

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146527.1 - phase: 0
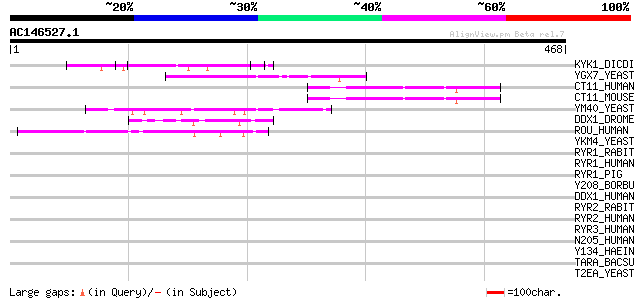
(468 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
KYK1_DICDI (P18160) Non-receptor tyrosine kinase spore lysis A (... 99 2e-20
YGX7_YEAST (P53076) Hypothetical 108.2 kDa protein in SAP4-OST5 ... 56 2e-07
CT11_HUMAN (Q9NWU2) Protein C20orf11 55 3e-07
CT11_MOUSE (Q9D7M1) Protein C20orf11 homolog 55 5e-07
YM40_YEAST (Q03212) Hypothetical 62.5 kDa protein in ALD2-DDR48 ... 54 9e-07
DDX1_DROME (Q9VNV3) ATP-dependent helicase DDX1 (DEAD box protei... 49 4e-05
ROU_HUMAN (Q00839) Heterogenous nuclear ribonucleoprotein U (hnR... 45 5e-04
YKM4_YEAST (P32343) Hypothetical 65.1 kDa protein in RRN3-SRP21 ... 42 0.003
RYR1_RABIT (P11716) Ryanodine receptor 1 (Skeletal muscle-type r... 41 0.006
RYR1_HUMAN (P21817) Ryanodine receptor 1 (Skeletal muscle-type r... 40 0.010
RYR1_PIG (P16960) Ryanodine receptor 1 (Skeletal muscle-type rya... 40 0.013
Y208_BORBU (O51226) Hypothetical protein BB0208 39 0.029
DDX1_HUMAN (Q92499) ATP-dependent helicase DDX1 (DEAD-box protei... 39 0.037
RYR2_RABIT (P30957) Ryanodine receptor 2 (Cardiac muscle-type ry... 37 0.11
RYR2_HUMAN (Q92736) Ryanodine receptor 2 (Cardiac muscle-type ry... 35 0.41
RYR3_HUMAN (Q15413) Ryanodine receptor 3 (Brain-type ryanodine r... 35 0.54
N205_HUMAN (Q92621) Nuclear pore complex protein Nup205 (Nucleop... 34 0.71
Y134_HAEIN (P43952) Hypothetical protein HI0134 34 0.92
TARA_BACSU (Q8RKI7) Putative N-acetylmannosaminyltransferase (EC... 33 1.6
T2EA_YEAST (P36100) Transcription initiation factor IIE, alpha s... 31 6.0
>KYK1_DICDI (P18160) Non-receptor tyrosine kinase spore lysis A (EC
2.7.1.112) (Tyrosine-protein kinase 1)
Length = 1584
Score = 99.0 bits (245), Expect = 2e-20
Identities = 65/203 (32%), Positives = 95/203 (46%), Gaps = 31/203 (15%)
Query: 49 ESPSELNTINSSGGFVVVATDKLSVKYT------SVNLHGHDVGVIQANKVA-------- 94
E PS+ +TI SGG ++ ++ L K S + H V+ N+ A
Sbjct: 184 EKPSKQSTIKDSGGSSIIPSEDLIPKEEFEVCRWSEKKNYHGKHVVVRNRTAFLPLDSPK 243
Query: 95 ----------PMKRIVYYFEIFVKDAGVKGQVSIGFTTESFKMRRQPGWEANSCGYHGDD 144
P YFE+ + D KGQ+SIG + GW S GYH DD
Sbjct: 244 DTIGGVRATQPFGEGFCYFEVII-DQLDKGQLSIGLANLEYPTFYHVGWMPRSYGYHNDD 302
Query: 145 GFLYR-----GQGKGEAFGPTYTTGDIVGAGINYAAQEFFFTKNGQVVGTVFKEMKGPLF 199
