI+ P + + + +LK+SLYGLKQ+ W+E +S L+ G E +
Sbjct: 1352 LYADIKEELYIRPPPHL--GMNDKLIRLKKSLYGLKQSGANWYETIKSYLIQQCGME--E 1407
Query: 694 SRYDPSLFLQRTPKGMVVLLVYVDDIVVTGSDQDAISRIKNLLHSTFHMKELG------R 747
R +F V + ++VDD+V+ + ++ RI L + K +
Sbjct: 1408 VRGWSCVF----KNSQVTICLFVDDMVLFSKNLNSNKRIIEKLKMQYDTKIINLGESDEE 1463
Query: 748 LTY-FLGLEVHYHHEGVFLNQQKYIQDLVQLAGLTNATLVDTP---MEVNVKYRR----- 798
+ Y LGLE+ Y + KY++ G+ N+ P + +N K R+
Sbjct: 1464 IQYDILGLEIKY-------QRGKYMK-----LGMENSLTEKIPKLNVPLNPKGRKLSAPG 1511
Query: 799 -------------DEGDHLDDPTQYRKLVGSLIYVTIT-RPDISFAVHTVSKFMQAPRHF 844
+E D+ + +KL+G YV R D+ + ++T+++ + P
Sbjct: 1512 QPGLYIDQQELELEEDDYKMKVHEMQKLIGLASYVGYKFRFDLLYYINTLAQHILFPSKQ 1571
Query: 845 HLSAVQQIIRYLLGTLKRGLFF----PVGSSIKLQAYSDADWAGCPDTRKSTTGWCMFLG 900
L ++I+++ T + L + PV + KL SDA + P KS G L
Sbjct: 1572 VLDMTYELIQFIWNTRDKQLIWHKSKPVKPTNKLVVISDASYGNQP-YYKSQIGNIYLLN 1630
Query: 901 NAPISWKCKKQDSVSKSSTEAEYRAMSAACSEIIWLRGLLTELGFSQDQPTPLHADNTSA 960
I K K S+TEAE A+S + + L L+ EL + L D+ S
Sbjct: 1631 GKVIGGKSTKASLTCTSTTEAEIHAISESVPLLNNLSYLIQELD-KKPITKGLLTDSKST 1689
Query: 961 IQ-IAANPVYHERTKHIEVDCHSIREAYDRRIINLPHVSTSVQTADIFTKSL 1011
I I +N R + +R+ +++ ++ T AD+ TK L
Sbjct: 1690 ISIIISNNEEKFRNRFFGTKAMRLRDEVSGNHLHVCYIETKKNIADVMTKPL 1741
Score = 55.1 bits (131), Expect = 1e-06
Identities = 51/175 (29%), Positives = 81/175 (46%), Gaps = 10/175 (5%)
Query: 266 FIKNQN------ISL*QWGEYMSHSFQEFLQSNGIISQRSCPSTP*QNGVAERKNRHLLD 319
FIKNQ I + + EY + + +FL+ NGI + + +GVAER NR LLD
Sbjct: 721 FIKNQFQASVLVIQMDRGSEYTNRTLHKFLEKNGITPCYTTTADSRAHGVAERLNRTLLD 780
Query: 320 VVRTLLLESHVPSRFWCEALSTAVHLINRMPSP-SIGNESPFTRLYGHPPNYSTLRVFGC 378
RT L S +P+ W A+ + + N + SP S + L G + STL FG
Sbjct: 781 DCRTQLQCSGLPNHLWFSAIEFSTIVRNSLASPKSKKSARQHAGLAG--LDISTLLPFGQ 838
Query: 379 VCYVHLPPQERTKFTAQSVECAFLGYSPHQKGFLCYDPNLRRIRVSRNVIFQENK 433
V+ +K + + L S + G++ Y P+L++ + N + + K
Sbjct: 839 PVIVN-DHNPNSKIHPRGIPGYALHPSRNSYGYIIYLPSLKKTVDTTNYVILQGK 892
>YJZ7_YEAST (P47098) Transposon Ty1 protein B
Length = 1755
Score = 98.2 bits (243), Expect = 1e-19
Identities = 129/532 (24%), Positives = 229/532 (42%), Gaps = 79/532 (14%)
Query: 527 KQAMENDCWQKAIESELLALEENQTWDIVPC--PSSVKP---LGSKFVFSIKLRSDGSID 581
K E + + +A E+ L + +TWD + P + S F+F+ K DG+
Sbjct: 1242 KDIKEKEKYIEAYHKEVNQLLKMKTWDTDEYYDRKEIDPKRVINSMFIFNKK--RDGT-- 1297
Query: 582 RYKAHLVVLGNKQEYGLDYDETFAPVAKMTTVR-----TILAIAASQAWPLHQMDVKNAF 636
+KA V G+ Q + +T+ + TV T L++A + + Q+D+ +A+
