

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146343.9 + phase: 0
(436 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
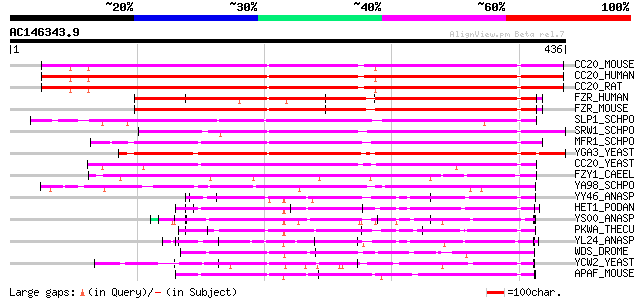
Score E
Sequences producing significant alignments: (bits) Value
CC20_MOUSE (Q9JJ66) Cell division cycle protein 20 homolog (p55C... 342 1e-93
CC20_HUMAN (Q12834) Cell division cycle protein 20 homolog (p55CDC) 341 2e-93
CC20_RAT (Q62623) Cell division cycle protein 20 homolog (p55CDC) 338 1e-92
FZR_HUMAN (Q9UM11) Fizzy-related protein homolog (Fzr) (Cdh1/Hct... 301 2e-81
FZR_MOUSE (Q9R1K5) Fizzy-related protein homolog (Fzr) (Cdh1/Hct... 301 3e-81
SLP1_SCHPO (P78972) WD-repeat containing protein slp1 298 1e-80
SRW1_SCHPO (O13286) WD-repeat containing protein srw1 (Suppresso... 291 2e-78
MFR1_SCHPO (O94423) Meiotic fizzy-related protein 1 284 4e-76
YGA3_YEAST (P53197) Hypothetical 62.8 kDa Trp-Asp repeats contai... 283 8e-76
CC20_YEAST (P26309) Cell division control protein 20 243 5e-64
FZY1_CAEEL (Q09373) WD-repeat containing protein fzy-1 (Fizzy pr... 149 1e-35
YA98_SCHPO (Q09786) Hypothetical WD-repeat protein C13G6.08 in c... 134 4e-31
YY46_ANASP (Q8YRI1) Hypothetical WD-repeat protein alr3466 110 7e-24
HET1_PODAN (Q00808) Vegetatible incompatibility protein HET-E-1 103 7e-22
YS00_ANASP (Q8YTC2) Hypothetical WD-repeat protein alr2800 97 8e-20
PKWA_THECU (P49695) Probable serine/threonine-protein kinase pkw... 96 2e-19
YL24_ANASP (Q8YV57) Hypothetical WD-repeat protein all2124 90 1e-17
WDS_DROME (Q9V3J8) Will die slowly protein 81 5e-15
YCW2_YEAST (P25382) Hypothetical WD-repeat protein YCR072C 75 3e-13
APAF_MOUSE (O88879) Apoptotic protease activating factor 1 (Apaf-1) 75 4e-13
>CC20_MOUSE (Q9JJ66) Cell division cycle protein 20 homolog (p55CDC)
(mmCdc20)
Length = 499
Score = 342 bits (877), Expect = 1e-93
Identities = 178/425 (41%), Positives = 258/425 (59%), Gaps = 22/425 (5%)
Query: 26 DRFIPNRSAIDWDHATTILST--------TTTNVTKENLWLAS----NVYQKKLAEAADL 73
DRFIP RSA + A+ +LS T T + W + +V + K+ +
Sbjct: 77 DRFIPQRSASQMEVASFLLSKENQPEDRGTPTKKEHQKAWSLNLNGFDVEEAKILRLSGK 136
Query: 74 PTRIL-AFRNKPRKRNVISPPPPPRSKPMRYIPKTCEGTFDLPDLSDDFSLNLLDWGSRN 132
P ++N+ + P K RYIP + D P++ +D+ LNL+DW S N
Sbjct: 137 PQNAPEGYQNRLKVLYSQKATPGSSRKTCRYIPSLPDRILDAPEIRNDYYLNLVDWSSGN 196
Query: 133 VLSIALDHTIYFWNASDSSGSEFVTVDEEEGPVTSVCWAPDGRHLAVGLTNSHVQLWDTA 192
VL++ALD+++Y WNA + + +++ ++SV W +G +LAVG +N+ VQLWD
Sbjct: 197 VLAVALDNSVYLWNAGSGDILQLLQMEQPGDYISSVAWIKEGNYLAVGTSNAEVQLWDVQ 256
Query: 193 ANKQLRTLKGGHRARVGSLAWNGHVLTTGGMDGKIVNNDVRLRSQIINTYRGHRREVCGL 252
K+LR + H ARV SL+WN ++L++G G I ++DVR+ + T GH +EVCGL
Sbjct: 257 QQKRLRNMTS-HSARVSSLSWNSYILSSGSRSGHIHHHDVRVAEHHVATLSGHSQEVCGL 315
Query: 253 KWSLDGKQLASGGNDNVVHIWDMSAVSSNSPTRW--LYRFDEHKAAVKALAWCPFQGNLL 310
+W+ DG+ LASGGNDN+V++W S + W L F +H+ AVKA+AWCP+Q N+L
Sbjct: 316 RWAPDGRHLASGGNDNIVNVWP----SGPGESGWAPLQTFTQHQGAVKAVAWCPWQSNIL 371
Query: 311 ASGGGGGDCCVKLWNTGMGERMNSVDTGSQVCALLWSKNERELLSSHGLTQNQLTLWKYP 370
A+GGG D +++WN G +++VD SQVC++LWS + +EL+S HG QNQL +WKYP
Sbjct: 372 ATGGGTSDRHIRIWNVCSGACLSAVDVHSQVCSILWSPHYKELISGHGFAQNQLVIWKYP 431
Query: 371 SMLKIAELHGHTSRVLHMTQSPDGSTVASAAAAADQTLRFWEVFGTPPAAPPKRNKMPFA 430
+M K+AEL GHT+RVL +T SPDG+TVAS AAAD+TLR W F PA +R K A
Sbjct: 432 TMAKVAELKGHTARVLGLTMSPDGATVAS--AAADETLRLWRCFEMDPALRREREKASVA 489
Query: 431 DSNRI 435
S+ I
Sbjct: 490 KSSLI 494
>CC20_HUMAN (Q12834) Cell division cycle protein 20 homolog (p55CDC)
Length = 499
Score = 341 bits (874), Expect = 2e-93
Identities = 178/425 (41%), Positives = 261/425 (60%), Gaps = 22/425 (5%)
Query: 26 DRFIPNRSAIDWDHATTILST--------TTTNVTKENLWLAS----NVYQKKLAEAADL 73
DR+IP+RSA + A+ +LS T T + W + +V + K+ +
Sbjct: 77 DRYIPHRSAAQMEVASFLLSKENQPENSQTPTKKEHQKAWALNLNGFDVEEAKILRLSGK 136
Query: 74 PTRIL-AFRNKPRKRNVISPPPPPRSKPMRYIPKTCEGTFDLPDLSDDFSLNLLDWGSRN 132
P ++N+ + P K RYIP + D P++ +D+ LNL+DW S N
Sbjct: 137 PQNAPEGYQNRLKVLYSQKATPGSSRKTCRYIPSLPDRILDAPEIRNDYYLNLVDWSSGN 196
Query: 133 VLSIALDHTIYFWNASDSSGSEFVTVDEEEGPVTSVCWAPDGRHLAVGLTNSHVQLWDTA 192
VL++ALD+++Y W+AS + + +++ ++SV W +G +LAVG +++ VQLWD
Sbjct: 197 VLAVALDNSVYLWSASSGDILQLLQMEQPGEYISSVAWIKEGNYLAVGTSSAEVQLWDVQ 256
Query: 193 ANKQLRTLKGGHRARVGSLAWNGHVLTTGGMDGKIVNNDVRLRSQIINTYRGHRREVCGL 252
K+LR + H ARVGSL+WN ++L++G G I ++DVR+ + T GH +EVCGL
Sbjct: 257 QQKRLRNMTS-HSARVGSLSWNSYILSSGSRSGHIHHHDVRVAEHHVATLSGHSQEVCGL 315
Query: 253 KWSLDGKQLASGGNDNVVHIWDMSAVSSNSPTRW--LYRFDEHKAAVKALAWCPFQGNLL 310
+W+ DG+ LASGGNDN+V++W S+ W L F +H+ AVKA+AWCP+Q N+L
Sbjct: 316 RWAPDGRHLASGGNDNLVNVWP----SAPGEGGWVPLQTFTQHQGAVKAVAWCPWQSNVL 371
Query: 311 ASGGGGGDCCVKLWNTGMGERMNSVDTGSQVCALLWSKNERELLSSHGLTQNQLTLWKYP 370
A+GGG D +++WN G +++VD SQVC++LWS + +EL+S HG QNQL +WKYP
Sbjct: 372 ATGGGTSDRHIRIWNVCSGACLSAVDAHSQVCSILWSPHYKELISGHGFAQNQLVIWKYP 431
Query: 371 SMLKIAELHGHTSRVLHMTQSPDGSTVASAAAAADQTLRFWEVFGTPPAAPPKRNKMPFA 430
+M K+AEL GHTSRVL +T SPDG+TVAS AAAD+TLR W F PA +R K A
Sbjct: 432 TMAKVAELKGHTSRVLSLTMSPDGATVAS--AAADETLRLWRCFELDPARRREREKASAA 489
Query: 431 DSNRI 435
S+ I
Sbjct: 490 KSSLI 494
>CC20_RAT (Q62623) Cell division cycle protein 20 homolog (p55CDC)
Length = 499
Score = 338 bits (868), Expect = 1e-92
Identities = 175/425 (41%), Positives = 258/425 (60%), Gaps = 22/425 (5%)
Query: 26 DRFIPNRSAIDWDHATTILST--------TTTNVTKENLWLAS----NVYQKKLAEAADL 73
+R+IP RSA + A+ +LS T T + W + +V + K+ +
Sbjct: 77 ERYIPQRSASQMEVASFLLSKENQPEDGGTPTKKEHQKAWARNLNGFDVEEAKILRLSGK 136
Query: 74 PTRIL-AFRNKPRKRNVISPPPPPRSKPMRYIPKTCEGTFDLPDLSDDFSLNLLDWGSRN 132
P ++N+ + P K RYIP + D P++ +D+ LNL+DW S N
Sbjct: 137 PQNAPEGYQNRLKVLYSQKATPGSSRKACRYIPSLPDRILDAPEIRNDYYLNLVDWSSGN 196
Query: 133 VLSIALDHTIYFWNASDSSGSEFVTVDEEEGPVTSVCWAPDGRHLAVGLTNSHVQLWDTA 192
VL++ALD+++Y WNA + + +++ ++SV W +G +LAVG +N+ VQLWD
Sbjct: 197 VLAVALDNSVYLWNAGSGDILQLLQMEQPGDYISSVAWIKEGNYLAVGTSNAEVQLWDVQ 256
Query: 193 ANKQLRTLKGGHRARVGSLAWNGHVLTTGGMDGKIVNNDVRLRSQIINTYRGHRREVCGL 252
K+LR + H ARV SL+WN ++L++G G I ++DVR+ + T GH +EVCGL
Sbjct: 257 QQKRLRNMTS-HSARVSSLSWNSYILSSGSRSGHIHHHDVRVAEHHVATLSGHSQEVCGL 315
Query: 253 KWSLDGKQLASGGNDNVVHIWDMSAVSSNSPTRW--LYRFDEHKAAVKALAWCPFQGNLL 310
+W+ DG+ LASGGNDN+V++W S + W L F +H+ AVKA+AWCP+Q N+L
Sbjct: 316 RWAPDGRHLASGGNDNIVNVWP----SGPGESGWVPLQTFTQHQGAVKAVAWCPWQSNIL 371
Query: 311 ASGGGGGDCCVKLWNTGMGERMNSVDTGSQVCALLWSKNERELLSSHGLTQNQLTLWKYP 370
A+GGG D +++WN G +++VD SQVC++LWS + +EL+S HG QNQL +WKYP
Sbjct: 372 ATGGGTSDRHIRIWNVCSGACLSAVDVHSQVCSILWSPHYKELISGHGFAQNQLVIWKYP 431
Query: 371 SMLKIAELHGHTSRVLHMTQSPDGSTVASAAAAADQTLRFWEVFGTPPAAPPKRNKMPFA 430
+M K+AEL GHT+RVL +T SPDG+TVAS AAAD+TLR W F PA +R K +
Sbjct: 432 TMAKVAELKGHTARVLSLTMSPDGATVAS--AAADETLRLWRCFELDPALRREREKASTS 489
Query: 431 DSNRI 435
S+ I
Sbjct: 490 KSSLI 494
>FZR_HUMAN (Q9UM11) Fizzy-related protein homolog (Fzr) (Cdh1/Hct1
homolog) (hCDH1) (CDC20-like protein 1)
Length = 496
Score = 301 bits (771), Expect = 2e-81
Identities = 153/317 (48%), Positives = 203/317 (63%), Gaps = 10/317 (3%)
Query: 99 KPMRYIPKTCEGTFDLPDLSDDFSLNLLDWGSRNVLSIALDHTIYFWNASDSSGSEFVTV 158
KP R I K D P+L DDF LNL+DW S NVLS+ L +Y W+A S + +
Sbjct: 166 KPTRKISKIPFKVLDAPELQDDFYLNLVDWSSLNVLSVGLGTCVYLWSACTSQVTRLCDL 225
Query: 159 DEEEGPVTSVCWAPDGRHLAVGLTNSHVQLWDTAANKQLRTLKGGHRARVGSLAWNGHVL 218
E VTSV W+ G +AVG VQ+WD AA K+L L+G H ARVG+LAWN L
Sbjct: 226 SVEGDSVTSVGWSERGNLVAVGTHKGFVQIWDAAAGKKLSMLEG-HTARVGALAWNAEQL 284
Query: 219 TTGGMDGKIVNNDVRLRS-QIINTYRGHRREVCGLKWSLDGKQLASGGNDNVVHIWDMSA 277
++G D I+ D+R Q +GHR+EVCGLKWS D + LASGGNDN + +W+ S+
Sbjct: 285 SSGSRDRMILQRDIRTPPLQSERRLQGHRQEVCGLKWSTDHQLLASGGNDNKLLVWNHSS 344