G +R G +GE++G +Y GDI+G G+++ ++E FFTKNG +GT F + G +
Sbjct: 303 GRKFRWREEPGVNEGESYGSSYKKGDIIGCGLSFTSREIFFTKNGMYLGTAFSNVYGVFY 362
Query: 200 PTVAVHSQNEEVHVNFGQKPFTF 222
P+VA + + FG PF F
Sbjct: 363 PSVAFNEPGISITGVFG-PPFKF 384
Score = 77.4 bits (189), Expect = 7e-14
Identities = 42/127 (33%), Positives = 68/127 (53%), Gaps = 3/127 (2%)
Query: 90 ANKVAPMKRIVYYFEIFVKDAGVKGQVSIGFTTESFKMRRQPGWEANSCGYHGDDGFLYR 149
A P YFE+F+ + G ++ G TT + PG S GY GD G Y
Sbjct: 49 AKSTQPFSSSFTYFELFITN-GNGDKICFGLTTNDHPIEVYPGNYQGSYGYSGD-GKCYF 106
Query: 150 GQGKGEAFGPTYTTGDIVGAGINYAAQEFFFTKNGQVVGTVFKEMK-GPLFPTVAVHSQN 208
G +G +GP++++GD+VG G + +++ +FTKNG +G +++ L+PTV + +
Sbjct: 107 GTNEGRVYGPSFSSGDVVGCGYDSSSKTLYFTKNGVYLGVAAQKVNLIGLYPTVGLQNPG 166
Query: 209 EEVHVNF 215
E V +NF
Sbjct: 167 ESVVINF 173
Score = 64.3 bits (155), Expect = 6e-10
Identities = 34/112 (30%), Positives = 61/112 (54%), Gaps = 9/112 (8%)
Query: 100 VYYFEIFVKDAGVKGQVSIGFTTESFKMRRQPGWEANSCGYHGDDGFLYRGQGKGEAFGP 159
+ YFE++++ KG +++G + ++ + G E S G+ +G Y G GE +GP
Sbjct: 749 ICYFEVYLEGHDKKGSITVGLSHSTYPFIKHIGREPKSYGF-SSEGEKYGGSEIGEPYGP 807
Query: 160 TYT-TGD------IVGAGINYAAQEFFFTKNGQVVGTVFKEM-KGPLFPTVA 203
+ GD ++G GIN + ++ FFTKNG +G F + PL+P+++
Sbjct: 808 FFFFDGDSIASSCVIGCGINTSTRDIFFTKNGHYLGVAFSRVTSDPLYPSIS 859
>YGX7_YEAST (P53076) Hypothetical 108.2 kDa protein in SAP4-OST5
intergenic region
Length = 958
Score = 56.2 bits (134), Expect = 2e-07
Identities = 49/175 (28%), Positives = 78/175 (44%), Gaps = 10/175 (5%)
Query: 132 GWEANSCGYHGDDGFLYRGQGKGEAFGPTYTTGDIVGAGINYAAQEFFFTKNGQVVGTVF 191
G++ N GY G DG + + + + + D++G GIN+ FFTKNG +G F
Sbjct: 504 GFDLNVFGYCGFDGLITNSTEQSKEYAKPFGRDDVIGCGINFIDGSIFFTKNGIHLGNAF 563
Query: 192 KEMKGPLF-PTVAVHSQNEEVHVNFG-QKPFTFDLKEYEAQERMKQQIKIEEVPVSPNAS 249
++ F P VA+ N + NFG + F FD+ Y Q++ K + E +
Sbjct: 564 TDLNDLEFVPYVALRPGN-SIKTNFGLNEDFVFDIIGY--QDKWK-SLAYEHICRGRQMD 619
Query: 250 YGIVRSYLLHYGYEDTLNSFDEASRST---IPPINIVQENGIDDQETTYALNHRK 301
I + ED + E ++ST ++I QE+G + T LN K
Sbjct: 620 VSI-EEFDSDESEEDETENGPEENKSTNVNEDLMDIDQEDGAAGNKDTKKLNDEK 673
Score = 35.4 bits (80), Expect = 0.32
Identities = 43/197 (21%), Positives = 77/197 (38%), Gaps = 29/197 (14%)
Query: 246 PNASYGIVRSYLLHYGYEDTLNSFDEASRSTIPPIN--------IVQENGIDDQETTYAL 297
PN ++ YL+H G D F + + +N +++ N + +
Sbjct: 711 PNTLNVMINDYLIHEGLVDVAKGFLKDLQKDAVNVNGQHSESKDVIRHNERQIMKEERMV 770