Sbjct: 1298 -HKARFVARGDIQ-----HPDTYDTGMQSNTVHHYALMTSLSLALDNNYYITQLDISSAY 1351
Query: 637 LHGDLQEEVYIKLPNGMPTPSPNTVCKLKRSLYGLKQAPRVWFEKFRSTLL---GFEFSQ 693
L+ D++EE+YI+ P + + + +LK+S YGLKQ+ W+E +S L+ G E +
Sbjct: 1352 LYADIKEELYIRPPPHL--GMNDKLIRLKKSHYGLKQSGANWYETIKSYLIKQCGME--E 1407
Query: 694 SRYDPSLFLQRTPKGMVVLLVYVDDIVVTGSDQDAISRIKNLLHSTFHMKEL------GR 747
R +F V + ++VDD+++ D +A +I L + K +
Sbjct: 1408 VRGWSCVF----KNSQVTICLFVDDMILFSKDLNANKKIITTLKKQYDTKIINLGESDNE 1463
Query: 748 LTY-FLGLEVHYHHEGVFLNQQKYIQDLVQLAGLTNATLVDTP---MEVNVKYRR----- 798
+ Y LGLE+ Y + KY++ G+ N+ P + +N K R+
Sbjct: 1464 IQYDILGLEIKY-------QRGKYMK-----LGMENSLTEKIPKLNVPLNPKGRKLSAPG 1511
Query: 799 -------------DEGDHLDDPTQYRKLVGSLIYVTIT-RPDISFAVHTVSKFMQAPRHF 844
DE ++ + + +KL+G YV R D+ + ++T+++ + P
Sbjct: 1512 QPGLYIDQDELEIDEDEYKEKVHEMQKLIGLASYVGYKFRFDLLYYINTLAQHILFPSRQ 1571
Query: 845 HLSAVQQIIRYLLGTLKRGLFF----PVGSSIKLQAYSDADWAGCPDTRKSTTGWCMFLG 900
L ++I+++ T + L + P KL A SDA + P KS G L
Sbjct: 1572 VLDMTYELIQFMWDTRDKQLIWHKNKPTEPDNKLVAISDASYGNQP-YYKSQIGNIFLLN 1630
Query: 901 NAPISWKCKKQDSVSKSSTEAEYRAMSAACSEIIWLRGLLTELGFSQDQPTPLHADNTSA 960
I K K S+TEAE A+S + + L L+ EL + L D+ S
Sbjct: 1631 GKVIGGKSTKASLTCTSTTEAEIHAISESVPLLNNLSYLIQELN-KKPIIKGLLTDSRST 1689
Query: 961 IQIAANPVYHE-RTKHIEVDCHSIREAYDRRIINLPHVSTSVQTADIFTKSL 1011
I I + + R + +R+ + + ++ T AD+ TK L
Sbjct: 1690 ISIIKSTNEEKFRNRFFGTKAMRLRDEVSGNNLYVYYIETKKNIADVMTKPL 1741
Score = 55.1 bits (131), Expect = 1e-06
Identities = 51/175 (29%), Positives = 81/175 (46%), Gaps = 10/175 (5%)
Query: 266 FIKNQN------ISL*QWGEYMSHSFQEFLQSNGIISQRSCPSTP*QNGVAERKNRHLLD 319
FIKNQ I + + EY + + +FL+ NGI + + +GVAER NR LLD
Sbjct: 721 FIKNQFQASVLVIQMDRGSEYTNRTLHKFLEKNGITPCYTTTADSRAHGVAERLNRTLLD 780
Query: 320 VVRTLLLESHVPSRFWCEALSTAVHLINRMPSP-SIGNESPFTRLYGHPPNYSTLRVFGC 378
RT L S +P+ W A+ + + N + SP S + L G + STL FG
Sbjct: 781 DCRTQLQCSGLPNHLWFSAIEFSTIVRNSLASPKSKKSARQHAGLAG--LDISTLLPFGQ 838
Query: 379 VCYVHLPPQERTKFTAQSVECAFLGYSPHQKGFLCYDPNLRRIRVSRNVIFQENK 433
V+ +K + + L S + G++ Y P+L++ + N + + K
Sbjct: 839 PVIVN-DHNPNSKIHPRGIPGYALHPSRNSYGYIIYLPSLKKTVDTTNYVILQGK 892
>M240_ARATH (P93290) Hypothetical mitochondrial protein AtMg00240
(ORF111a)
Length = 111
Score = 92.4 bits (228), Expect = 5e-18
Identities = 38/79 (48%), Positives = 61/79 (77%)
Query: 818 IYVTITRPDISFAVHTVSKFMQAPRHFHLSAVQQIIRYLLGTLKRGLFFPVGSSIKLQAY 877
+Y+TITRPD++FAV+ +S+F A R + AV +++ Y+ GT+ +GLF+ S ++L+A+
Sbjct: 1 MYLTITRPDLTFAVNRLSQFSSASRTAQMQAVYKVLHYVKGTVGQGLFYSATSDLQLKAF 60