Query: 278 VSSNSPTRWLYRFDEHKAAVKALAWCPFQGNLLASGGGGGDCCVKLWNTGMGERMNSVDT 337
+S + ++ EH AAVKA+AW P Q LLASGGG D C++ WNT G+ + +DT
Sbjct: 345 LSP------VQQYTEHLAAVKAIAWSPHQHGLLASGGGTADRCIRFWNTLTGQPLQCIDT 398
Query: 338 GSQVCALLWSKNERELLSSHGLTQNQLTLWKYPSMLKIAELHGHTSRVLHMTQSPDGSTV 397
GSQVC L WSK+ EL+S+HG +QNQ+ +WKYPS+ ++A+L GH+ RVL++ SPDG +
Sbjct: 399 GSQVCNLAWSKHANELVSTHGYSQNQILVWKYPSLTQVAKLTGHSYRVLYLAMSPDGEAI 458
Query: 398 ASAAAAADQTLRFWEVF 414
+ A D+TLRFW VF
Sbjct: 459 VT--GAGDETLRFWNVF 473
Score = 55.5 bits (132), Expect = 3e-07
Identities = 50/172 (29%), Positives = 73/172 (42%), Gaps = 13/172 (7%)
Query: 249 VCGLKWSLDGKQLASGGNDNVVHIWDMSAVSSNSPTRWLYRFDEHKAAVKALAWCPFQGN 308
V + WS G +A G + V IWD +A L + H A V ALAW Q
Sbjct: 232 VTSVGWSERGNLVAVGTHKGFVQIWDAAAGKK------LSMLEGHTARVGALAWNAEQ-- 283
Query: 309 LLASGGGGGDCCVKLWNTGMGERMNSVDTGSQ-VCALLWSKNERELLSSHGLTQNQLTLW 367
L+SG + T + + Q VC L WS + +LL+S G N+L +W
Sbjct: 284 -LSSGSRDRMILQRDIRTPPLQSERRLQGHRQEVCGLKWS-TDHQLLASGG-NDNKLLVW 340
Query: 368 KYPSMLKIAELHGHTSRVLHMTQSP-DGSTVASAAAAADQTLRFWEVFGTPP 418
+ S+ + + H + V + SP +AS AD+ +RFW P
Sbjct: 341 NHSSLSPVQQYTEHLAAVKAIAWSPHQHGLLASGGGTADRCIRFWNTLTGQP 392
Score = 48.9 bits (115), Expect = 3e-05
Identities = 37/155 (23%), Positives = 69/155 (43%), Gaps = 12/155 (7%)
Query: 139 DHTIYFWNASDSSGSEFVTVDEEEGPVTSVCWAPDGRHLAV---GLTNSHVQLWDTAANK 195
D+ + WN S S + T E V ++ W+P L G + ++ W+T +
Sbjct: 334 DNKLLVWNHSSLSPVQQYT--EHLAAVKAIAWSPHQHGLLASGGGTADRCIRFWNTLTGQ 391
Query: 196 QLRTLKGGHRARVGSLAWNGH----VLTTGGMDGKIVNNDVRLRSQIINTYRGHRREVCG 251
L+ + G ++V +LAW+ H V T G +I+ +Q+ GH V
Sbjct: 392 PLQCIDTG--SQVCNLAWSKHANELVSTHGYSQNQILVWKYPSLTQVAKL-TGHSYRVLY 448
Query: 252 LKWSLDGKQLASGGNDNVVHIWDMSAVSSNSPTRW 286
L S DG+ + +G D + W++ + + ++ +W
Sbjct: 449 LAMSPDGEAIVTGAGDETLRFWNVFSKTRSTKVKW 483
>FZR_MOUSE (Q9R1K5) Fizzy-related protein homolog (Fzr) (Cdh1/Hct1
homolog)
Length = 493
Score = 301 bits (770), Expect = 3e-81
Identities = 153/317 (48%), Positives = 203/317 (63%), Gaps = 10/317 (3%)
Query: 99 KPMRYIPKTCEGTFDLPDLSDDFSLNLLDWGSRNVLSIALDHTIYFWNASDSSGSEFVTV 158
KP R I K D P+L DDF LNL+DW S NVLS+ L +Y W+A S + +
Sbjct: 166 KPTRKISKIPFKVLDAPELQDDFYLNLVDWSSLNVLSVGLGTCVYLWSACTSQVTRLCDL 225
Query: 159 DEEEGPVTSVCWAPDGRHLAVGLTNSHVQLWDTAANKQLRTLKGGHRARVGSLAWNGHVL 218
E VTSV W+ G +AVG VQ+WD AA K+L L+G H ARVG+LAWN L
Sbjct: 226 SVEGDSVTSVGWSERGNLVAVGTHKGFVQIWDAAAGKKLSMLEG-HTARVGALAWNADQL 284
Query: 219 TTGGMDGKIVNNDVRLRS-QIINTYRGHRREVCGLKWSLDGKQLASGGNDNVVHIWDMSA 277
++G D I+ D+R Q +GHR+EVCGLKWS D + LASGGNDN + +W+ S+
Sbjct: 285 SSGSRDRMILQRDIRTPPLQSERRLQGHRQEVCGLKWSTDHQLLASGGNDNKLLVWNHSS 344
Query: 278 VSSNSPTRWLYRFDEHKAAVKALAWCPFQGNLLASGGGGGDCCVKLWNTGMGERMNSVDT 337
+S + ++ EH AAVKA+AW P Q LLASGGG D C++ WNT G+ + +DT
Sbjct: 345 LSP------VQQYTEHLAAVKAIAWSPHQHGLLASGGGTADRCIRFWNTLTGQPLQCIDT 398
Query: 338 GSQVCALLWSKNERELLSSHGLTQNQLTLWKYPSMLKIAELHGHTSRVLHMTQSPDGSTV 397
GSQVC L WSK+ EL+S+HG +QNQ+ +WKYPS+ ++A+L GH+ RVL++ SPDG +
Sbjct: 399 GSQVCNLAWSKHANELVSTHGYSQNQILVWKYPSLTQVAKLTGHSYRVLYLAMSPDGEAI 458
Query: 398 ASAAAAADQTLRFWEVF 414
+ A D+TLRFW VF
Sbjct: 459 VT--GAGDETLRFWNVF 473
Score = 55.8 bits (133), Expect = 2e-07
Identities = 49/172 (28%), Positives = 73/172 (41%), Gaps = 13/172 (7%)
Query: 249 VCGLKWSLDGKQLASGGNDNVVHIWDMSAVSSNSPTRWLYRFDEHKAAVKALAWCPFQGN 308
V + WS G +A G + V IWD +A L + H A V ALAW +
Sbjct: 232 VTSVGWSERGNLVAVGTHKGFVQIWDAAAGKK------LSMLEGHTARVGALAW---NAD 282
Query: 309 LLASGGGGGDCCVKLWNTGMGERMNSVDTGSQ-VCALLWSKNERELLSSHGLTQNQLTLW 367
L+SG + T + + Q VC L WS + +LL+S G N+L +W
Sbjct: 283 QLSSGSRDRMILQRDIRTPPLQSERRLQGHRQEVCGLKWS-TDHQLLASGG-NDNKLLVW 340
Query: 368 KYPSMLKIAELHGHTSRVLHMTQSP-DGSTVASAAAAADQTLRFWEVFGTPP 418
+ S+ + + H + V + SP +AS AD+ +RFW P
Sbjct: 341 NHSSLSPVQQYTEHLAAVKAIAWSPHQHGLLASGGGTADRCIRFWNTLTGQP 392
>SLP1_SCHPO (P78972) WD-repeat containing protein slp1
Length = 488
Score = 298 bits (764), Expect = 1e-80
Identities = 170/411 (41%), Positives = 241/411 (58%), Gaps = 36/411 (8%)
Query: 17 NNSRWHQNIDRFIPNRSAIDWDHATTILSTTTTNVTKENLWLASNVYQKKLAEAA--DLP 74
N SR DRFIP+R + A +++ +++V + Y + +AEA DL
Sbjct: 79 NKSRPASRSDRFIPSRP----NTANAFVNSISSDVPFD--------YSESVAEACGFDLN 126
Query: 75 TRILAFR-NKPRKRNVIS-------PPPPPRSKPMRYIPKTCEGTFDLPDLSDDFSLNLL 126
TR+LAF+ + P + + P P + R T E D P + DD+ LNLL
Sbjct: 127 TRVLAFKLDAPEAKKPVDLRTQHNRPQRPVVTPAKRRFNTTPERVLDAPGIIDDYYLNLL 186
Query: 127 DWGSRNVLSIALDHTIYFWNASDSSGSEFVTVDEEEGPVTSVCWAPDGRHLAVGLTNSHV 186
DW + NV+++AL+ +Y WNA S S DE V SV W+ DG L+VGL N V
Sbjct: 187 DWSNLNVVAVALERNVYVWNADSGSVSALAETDESTY-VASVKWSHDGSFLSVGLGNGLV 245
Query: 187 QLWDTAANKQLRTLKGGHRARVGSLAWNGHVLTTGGMDGKIVNNDVRLRSQIINTYRGHR 246
++D + +LRT+ GH+ARVG L+WN HVL++G G I ++DVR+ + I T +GH
Sbjct: 246 DIYDVESQTKLRTM-AGHQARVGCLSWNRHVLSSGSRSGAIHHHDVRIANHQIGTLQGHS 304
Query: 247 REVCGLKWSLDGKQLASGGNDNVVHIWDM-SAVSSNSPTRWLYRFDEHKAAVKALAWCPF 305
EVCGL W DG QLASGGNDNVV IWD S++ + T H AAVKA+AWCP+
Sbjct: 305 SEVCGLAWRSDGLQLASGGNDNVVQIWDARSSIPKFTKTN-------HNAAVKAVAWCPW 357
Query: 306 QGNLLASGGGGGDCCVKLWNTGMGERMNSVDTGSQVCALLWSKNERELLSSHGLTQNQLT 365
Q NLLA+GGG D + WN G R+N+VD GSQV +L+WS + +E++S+HG N L+
Sbjct: 358 QSNLLATGGGTMDKQIHFWNAATGARVNTVDAGSQVTSLIWSPHSKEIMSTHGFPDNNLS 417
Query: 366 LWKYPS--MLKIAELHGHTSRVLHMTQSPDGSTVASAAAAADQTLRFWEVF 414
+W Y S + K ++ H +RVL+ SPDG +++ AA+D+ L+FW V+
Sbjct: 418 IWSYSSSGLTKQVDIPAHDTRVLYSALSPDGRILST--AASDENLKFWRVY 466
>SRW1_SCHPO (O13286) WD-repeat containing protein srw1 (Suppressor
of rad/wee1)
Length = 556
Score = 291 bits (746), Expect = 2e-78
Identities = 149/338 (44%), Positives = 200/338 (59%), Gaps = 16/338 (4%)
Query: 102 RYIPKTCEGTFDLPDLSDDFSLNLLDWGSRNVLSIALDHTIYFWNASDSSGSEFVTVDEE 161
R +P D P L+ DF LNLLDWG N+L++AL +Y W+ S VTV
Sbjct: 232 RELPSIPYRVLDAPGLAGDFYLNLLDWGQCNMLAVALASRVYLWSGISSE----VTVMHN 287
Query: 162 EGP---VTSVCWAPDGRHLAVGLTNSHVQLWDTAANKQLRTLKGGHRARVGSLAWNGHVL 218
P VTS+ W G HLAVG N V++WD A K+ RT+ G H RVG+L+WN HVL
Sbjct: 288 FYPTDTVTSLRWVQRGTHLAVGTHNGSVEIWDAATCKKTRTMSG-HTERVGALSWNDHVL 346
Query: 219 TTGGMDGKIVNNDVRLRSQIINTYRGHRREVCGLKWSLDGKQLASGGNDNVVHIWDMSAV 278
++GG D I++ DVR HR+EVCGL+W+ + LASGGNDN + +WD
Sbjct: 347 SSGGRDNHILHRDVRAPEHYFRVLTAHRQEVCGLEWNSNENLLASGGNDNALMVWD---- 402
Query: 279 SSNSPTRWLYRFDEHKAAVKALAWCPFQGNLLASGGGGGDCCVKLWNTGMGERMNSVDTG 338
+ LY F H AAVKA+ W P Q +LASGGG D +KLWNT G ++++DTG
Sbjct: 403 --KFEEKPLYSFHNHIAAVKAITWSPHQRGILASGGGTADRTIKLWNTQRGSMLHNIDTG 460
Query: 339 SQVCALLWSKNERELLSSHGLTQNQLTLWKYPSMLKIAELHGHTSRVLHMTQSPDGSTVA 398
SQVC LLWSK E +S+HG +N++ LW YPS+ ++ L GHT RVL++ SP+G +
Sbjct: 461 SQVCNLLWSKQTNEFISTHGFMENEVALWNYPSVSRVGTLKGHTDRVLYLAMSPNGENIV 520
Query: 399 SAAAAADQTLRFWEVFGTPPAAPPKRNKMPFADSNRIR 436
+ AAD+TLRFW++F + PF + +IR
Sbjct: 521 T--GAADETLRFWKLFDSKSKHSASTMSSPFDPTMKIR 556
>MFR1_SCHPO (O94423) Meiotic fizzy-related protein 1
Length = 421
Score = 284 bits (726), Expect = 4e-76
Identities = 151/356 (42%), Positives = 215/356 (59%), Gaps = 17/356 (4%)
Query: 64 QKKLAEAADLPTRILAFRNKPRKRNVISPPPPPRSKPMRYIPKTCEGTFDLPDLSDDFSL 123
+KK+ + D + L+ P+ ++++ P KP R PKT D P L +DF L
Sbjct: 74 EKKMLDTPDRKSYSLS-PISPQSQDMLRQP----QKPKRAFPKTPYKILDAPYLKNDFYL 128
Query: 124 NLLDWGSRNVLSIALDHTIYFWNASDSSGSEFVTVD-EEEGPVTSVCWAPDGRHLAVGLT 182
NLLDWG NVL++ L +IY W+A+ SG D VTSV W G LAVG
Sbjct: 129 NLLDWGQSNVLAVGLASSIYLWSAA--SGKVVQLHDFGATNHVTSVLWTGKGTQLAVGTD 186
Query: 183 NSHVQLWDTAANKQLRTLKGGHRARVGSLAWNGHVLTTGGMDGKIVNNDVRLRSQIINTY 242
+ + +WD + K +R+LKG H RV +LAWN + LT+GG D I+++D+R
Sbjct: 187 SGVIYIWDIESTKSVRSLKG-HSERVAALAWNDNTLTSGGKDEVILHHDLRAPGCCAEMM 245
Query: 243 RGHRREVCGLKWSLDGKQLASGGNDNVVHIWDMSAVSSNSPTRWLYRFDEHKAAVKALAW 302