Query: 298 NHRKTLRQLIRDGEIDAAFGKLREWYPQIAEDNTSAMCFLLHCQKFIELVRVGA------ 351
R+ LR LI G+I + P + ++N + F L ++ +++ +
Sbjct: 771 KIRQELRYLINKGQISKCINYIDNEIPDLLKNNLE-LVFELKLANYLVMIKKSSSKDDDE 829
Query: 352 LEEAVKYGRIELSSFFGLSL---------FEDIVQDCVALLAYERPLESA----VGYLLK 398
+E + G+ ELS+ F F + + ALLAY PL A GYL
Sbjct: 830 IENLILKGQ-ELSNEFIYDTKIPQSLRDRFSGQLSNVSALLAYSNPLVEAPKEISGYLSD 888
Query: 399 DSQREVVADAVNAMILS 415
+ +E + N IL+
Sbjct: 889 EYLQERLFQVSNNTILT 905
>CT11_HUMAN (Q9NWU2) Protein C20orf11
Length = 228
Score = 55.5 bits (132), Expect = 3e-07
Identities = 44/165 (26%), Positives = 79/165 (47%), Gaps = 17/165 (10%)
Query: 252 IVRSYLLHYGYEDTLNSFDEASRSTIPPINIVQENGIDDQETTYALNHRKTLRQLIRDGE 311
++ +YL+ G+++ F E+GI+ L+ R +R++I G+
Sbjct: 32 LIMNYLVTEGFKEAAEKFR-------------MESGIEPSVDLETLDERIKIREMILKGQ 78
Query: 312 IDAAFGKLREWYPQIAEDNTSAMCFLLHCQKFIELVRVGALEEAVKYGRIELSSFFGLSL 371
I A + +P++ + N + F L Q IEL+R E A+++ + +L+ G
Sbjct: 79 IQEAIALINSLHPELLDTNRY-LYFHLQQQHLIELIRQRETEAALEFAQTQLAEQ-GEES 136
Query: 372 FEDI--VQDCVALLAYERPLESAVGYLLKDSQREVVADAVNAMIL 414
E + ++ +ALLA++ P ES G LL QR+ V VN +L
Sbjct: 137 RECLTEMERTLALLAFDSPEESPFGDLLHTMQRQKVWSEVNQAVL 181
>CT11_MOUSE (Q9D7M1) Protein C20orf11 homolog
Length = 228
Score = 54.7 bits (130), Expect = 5e-07
Identities = 44/165 (26%), Positives = 79/165 (47%), Gaps = 17/165 (10%)
Query: 252 IVRSYLLHYGYEDTLNSFDEASRSTIPPINIVQENGIDDQETTYALNHRKTLRQLIRDGE 311
++ +YL+ G+++ F E+GI+ L+ R +R++I G+
Sbjct: 32 LIMNYLVTEGFKEAAEKFR-------------MESGIEPSVDLETLDERIKIREMILKGQ 78
Query: 312 IDAAFGKLREWYPQIAEDNTSAMCFLLHCQKFIELVRVGALEEAVKYGRIELSSFFGLSL 371
I A + +P++ + N + F L Q IEL+R E A+++ + +L+ G
Sbjct: 79 IQEAIALINSLHPELLDTNRY-LYFHLQQQHLIELIRQRETEAALEFAQTQLAEQ-GEES 136
Query: 372 FEDI--VQDCVALLAYERPLESAVGYLLKDSQREVVADAVNAMIL 414
E + ++ +ALLA++ P ES G LL QR+ V VN +L
Sbjct: 137 RECLTEMERTLALLAFDSPEESPFGDLLHMMQRQKVWSEVNQAVL 181
>YM40_YEAST (Q03212) Hypothetical 62.5 kDa protein in ALD2-DDR48
intergenic region
Length = 550
Score = 53.9 bits (128), Expect = 9e-07
Identities = 62/228 (27%), Positives = 93/228 (40%), Gaps = 34/228 (14%)
Query: 65 VVATDKLSVKYTSVNLHGHDVGVIQANKVAPMKRIVYY--FEIFVKDAGV-------KGQ 115
V+ +K V + + N I N P VYY F+IF D + KG
Sbjct: 206 VIVENKTEVSFLNYNYDAS----ISTNLPIPCINKVYYCEFKIFETDGPLNSDENVSKGV 261
Query: 116 VSIGFTTESFKMRRQPGWEANSCGYHGD------DGFLYRGQGKGEAFGPTYTTGDIVGA 169
