

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146343.8 + phase: 0 /pseudo
(379 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
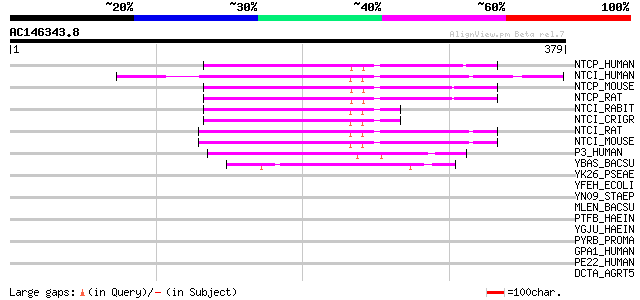
Score E
Sequences producing significant alignments: (bits) Value
NTCP_HUMAN (Q14973) Sodium/bile acid cotransporter (Na(+)/bile a... 70 1e-11
NTCI_HUMAN (Q12908) Ileal sodium/bile acid cotransporter (Ileal ... 66 1e-10
NTCP_MOUSE (O08705) Sodium/bile acid cotransporter (Na(+)/bile a... 62 2e-09
NTCP_RAT (P26435) Sodium/bile acid cotransporter (Na(+)/bile aci... 62 3e-09
NTCI_RABIT (Q28727) Ileal sodium/bile acid cotransporter (Ileal ... 62 3e-09
NTCI_CRIGR (Q60414) Ileal sodium/bile acid cotransporter (Ileal ... 60 9e-09
NTCI_RAT (Q62633) Ileal sodium/bile acid cotransporter (Ileal Na... 59 2e-08
NTCI_MOUSE (P70172) Ileal sodium/bile acid cotransporter (Ileal ... 59 2e-08
P3_HUMAN (P09131) P3 protein 58 3e-08
YBAS_BACSU (P55190) Hypothetical protein ybaS 57 1e-07
YK26_PSEAE (P39879) Hypothetical protein PA2026 43 0.002
YFEH_ECOLI (P39836) Hypothetical protein yfeH 39 0.017
YN09_STAEP (Q8CQT3) Hypothetical UPF0324 membrane protein SE2309 37 0.063
MLEN_BACSU (P54571) Malate-2H(+)/Na(+)-lactate antiporter 37 0.11
PTFB_HAEIN (P44714) PTS system, fructose-specific IIBC component... 36 0.18
YGJU_HAEIN (P45246) Hypothetical symporter HI1545 34 0.54
PYRB_PROMA (Q7VDV6) Aspartate carbamoyltransferase (EC 2.1.3.2) ... 33 0.92
GPA1_HUMAN (O43292) Glycosylphosphatidylinositol anchor attachme... 33 1.2
PE22_HUMAN (P43116) Prostaglandin E2 receptor, EP2 subtype (Pros... 32 2.0
DCTA_AGRT5 (P58734) C4-dicarboxylate transport protein 32 2.0
>NTCP_HUMAN (Q14973) Sodium/bile acid cotransporter (Na(+)/bile acid
cotransporter) (Na(+)/taurocholate transport protein)
(Sodium/taurocholate cotransporting polypeptide)
Length = 349
Score = 69.7 bits (169), Expect = 1e-11
Identities = 55/210 (26%), Positives = 100/210 (47%), Gaps = 13/210 (6%)
Query: 133 IMLSIGIGLSMEDFALAFKRPLPLSIGFIAQYVLKPVLGVLIAKAFGLPRMFYAGFVLTA 192
IMLS+G + +P L+I +AQY + P+ ++ K F L + ++
Sbjct: 38 IMLSLGCTMEFSKIKAHLWKPKGLAIALVAQYGIMPLTAFVLGKVFRLKNIEALAILVCG 97
Query: 193 CVSGAQLSSYASFISKGDVALCIVLTSYTTIASVIVTPLL-----TGLLIGSV---VPVD 244
C G LS+ S KGD+ L IV+T+ +T ++ + PLL G+ G + VP
Sbjct: 98 CSPGGNLSNVFSLAMKGDMNLSIVMTTCSTFCALGMMPLLLYIYSRGIYDGDLKDKVPYK 157
Query: 245 AVAMSKSILQVVLAPVTLGLLLNTYAKPVVS-ILRPVMPFVAMICTSLCIGSPLAINRSQ 303
+ +S L +VL P T+G++L + + +++ M + + ++ + S + + +S
Sbjct: 158 GIVIS---LVLVLIPCTIGIVLKSKRPQYMRYVIKGGMIIILLCSVAVTVLSAINVGKSI 214
Query: 304 ILSGEGLRLVAPVLIFHAAAFTLGYWFSNL 333