+ H +E+CGL+W QLASGGNDN + +WD + +R L++F+EH AAVKA+ W
Sbjct: 246 KVHEQEICGLQWDRSLGQLASGGNDNNLFVWDYRS------SRPLHKFEEHTAAVKAIGW 299
Query: 303 CPFQGNLLASGGGGGDCCVKLWNTGMGERMNSVDTGSQVCALLWSKNERELLSSHGLTQN 362
P Q +LASGGG D C+ + NT G N +DTGSQVC + WSK E++++HG +N
Sbjct: 300 SPHQRGILASGGGTIDRCLTIHNTLTGRLQNKLDTGSQVCNMAWSKTSNEIVTTHGFAKN 359
Query: 363 QLTLWKYPSMLKIAELHGHTSRVLHMTQSPDGSTVASAAAAADQTLRFWEVFGTPP 418
Q++LWKYPS+ IA L HT+RVL+++ SPDG ++ + A D+TLRFW++F P
Sbjct: 360 QVSLWKYPSLKNIANLTAHTNRVLYLSMSPDGQSIVT--GAGDETLRFWKLFNKKP 413
>YGA3_YEAST (P53197) Hypothetical 62.8 kDa Trp-Asp repeats
containing protein in PMC1-TFG2 intergenic region
Length = 566
Score = 283 bits (723), Expect = 8e-76
Identities = 154/351 (43%), Positives = 218/351 (61%), Gaps = 18/351 (5%)
Query: 86 KRNVISPPPPPRSKPMRYIPKTCEGTFDLPDLSDDFSLNLLDWGSRNVLSIALDHTIYFW 145
K+ ++SP K R I K D P L+DDF +L+DW S +VL++AL +I+
Sbjct: 234 KQLLLSP-----GKQFRQIAKVPYRVLDAPSLADDFYYSLIDWSSTDVLAVALGKSIFL- 287
Query: 146 NASDSSGSEFVTVDEEEGPVTSVCWAPDGRHLAVGLTNSHVQLWDTAANKQLRTLKGGHR 205
+D++ + V + + E TS+ W G HLAVG N V+++D K +RTL G H
Sbjct: 288 --TDNNTGDVVHLCDTENEYTSLSWIGAGSHLAVGQANGLVEIYDVMKRKCIRTLSG-HI 344
Query: 206 ARVGSLAWNGHVLTTGGMDGKIVNNDVRLRSQIINTYRGHRREVCGLKWSLDGKQLASGG 265
RV L+WN HVLT+G D +I++ DVR+ T H +EVCGLKW++ +LASGG
Sbjct: 345 DRVACLSWNNHVLTSGSRDHRILHRDVRMPDPFFETIESHTQEVCGLKWNVADNKLASGG 404
Query: 266 NDNVVHIWDMSAVSSNSPTRWLYRFDEHKAAVKALAWCPFQGNLLASGGGGGDCCVKLWN 325
NDNVVH+++ + S SP + FDEHKAAVKA+AW P + +LA+GGG D +K+WN
Sbjct: 405 NDNVVHVYEGT---SKSP---ILTFDEHKAAVKAMAWSPHKRGVLATGGGTADRRLKIWN 458
Query: 326 TGMGERMNSVDTGSQVCALLWSKNERELLSSHGLTQNQLTLWKYPSMLKIAELHGHTSRV 385
+M+ +D+GSQ+C ++WSKN EL++SHG ++ LTLW SM IA L GH+ RV
Sbjct: 459 VNTSIKMSDIDSGSQICNMVWSKNTNELVTSHGYSKYNLTLWDCNSMDPIAILKGHSFRV 518
Query: 386 LHMTQSPDGSTVASAAAAADQTLRFWEVFGTPPAAPPKRNKMPFADSNRIR 436
LH+T S DG+TV S A D+TLR+W++F P A + N + F N+IR
Sbjct: 519 LHLTLSNDGTTVVS--GAGDETLRYWKLF-DKPKAKVQPNSLIFDAFNQIR 566
>CC20_YEAST (P26309) Cell division control protein 20
Length = 610
Score = 243 bits (621), Expect = 5e-64
Identities = 136/381 (35%), Positives = 211/381 (54%), Gaps = 38/381 (9%)
Query: 62 VYQKKLAEAA--DLPTRILAFRNKPRKRNVISPPPPPRSKPMRY---------------- 103
V+++ +AEA D+ RIL + +P K + + K Y
Sbjct: 185 VFKQNVAEACGLDMNKRILQYMPEPPKCSSLRQKSYIMKKRTHYSYQQEQKIPDLIKLRK 244
Query: 104 IPKTCEGTFDLPDLSDDFSLNLLDWGSRNVLSIALDHTIYFWNASDSSGSEFVTVDEEEG 163
I E D P DDF LNLL W +NVL+IALD +Y WNA+ +G + D E
Sbjct: 245 INTNPERILDAPGFQDDFYLNLLSWSKKNVLAIALDTALYLWNAT--TGDVSLLTDFENT 302
Query: 164 PVTSVCWAPDGRHLAVGLTNSHVQLWDTAANKQLRTLKGGHRARVGSLAWNGHVLTTGGM 223
+ SV W+ D H+++G + + ++WD +RT++ G R+GSL+W ++ TG
Sbjct: 303 TICSVTWSDDDCHISIGKEDGNTEIWDVETMSLIRTMRSGLGVRIGSLSWLDTLIATGSR 362
Query: 224 DGKIVNNDVRLRSQIINTYRGHRREVCGLKWSLDGKQLASGGNDNVVHIWDMSAVSSNSP 283
G+I NDVR++ I++T+ H EVCGL + DG QLASGGNDN V IWD ++ P
Sbjct: 363 SGEIQINDVRIKQHIVSTWAEHTGEVCGLSYKSDGLQLASGGNDNTVMIWD---TRTSLP 419
Query: 284 TRWLYRFDEHKAAVKALAWCPFQGNLLASGGGGGDCCVKLWNTGMGERMNSVDTGSQVCA 343
+ H AAVKAL+WCP+ N+LASGGG D + WN+ G R+ S++TGSQV +
Sbjct: 420 Q---FSKKTHTAAVKALSWCPYSPNILASGGGQTDKHIHFWNSITGARVGSINTGSQVSS 476
Query: 344 LLWSKN---------ERELLSSHGLTQNQLTLWKYPSMLKIAE-LHGHTSRVLHMTQSPD 393
L W ++ +E++++ G +N ++++ Y + K+AE +H H +R+ SPD
Sbjct: 477 LHWGQSHTSTNGGMMNKEIVATGGNPENAISVYNYETKFKVAEVVHAHEARICCSQLSPD 536
Query: 394 GSTVASAAAAADQTLRFWEVF 414
G+T+A+ D+ L+F+++F
Sbjct: 537 GTTLAT--VGGDENLKFYKIF 555
>FZY1_CAEEL (Q09373) WD-repeat containing protein fzy-1 (Fizzy
protein 1)
Length = 507
Score = 149 bits (376), Expect = 1e-35
Identities = 102/375 (27%), Positives = 181/375 (48%), Gaps = 34/375 (9%)
Query: 63 YQKKLAEAADLPTRILAFRNKPR---KRNVISPPPPPRSKPMRYIPKTCEGTFDLPDLSD 119
Y+K LA P + + N+ + N + P K R++ +T D P L+
Sbjct: 122 YKKNLA-----PPPAIGYINQAKVLYSTNSVINPASSVKKSTRHVKETATKVLDGPGLTK 176
Query: 120 DFSLNLLDWGSRNVLSIALDHTIYFWNASDSSGSEFV--TVDEEEGPVTSVCWAPDGRHL 177
D LDWG N +++AL H +Y WN EG +TSV W+ +GR++
Sbjct: 177 DLYSRHLDWGCHNWVAVALGHELYLWNTETCVIKNLFEDNAPTNEGLITSVRWSQEGRYI 236
Query: 178 AVGLTNSHVQLWD------TAANKQLRTLKGGHRARVGSLAWNGH-VLTTGGMDGKIVNN 230
++G + V+++D T ++LRTL+ G +R S+AW V+T G G IVN+
Sbjct: 237 SLGYASGAVKIYDPNRPKTTEYVRELRTLRVGGASRCASIAWRKQGVMTCGYKSGDIVNH 296
Query: 231 DVRLRSQIINTY---RGHRREVCGLKWSLDGKQLASGGNDNVVHIWDMSAVSSNS---PT 284
DVR+ +++++ GH R+V L+WS D SG +D IWD V ++
Sbjct: 297 DVRISQHVVSSWGGDNGHCRDVTALEWSADENMCVSGSSDRTAKIWDGRHVRGSTVIQDP 356
Query: 285 RWLYRFDEHKAAVKALAWCPFQGNLLASGGGGGDCCVKLWNTGMG----ERMNSVDTGSQ 340
++ DEH V+ +C F+ +LA+GGG D VKLW+ +N +TG
Sbjct: 357 EPMFTIDEHTGQVRTAQFCSFRDGILATGGGINDGTVKLWDVKRQFQKVRELNVCETGG- 415
Query: 341 VCALLWSKNERELLSSHGLTQNQLTLWKYPSMLKIA-ELHGHTSRVLHMTQSPDGSTVAS 399
V +++++ E+L++ L ++++ + K++ E+ ++ + SP +
Sbjct: 416 VGGIVFNRPYSEMLTAS--DDGFLRIYRFNANYKLSHEIQASNEPIMDLVGSPFDEVL-- 471
Query: 400 AAAAADQTLRFWEVF 414
++TL+ +++F
Sbjct: 472 -IGDMEETLKVFQLF 485
>YA98_SCHPO (Q09786) Hypothetical WD-repeat protein C13G6.08 in
chromosome I
Length = 535
Score = 134 bits (338), Expect = 4e-31
Identities = 120/437 (27%), Positives = 204/437 (46%), Gaps = 72/437 (16%)
Query: 25 IDRFIP---NRSAIDWDHATTILSTTTTNVTKENLWLASNVYQKKLAEAADL-------- 73
+DRFIP N+ I ++ LS N TK AS YQ+ L A ++
Sbjct: 85 LDRFIPMTSNKDTISLGRHSS-LSRNLVNKTKN----ASETYQQLLEYALEVERDDNVFT 139
Query: 74 -------------PTRILAFRNKPRKRNVISPPPPPRSKPMRYIPKTCEGTFDLPDLSDD 120
PT + + ++ K N P P R + D P+L DD
Sbjct: 140 YAKLQKSDMTQKCPTMVASEKDNKGKLNEKKNRSPENLLPFRIL--------DAPELRDD 191
Query: 121 FSLNLLDWGSRNVLSIALDHTIYFWNASDSSGSEFVTVDEEEGPVTSVCWAPDGRHLAVG 180
F +LL W + L+I L IY W S G V ++E V+SV ++ +G LAVG
Sbjct: 192 FYTSLLSWSPKGDLAIGLAENIYLW--SKELGPTRV-LEESIYDVSSVAYSYNGDILAVG 248
Query: 181 LTNSHVQLWDTAANKQLRTLKGGHRARVGSLAWNGHVLTTGGMDGK---------IVNND 231
+ +Q W N+++ + H +G LAW + T + GK I+ ++
Sbjct: 249 RVDGTLQFWQD--NERVPRISIHHPGDIGVLAWKPVLETNRLLVGKGNGNIFVYDIIWSE 306
Query: 232 VRLRSQIINTY-RGHRREVCGLKWSLDGKQLASGGNDNVVHIWDMSAVSSNSPTRWLYRF 290
++ ++ T H +VCGL W+ DG Q ASGGNDN V ++ S + LY +
Sbjct: 307 STSKAVLVATITNAHDEQVCGLTWNHDGSQFASGGNDNRVCLFKGSDLRQP-----LYVW 361
Query: 291 DEHKAAVKALAWCPFQGNLLASGGGGGDCCVKLWNTGMGERMNSVDTGSQVCALLWSKNE 350
++ AAVKAL++CP+Q +LLA+G G D ++ +N G++++ + G+Q+ ++ WS
Sbjct: 362 QQN-AAVKALSFCPWQRSLLATGAGSHDKHIRFYNCFNGKKIDELYCGAQITSIHWSPKY 420
Query: 351 RELLSSHGLT----QNQLTLWKY----------PSMLKIAELHGHTSRVLHMTQSPDGST 396
RE + G + Q++ ++ + PS+ I +H + L+ T T
Sbjct: 421 REFSVTFGYSLETVQHRFAVYSWPQLECLVSVLPSVPDIRCVHSVLTSQLNETTGRCEMT 480
Query: 397 VASAAAAADQTLRFWEV 413
+ A++++T++F+++
Sbjct: 481 DSIIIASSNETIKFFDL 497
>YY46_ANASP (Q8YRI1) Hypothetical WD-repeat protein alr3466
Length = 1526
Score = 110 bits (275), Expect = 7e-24
Identities = 94/316 (29%), Positives = 147/316 (45%), Gaps = 60/316 (18%)
Query: 142 IYFWNASDSSGSEFVTVDEEEGPVTSVCWAPDGRHLAVGLTNSHVQLWDTAANKQLRTLK 201
+ FW A+ +G E +T V SV ++ DG+ LA G + V+LWD ++ + L+T K
Sbjct: 888 VRFWEAA--TGKELLTCKGHNSWVNSVGFSQDGKMLASGSDDQTVRLWDISSGQCLKTFK 945
Query: 202 G-----------------------------------------GHRARVGSLAWN--GHVL 218
G GH V S+A+N G +L
Sbjct: 946 GHTSRVRSVVFSPNSLMLASGSSDQTVRLWDISSGECLYIFQGHTGWVYSVAFNLDGSML 1005
Query: 219 TTGGMDGKIVNNDVRLRSQIINTYRGHRREVCGLKWSLDGKQLASGGNDNVVHIWDMSAV 278
TG D + D+ SQ ++GH V + +S DG LASG +D V +WD+S+
Sbjct: 1006 ATGSGDQTVRLWDIS-SSQCFYIFQGHTSCVRSVVFSSDGAMLASGSDDQTVRLWDISSG 1064
Query: 279 SSNSPTRWLYRFDEHKAAVKALAWCPFQGNLLASGGGGGDCCVKLWNTGMGERMNSVD-T 337
+ LY H + V+++ + P G +LAS GG D V+LW+ G + ++
Sbjct: 1065 NC------LYTLQGHTSCVRSVVFSP-DGAMLAS--GGDDQIVRLWDISSGNCLYTLQGY 1115
Query: 338 GSQVCALLWSKNERELLSSHGLTQNQLTLWKYPSMLKIAELHGHTSRVLHMTQSPDGSTV 397