+S G +T+ + R PG +S Y + D F Q + P GDIVG
Sbjct: 262 ISFGLSTQPYPYFRLPGRHHHSIAYDSNGARRFNDSFKLNEQLR--TLFPQCEKGDIVGI 319
Query: 170 GINYAAQEFFFTKNGQVVG--TVFKEMKG----PLFPTVAVHSQNEEVHVNFGQKPFTFD 223
G + FFT+NG+ + +V ++G L+P + + ++HVNFG F +
Sbjct: 320 GYRSRSGTVFFTRNGKKLNEKSVGGHIRGWKFQYLYPIIGSNVPC-QIHVNFGTYGFVY- 377
Query: 224 LKEYEAQERMKQQIKIEEVPVSPNASYGIVRSYLLHYGYEDTLNSFDE 271
EA + K + + P + + LL G ED N FDE
Sbjct: 378 ---IEANVKKWGYAKSNGIKLPPPSYEDYGKDTLLESGGED--NDFDE 420
>DDX1_DROME (Q9VNV3) ATP-dependent helicase DDX1 (DEAD box protein
1)
Length = 727
Score = 48.5 bits (114), Expect = 4e-05
Identities = 41/136 (30%), Positives = 62/136 (45%), Gaps = 33/136 (24%)
Query: 101 YYFEIFVKDAGVKGQVSIGFTTESFKMRRQPGWEANSCGYHGDDGFLYRGQGK------- 153
+YFE V D G+ +G++T+ Q + +C GF + G GK
Sbjct: 132 FYFEATVTDEGL---CRVGWSTQ------QANLDLGTCRM----GFGFGGTGKKSNNRQF 178
Query: 154 ---GEAFGPTYTTGDIVGAGINYAAQEFFFTKNGQVVGTVFK----EMKGPLFPTVAVHS 206
GEAFG D++G ++ QE FTKNGQ +G F+ K +P V +
Sbjct: 179 DDYGEAFGKA----DVIGCLLDLKNQEVSFTKNGQNLGVAFRLPDNLAKETFYPAVVL-- 232
Query: 207 QNEEVHVNFGQKPFTF 222
+N E+ NFG+ F +
Sbjct: 233 KNAEMQFNFGKTDFKY 248
>ROU_HUMAN (Q00839) Heterogenous nuclear ribonucleoprotein U (hnRNP
U) (Scaffold attachment factor A) (SAF-A) (pp120)
Length = 824
Score = 44.7 bits (104), Expect = 5e-04
Identities = 57/228 (25%), Positives = 94/228 (41%), Gaps = 24/228 (10%)
Query: 7 KNTIEKPDQDQNQDLGFHFLELSRRKAKLDSEDFEVDDEEKEESPSELNTINSSGGFVVV 66
K +++P +D + + E +AK E +DE +++ L+T N F +
Sbjct: 242 KRGVKRPREDHGRGYFEYIEENKYSRAKSPQPPVEEEDEHFDDTVVCLDTYNCDLHF-KI 300
Query: 67 ATDKLSVKYTSVNLHGHDVGVIQANKVAPMKRIVYYFEIFVKDAGVKGQVSIGFTTESFK 126
+ D+LS ++ +A+ ++ FE+ V + K V +T +
Sbjct: 301 SRDRLSASSLTMESFAFLWAGGRASYGVSKGKVC--FEMKVTE---KIPVRHLYTKDIDI 355
Query: 127 MRRQPGWEANSCG-YHGDDGFLYRGQGKG--------EAFGPTYTTGDIVGAGINYAAQ- 176
+ GW + G G++ F Y KG E +G + D++ N+ +
Sbjct: 356 HEVRIGWSLTTSGMLLGEEEFSYGYSLKGIKTCNCETEDYGEKFDENDVITCFANFESDE 415
Query: 177 -EFFFTKNGQVVGTVFKEMK-----GPLFPTVAVHSQNEEVHVNFGQK 218
E + KNGQ +G FK K PLFP V H N V NFGQK
Sbjct: 416 VELSYAKNGQDLGVAFKISKEVLAGRPLFPHVLCH--NCAVEFNFGQK 461
>YKM4_YEAST (P32343) Hypothetical 65.1 kDa protein in RRN3-SRP21
intergenic region
Length = 579
Score = 42.4 bits (98), Expect = 0.003
Identities = 46/174 (26%), Positives = 65/174 (36%), Gaps = 13/174 (7%)