+ + L L+A + F LGY S L
Sbjct: 215 MFAMTPL-LIATSSLMPFIGFLLGYVLSAL 243
>NTCI_HUMAN (Q12908) Ileal sodium/bile acid cotransporter (Ileal
Na(+)/bile acid cotransporter) (Na(+) dependent ileal
bile acid transporter) (Ileal sodium-dependent bile acid
transporter) (ISBT) (Sodium/taurocholate cotransporting
polypeptide, ileal)
Length = 348
Score = 66.2 bits (160), Expect = 1e-10
Identities = 66/312 (21%), Positives = 132/312 (42%), Gaps = 39/312 (12%)
Query: 74 VDESVVGGGGNGDVGERDWSQILSALLPFVVAVTAVAALSQPSTFTWVSKELYAPALGGI 133
VD + V G + V E +++ ILS +L V+ + L +
Sbjct: 8 VDNATVCSGASCVVPESNFNNILSVVLSTVLTIL----------------------LALV 45
Query: 134 MLSIGIGLSMEDFALAFKRPLPLSIGFIAQYVLKPVLGVLIAKAFGLPRMFYAGFVLTAC 193
M S+G + ++ F KRP + +GF+ Q+ + P+ G +++ AF + + ++ C
Sbjct: 46 MFSMGCNVEIKKFLGHIKRPWGICVGFLCQFGIMPLTGFILSVAFDILPLQAVVVLIIGC 105
Query: 194 VSGAQLSSYASFISKGDVALCIVLTSYTTIASVIVTPL----LTGLLIGS---VVPVDAV 246
G S+ ++ GD+ L + +T+ +T+ ++ + PL T + + S V+P D +
Sbjct: 106 CPGGTASNILAYWVDGDMDLSVSMTTCSTLLALGMMPLCLLIYTKMWVDSGSIVIPYDNI 165
Query: 247 AMSKSILQVVLAPVTLGLLLNTYAKPVVSILRPVMPFVAMICTSLCIGSPLAINRSQILS 306
S L ++ PV++G+ +N I+ + I L + +S +
Sbjct: 166 GTS---LVALVVPVSIGMFVNHKWPQKAKIILKIGSIAGAILIVLIAVVGGILYQSAWII 222
Query: 307 GEGLRLVAPVLIFHAAAFTLGYWFSNLPSLRFLTSITINTLSQAGRTGEQDNFIMHWNAE 366
L ++ IF A ++LG+ + + L + T+ A TG Q+ + +
Sbjct: 223 APKLWIIG--TIFPVAGYSLGFLLARIAGLPWYRCRTV-----AFETGMQNTQLCSTIVQ 275
Query: 367 LHTCRPSSNTIF 378
L N +F
Sbjct: 276 LSFTPEELNVVF 287
>NTCP_MOUSE (O08705) Sodium/bile acid cotransporter (Na(+)/bile acid
cotransporter) (Na(+)/taurocholate transport protein)
(Sodium/taurocholate cotransporting polypeptide)
Length = 362
Score = 62.4 bits (150), Expect = 2e-09
Identities = 54/210 (25%), Positives = 95/210 (44%), Gaps = 13/210 (6%)
Query: 133 IMLSIGIGLSMEDFALAFKRPLPLSIGFIAQYVLKPVLGVLIAKAFGLPRMFYAGFVLTA 192
IMLS+G + F +P + I +AQY + P+ L+ K F L + ++
Sbjct: 38 IMLSLGCTMEFSKIKAHFWKPKGVIIAIVAQYGIMPLSAFLLGKVFHLTSIEALAILICG 97
Query: 193 CVSGAQLSSYASFISKGDVALCIVLTSYTTIASVIVTPLL-----TGLLIGSV---VPVD 244
C G LS+ + KGD+ L IV+T+ ++ ++ + PLL G+ G + VP
Sbjct: 98 CSPGGNLSNLFTLAMKGDMNLSIVMTTCSSFTALGMMPLLLYIYSKGIYDGDLKDKVPYK 157
Query: 245 AVAMSKSILQVVLAPVTLGLLLNTYAKPVVS-ILRPVMPFVAMICTSLCIGSPLAINRSQ 303
+ +S L +VL P +G+ L + V +L+ M + ++ + S + + S
Sbjct: 158 GIMLS---LVMVLIPCAIGIFLKSKRPHYVPYVLKAGMIITFSLSVAVTVLSVINVGNS- 213
Query: 304 ILSGEGLRLVAPVLIFHAAAFTLGYWFSNL 333
I+ L+A + F +GY S L
Sbjct: 214 IMFVMTPHLLATSSLMPFTGFLMGYILSAL 243
>NTCP_RAT (P26435) Sodium/bile acid cotransporter (Na(+)/bile acid
cotransporter) (Na(+)/taurocholate transport protein)