S V L++S N L ++G + + LW S + L GHT+ V + SPDG+T+
Sbjct: 1116 TSWVRFLVFSPNGVTL--ANGSSDQIVRLWDISSKKCLYTLQGHTNWVNAVAFSPDGATL 1173
Query: 398 ASAAAAADQTLRFWEV 413
AS + DQT+R W++
Sbjct: 1174 AS--GSGDQTVRLWDI 1187
Score = 108 bits (271), Expect = 2e-23
Identities = 84/278 (30%), Positives = 142/278 (50%), Gaps = 20/278 (7%)
Query: 139 DHTIYFWNASDSSGSEFVTVDEEEGPVTSVCWAPDGRHLAVGLTNSHVQLWDTAANKQLR 198
D T+ W+ S S + V SV + PDG LA G ++ V+LW+ ++K L
Sbjct: 1179 DQTVRLWDISSSKC--LYILQGHTSWVNSVVFNPDGSTLASGSSDQTVRLWEINSSKCLC 1236
Query: 199 TLKGGHRARVGSLAWN--GHVLTTGGMDGKIVNNDVRLRSQIINTYRGHRREVCGLKWSL 256
T + GH + V S+ +N G +L +G D + D+ S+ ++T++GH V + ++
Sbjct: 1237 TFQ-GHTSWVNSVVFNPDGSMLASGSSDKTVRLWDIS-SSKCLHTFQGHTNWVNSVAFNP 1294
Query: 257 DGKQLASGGNDNVVHIWDMSAVSSNSPTRWLYRFDEHKAAVKALAWCPFQGNLLASGGGG 316
DG LASG D V +W++S+ ++ L+ F H + V ++ + P G +LAS G
Sbjct: 1295 DGSMLASGSGDQTVRLWEISS------SKCLHTFQGHTSWVSSVTFSP-DGTMLAS--GS 1345
Query: 317 GDCCVKLWNTGMGERMNS-VDTGSQVCALLWSKNERELLSSHGLTQNQLTLWKYPSMLKI 375
D V+LW+ GE + + + + V ++++S + L S G + LW S +
Sbjct: 1346 DDQTVRLWSISSGECLYTFLGHTNWVGSVIFSPDGAILASGSG--DQTVRLWSISSGKCL 1403
Query: 376 AELHGHTSRVLHMTQSPDGSTVASAAAAADQTLRFWEV 413
L GH + V + SPDG+ +AS + DQT+R W +
Sbjct: 1404 YTLQGHNNWVGSIVFSPDGTLLAS--GSDDQTVRLWNI 1439
Score = 108 bits (270), Expect = 3e-23
Identities = 85/277 (30%), Positives = 137/277 (48%), Gaps = 18/277 (6%)
Query: 139 DHTIYFWNASDSSGSEFVTVDEEEGPVTSVCWAPDGRHLAVGLTNSHVQLWDTAANKQLR 198
D T+ W + S T V SV + PDG LA G ++ V+LWD +++K L
Sbjct: 1221 DQTVRLWEINSSKC--LCTFQGHTSWVNSVVFNPDGSMLASGSSDKTVRLWDISSSKCLH 1278
Query: 199 TLKGGHRARVGSLAWN--GHVLTTGGMDGKIVNNDVRLRSQIINTYRGHRREVCGLKWSL 256
T +G H V S+A+N G +L +G D + ++ S+ ++T++GH V + +S
Sbjct: 1279 TFQG-HTNWVNSVAFNPDGSMLASGSGDQTVRLWEIS-SSKCLHTFQGHTSWVSSVTFSP 1336
Query: 257 DGKQLASGGNDNVVHIWDMSAVSSNSPTRWLYRFDEHKAAVKALAWCPFQGNLLASGGGG 316
DG LASG +D V +W +S+ LY F H V ++ + P G +LAS G
Sbjct: 1337 DGTMLASGSDDQTVRLWSISS------GECLYTFLGHTNWVGSVIFSP-DGAILAS--GS 1387
Query: 317 GDCCVKLWNTGMGERMNSVDTGSQVCALLWSKNERELLSSHGLTQNQLTLWKYPSMLKIA 376
GD V+LW+ G+ + ++ + + + LL+S G + LW S +
Sbjct: 1388 GDQTVRLWSISSGKCLYTLQGHNNWVGSIVFSPDGTLLAS-GSDDQTVRLWNISSGECLY 1446
Query: 377 ELHGHTSRVLHMTQSPDGSTVASAAAAADQTLRFWEV 413
LHGH + V + S DG +AS + D+T++ W+V
Sbjct: 1447 TLHGHINSVRSVAFSSDGLILAS--GSDDETIKLWDV 1481
Score = 102 bits (253), Expect = 3e-21
Identities = 76/253 (30%), Positives = 125/253 (49%), Gaps = 16/253 (6%)
Query: 163 GPVTSVCWAPDGRHLAVGLTNSHVQLWDTAANKQLRTLKGGHRARVGSLAW--NGHVLTT 220
G V +V ++PDG+ A G + V+ W+ A K+L T K GH + V S+ + +G +L +
Sbjct: 865 GSVLTVAFSPDGKLFATGDSGGIVRFWEAATGKELLTCK-GHNSWVNSVGFSQDGKMLAS 923
Query: 221 GGMDGKIVNNDVRLRSQIINTYRGHRREVCGLKWSLDGKQLASGGNDNVVHIWDMSAVSS 280
G D + D+ Q + T++GH V + +S + LASG +D V +WD+S+
Sbjct: 924 GSDDQTVRLWDIS-SGQCLKTFKGHTSRVRSVVFSPNSLMLASGSSDQTVRLWDISS--- 979
Query: 281 NSPTRWLYRFDEHKAAVKALAWCPFQGNLLASGGGGGDCCVKLWNTGMGERMNSVDTGSQ 340
LY F H V ++A+ G++LA+ G GD V+LW+ + +
Sbjct: 980 ---GECLYIFQGHTGWVYSVAF-NLDGSMLAT--GSGDQTVRLWDISSSQCFYIFQGHTS 1033
Query: 341 VCALLWSKNERELLSSHGLTQNQLTLWKYPSMLKIAELHGHTSRVLHMTQSPDGSTVASA 400
+ ++ +L+S G + LW S + L GHTS V + SPDG+ +AS
Sbjct: 1034 CVRSVVFSSDGAMLAS-GSDDQTVRLWDISSGNCLYTLQGHTSCVRSVVFSPDGAMLAS- 1091
Query: 401 AAAADQTLRFWEV 413
DQ +R W++
Sbjct: 1092 -GGDDQIVRLWDI 1103
Score = 101 bits (252), Expect = 3e-21
Identities = 84/278 (30%), Positives = 136/278 (48%), Gaps = 20/278 (7%)
Query: 139 DHTIYFWNASDSSGSEFVTVDEEEGPVTSVCWAPDGRHLAVGLTNSHVQLWDTAANKQLR 198
D T+ W+ S SG G V SV + DG LA G + V+LWD ++++
Sbjct: 969 DQTVRLWDIS--SGECLYIFQGHTGWVYSVAFNLDGSMLATGSGDQTVRLWDISSSQCFY 1026
Query: 199 TLKGGHRARVGSLAWN--GHVLTTGGMDGKIVNNDVRLRSQIINTYRGHRREVCGLKWSL 256
+G H + V S+ ++ G +L +G D + D+ + T +GH V + +S
Sbjct: 1027 IFQG-HTSCVRSVVFSSDGAMLASGSDDQTVRLWDIS-SGNCLYTLQGHTSCVRSVVFSP 1084
Query: 257 DGKQLASGGNDNVVHIWDMSAVSSNSPTRWLYRFDEHKAAVKALAWCPFQGNLLASGGGG 316
DG LASGG+D +V +WD+S+ + LY + + V+ L + P N + G
Sbjct: 1085 DGAMLASGGDDQIVRLWDISSGNC------LYTLQGYTSWVRFLVFSP---NGVTLANGS 1135
Query: 317 GDCCVKLWNTGMGERMNSVDTGSQ-VCALLWSKNERELLSSHGLTQNQLTLWKYPSMLKI 375
D V+LW+ + + ++ + V A+ +S + L S G + LW S +
Sbjct: 1136 SDQIVRLWDISSKKCLYTLQGHTNWVNAVAFSPDGATLASGSG--DQTVRLWDISSSKCL 1193
Query: 376 AELHGHTSRVLHMTQSPDGSTVASAAAAADQTLRFWEV 413
L GHTS V + +PDGST+AS ++DQT+R WE+
Sbjct: 1194 YILQGHTSWVNSVVFNPDGSTLAS--GSSDQTVRLWEI 1229
Score = 74.3 bits (181), Expect = 6e-13
Identities = 64/198 (32%), Positives = 96/198 (48%), Gaps = 23/198 (11%)
Query: 139 DHTIYFWNASDSSGSEFVTVDEEEGPVTSVCWAPDGRHLAVGLTNSHVQLWDTAANKQLR 198
D T+ W S S T V+SV ++PDG LA G + V+LW ++ + L
Sbjct: 1305 DQTVRLWEISSSKCLH--TFQGHTSWVSSVTFSPDGTMLASGSDDQTVRLWSISSGECLY 1362
Query: 199 TLKGGHRARVGSLAWN--GHVLTTGGMDGKIVNNDVRLRS----QIINTYRGHRREVCGL 252
T G H VGS+ ++ G +L +G D VRL S + + T +GH V +
Sbjct: 1363 TFLG-HTNWVGSVIFSPDGAILASGSGD-----QTVRLWSISSGKCLYTLQGHNNWVGSI 1416
Query: 253 KWSLDGKQLASGGNDNVVHIWDMSAVSSNSPTRWLYRFDEHKAAVKALAWCPFQGNLLAS 312
+S DG LASG +D V +W++S+ LY H +V+++A+ G +LAS
Sbjct: 1417 VFSPDGTLLASGSDDQTVRLWNISSGEC------LYTLHGHINSVRSVAFSS-DGLILAS 1469
Query: 313 GGGGGDCCVKLWNTGMGE 330
G D +KLW+ GE
Sbjct: 1470 GSD--DETIKLWDVKTGE 1485
Score = 39.3 bits (90), Expect = 0.020
Identities = 22/63 (34%), Positives = 32/63 (49%), Gaps = 2/63 (3%)
Query: 139 DHTIYFWNASDSSGSEFVTVDEEEGPVTSVCWAPDGRHLAVGLTNSHVQLWDTAANKQLR 198
D T+ WN S SG T+ V SV ++ DG LA G + ++LWD + ++
Sbjct: 1431 DQTVRLWNIS--SGECLYTLHGHINSVRSVAFSSDGLILASGSDDETIKLWDVKTGECIK 1488
Query: 199 TLK 201
TLK
Sbjct: 1489 TLK 1491
>HET1_PODAN (Q00808) Vegetatible incompatibility protein HET-E-1
Length = 1356
Score = 103 bits (258), Expect = 7e-22
Identities = 83/285 (29%), Positives = 143/285 (50%), Gaps = 20/285 (7%)
Query: 131 RNVLSIALDHTIYFWNASDSSGSEFVTVDEEEGPVTSVCWAPDGRHLAVGLTNSHVQLWD 190
+ V S + D TI W+ + +G++ T++ G V SV ++PD +A G + +++WD
Sbjct: 854 QRVASGSDDKTIKIWDTASGTGTQ--TLEGHGGSVWSVAFSPDRERVASGSDDKTIKIWD 911
Query: 191 TAANKQLRTLKGGHRARVGSLAW--NGHVLTTGGMDGKIVNNDVRLRSQIINTYRGHRRE 248
A+ +TL+ GH RV S+A+ +G + +G D I D T GH
Sbjct: 912 AASGTCTQTLE-GHGGRVQSVAFSPDGQRVASGSDDHTIKIWDA-ASGTCTQTLEGHGSS 969
Query: 249 VCGLKWSLDGKQLASGGNDNVVHIWDMSAVSSNSPTRWLYRFDEHKAAVKALAWCPFQGN 308
V + +S DG+++ASG D + IWD +S + T+ L + H +V ++A+ P G
Sbjct: 970 VLSVAFSPDGQRVASGSGDKTIKIWD---TASGTCTQTL---EGHGGSVWSVAFSP-DGQ 1022
Query: 309 LLASGGGGGDCCVKLWNTGMGERMNSVD-TGSQVCALLWSKNERELLSSHGLTQNQLTLW 367
+AS G D +K+W+T G +++ G V ++++S + + + S G + + +W
Sbjct: 1023 RVAS--GSDDKTIKIWDTASGTCTQTLEGHGGWVQSVVFSPDGQRVAS--GSDDHTIKIW 1078
Query: 368 KYPSMLKIAELHGHTSRVLHMTQSPDGSTVASAAAAADQTLRFWE 412
S L GH V + SPDG VAS + D T++ W+
Sbjct: 1079 DAVSGTCTQTLEGHGDSVWSVAFSPDGQRVAS--GSIDGTIKIWD 1121
Score = 101 bits (251), Expect = 4e-21
Identities = 83/284 (29%), Positives = 140/284 (49%), Gaps = 18/284 (6%)
Query: 131 RNVLSIALDHTIYFWNASDSSGSEFVTVDEEEGPVTSVCWAPDGRHLAVGLTNSHVQLWD 190
+ V S + DHTI W+A+ SG+ T++ V SV ++PDG+ +A G + +++WD
Sbjct: 938 QRVASGSDDHTIKIWDAA--SGTCTQTLEGHGSSVLSVAFSPDGQRVASGSGDKTIKIWD 995
Query: 191 TAANKQLRTLKGGHRARVGSLAWN-GHVLTTGGMDGKIVNNDVRLRSQIINTYRGHRREV 249
TA+ +TL+ GH V S+A++ G D K + T GH V
Sbjct: 996 TASGTCTQTLE-GHGGSVWSVAFSPDGQRVASGSDDKTIKIWDTASGTCTQTLEGHGGWV 1054
Query: 250 CGLKWSLDGKQLASGGNDNVVHIWDMSAVSSNSPTRWLYRFDEHKAAVKALAWCPFQGNL 309
+ +S DG+++ASG +D+ + IWD AVS + H +V ++A+ P G
Sbjct: 1055 QSVVFSPDGQRVASGSDDHTIKIWD--AVSGTC----TQTLEGHGDSVWSVAFSP-DGQR 1107
Query: 310 LASGGGGGDCCVKLWNTGMGERMNSVD-TGSQVCALLWSKNERELLSSHGLTQNQLTLWK 368
+ASG G +K+W+ G +++ G V ++ +S + + + S G + +W
Sbjct: 1108 VASGSIDG--TIKIWDAASGTCTQTLEGHGGWVHSVAFSPDGQRVAS--GSIDGTIKIWD 1163