Query: 878 SDADWAGCPDTRKSTTGWC 896
+D+DWA CPDTR+S TG+C
Sbjct: 61 ADSDWASCPDTRRSVTGFC 79
>M820_ARATH (P92520) Hypothetical mitochondrial protein AtMg00820
(ORF170)
Length = 170
Score = 89.0 bits (219), Expect = 6e-17
Identities = 47/108 (43%), Positives = 68/108 (62%)
Query: 513 MTATLSSIPIPSSYKQAMENDCWQKAIESELLALEENQTWDIVPCPSSVKPLGSKFVFSI 572
+T T + P S A+++ W +A++ EL AL N+TW +VP P + LG K+VF
Sbjct: 18 LTITTTIKKEPKSVIFALKDPGWCQAMQEELDALSRNKTWILVPPPVNQNILGCKWVFKT 77
Query: 573 KLRSDGSIDRYKAHLVVLGNKQEYGLDYDETFAPVAKMTTVRTILAIA 620
KL SDG++DR KA LV G QE G+ + ET++PV + T+RTIL +A
Sbjct: 78 KLHSDGTLDRLKARLVAKGFHQEEGIYFVETYSPVVRTATIRTILNVA 125
>M710_ARATH (P92512) Hypothetical mitochondrial protein AtMg00710
(ORF120)
Length = 120
Score = 62.8 bits (151), Expect = 5e-09
Identities = 30/70 (42%), Positives = 43/70 (60%)
Query: 314 NRHLLDVVRTLLLESHVPSRFWCEALSTAVHLINRMPSPSIGNESPFTRLYGHPPNYSTL 373
NR +++ VR++L E +P F +A +TAVH+IN+ PS +I P + P YS L
Sbjct: 2 NRTIIEKVRSMLCECGLPKTFRADAANTAVHIINKYPSTAINFHVPDEVWFQSVPTYSYL 61
Query: 374 RVFGCVCYVH 383
R FGCV Y+H
Sbjct: 62 RRFGCVAYIH 71
>POL_FENV1 (P31792) Pol polyprotein [Contains: Reverse
transcriptase/ribonuclease H (EC 2.7.7.49) (EC 3.1.26.4)
(RT); Integrase (IN)] (Fragment)
Length = 1046
Score = 39.3 bits (90), Expect = 0.054
Identities = 29/94 (30%), Positives = 45/94 (47%), Gaps = 4/94 (4%)
Query: 280 YMSHSFQEFLQSNGIISQRSCPSTP*QNGVAERKNRHLLDVVRTLLLESHVPSRFWCEAL 339
++S Q ++ GI + C P +G ER NR + + + L LE+ + W L
Sbjct: 832 FVSQVSQGLARTLGINWKLHCAYRPQSSGQVERMNRTIKETLTKLTLETGLKD--WRRLL 889
Query: 340 STAVHLINRMPSPSIGNESPFTRLYGHPPNYSTL 373
S A L+ +P+ +P+ LYG PP STL
Sbjct: 890 SLA--LLRARNTPNRFGLTPYEILYGGPPPLSTL 921
>POL_BAEVM (P10272) Pol polyprotein [Contains: Protease (EC 3.4.23.-);
Reverse transcriptase/ribonuclease H (EC 2.7.7.49) (EC
3.1.26.4) (RT); Integrase (IN)]
Length = 1189
Score = 38.1 bits (87), Expect = 0.12
Identities = 29/94 (30%), Positives = 44/94 (45%), Gaps = 4/94 (4%)
Query: 280 YMSHSFQEFLQSNGIISQRSCPSTP*QNGVAERKNRHLLDVVRTLLLESHVPSRFWCEAL 339
++S Q + GI + C P +G ER NR + + + L LE+ + W L
Sbjct: 975 FVSQVSQGLARILGINWKLHCAYRPQSSGQVERMNRTIKETLTKLTLETGLKD--WRRLL 1032
Query: 340 STAVHLINRMPSPSIGNESPFTRLYGHPPNYSTL 373
S A L+ +P+ +P+ LYG PP STL
Sbjct: 1033 SLA--LLRARNTPNRFGLTPYEILYGGPPPLSTL 1064
>KCD1_MOUSE (Q03719) Potassium voltage-gated channel subfamily D
member 1 (Voltage-gated potassium channel subunit Kv4.1)
(mShal)
Length = 651