Query: 35 LDSEDFEVDDEEKEES----PSELNTINSSGGFV--VVATDKLSVKYTSVNLHGHDVGVI 88
LDS+D + + +S PS + I+ G F+ + DKL ++++ N V
Sbjct: 162 LDSDDQQFIKDRGIQSYFLLPSINDNIDEYGNFLPSFIVQDKLDIQFSKFNKSSSTV--- 218
Query: 89 QANKVAPMKRI-VYYFEIFVKDAGVKGQ--VSIGFTTESFKMRRQPGWEANSCGYHGDDG 145
N P R YFE+ + K SIG TT + R PG S Y
Sbjct: 219 -MNYPLPHNRKDAVYFEVKIFRHIQKSNSIFSIGLTTVPYPYFRVPGMAKYSIAYESTGK 277
Query: 146 FLYRGQGKGEAFGPTYTTGDIVGAGINYAAQEFFFTKNGQVVGTVFKEMKGPLF 199
P GD VG G Y F T NG+ + V + + LF
Sbjct: 278 LRINNPFTASTLLPKLEEGDTVGFGYRYKTGTIFITHNGKKLMDVTQNIGIDLF 331
>RYR1_RABIT (P11716) Ryanodine receptor 1 (Skeletal muscle-type
ryanodine receptor) (RyR1) (RYR-1) (Skeletal muscle
calcium release channel)
Length = 5037
Score = 41.2 bits (95), Expect = 0.006
Identities = 36/140 (25%), Positives = 63/140 (44%), Gaps = 12/140 (8%)
Query: 85 VGVIQANKVAPMKRIVYYFEIFVKDAGVKGQVSIGFTTESFKMRRQPGWEANSCGYHGDD 144
V + +A K ++ +YFE +A G++ +G+ + + G + + ++G
Sbjct: 1072 VRIFRAEKSYTVQSGRWYFEF---EAVTTGEMRVGWARPELRPDVELGADELAYVFNGHR 1128
Query: 145 GFLYRGQGKGEAFGPTYTTGDIVGAGINYAAQEFFFTKNGQVV------GTVFKEMK-GP 197
G R E FG + +GD+VG I+ FT NG+V+ T F+E++ G
Sbjct: 1129 G--QRWHLGSEPFGRPWQSGDVVGCMIDLTENTIIFTLNGEVLMSDSGSETAFREIEIGD 1186
Query: 198 LFPTVAVHSQNEEVHVNFGQ 217
F V + H+N GQ
Sbjct: 1187 GFLPVCSLGPGQVGHLNLGQ 1206
>RYR1_HUMAN (P21817) Ryanodine receptor 1 (Skeletal muscle-type
ryanodine receptor) (RyR1) (RYR-1) (Skeletal muscle
calcium release channel)
Length = 5038
Score = 40.4 bits (93), Expect = 0.010
Identities = 36/140 (25%), Positives = 62/140 (43%), Gaps = 12/140 (8%)
Query: 85 VGVIQANKVAPMKRIVYYFEIFVKDAGVKGQVSIGFTTESFKMRRQPGWEANSCGYHGDD 144
V + +A K ++ +YFE +A G++ +G+ + + G + + ++G
Sbjct: 1071 VRIFRAEKSYTVQSGRWYFEF---EAVTTGEMRVGWARPELRPDVELGADELAYVFNGHR 1127
Query: 145 GFLYRGQGKGEAFGPTYTTGDIVGAGINYAAQEFFFTKNGQVV------GTVFKEMK-GP 197
G R E FG + GD+VG I+ FT NG+V+ T F+E++ G
Sbjct: 1128 G--QRWHLGSEPFGRPWQPGDVVGCMIDLTENTIIFTLNGEVLMSDSGSETAFREIEIGD 1185
Query: 198 LFPTVAVHSQNEEVHVNFGQ 217
F V + H+N GQ
Sbjct: 1186 GFLPVCSLGPGQVGHLNLGQ 1205
>RYR1_PIG (P16960) Ryanodine receptor 1 (Skeletal muscle-type
ryanodine receptor) (RyR1) (RYR-1) (Skeletal muscle
calcium release channel)
Length = 5035
Score = 40.0 bits (92), Expect = 0.013
Identities = 35/140 (25%), Positives = 63/140 (45%), Gaps = 12/140 (8%)