Query: 369 YPSMLKIAELHGHTSRVLHMTQSPDGSTVASAAAAADQTLRFWE 412
S L GH V + SPDG VAS ++D+T++ W+
Sbjct: 1164 AASGTCTQTLEGHGGWVQSVAFSPDGQRVAS--GSSDKTIKIWD 1205
Score = 98.6 bits (244), Expect = 3e-20
Identities = 83/285 (29%), Positives = 142/285 (49%), Gaps = 20/285 (7%)
Query: 131 RNVLSIALDHTIYFWNASDSSGSEFVTVDEEEGPVTSVCWAPDGRHLAVGLTNSHVQLWD 190
+ V S + D TI W+ + SG+ T++ G V SV ++PDG+ +A G + +++WD
Sbjct: 980 QRVASGSGDKTIKIWDTA--SGTCTQTLEGHGGSVWSVAFSPDGQRVASGSDDKTIKIWD 1037
Query: 191 TAANKQLRTLKGGHRARVGSLAW--NGHVLTTGGMDGKIVNNDVRLRSQIINTYRGHRRE 248
TA+ +TL+ GH V S+ + +G + +G D I D + T GH
Sbjct: 1038 TASGTCTQTLE-GHGGWVQSVVFSPDGQRVASGSDDHTIKIWDA-VSGTCTQTLEGHGDS 1095
Query: 249 VCGLKWSLDGKQLASGGNDNVVHIWDMSAVSSNSPTRWLYRFDEHKAAVKALAWCPFQGN 308
V + +S DG+++ASG D + IWD +S + T+ L + H V ++A+ P G
Sbjct: 1096 VWSVAFSPDGQRVASGSIDGTIKIWD---AASGTCTQTL---EGHGGWVHSVAFSP-DGQ 1148
Query: 309 LLASGGGGGDCCVKLWNTGMGERMNSVD-TGSQVCALLWSKNERELLSSHGLTQNQLTLW 367
+ASG G +K+W+ G +++ G V ++ +S + + + S G + + +W
Sbjct: 1149 RVASGSIDG--TIKIWDAASGTCTQTLEGHGGWVQSVAFSPDGQRVAS--GSSDKTIKIW 1204
Query: 368 KYPSMLKIAELHGHTSRVLHMTQSPDGSTVASAAAAADQTLRFWE 412
S L GH V + SPDG VAS ++D T++ W+
Sbjct: 1205 DTASGTCTQTLEGHGGWVQSVAFSPDGQRVAS--GSSDNTIKIWD 1247
Score = 98.6 bits (244), Expect = 3e-20
Identities = 80/275 (29%), Positives = 134/275 (48%), Gaps = 23/275 (8%)
Query: 145 WNASDSSGSEFVTVDEEEGPVTSVCWAPDGRHLAVGLTNSHVQLWDTAANKQLRTLKGGH 204
WNA T++ V SV ++ DG+ +A G + +++WDTA+ +TL+ GH
Sbjct: 830 WNACTQ------TLEGHGSSVLSVAFSADGQRVASGSDDKTIKIWDTASGTGTQTLE-GH 882
Query: 205 RARVGSLAWN-GHVLTTGGMDGKIVNNDVRLRSQIINTYRGHRREVCGLKWSLDGKQLAS 263
V S+A++ G D K + T GH V + +S DG+++AS
Sbjct: 883 GGSVWSVAFSPDRERVASGSDDKTIKIWDAASGTCTQTLEGHGGRVQSVAFSPDGQRVAS 942
Query: 264 GGNDNVVHIWDMSAVSSNSPTRWLYRFDEHKAAVKALAWCPFQGNLLASGGGGGDCCVKL 323
G +D+ + IWD +S + T+ L + H ++V ++A+ P G +AS G GD +K+
Sbjct: 943 GSDDHTIKIWD---AASGTCTQTL---EGHGSSVLSVAFSP-DGQRVAS--GSGDKTIKI 993
Query: 324 WNTGMGERMNSVD-TGSQVCALLWSKNERELLSSHGLTQNQLTLWKYPSMLKIAELHGHT 382
W+T G +++ G V ++ +S + + + S G + +W S L GH
Sbjct: 994 WDTASGTCTQTLEGHGGSVWSVAFSPDGQRVAS--GSDDKTIKIWDTASGTCTQTLEGHG 1051
Query: 383 SRVLHMTQSPDGSTVASAAAAADQTLRFWE-VFGT 416
V + SPDG VAS + D T++ W+ V GT
Sbjct: 1052 GWVQSVVFSPDGQRVAS--GSDDHTIKIWDAVSGT 1084
Score = 97.1 bits (240), Expect = 8e-20
Identities = 81/277 (29%), Positives = 138/277 (49%), Gaps = 20/277 (7%)
Query: 139 DHTIYFWNASDSSGSEFVTVDEEEGPVTSVCWAPDGRHLAVGLTNSHVQLWDTAANKQLR 198
D TI W+A+ SG+ T++ G V SV ++PDG+ +A G + +++WD A+ +
Sbjct: 904 DKTIKIWDAA--SGTCTQTLEGHGGRVQSVAFSPDGQRVASGSDDHTIKIWDAASGTCTQ 961
Query: 199 TLKGGHRARVGSLAW--NGHVLTTGGMDGKIVNNDVRLRSQIINTYRGHRREVCGLKWSL 256
TL+ GH + V S+A+ +G + +G D I D T GH V + +S
Sbjct: 962 TLE-GHGSSVLSVAFSPDGQRVASGSGDKTIKIWDT-ASGTCTQTLEGHGGSVWSVAFSP 1019
Query: 257 DGKQLASGGNDNVVHIWDMSAVSSNSPTRWLYRFDEHKAAVKALAWCPFQGNLLASGGGG 316
DG+++ASG +D + IWD +S + T+ L + H V+++ + P G +AS G
Sbjct: 1020 DGQRVASGSDDKTIKIWD---TASGTCTQTL---EGHGGWVQSVVFSP-DGQRVAS--GS 1070
Query: 317 GDCCVKLWNTGMGERMNSVD-TGSQVCALLWSKNERELLSSHGLTQNQLTLWKYPSMLKI 375
D +K+W+ G +++ G V ++ +S + + + S G + +W S
Sbjct: 1071 DDHTIKIWDAVSGTCTQTLEGHGDSVWSVAFSPDGQRVAS--GSIDGTIKIWDAASGTCT 1128
Query: 376 AELHGHTSRVLHMTQSPDGSTVASAAAAADQTLRFWE 412
L GH V + SPDG VAS + D T++ W+
Sbjct: 1129 QTLEGHGGWVHSVAFSPDGQRVAS--GSIDGTIKIWD 1163
Score = 72.4 bits (176), Expect = 2e-12
Identities = 49/149 (32%), Positives = 82/149 (54%), Gaps = 6/149 (4%)
Query: 131 RNVLSIALDHTIYFWNASDSSGSEFVTVDEEEGPVTSVCWAPDGRHLAVGLTNSHVQLWD 190
+ V S ++D TI W+A+ SG+ T++ G V SV ++PDG+ +A G + +++WD
Sbjct: 1106 QRVASGSIDGTIKIWDAA--SGTCTQTLEGHGGWVHSVAFSPDGQRVASGSIDGTIKIWD 1163
Query: 191 TAANKQLRTLKGGHRARVGSLAWN--GHVLTTGGMDGKIVNNDVRLRSQIINTYRGHRRE 248
A+ +TL+G H V S+A++ G + +G D I D T GH
Sbjct: 1164 AASGTCTQTLEG-HGGWVQSVAFSPDGQRVASGSSDKTIKIWDTA-SGTCTQTLEGHGGW 1221
Query: 249 VCGLKWSLDGKQLASGGNDNVVHIWDMSA 277
V + +S DG+++ASG +DN + IWD ++
Sbjct: 1222 VQSVAFSPDGQRVASGSSDNTIKIWDTAS 1250
Score = 47.0 bits (110), Expect = 1e-04
Identities = 28/90 (31%), Positives = 49/90 (54%), Gaps = 2/90 (2%)
Query: 131 RNVLSIALDHTIYFWNASDSSGSEFVTVDEEEGPVTSVCWAPDGRHLAVGLTNSHVQLWD 190
+ V S + D TI W+ + SG+ T++ G V SV ++PDG+ +A G +++ +++WD
Sbjct: 1190 QRVASGSSDKTIKIWDTA--SGTCTQTLEGHGGWVQSVAFSPDGQRVASGSSDNTIKIWD 1247
Query: 191 TAANKQLRTLKGGHRARVGSLAWNGHVLTT 220
TA+ +TL G A S + + T
Sbjct: 1248 TASGTCTQTLNVGSTATCLSFDYTNAYINT 1277
>YS00_ANASP (Q8YTC2) Hypothetical WD-repeat protein alr2800
Length = 1258
Score = 97.1 bits (240), Expect = 8e-20
Identities = 82/279 (29%), Positives = 126/279 (44%), Gaps = 22/279 (7%)
Query: 139 DHTIYFWNASDSSGSEFVTVDEEEGPVTSVCWAPDGRHLAVGLTNSHVQLWDTAANKQLR 198
D TI W+ D G+ T+ V V ++PDG LA + ++LWD + K LR
Sbjct: 747 DKTIKLWDIQD--GTCLQTLTGHTDWVRCVAFSPDGNTLASSAADHTIKLWDVSQGKCLR 804
Query: 199 TLKGGHRARVGSLAWN--GHVLTTGGMDG--KIVNNDVRLRSQIINTYRGHRREVCGLKW 254
TLK H V S+A++ G L +G D KI N + + TY GH V + +
Sbjct: 805 TLKS-HTGWVRSVAFSADGQTLASGSGDRTIKIWNYHT---GECLKTYIGHTNSVYSIAY 860
Query: 255 SLDGKQLASGGNDNVVHIWDMSAVSSNSPTRWLYRFDEHKAAVKALAWCPFQGNLLASGG 314
S D K L SG D + +WD + H V ++A+ P G LA
Sbjct: 861 SPDSKILVSGSGDRTIKLWDCQTHIC------IKTLHGHTNEVCSVAFSP-DGQTLAC-- 911
Query: 315 GGGDCCVKLWNTGMGERMNSVDTGSQVCALLWSKNERELLSSHGLTQNQLTLWKYPSMLK 374
D V+LWN G+ + + + + +R++L+S G + LW + +
Sbjct: 912 VSLDQSVRLWNCRTGQCLKAWYGNTDWALPVAFSPDRQILAS-GSNDKTVKLWDWQTGKY 970
Query: 375 IAELHGHTSRVLHMTQSPDGSTVASAAAAADQTLRFWEV 413
I+ L GHT + + SPD T+AS A+ D ++R W +
Sbjct: 971 ISSLEGHTDFIYGIAFSPDSQTLAS--ASTDSSVRLWNI 1007
Score = 95.5 bits (236), Expect = 2e-19
Identities = 84/310 (27%), Positives = 137/310 (44%), Gaps = 50/310 (16%)
Query: 140 HTIYFWNASDSSGSEFVTVDEEEGPVTSVCWAPDGRHLAVGLTNSHVQLWDTAANKQLRT 199
H + F N SD S F E G + S ++P+G+ LA T+ HV++W+ + K L
Sbjct: 624 HDVDFAN-SDLSCCVFT---ETLGNILSAAFSPEGQLLATCDTDCHVRVWEVKSGKLLLI 679
Query: 200 LKG-GHRARVGSLAWNGHVLTTGGMDGKIVNNDVRLRSQIINTYRGHRREVCGLKWSLDG 258
+G + R + +G +L + G D + VR I T GH EV + + DG
Sbjct: 680 CRGHSNWVRFVVFSPDGEILASCGADENVKLWSVR-DGVCIKTLTGHEHEVFSVAFHPDG 738
Query: 259 KQLASGGNDNVVHIWDM-----------------------------SAVSSNSPTRW--- 286
+ LAS D + +WD+ S+ + ++ W
Sbjct: 739 ETLASASGDKTIKLWDIQDGTCLQTLTGHTDWVRCVAFSPDGNTLASSAADHTIKLWDVS 798
Query: 287 ----LYRFDEHKAAVKALAWCPFQGNLLASGGGGGDCCVKLWNTGMGERMNS-VDTGSQV 341
L H V+++A+ G LAS G GD +K+WN GE + + + + V
Sbjct: 799 QGKCLRTLKSHTGWVRSVAFSA-DGQTLAS--GSGDRTIKIWNYHTGECLKTYIGHTNSV 855
Query: 342 CALLWSKNERELLSSHGLTQNQLTLWKYPSMLKIAELHGHTSRVLHMTQSPDGSTVASAA 401
++ +S + + L+S G + LW + + I LHGHT+ V + SPDG T+ A
Sbjct: 856 YSIAYSPDSKILVSGSG--DRTIKLWDCQTHICIKTLHGHTNEVCSVAFSPDGQTL--AC 911
Query: 402 AAADQTLRFW 411
+ DQ++R W
Sbjct: 912 VSLDQSVRLW 921
Score = 90.5 bits (223), Expect = 8e-18
Identities = 77/285 (27%), Positives = 139/285 (48%), Gaps = 18/285 (6%)
Query: 130 SRNVLSIALDHTIYFWNASDSSGSEFVTVDEEEGPVTSVCWAPDGRHLAVGLTNSHVQLW 189
S+ ++S + D TI W+ + T+ V SV ++PDG+ LA + V+LW
Sbjct: 864 SKILVSGSGDRTIKLWDCQTHICIK--TLHGHTNEVCSVAFSPDGQTLACVSLDQSVRLW 921
Query: 190 DTAANKQLRTLKGGHR-ARVGSLAWNGHVLTTGGMDGKIVNNDVRLRSQIINTYRGHRRE 248
+ + L+ G A + + + +L +G D + D + + I++ GH
Sbjct: 922 NCRTGQCLKAWYGNTDWALPVAFSPDRQILASGSNDKTVKLWDWQT-GKYISSLEGHTDF 980
Query: 249 VCGLKWSLDGKQLASGGNDNVVHIWDMSAVSSNSPTRWLYRFDEHKAAVKALAWCPFQGN 308
+ G+ +S D + LAS D+ V +W++S + EH V A+ + P QG
Sbjct: 981 IYGIAFSPDSQTLASASTDSSVRLWNIST------GQCFQILLEHTDWVYAVVFHP-QGK 1033
Query: 309 LLASGGGGGDCCVKLWNTGMGERMNSVDTGS-QVCALLWSKNERELLSSHGLTQNQLTLW 367
++A+ G DC VKLWN G+ + ++ S ++ + WS + +LL+S Q+ + LW