(Sodium/taurocholate cotransporting polypeptide)
Length = 362
Score = 61.6 bits (148), Expect = 3e-09
Identities = 53/210 (25%), Positives = 98/210 (46%), Gaps = 13/210 (6%)
Query: 133 IMLSIGIGLSMEDFALAFKRPLPLSIGFIAQYVLKPVLGVLIAKAFGLPRMFYAGFVLTA 192
IMLS+G + +P + + +AQ+ + P+ L+ K F L + ++
Sbjct: 38 IMLSLGCTMEFSKIKAHLWKPKGVIVALVAQFGIMPLAAFLLGKIFHLSNIEALAILICG 97
Query: 193 CVSGAQLSSYASFISKGDVALCIVLTSYTTIASVIVTPLL-----TGLLIGSV---VPVD 244
C G LS+ + KGD+ L IV+T+ ++ +++ + PLL G+ G + VP
Sbjct: 98 CSPGGNLSNLFTLAMKGDMNLSIVMTTCSSFSALGMMPLLLYVYSKGIYDGDLKDKVPYK 157
Query: 245 AVAMSKSILQVVLAPVTLGLLLNTYAKPVVS-ILRPVMPFVAMICTSLCIGSPLAINRSQ 303
+ +S L +VL P T+G++L + V IL+ M ++ ++ S + + S
Sbjct: 158 GIMIS---LVIVLIPCTIGIVLKSKRPHYVPYILKGGMIITFLLSVAVTALSVINVGNS- 213
Query: 304 ILSGEGLRLVAPVLIFHAAAFTLGYWFSNL 333
I+ L+A + + F +GY S L
Sbjct: 214 IMFVMTPHLLATSSLMPFSGFLMGYILSAL 243
>NTCI_RABIT (Q28727) Ileal sodium/bile acid cotransporter (Ileal
Na(+)/bile acid cotransporter) (Na(+) dependent ileal
bile acid transporter) (Ileal sodium-dependent bile acid
transporter) (ISBT) (Sodium/taurocholate cotransporting
polypeptide, ileal)
Length = 347
Score = 61.6 bits (148), Expect = 3e-09
Identities = 37/142 (26%), Positives = 76/142 (53%), Gaps = 10/142 (7%)
Query: 133 IMLSIGIGLSMEDFALAFKRPLPLSIGFIAQYVLKPVLGVLIAKAFGLPRMFYAGFVLTA 192
+M S+G + ++ F +RP + IGF+ Q+ + P+ G ++A AFG+ + ++
Sbjct: 46 VMFSMGCNVEIKKFLGHIRRPWGIFIGFLCQFGIMPLTGFVLAVAFGIMPIQAVVVLIMG 105
Query: 193 CVSGAQLSSYASFISKGDVALCIVLTSYTTIASVIVTPL----LTGLLIGS---VVPVDA 245
C G S+ ++ GD+ L + +T+ +T+ ++ + PL T + + S V+P D
Sbjct: 106 CCPGGTASNILAYWVDGDMDLSVSMTTCSTLLALGMMPLCLYVYTKMWVDSGTIVIPYDN 165
Query: 246 VAMSKSILQVVLAPVTLGLLLN 267
+ S L ++ PV++G+ +N
Sbjct: 166 IGTS---LVALVVPVSIGMFVN 184
>NTCI_CRIGR (Q60414) Ileal sodium/bile acid cotransporter (Ileal
Na(+)/bile acid cotransporter) (Na(+) dependent ileal
bile acid transporter) (Ileal sodium-dependent bile acid
transporter) (ISBT) (Sodium/taurocholate cotransporting
polypeptide, ileal)
Length = 348
Score = 60.1 bits (144), Expect = 9e-09
Identities = 35/142 (24%), Positives = 76/142 (52%), Gaps = 10/142 (7%)
Query: 133 IMLSIGIGLSMEDFALAFKRPLPLSIGFIAQYVLKPVLGVLIAKAFGLPRMFYAGFVLTA 192
+M S+G + + F +RP + +GF+ Q+ + P+ G +++ AFG+ + ++
Sbjct: 45 VMFSMGCNVELHKFLGHLRRPWGIVVGFLCQFGIMPLTGFVLSVAFGILPVQAVVVLIQG 104
Query: 193 CVSGAQLSSYASFISKGDVALCIVLTSYTTIASVIVTPL----LTGLLIGS---VVPVDA 245
C G S+ ++ GD+ L + +T+ +T+ ++ + PL T + + S V+P D+
Sbjct: 105 CCPGGTASNILAYWVDGDMDLSVSMTTCSTLLALGMMPLCLFIYTKMWVDSGTIVIPYDS 164
Query: 246 VAMSKSILQVVLAPVTLGLLLN 267
+ S L ++ PV++G+ +N