Query: 85 VGVIQANKVAPMKRIVYYFEIFVKDAGVKGQVSIGFTTESFKMRRQPGWEANSCGYHGDD 144
V + +A K ++ +YFE +A G++ +G+ + + G + + ++G
Sbjct: 1072 VRIFRAEKSYAVQSGRWYFEF---EAVTTGEMRVGWARPELRPDVELGADELAYVFNGHR 1128
Query: 145 GFLYRGQGKGEAFGPTYTTGDIVGAGINYAAQEFFFTKNGQVV------GTVFKEMK-GP 197
G R E FG + +GD+VG I+ FT NG+V+ T F++++ G
Sbjct: 1129 G--QRWHLGSELFGRPWQSGDVVGCMIDLTENTIIFTLNGEVLMSDSGSETAFRDIEVGD 1186
Query: 198 LFPTVAVHSQNEEVHVNFGQ 217
F V + H+N GQ
Sbjct: 1187 GFLPVCSLGPGQVGHLNLGQ 1206
>Y208_BORBU (O51226) Hypothetical protein BB0208
Length = 581
Score = 38.9 bits (89), Expect = 0.029
Identities = 35/168 (20%), Positives = 73/168 (42%), Gaps = 13/168 (7%)
Query: 214 NFGQKPFTFDLKEYEAQE--RMKQQIKIEEVPVSPNASYGIVRSYLLHYGYEDTLNSFDE 271
N +K F +KE+ + ++ QI + + PN Y H+ + + + +
Sbjct: 409 NKDKKIFCQRMKEFFKSKTFEIQNQIFVYFLSFYPNIEYK-------HFKFISDILLYLD 461
Query: 272 ASRSTIPP----INIVQENGIDDQETTYALNHRKTLRQLIRDGEIDAAFGKLREWYPQIA 327
++ TIP I + N +D Q++ + KT ++++ + F + + +
Sbjct: 462 ENKITIPKKIIKIQKINPNQLDYQKSYLLIKSLKTRSRILKIIIKNIRFNLIEKLEDENE 521
Query: 328 EDNTSAMCFLLHCQKFIELVRVGALEEAVKYGRIELSSFFGLSLFEDI 375
+ AMC+L++C+ +EL ++ K +E SFF L + I
Sbjct: 522 KKLLPAMCYLIYCENLVELKNNRKIKSNEKNSLLEFISFFKTKLKQKI 569
>DDX1_HUMAN (Q92499) ATP-dependent helicase DDX1 (DEAD-box protein
1) (DEAD-box protein-retinoblastoma) (DBP-RB)
Length = 740
Score = 38.5 bits (88), Expect = 0.037
Identities = 28/92 (30%), Positives = 45/92 (48%), Gaps = 12/92 (13%)
Query: 145 GFLYRGQGKG------EAFGPTYTTGDIVGAGINYAAQEFFFTKNGQVVGTVFK---EMK 195
GF + G GK + +G +T D +G ++ F+KNG+ +G F+ MK
Sbjct: 164 GFGFGGTGKKSHNKQFDNYGEEFTMHDTIGCYLDIDKGHVKFSKNGKDLGLAFEIPPHMK 223
Query: 196 G-PLFPTVAVHSQNEEVHVNFGQKPFTFDLKE 226
LFP + +N E+ NFG++ F F K+
Sbjct: 224 NQALFPACVL--KNAELKFNFGEEEFKFPPKD 253
>RYR2_RABIT (P30957) Ryanodine receptor 2 (Cardiac muscle-type
ryanodine receptor) (RyR2) (RYR-2) (Cardiac muscle
ryanodine receptor-calcium release channel)
Length = 4969
Score = 37.0 bits (84), Expect = 0.11
Identities = 25/107 (23%), Positives = 51/107 (47%), Gaps = 7/107 (6%)
Query: 82 GHDVGVIQANKVAPMKRIVYYFEIFVKDAGVKGQVSIGFTTESFKMRRQPGWEANSCGYH 141
G + +A K +K +YFE +A G + +G++ + ++ G + + +
Sbjct: 1082 GERFRIFRAEKTYAVKAGRWYFEF---EAVTSGDMRVGWSRPGCQPDQELGSDERAFAF- 1137
Query: 142 GDDGFLYRGQGKG-EAFGPTYTTGDIVGAGINYAAQEFFFTKNGQVV 187
DGF + +G E +G ++ GD+VG ++ FT NG+++