Sbjct: 1034 IIAT--GSADCTVKLWNISTGQCLKTLSEHSDKILGMAWSP-DGQLLASASADQS-VRLW 1089
Query: 368 KYPSMLKIAELHGHTSRVLHMTQSPDGSTVASAAAAADQTLRFWE 412
+ + L GH++RV SP+G +A+ + DQT++ W+
Sbjct: 1090 DCCTGRCVGILRGHSNRVYSAIFSPNGEIIAT--CSTDQTVKIWD 1132
Score = 87.0 bits (214), Expect = 8e-17
Identities = 90/350 (25%), Positives = 140/350 (39%), Gaps = 56/350 (16%)
Query: 111 TFDLPDLSDDFSLNLL----DW--------GSRNVLSIALDHTIYFWNASDSSGSEFVTV 158
T L D+ D L L DW + S A DHTI W+ S G T+
Sbjct: 749 TIKLWDIQDGTCLQTLTGHTDWVRCVAFSPDGNTLASSAADHTIKLWDV--SQGKCLRTL 806
Query: 159 DEEEGPVTSVCWAPDGRHLAVGLTNSHVQLWDTAANKQLRTLKGGHRARVGSLAW--NGH 216
G V SV ++ DG+ LA G + +++W+ + L+T GH V S+A+ +
Sbjct: 807 KSHTGWVRSVAFSADGQTLASGSGDRTIKIWNYHTGECLKTYI-GHTNSVYSIAYSPDSK 865
Query: 217 VLTTGGMDGKIVNNDVRLRSQIINTYRGHRREVCGLKWSLDGKQLASGGNDNVVHIW--- 273
+L +G D I D + I T GH EVC + +S DG+ LA D V +W
Sbjct: 866 ILVSGSGDRTIKLWDCQTHI-CIKTLHGHTNEVCSVAFSPDGQTLACVSLDQSVRLWNCR 924
Query: 274 ------------DMSAVSSNSPTRWLYRFDEHKAAVKALAW------------------C 303
D + + SP R + + VK W
Sbjct: 925 TGQCLKAWYGNTDWALPVAFSPDRQILASGSNDKTVKLWDWQTGKYISSLEGHTDFIYGI 984
Query: 304 PFQGNLLASGGGGGDCCVKLWNTGMGERMN-SVDTGSQVCALLWSKNERELLSSHGLTQN 362
F + D V+LWN G+ ++ V A+++ + + + G
Sbjct: 985 AFSPDSQTLASASTDSSVRLWNISTGQCFQILLEHTDWVYAVVFHPQGK--IIATGSADC 1042
Query: 363 QLTLWKYPSMLKIAELHGHTSRVLHMTQSPDGSTVASAAAAADQTLRFWE 412
+ LW + + L H+ ++L M SPDG +AS A+ADQ++R W+
Sbjct: 1043 TVKLWNISTGQCLKTLSEHSDKILGMAWSPDGQLLAS--ASADQSVRLWD 1090
Score = 79.7 bits (195), Expect = 1e-14
Identities = 59/194 (30%), Positives = 93/194 (47%), Gaps = 15/194 (7%)
Query: 139 DHTIYFWNASDSSGSEFVTVDEEEGPVTSVCWAPDGRHLAVGLTNSHVQLWDTAANKQLR 198
D T+ WN S +G T+ E + + W+PDG+ LA + V+LWD + +
Sbjct: 1041 DCTVKLWNIS--TGQCLKTLSEHSDKILGMAWSPDGQLLASASADQSVRLWDCCTGRCVG 1098
Query: 199 TLKGGHRARVGSLAW--NGHVLTTGGMDGKIVNNDVRLRSQIINTYRGHRREVCGLKWSL 256
L+ GH RV S + NG ++ T D + D + + + + T GH V + +S
Sbjct: 1099 ILR-GHSNRVYSAIFSPNGEIIATCSTDQTVKIWDWQ-QGKCLKTLTGHTNWVFDIAFSP 1156
Query: 257 DGKQLASGGNDNVVHIWDMSAVSSNSPTRWLYRFDEHKAAVKALAWCPFQGNLLASGGGG 316
DGK LAS +D V IWD++ + + H V ++A+ P G ++AS G
Sbjct: 1157 DGKILASASHDQTVRIWDVNTGKCH------HICIGHTHLVSSVAFSP-DGEVVAS--GS 1207
Query: 317 GDCCVKLWNTGMGE 330
D V++WN GE
Sbjct: 1208 QDQTVRIWNVKTGE 1221
Score = 71.2 bits (173), Expect = 5e-12
Identities = 77/301 (25%), Positives = 132/301 (43%), Gaps = 43/301 (14%)
Query: 118 SDDFSLNLLDWGSRNVLSIALDHTIYFWNASDSSGSEFVTVDEEEGPVTSVCWAPDGRHL 177
S+D ++ L DW + +S HT + + + ++PD + L
Sbjct: 955 SNDKTVKLWDWQTGKYISSLEGHTDFIY---------------------GIAFSPDSQTL 993
Query: 178 AVGLTNSHVQLWDTAANKQLRTLKGGHRARVGSLAWN--GHVLTTGGMDGKIVNNDVRLR 235
A T+S V+LW+ + + + L H V ++ ++ G ++ TG D + ++
Sbjct: 994 ASASTDSSVRLWNISTGQCFQILL-EHTDWVYAVVFHPQGKIIATGSADCTVKLWNIS-T 1051
Query: 236 SQIINTYRGHRREVCGLKWSLDGKQLASGGNDNVVHIWDMSAVSSNSPTRWLYRFDEHKA 295
Q + T H ++ G+ WS DG+ LAS D V +WD R + H
Sbjct: 1052 GQCLKTLSEHSDKILGMAWSPDGQLLASASADQSVRLWDCCT------GRCVGILRGHSN 1105
Query: 296 AVKALAWCPFQGNLLASGGGGGDCCVKLWNTGMGERMNSVDTG--SQVCALLWSKNEREL 353
V + + P G ++A+ D VK+W+ G+ + ++ TG + V + +S + + L
Sbjct: 1106 RVYSAIFSP-NGEIIAT--CSTDQTVKIWDWQQGKCLKTL-TGHTNWVFDIAFSPDGKIL 1161
Query: 354 LS-SHGLTQNQLTLWKYPSMLKIAELHGHTSRVLHMTQSPDGSTVASAAAAADQTLRFWE 412
S SH T + +W + GHT V + SPDG VAS + DQT+R W
Sbjct: 1162 ASASHDQT---VRIWDVNTGKCHHICIGHTHLVSSVAFSPDGEVVAS--GSQDQTVRIWN 1216
Query: 413 V 413
V
Sbjct: 1217 V 1217
Score = 48.5 bits (114), Expect = 3e-05
Identities = 36/124 (29%), Positives = 60/124 (48%), Gaps = 6/124 (4%)
Query: 290 FDEHKAAVKALAWCPFQGNLLASGGGGGDCCVKLWNTGMGERMNSVDTGSQVCALLWSKN 349
F E + + A+ P +G LLA+ DC V++W G+ + S +
Sbjct: 638 FTETLGNILSAAFSP-EGQLLATCDT--DCHVRVWEVKSGKLLLICRGHSNWVRFVVFSP 694
Query: 350 ERELLSSHGLTQNQLTLWKYPSMLKIAELHGHTSRVLHMTQSPDGSTVASAAAAADQTLR 409
+ E+L+S G +N + LW + I L GH V + PDG T+ASA+ D+T++
Sbjct: 695 DGEILASCGADEN-VKLWSVRDGVCIKTLTGHEHEVFSVAFHPDGETLASASG--DKTIK 751
Query: 410 FWEV 413
W++
Sbjct: 752 LWDI 755
>PKWA_THECU (P49695) Probable serine/threonine-protein kinase pkwA
(EC 2.7.1.37)
Length = 742
Score = 95.5 bits (236), Expect = 2e-19
Identities = 77/277 (27%), Positives = 131/277 (46%), Gaps = 19/277 (6%)
Query: 139 DHTIYFWNASDSSGSEFVTVDEEEGPVTSVCWAPDGRHLAVGLTNSHVQLWDTAANKQLR 198
D I+ W+ + SG E T++ V +V ++PDG LA G ++ V+LWD AA ++ R
Sbjct: 480 DKLIHVWDVA--SGDELHTLEGHTDWVRAVAFSPDGALLASGSDDATVRLWDVAAAEE-R 536
Query: 199 TLKGGHRARVGSLAW--NGHVLTTGGMDGKIVNNDVRLRSQIINTYRGHRREVCGLKWSL 256
+ GH V +A+ +G ++ +G DG +V ++ +GH V + +S
Sbjct: 537 AVFEGHTHYVLDIAFSPDGSMVASGSRDGTARLWNVATGTEHA-VLKGHTDYVYAVAFSP 595
Query: 257 DGKQLASGGNDNVVHIWDMSAVSSNSPTRWLYRFDEHKAAVKALAWCPFQGNLLASGGGG 316
DG +ASG D + +WD++ V +LA+ P G++L G
Sbjct: 596 DGSMVASGSRDGTIRLWDVATGKERDV------LQAPAENVVSLAFSP-DGSMLVH---G 645
Query: 317 GDCCVKLWNTGMGERMNSVDTGSQVCALLWSKNERELLSSHGLTQNQLTLWKYPSMLKIA 376
D V LW+ GE +++ + + + + LL+S G + LW + +
Sbjct: 646 SDSTVHLWDVASGEALHTFEGHTDWVRAVAFSPDGALLAS-GSDDRTIRLWDVAAQEEHT 704
Query: 377 ELHGHTSRVLHMTQSPDGSTVASAAAAADQTLRFWEV 413
L GHT V + P+G+T+AS A+ D T+R W +
Sbjct: 705 TLEGHTEPVHSVAFHPEGTTLAS--ASEDGTIRIWPI 739
Score = 90.5 bits (223), Expect = 8e-18
Identities = 73/261 (27%), Positives = 131/261 (49%), Gaps = 22/261 (8%)
Query: 156 VTVDEEEGPVTSVCWAPDGRHLAVGLTNSHVQLWDTAANKQLRTLKGGHRARVGSLAW-- 213
+T D E +V ++P G LA G + + +WD A+ +L TL+ GH V ++A+
Sbjct: 456 LTTDRE---AVAVAFSPGGSLLAGGSGDKLIHVWDVASGDELHTLE-GHTDWVRAVAFSP 511
Query: 214 NGHVLTTGGMDGKIVNNDVRLRSQIINTYRGHRREVCGLKWSLDGKQLASGGNDNVVHIW 273
+G +L +G D + DV + + GH V + +S DG +ASG D +W
Sbjct: 512 DGALLASGSDDATVRLWDVAAAEERA-VFEGHTHYVLDIAFSPDGSMVASGSRDGTARLW 570
Query: 274 DMSAVSSNSPTRWLYRFDEHKAAVKALAWCPFQGNLLASGGGGGDCCVKLWNTGMGERMN 333
+++ + ++ + H V A+A+ P G+++ASG G ++LW+ G+ +
Sbjct: 571 NVATGTEHAVLK------GHTDYVYAVAFSP-DGSMVASGSRDG--TIRLWDVATGKERD 621
Query: 334 SVDTGSQ-VCALLWSKNERELLSSHGLTQNQLTLWKYPSMLKIAELHGHTSRVLHMTQSP 392
+ ++ V +L +S + L+ HG + + + LW S + GHT V + SP
Sbjct: 622 VLQAPAENVVSLAFSPDGSMLV--HG-SDSTVHLWDVASGEALHTFEGHTDWVRAVAFSP 678
Query: 393 DGSTVASAAAAADQTLRFWEV 413
DG+ +AS + D+T+R W+V
Sbjct: 679 DGALLAS--GSDDRTIRLWDV 697
Score = 68.9 bits (167), Expect = 2e-11
Identities = 56/194 (28%), Positives = 95/194 (48%), Gaps = 16/194 (8%)
Query: 133 VLSIALDHTIYFWNASDSSGSEFVTVDEEEGPVTSVCWAPDGRHLAVGLTNSHVQLWDTA 192
V S + D T WN + +G+E + V +V ++PDG +A G + ++LWD A
Sbjct: 558 VASGSRDGTARLWNVA--TGTEHAVLKGHTDYVYAVAFSPDGSMVASGSRDGTIRLWDVA 615
Query: 193 ANKQLRTLKGGHRARVGSLAWN--GHVLTTGGMDGKIVNNDVRLRSQIINTYRGHRREVC 250
K+ L+ V SLA++ G +L G D + DV + ++T+ GH V
Sbjct: 616 TGKERDVLQAPAE-NVVSLAFSPDGSMLVHGS-DSTVHLWDVA-SGEALHTFEGHTDWVR 672
Query: 251 GLKWSLDGKQLASGGNDNVVHIWDMSAVSSNSPTRWLYRFDEHKAAVKALAWCPFQGNLL 310
+ +S DG LASG +D + +WD++A ++ + H V ++A+ P +G L
Sbjct: 673 AVAFSPDGALLASGSDDRTIRLWDVAAQEEHT------TLEGHTEPVHSVAFHP-EGTTL 725
Query: 311 ASGGGGGDCCVKLW 324
AS G +++W
Sbjct: 726 ASASEDG--TIRIW 737
Score = 56.6 bits (135), Expect = 1e-07
Identities = 44/146 (30%), Positives = 71/146 (48%), Gaps = 7/146 (4%)
Query: 133 VLSIALDHTIYFWNASDSSGSEFVTVDEEEGPVTSVCWAPDGRHLAVGLTNSHVQLWDTA 192
V S + D TI W+ + + + E V S+ ++PDG L G ++S V LWD A
Sbjct: 600 VASGSRDGTIRLWDVATGKERDVLQAPAEN--VVSLAFSPDGSMLVHG-SDSTVHLWDVA 656
Query: 193 ANKQLRTLKGGHRARVGSLAWN--GHVLTTGGMDGKIVNNDVRLRSQIINTYRGHRREVC 250
+ + L T +G H V ++A++ G +L +G D I DV + + T GH V
Sbjct: 657 SGEALHTFEG-HTDWVRAVAFSPDGALLASGSDDRTIRLWDVAAQEEH-TTLEGHTEPVH 714
Query: 251 GLKWSLDGKQLASGGNDNVVHIWDMS 276
+ + +G LAS D + IW ++
Sbjct: 715 SVAFHPEGTTLASASEDGTIRIWPIA 740
>YL24_ANASP (Q8YV57) Hypothetical WD-repeat protein all2124
Length = 1683
Score = 90.1 bits (222), Expect = 1e-17