Score = 33.5 bits (75), Expect = 3.0
Identities = 25/85 (29%), Positives = 38/85 (44%), Gaps = 5/85 (5%)
Query: 444 SSPISILPLFYDSHSRQQPSKPLLTYKRRSTATHGPPQDNSLVAGPVEEPAPLRRSSRES 503
S P+ LF SR+ + + ++ + G Q+ +AG PAP RSS +
Sbjct: 520 SQPMGPGSLFSSCCSRRVNRRAIRLANSTASVSRGSMQELDTLAGLRRSPAPQTRSSLNA 579
Query: 504 KPPERY-INC----MTATLSSIPIP 523
KP + +NC A + SIP P
Sbjct: 580 KPHDSLDLNCDSRDFVAAIISIPTP 604
>Y624_MYCPU (Q98PU5) Hypothetical UPF0082 protein MYPU_6240
Length = 241
Score = 33.1 bits (74), Expect = 3.9
Identities = 31/143 (21%), Positives = 58/143 (39%), Gaps = 10/143 (6%)
Query: 631 DVKNAFLHGDLQEEVYIKLPNGMPTPSPNTVCKLKRSLYGLKQAPRVWFEKFRSTLLGFE 690
D A + +E+Y+ G P N +L + K P+V EK + G
Sbjct: 18 DAARAKIFQKFAKEIYVAAVAGGPDVDSNPALRLAVNKAKSKSMPKVNIEKAIAKASGNS 77
Query: 691 FSQSRYDPSLFLQRTPKGMVVLLVYVDDIVVTGSDQDAISRIKNLLHSTFHMKELGRLTY 750
S S Y ++ KG+ +L++ + D +R+ + + S F+ K G+L+
Sbjct: 78 KSSSTYSEYIYSGTVSKGVQILVICL---------SDNFNRLSSNIKSYFN-KAGGQLSK 127
Query: 751 FLGLEVHYHHEGVFLNQQKYIQD 773
+ Y +G+ +K I +
Sbjct: 128 QGSIPYVYQQKGLIEIDKKLISE 150
>POL_SMSAV (P03359) Pol polyprotein [Contains: Reverse
transcriptase/ribonuclease H (EC 2.7.7.49) (EC 3.1.26.4)
(RT); Integrase (IN)] (Fragment)
Length = 294
Score = 32.7 bits (73), Expect = 5.1
Identities = 24/76 (31%), Positives = 35/76 (45%), Gaps = 4/76 (5%)
Query: 293 GIISQRSCPSTP*QNGVAERKNRHLLDVVRTLLLESHVPSRFWCEALSTAVHLINRMPSP 352
GI + C P +G ER NR + + + L LE+ + W L A L+ +P
Sbjct: 89 GINWKLHCAYRPQSSGQVERMNRTIKETLTKLALETG--GKDWVALLPLA--LLRAKNTP 144
Query: 353 SIGNESPFTRLYGHPP 368
S +P+ LYG PP
Sbjct: 145 SRFGLTPYEILYGGPP 160
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.339 0.147 0.496
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 113,612,085
Number of Sequences: 164201
Number of extensions: 4716198
Number of successful extensions: 14659
Number of sequences better than 10.0: 22
Number of HSP's better than 10.0 without gapping: 17
Number of HSP's successfully gapped in prelim test: 5
Number of HSP's that attempted gapping in prelim test: 14587
Number of HSP's gapped (non-prelim): 38
length of query: 1019
length of database: 59,974,054
effective HSP length: 120
effective length of query: 899
effective length of database: 40,269,934
effective search space: 36202670666
effective search space used: 36202670666
T: 11
A: 40
X1: 15 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (21.8 bits)
S2: 71 (32.0 bits)
Medicago: description of AC146527.6