Sbjct: 1138 --DGFKAQRWHQGNEHYGRSWQAGDVVGCMVDMNEHTMMFTLNGEIL 1182
>RYR2_HUMAN (Q92736) Ryanodine receptor 2 (Cardiac muscle-type
ryanodine receptor) (RyR2) (RYR-2) (Cardiac muscle
ryanodine receptor-calcium release channel) (hRYR-2)
Length = 4967
Score = 35.0 bits (79), Expect = 0.41
Identities = 25/107 (23%), Positives = 50/107 (46%), Gaps = 7/107 (6%)
Query: 82 GHDVGVIQANKVAPMKRIVYYFEIFVKDAGVKGQVSIGFTTESFKMRRQPGWEANSCGYH 141
G + +A K +K +YFE AG + +G++ + ++ G + + +
Sbjct: 1082 GERFRIFRAEKTYAVKAGRWYFEFETVTAG---DMRVGWSRPGCQPDQELGSDERAFAF- 1137
Query: 142 GDDGFLYRGQGKG-EAFGPTYTTGDIVGAGINYAAQEFFFTKNGQVV 187
DGF + +G E +G ++ GD+VG ++ FT NG+++
Sbjct: 1138 --DGFKAQRWHQGNEHYGRSWQAGDVVGCMVDMNEHTMMFTLNGEIL 1182
>RYR3_HUMAN (Q15413) Ryanodine receptor 3 (Brain-type ryanodine
receptor) (RyR3) (RYR-3) (Brain ryanodine
receptor-calcium release channel)
Length = 4870
Score = 34.7 bits (78), Expect = 0.54
Identities = 26/89 (29%), Positives = 42/89 (46%), Gaps = 5/89 (5%)
Query: 101 YYFEIFVKDAGVKGQVSIGFTTESFKMRRQPGWEANSCGYHGDDGFLYRGQGKGEAFGPT 160
+YFE V G + +G+ + + G + + + G+ G + QG G FG T
Sbjct: 1087 WYFEFEVVTGG---DMRVGWARPGCRPDVELGADDQAFVFEGNRGQRWH-QGSGY-FGRT 1141
Query: 161 YTTGDIVGAGINYAAQEFFFTKNGQVVGT 189
+ GD+VG IN FT NG+++ T
Sbjct: 1142 WQPGDVVGCMINLDDASMIFTLNGELLIT 1170
>N205_HUMAN (Q92621) Nuclear pore complex protein Nup205
(Nucleoporin Nup205) (205 kDa nucleoporin)
Length = 2012
Score = 34.3 bits (77), Expect = 0.71
Identities = 32/104 (30%), Positives = 47/104 (44%), Gaps = 4/104 (3%)
Query: 332 SAMCFLLHCQKFIELVRVGALEEAVKYGRIELSSFFGLSLFEDIVQDCVALLAYERPLES 391
S M LL+ +EL + LEE VK + F G E VQ C+ALL E+
Sbjct: 804 SLMYHLLNESPMLELA-LSLLEEGVKQ-LDTYAPFPGKKHLEKAVQHCLALLNLTLQKEN 861
Query: 392 AVGYLLKDSQREVVADAVNAMILSTNPNIKVTKNCLHSNLERLL 435
LL++SQ ++ + ++ NP K N + N+ R L
Sbjct: 862 LFMDLLRESQLALIVCPLEQLLQGINPRTKKADNVV--NIARYL 903
>Y134_HAEIN (P43952) Hypothetical protein HI0134
Length = 391
Score = 33.9 bits (76), Expect = 0.92
Identities = 40/164 (24%), Positives = 71/164 (42%), Gaps = 22/164 (13%)
Query: 160 TYTTGDIVGAGINYAAQEFFFTKNGQVVGTVFKEMKGPLFPTVAVHS--QNEEVHVNFGQ 217
++++ ++ +N+ AQ NG+++G +M FP + QN+E++ F Q
Sbjct: 247 SFSSSSLIQGKLNFLAQ------NGEILGVNLLDMVAQYFPINYNNDLLQNKELNTRFEQ 300
Query: 218 KPFTFDLKEYEAQERMKQQIKIE-EVPVSPNASYGIVRSYLLHYGYEDTLNSFDEASRST 276