Identities = 76/285 (26%), Positives = 135/285 (46%), Gaps = 22/285 (7%)
Query: 131 RNVLSIALDHTIYFWNASDSSGSEFVTVDEEEGPVTSVCWAPDGRHLAVGLTNSHVQLWD 190
+ + S +LD TI W+ G F T++ E V SV ++PDG+ +A G ++ ++LW
Sbjct: 1085 QTIASGSLDKTIKLWSRD---GRLFRTLNGHEDAVYSVSFSPDGQTIASGGSDKTIKLWQ 1141
Query: 191 TAANKQLRTLKGGHRARVGSLAW--NGHVLTTGGMDGKIVNNDVRLRSQIINTYRGHRRE 248
T+ L+T+ GH V ++ + +G L + D I D Q++ T GH
Sbjct: 1142 TSDGTLLKTIT-GHEQTVNNVYFSPDGKNLASASSDHSIKLWDT-TSGQLLMTLTGHSAG 1199
Query: 249 VCGLKWSLDGKQLASGGNDNVVHIWDMSAVSSNSPTRWLYRFDEHKAAVKALAWCPFQGN 308
V +++S DG+ +A+G D V +W + L + H+ V +L++ P G
Sbjct: 1200 VITVRFSPDGQTIAAGSEDKTVKLW------HRQDGKLLKTLNGHQDWVNSLSFSP-DGK 1252
Query: 309 LLASGGGGGDCCVKLWNTGMGERMNSV-DTGSQVCALLWSKNERELLSSHGLTQNQLTLW 367
LAS D +KLW G+ + ++ V + +S + + + S+ N + LW
Sbjct: 1253 TLAS--ASADKTIKLWRIADGKLVKTLKGHNDSVWDVNFSSDGKAIASAS--RDNTIKLW 1308
Query: 368 KYPSMLKIAELHGHTSRVLHMTQSPDGSTVASAAAAADQTLRFWE 412
+++ GH+ V + PD + +AS A+ D T+R W+
Sbjct: 1309 NRHG-IELETFTGHSGGVYAVNFLPDSNIIAS--ASLDNTIRLWQ 1350
Score = 84.3 bits (207), Expect = 5e-16
Identities = 78/289 (26%), Positives = 129/289 (43%), Gaps = 32/289 (11%)
Query: 131 RNVLSIALDHTIYFWNASDSSGSEFVTVDEEEGPVTSVCWAPDGRHLAVGLTNSHVQLWD 190
+ + S D TI W SD G+ T+ E V +V ++PDG++LA ++ ++LWD
Sbjct: 1126 QTIASGGSDKTIKLWQTSD--GTLLKTITGHEQTVNNVYFSPDGKNLASASSDHSIKLWD 1183
Query: 191 TAANKQLRTLKGGHRARVGSLAW--NGHVLTTGGMDGKIVNNDVRLRSQIINTYRGHRRE 248
T + + L TL GH A V ++ + +G + G D K V R +++ T GH+
Sbjct: 1184 TTSGQLLMTLT-GHSAGVITVRFSPDGQTIAAGSED-KTVKLWHRQDGKLLKTLNGHQDW 1241
Query: 249 VCGLKWSLDGKQLASGGNDNVVHIWDMS------AVSSNSPTRWLYRFDEHKAAVKALAW 302
V L +S DGK LAS D + +W ++ + ++ + W F
Sbjct: 1242 VNSLSFSPDGKTLASASADKTIKLWRIADGKLVKTLKGHNDSVWDVNFSS---------- 1291
Query: 303 CPFQGNLLASGGGGGDCCVKLWNTGMGERMNSVDTGSQVCALLWSKNERELLSSHGLTQN 362
G +AS D +KLWN E V A+ + + + S+ N
Sbjct: 1292 ---DGKAIAS--ASRDNTIKLWNRHGIELETFTGHSGGVYAVNFLPDSNIIASAS--LDN 1344
Query: 363 QLTLWKYPSMLKIAELHGHTSRVLHMTQSPDGSTVASAAAAADQTLRFW 411
+ LW+ P + + L G+ S V ++ DGS +A+ A AD ++ W
Sbjct: 1345 TIRLWQRPLISPLEVLAGN-SGVYAVSFLHDGSIIAT--AGADGNIQLW 1390
Score = 82.8 bits (203), Expect = 2e-15
Identities = 74/295 (25%), Positives = 133/295 (45%), Gaps = 17/295 (5%)
Query: 121 FSLNLLDWGSRNVLSIALDHTIYFWNASDSSGSEFVTVDEEEGPVTSVCWAPDGRHLAVG 180
+++N L S + S +LD+TI W S E + + V +V + DG +A
Sbjct: 1326 YAVNFLP-DSNIIASASLDNTIRLWQRPLISPLEVLAGNSG---VYAVSFLHDGSIIATA 1381
Query: 181 LTNSHVQLWDTAANKQLRTLKGGHRARVGSLAWNGHVLTTGGMDGKIVNNDVRLRSQIIN 240
+ ++QLW + L+TL G S G ++ + D + VR + +
Sbjct: 1382 GADGNIQLWHSQDGSLLKTLPGNKAIYGISFTPQGDLIASANADKTVKIWRVR-DGKALK 1440
Query: 241 TYRGHRREVCGLKWSLDGKQLASGGNDNVVHIWDMSAVSSNSPTRWLYRFDEHKAAVKAL 300
T GH EV + +S DGK LAS DN V +W++ S ++ H V +
Sbjct: 1441 TLIGHDNEVNKVNFSPDGKTLASASRDNTVKLWNV------SDGKFKKTLKGHTDEVFWV 1494
Query: 301 AWCPFQGNLLASGGGGGDCCVKLWNTGMGERMNSVDTGSQVCALLWSKNERELLSSHGLT 360
++ P G ++AS D ++LW++ G + S+ + + + + +L+S
Sbjct: 1495 SFSP-DGKIIAS--ASADKTIRLWDSFSGNLIKSLPAHNDLVYSVNFNPDGSMLASTS-A 1550
Query: 361 QNQLTLWKYPSMLKIAELHGHTSRVLHMTQSPDGSTVASAAAAADQTLRFWEVFG 415
+ LW+ + GH++ V + SPDG +AS A+ D+T++ W++ G
Sbjct: 1551 DKTVKLWRSHDGHLLHTFSGHSNVVYSSSFSPDGRYIAS--ASEDKTVKIWQIDG 1603
Score = 81.3 bits (199), Expect = 5e-15
Identities = 71/255 (27%), Positives = 122/255 (47%), Gaps = 21/255 (8%)
Query: 165 VTSVCWAPDGRHLAVGLTNSHVQLWDTAANKQLRTLKGGHRARVGSLAW--NGHVLTTGG 222
V S+ + DG+ +A G + ++LW + + RTL GH V S+++ +G + +GG
Sbjct: 1075 VISISISRDGQTIASGSLDKTIKLW-SRDGRLFRTL-NGHEDAVYSVSFSPDGQTIASGG 1132
Query: 223 MDGKIVNNDVRLRSQIINTYRGHRREVCGLKWSLDGKQLASGGNDNVVHIWDMSAVSSNS 282
D K + ++ T GH + V + +S DGK LAS +D+ + +WD ++
Sbjct: 1133 SD-KTIKLWQTSDGTLLKTITGHEQTVNNVYFSPDGKNLASASSDHSIKLWDTTS----- 1186
Query: 283 PTRWLYRFDEHKAAVKALAWCPFQGNLLASGGGGGDCCVKLWNTGMGERMNSVDTGSQ-- 340
+ L H A V + + P G +A+ G D VKLW+ G+ + +++ G Q
Sbjct: 1187 -GQLLMTLTGHSAGVITVRFSP-DGQTIAA--GSEDKTVKLWHRQDGKLLKTLN-GHQDW 1241
Query: 341 VCALLWSKNERELLSSHGLTQNQLTLWKYPSMLKIAELHGHTSRVLHMTQSPDGSTVASA 400
V +L +S + + L S+ + LW+ + L GH V + S DG +AS
Sbjct: 1242 VNSLSFSPDGKTLASAS--ADKTIKLWRIADGKLVKTLKGHNDSVWDVNFSSDGKAIAS- 1298
Query: 401 AAAADQTLRFWEVFG 415
A+ D T++ W G
Sbjct: 1299 -ASRDNTIKLWNRHG 1312
Score = 71.2 bits (173), Expect = 5e-12
Identities = 48/172 (27%), Positives = 83/172 (47%), Gaps = 13/172 (7%)
Query: 240 NTYRGHRREVCGLKWSLDGKQLASGGNDNVVHIWDMSAVSSNSPTRWLYRFDEHKAAVKA 299
N GH+ V + S DG+ +ASG D + +W R + H+ AV +
Sbjct: 1066 NRLEGHKDGVISISISRDGQTIASGSLDKTIKLWSRDG-------RLFRTLNGHEDAVYS 1118
Query: 300 LAWCPFQGNLLASGGGGGDCCVKLWNTGMGERMNSVDTGSQVCALLWSKNERELLSSHGL 359
+++ P G +AS GG D +KLW T G + ++ Q ++ + + L+S
Sbjct: 1119 VSFSP-DGQTIAS--GGSDKTIKLWQTSDGTLLKTITGHEQTVNNVYFSPDGKNLAS-AS 1174
Query: 360 TQNQLTLWKYPSMLKIAELHGHTSRVLHMTQSPDGSTVASAAAAADQTLRFW 411
+ + + LW S + L GH++ V+ + SPDG T+ AA + D+T++ W
Sbjct: 1175 SDHSIKLWDTTSGQLLMTLTGHSAGVITVRFSPDGQTI--AAGSEDKTVKLW 1224
Score = 43.9 bits (102), Expect = 8e-04
Identities = 37/142 (26%), Positives = 60/142 (42%), Gaps = 5/142 (3%)
Query: 133 VLSIALDHTIYFWNASDSSGSEFVTVDEEEGPVTSVCWAPDGRHLAVGLTNSHVQLWDTA 192
+ S + D TI W++ SG+ ++ V SV + PDG LA + V+LW +
Sbjct: 1503 IASASADKTIRLWDSF--SGNLIKSLPAHNDLVYSVNFNPDGSMLASTSADKTVKLWRSH 1560
Query: 193 ANKQLRTLKG-GHRARVGSLAWNGHVLTTGGMDGKIVNNDVRLRSQIINTYRGHRREVCG 251
L T G + S + +G + + D + ++ ++ T H+ V
Sbjct: 1561 DGHLLHTFSGHSNVVYSSSFSPDGRYIASASEDKTV--KIWQIDGHLLTTLPQHQAGVMS 1618
Query: 252 LKWSLDGKQLASGGNDNVVHIW 273
+S DGK L SG D IW
Sbjct: 1619 AIFSPDGKTLISGSLDTTTKIW 1640
>WDS_DROME (Q9V3J8) Will die slowly protein
Length = 361
Score = 81.3 bits (199), Expect = 5e-15
Identities = 73/291 (25%), Positives = 126/291 (43%), Gaps = 34/291 (11%)
Query: 135 SIALDHTIYFWNASDSSGSEFVTVDEEEGPVTSVCWAPDGRHLAVGLTNSHVQLWDTAAN 194
S + D I W A D + T+ + ++ V W+ D R L G + +++W+ +
Sbjct: 89 SSSADKLIKIWGAYDGKFEK--TISGHKLGISDVAWSSDSRLLVSGSDDKTLKVWELSTG 146
Query: 195 KQLRTLKGGHRARVGSLAWN--GHVLTTGGMDGKIVNNDVRLRSQIINTYRGHRREVCGL 252
K L+TLKG H V +N +++ +G D + DVR + + T H V +
Sbjct: 147 KSLKTLKG-HSNYVFCCNFNPQSNLIVSGSFDESVRIWDVRT-GKCLKTLPAHSDPVSAV 204
Query: 253 KWSLDGKQLASGGNDNVVHIWDMSAVSSNSPTRWLYRFDEHKAAVKALAWCPFQGNLLAS 312
++ DG + S D + IWD +S + L D+ V + + P +LA+
Sbjct: 205 HFNRDGSLIVSSSYDGLCRIWD---TASGQCLKTL--IDDDNPPVSFVKFSPNGKYILAA 259
Query: 313 GGGGGDCCVKLWNTGMGERMNSVDTGSQVCALLWSKNERELLSSH-----------GLTQ 361
D +KLW+ G+ + + TG KNE+ + ++ G
Sbjct: 260 TL---DNTLKLWDYSKGKCLKTY-TGH--------KNEKYCIFANFSVTGGKWIVSGSED 307
Query: 362 NQLTLWKYPSMLKIAELHGHTSRVLHMTQSPDGSTVASAAAAADQTLRFWE 412
N + +W S + +L GHT VL P + +ASAA D+T++ W+
Sbjct: 308 NMVYIWNLQSKEVVQKLQGHTDTVLCTACHPTENIIASAALENDKTIKLWK 358
Score = 45.8 bits (107), Expect = 2e-04
Identities = 40/172 (23%), Positives = 70/172 (40%), Gaps = 12/172 (6%)
Query: 241 TYRGHRREVCGLKWSLDGKQLASGGNDNVVHIWDMSAVSSNSPTRWLYRFDEHKAAVKAL 300
T GH + V +K+S +G+ LAS D ++ IW ++ HK + +
Sbjct: 67 TLAGHTKAVSAVKFSPNGEWLASSSADKLIKIW------GAYDGKFEKTISGHKLGISDV 120
Query: 301 AWCPFQGNLLASGGGGGDCCVKLWNTGMGERMNSVDTGSQVCALLWSKNERELLSSHGLT 360
AW LL S G D +K+W G+ + ++ G + N + L G
Sbjct: 121 AWSS-DSRLLVS--GSDDKTLKVWELSTGKSLKTL-KGHSNYVFCCNFNPQSNLIVSGSF 176
Query: 361 QNQLTLWKYPSMLKIAELHGHTSRVLHMTQSPDGSTVASAAAAADQTLRFWE 412
+ +W + + L H+ V + + DGS + S ++ D R W+
Sbjct: 177 DESVRIWDVRTGKCLKTLPAHSDPVSAVHFNRDGSLIVS--SSYDGLCRIWD 226
>YCW2_YEAST (P25382) Hypothetical WD-repeat protein YCR072C
Length = 515
Score = 75.1 bits (183), Expect = 3e-13
Identities = 55/209 (26%), Positives = 99/209 (47%), Gaps = 23/209 (11%)
Query: 67 LAEAADLPTRILAFRNKPRKRNVISPPPPPRSKPMRYIPKTCEGTFDLPDLSDDFSLNLL 126
L+ + D RI AF + +K S P + K + K C+ + ++
Sbjct: 326 LSLSTDYALRIGAFDHTGKKP---STPEEAQKKALENYEKICKKNGNSEEM--------- 373
Query: 127 DWGSRNVLSIALDHTIYFWNASDSSGSEFVTVDEEEGPVTSVCWAPDGRHLAVGLTNSHV 186
+++ + D+T++ WN S+ + + V V ++PDGR++ ++ +
Sbjct: 374 ------MVTASDDYTMFLWNPLKST-KPIARMTGHQKLVNHVAFSPDGRYIVSASFDNSI 426
Query: 187 QLWDTAANKQLRTLKGGHRARVGSLAWNG--HVLTTGGMDGKIVNNDVRLRSQIINTYRG 244
+LWD K + T +G H A V +AW+ +L + D + DVR R ++ G
Sbjct: 427 KLWDGRDGKFISTFRG-HVASVYQVAWSSDCRLLVSCSKDTTLKVWDVRTRKLSVDL-PG 484
Query: 245 HRREVCGLKWSLDGKQLASGGNDNVVHIW 273
H+ EV + WS+DGK++ SGG D +V +W
Sbjct: 485 HKDEVYTVDWSVDGKRVCSGGKDKMVRLW 513
Score = 72.8 bits (177), Expect = 2e-12
Identities = 71/292 (24%), Positives = 121/292 (41%), Gaps = 26/292 (8%)
Query: 130 SRNVLSIALDHTIYFWNASDSSGSEFVTVDEEEGPVTSVCWAPDGRHLAVGLTNSHVQLW 189
S +++ A D+T W+ + T+ V V W+PDG +A G ++ ++LW
Sbjct: 156 SSRMVTGAGDNTARIWDCDTQTPMH--TLKGHYNWVLCVSWSPDGEVIATGSMDNTIRLW 213
Query: 190 DTAANKQLRTLKGGHRARVGSLAWNGHVLTTGGMDGKIVNND----VRLRSQIIN----T 241
D + + L GH + SL+W L G ++ ++ +++ + T
Sbjct: 214 DPKSGQCLGDALRGHSKWITSLSWEPIHLVKPGSKPRLASSSKDGTIKIWDTVSRVCQYT 273
Query: 242 YRGHRREVCGLKWSLDGKQLASGGNDNVVHIWDMSAVSSNSPTRWLYRFDEHKAAVKALA 301
GH V +KW G L SG +D V +WD+ NS R + H V L+
Sbjct: 274 MSGHTNSVSCVKWGGQG-LLYSGSHDRTVRVWDI-----NSQGRCINILKSHAHWVNHLS 327
Query: 302 WCPFQGNLLASGGGGGDCCVKLWNTGMGERMNSVDTGSQVCALLWSKNERELLSSHGLTQ 361
A G D K +T + +++ ++C + E + +S T
Sbjct: 328 ----LSTDYALRIGAFDHTGKKPSTPEEAQKKALENYEKICKKNGNSEEMMVTASDDYT- 382
Query: 362 NQLTLWK-YPSMLKIAELHGHTSRVLHMTQSPDGSTVASAAAAADQTLRFWE 412
+ LW S IA + GH V H+ SPDG + S A+ D +++ W+
Sbjct: 383 --MFLWNPLKSTKPIARMTGHQKLVNHVAFSPDGRYIVS--ASFDNSIKLWD 430
Score = 46.2 bits (108), Expect = 2e-04
Identities = 67/296 (22%), Positives = 109/296 (36%), Gaps = 63/296 (21%)
Query: 165 VTSVCWAP-------DGRHLAVGLTNSHVQLWDTAANKQLRTLKGGHRARVGSLAWNGH- 216
+TS+ W P LA + +++WDT + T+ G H V + W G
Sbjct: 232 ITSLSWEPIHLVKPGSKPRLASSSKDGTIKIWDTVSRVCQYTMSG-HTNSVSCVKWGGQG 290
Query: 217 VLTTGGMDGKIVNNDVRLRSQIINTYRGHRREVCGLKWSLD-----------GKQ----- 260
+L +G D + D+ + + IN + H V L S D GK+
Sbjct: 291 LLYSGSHDRTVRVWDINSQGRCINILKSHAHWVNHLSLSTDYALRIGAFDHTGKKPSTPE 350
Query: 261 -----------------------LASGGNDNVVHIWDMSAVSSNSPTRWLYRFDEHKAAV 297
+ + +D + +W+ + S P + R H+ V
Sbjct: 351 EAQKKALENYEKICKKNGNSEEMMVTASDDYTMFLWN--PLKSTKP---IARMTGHQKLV 405
Query: 298 KALAWCPFQGNLLASGGGGGDCCVKLWNTGMGERMNSVDTG--SQVCALLWSKNERELLS 355
+A+ P G + S D +KLW+ G + S G + V + WS + R L+S
Sbjct: 406 NHVAFSP-DGRYIVSASF--DNSIKLWD-GRDGKFISTFRGHVASVYQVAWSSDCRLLVS 461
Query: 356 SHGLTQNQLTLWKYPSMLKIAELHGHTSRVLHMTQSPDGSTVASAAAAADQTLRFW 411
T L +W + +L GH V + S DG V S D+ +R W
Sbjct: 462 CSKDTT--LKVWDVRTRKLSVDLPGHKDEVYTVDWSVDGKRVCS--GGKDKMVRLW 513
>APAF_MOUSE (O88879) Apoptotic protease activating factor 1 (Apaf-1)
Length = 1249
Score = 74.7 bits (182), Expect = 4e-13
Identities = 70/294 (23%), Positives = 126/294 (42%), Gaps = 23/294 (7%)
Query: 131 RNVLSIALDHTIYFWNASDSSGSEFVTVDEEEGPVTSVCWAPDGRHLAVGLTNSHVQLWD 190
+ + S D T+ + A +G + + + E V ++ D ++A + V++WD
Sbjct: 628 QRIASCGADKTLQVFKAE--TGEKLLDIKAHEDEVLCCAFSSDDSYIATCSADKKVKIWD 685
Query: 191 TAANKQLRTLKGGHRARVGSLAW----NGHVLTTGGMDGKIVNNDVRLRSQIINTYRGHR 246
+A K + T H +V + N +L TG D + D+ + + NT GH
Sbjct: 686 SATGKLVHTYDE-HSEQVNCCHFTNSSNHLLLATGSNDFFLKLWDLN-QKECRNTMFGHT 743
Query: 247 REVCGLKWSLDGKQLASGGNDNVVHIWDMSAVSSNSPTRWLYRF--------DEHKAAVK 298
V ++S D + LAS D + +WD+ + + + RF ++ + VK
Sbjct: 744 NSVNHCRFSPDDELLASCSADGTLRLWDVRSANERKSIN-VKRFFLSSEDPPEDVEVIVK 802
Query: 299 ALAWCPFQGNLLASGGGGGDCCVKLWNTGMGERMNSVDTGSQVCALLWSKNERELLSSHG 358
+W ++ + V L++ + + TG + + L+
Sbjct: 803 CCSWSADGDKIIVAAKNK----VLLFDIHTSGLLAEIHTGHHSTIQYCDFSPYDHLAVIA 858
Query: 359 LTQNQLTLWKYPSMLKIAELHGHTSRVLHMTQSPDGSTVASAAAAADQTLRFWE 412
L+Q + LW S LK+A+ GH S V + SPDGS+ + A+ DQT+R WE
Sbjct: 859 LSQYCVELWNIDSRLKVADCRGHLSWVHGVMFSPDGSSFLT--ASDDQTIRVWE 910
Score = 52.4 bits (124), Expect = 2e-06
Identities = 46/172 (26%), Positives = 73/172 (41%), Gaps = 12/172 (6%)
Query: 243 RGHRREVCGLKWSLDGKQLASGGNDNVVHIWDMSAVSSNSPTRWLYRFDEHKAAVKALAW 302
R H V +S DG+++AS G D + ++ + + KA +
Sbjct: 612 RPHTDAVYHACFSQDGQRIASCGADKTLQVFKAETGE---------KLLDIKAHEDEVLC 662
Query: 303 CPFQGNLLASGGGGGDCCVKLWNTGMGERMNSVDTGS-QVCALLWSKNERELLSSHGLTQ 361
C F + D VK+W++ G+ +++ D S QV ++ + LL + G
Sbjct: 663 CAFSSDDSYIATCSADKKVKIWDSATGKLVHTYDEHSEQVNCCHFTNSSNHLLLATGSND 722
Query: 362 NQLTLWKYPSMLKIAELHGHTSRVLHMTQSPDGSTVASAAAAADQTLRFWEV 413
L LW + GHT+ V H SPD +AS +A D TLR W+V
Sbjct: 723 FFLKLWDLNQKECRNTMFGHTNSVNHCRFSPDDELLASCSA--DGTLRLWDV 772
Score = 43.5 bits (101), Expect = 0.001
Identities = 45/180 (25%), Positives = 76/180 (42%), Gaps = 18/180 (10%)
Query: 236 SQIINTYRGHRREVCGLKWSLDGKQLASGGNDNVVHIWDMSAVSSNSPTRWLYRFDEHKA 295
+++ ++ GH++ V ++++ DGK L S D+V+ +W N T H+
Sbjct: 993 NRVFSSGVGHKKAVRHIQFTADGKTLISSSEDSVIQVW-------NWQTGDYVFLQAHQE 1045
Query: 296 AVKALAWCPFQGNLLASGGGGGDCCVKLWN--TGMGERMNSVDTGSQVCALLWSKNEREL 353
VK Q + L S G VK+WN TG ER + G+ + + S +
Sbjct: 1046 TVKDFRL--LQDSRLLSWSFDG--TVKVWNVITGRIERDFTCHQGTVLSCAISSDATKFS 1101
Query: 354 LSSHGLTQNQLTLWKYPSMLKIAELHGHTSRVLHMTQSPDGSTVASAAAAADQTLRFWEV 413
+S T +W + + + EL GH V S DG +A+ + +R W V
Sbjct: 1102 STSADKTAK---IWSFDLLSPLHELKGHNGCVRCSAFSLDGILLATGDDNGE--IRIWNV 1156
Score = 42.4 bits (98), Expect = 0.002
Identities = 36/163 (22%), Positives = 67/163 (41%), Gaps = 19/163 (11%)
Query: 135 SIALDHTIYFWNASDSSGSEFVTV-----------DEEEGPVTSVCWAPDGRHLAVGLTN 183
S + D T+ W+ ++ + + V ++ E V W+ DG + V N
Sbjct: 760 SCSADGTLRLWDVRSANERKSINVKRFFLSSEDPPEDVEVIVKCCSWSADGDKIIVAAKN 819
Query: 184 SHVQLWDTAANKQLRTLKGGHRARVGSLAWNGH----VLTTGGMDGKIVNNDVRLRSQII 239
V L+D + L + GH + + ++ + V+ ++ N D RL+ +
Sbjct: 820 K-VLLFDIHTSGLLAEIHTGHHSTIQYCDFSPYDHLAVIALSQYCVELWNIDSRLK---V 875
Query: 240 NTYRGHRREVCGLKWSLDGKQLASGGNDNVVHIWDMSAVSSNS 282
RGH V G+ +S DG + +D + +W+ V NS
Sbjct: 876 ADCRGHLSWVHGVMFSPDGSSFLTASDDQTIRVWETKKVCKNS 918
Score = 33.5 bits (75), Expect = 1.1
Identities = 46/207 (22%), Positives = 84/207 (40%), Gaps = 20/207 (9%)
Query: 131 RNVLSIALDHTIYFWNASDSSGSEFVTVDEEEGPVTSVCWAPDGRHLAVGLTNSHVQLWD 190
+ ++S + D I WN ++V + + V D R L+ + V++W+
Sbjct: 1016 KTLISSSEDSVIQVWNWQTG---DYVFLQAHQETVKDFRLLQDSRLLSWSFDGT-VKVWN 1071
Query: 191 TAANKQLRTLKGGHRARVGSLAWNGHVL----TTGGMDGKIVNNDVRLRSQIINTYRGHR 246
+ R H+ V S A + T+ KI + D+ ++ +GH
Sbjct: 1072 VITGRIERDFTC-HQGTVLSCAISSDATKFSSTSADKTAKIWSFDLL---SPLHELKGHN 1127
Query: 247 REVCGLKWSLDGKQLASGGNDNVVHIWDMS---AVSSNSPTRWLYRFDEHKAAVKALAWC 303
V +SLDG LA+G ++ + IW++S + S +P H V + +
Sbjct: 1128 GCVRCSAFSLDGILLATGDDNGEIRIWNVSDGQLLHSCAPISVEEGTATHGGWVTDVCFS 1187
Query: 304 PFQGNLLASGGGGGDCCVKLWNTGMGE 330
P L+++GG +K WN G+
Sbjct: 1188 PDSKTLVSAGG-----YLKWWNVATGD 1209
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.318 0.133 0.424
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 56,469,942
Number of Sequences: 164201
Number of extensions: 2463818
Number of successful extensions: 10789
Number of sequences better than 10.0: 474
Number of HSP's better than 10.0 without gapping: 259
Number of HSP's successfully gapped in prelim test: 215
Number of HSP's that attempted gapping in prelim test: 7679
Number of HSP's gapped (non-prelim): 1782
length of query: 436
length of database: 59,974,054
effective HSP length: 113
effective length of query: 323
effective length of database: 41,419,341
effective search space: 13378447143
effective search space used: 13378447143
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 67 (30.4 bits)
Medicago: description of AC146343.9