F L+ + Q ++ + KIE + P GI+ + + LNS D+ ++
Sbjct: 301 ----FYLQLFLQQNQLIAE-KIELKTPALLGQGKGIIDLNRMECNVDINLNSTDQRYQNL 355
Query: 277 IPPINIVQENGIDDQETTYALNHRKTLRQLIRDGEIDAAFGKLR 320
PIN + Y +N K R + IDA KLR
Sbjct: 356 TLPINFFG----NCNSPQYKINFTKKFRHQL----IDAIKEKLR 391
>TARA_BACSU (Q8RKI7) Putative N-acetylmannosaminyltransferase (EC
2.4.1.187) (UDP-N-acetylmannosamine transferase)
(N-acetylglucosaminyldiphospho-undecaprenol
N-acetyl-beta-D-mannosaminyltransferase)
Length = 257
Score = 33.1 bits (74), Expect = 1.6
Identities = 44/165 (26%), Positives = 65/165 (38%), Gaps = 32/165 (19%)
Query: 18 NQDLGFHFLELSRRKAKLDSEDFEVDDEEKEESPSELNTINSSGGFVVVATDKLSVKYTS 77
N ++GF ++ R +A L S DF + D G VV KL K
Sbjct: 40 NPEIGFEAMQNPRYEAVLSSADFILPD-----------------GIGVVMVSKLIGKPLQ 82
Query: 78 VNLHGHDVGVIQANKVAPMKRIVYYFEIFVKDAGVKGQVSIGFTTESFKMRRQPGWEANS 137
+ G+D+ +K K+ V+++ G V I T E K R PG E
Sbjct: 83 SRIAGYDLFTSLLDKADQKKKRVFFY-------GAAKHV-IAETIERVK-RDFPGIEI-- 131
Query: 138 CGYHGDDGFLYRGQGKGEAFGPTYTTGDIVGAGINYAAQEFFFTK 182
GY DG++ + + +T D+V + Y QEFF K
Sbjct: 132 AGY--SDGYVKDQREVADKIAA--STPDMVFVALGYPNQEFFIHK 172
>T2EA_YEAST (P36100) Transcription initiation factor IIE, alpha
subunit (TFIIE-alpha) (Transcription factor A large
subunit) (Factor A 66 kDa subunit)
Length = 482
Score = 31.2 bits (69), Expect = 6.0
Identities = 12/43 (27%), Positives = 28/43 (64%)
Query: 11 EKPDQDQNQDLGFHFLELSRRKAKLDSEDFEVDDEEKEESPSE 53
E +++ + L ++ L++++AKL+ E+ E ++EE++E E
Sbjct: 347 EMEERENEKTLNDYYAALAKKQAKLNKEEEEEEEEEEDEEEEE 389
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.317 0.135 0.388
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 55,679,322
Number of Sequences: 164201
Number of extensions: 2444191
Number of successful extensions: 7251
Number of sequences better than 10.0: 23
Number of HSP's better than 10.0 without gapping: 5
Number of HSP's successfully gapped in prelim test: 18
Number of HSP's that attempted gapping in prelim test: 7212
Number of HSP's gapped (non-prelim): 47
length of query: 468
length of database: 59,974,054
effective HSP length: 114
effective length of query: 354
effective length of database: 41,255,140
effective search space: 14604319560
effective search space used: 14604319560
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 68 (30.8 bits)
Medicago: description of AC146527.1