Sbjct: 165 IGTS---LVALVIPVSIGMYVN 183
>NTCI_RAT (Q62633) Ileal sodium/bile acid cotransporter (Ileal
Na(+)/bile acid cotransporter) (Na(+) dependent ileal
bile acid transporter) (Ileal sodium-dependent bile acid
transporter) (ISBT) (Sodium/taurocholate cotransporting
polypeptide, ileal)
Length = 348
Score = 59.3 bits (142), Expect = 2e-08
Identities = 47/211 (22%), Positives = 100/211 (47%), Gaps = 12/211 (5%)
Query: 130 LGGIMLSIGIGLSMEDFALAFKRPLPLSIGFIAQYVLKPVLGVLIAKAFGLPRMFYAGFV 189
L +M S+G + + F KRP + +GF+ Q+ + P+ G +++ A G+ + +
Sbjct: 42 LAMVMFSMGCNVEINKFLGHIKRPWGIFVGFLCQFGIMPLTGFILSVASGILPVQAVVVL 101
Query: 190 LTACVSGAQLSSYASFISKGDVALCIVLTSYTTIASVIVTPL----LTGLLIGS---VVP 242
+ C G S+ ++ GD+ L + +T+ +T+ ++ + PL T + + S V+P
Sbjct: 102 IMGCCPGGTGSNILAYWIDGDMDLSVSMTTCSTLLALGMMPLCLFIYTKMWVDSGTIVIP 161
Query: 243 VDAVAMSKSILQVVLAPVTLGLLLNTYAKPVVSILRPVMPFVAMICTSLCIGSPLAINRS 302
D++ +S L ++ PV++G+ +N I+ + I L + +S
Sbjct: 162 YDSIGIS---LVALVIPVSIGMFVNHKWPQKAKIILKIGSIAGAILIVLIAVVGGILYQS 218
Query: 303 QILSGEGLRLVAPVLIFHAAAFTLGYWFSNL 333
+ L ++ IF A ++LG++ + L
Sbjct: 219 AWIIEPKLWIIG--TIFPIAGYSLGFFLARL 247
>NTCI_MOUSE (P70172) Ileal sodium/bile acid cotransporter (Ileal
Na(+)/bile acid cotransporter) (Na+ dependent ileal bile
acid transporter) (Ileal sodium-dependent bile acid
transporter) (ISBT) (Sodium/taurocholate cotransporting
polypeptide, ileal)
Length = 348
Score = 59.3 bits (142), Expect = 2e-08
Identities = 47/211 (22%), Positives = 100/211 (47%), Gaps = 12/211 (5%)
Query: 130 LGGIMLSIGIGLSMEDFALAFKRPLPLSIGFIAQYVLKPVLGVLIAKAFGLPRMFYAGFV 189
L +M S+G + + F KRP + +GF+ Q+ + P+ G +++ A G+ + +
Sbjct: 42 LAMVMFSMGCNVEVHKFLGHIKRPWGIFVGFLCQFGIMPLTGFILSVASGILPVQAVVVL 101
Query: 190 LTACVSGAQLSSYASFISKGDVALCIVLTSYTTIASVIVTPL----LTGLLIGS---VVP 242
+ C G S+ ++ GD+ L + +T+ +T+ ++ + PL T + + S V+P
Sbjct: 102 IMGCCPGGTGSNILAYWIDGDMDLSVSMTTCSTLLALGMMPLCLFVYTKMWVDSGTIVIP 161
Query: 243 VDAVAMSKSILQVVLAPVTLGLLLNTYAKPVVSILRPVMPFVAMICTSLCIGSPLAINRS 302
D++ +S L ++ PV+ G+ +N I+ + +I L + +S
Sbjct: 162 YDSIGIS---LVALVIPVSFGMFVNHKWPQKAKIILKIGSITGVILIVLIAVIGGILYQS 218
Query: 303 QILSGEGLRLVAPVLIFHAAAFTLGYWFSNL 333
+ L ++ IF A ++LG++ + L
Sbjct: 219 AWIIEPKLWIIG--TIFPIAGYSLGFFLARL 247
>P3_HUMAN (P09131) P3 protein
Length = 477
Score = 58.2 bits (139), Expect = 3e-08
Identities = 48/182 (26%), Positives = 85/182 (46%), Gaps = 9/182 (4%)
Query: 136 SIGIGLSMEDFALAFKRPLPLSIGFIAQYVLKPVLGVLIAKAFGLPRMFYAGFVLTACVS 195
S G + +E + P P+ +G + Q+++ P+ L+AK F LP+ G ++T
Sbjct: 206 SFGCKVELEVLKGLMQSPQPMLLGLLGQFLVMPLYAFLMAKVFMLPKALALGLIITCSSP 265
Query: 196 GAQLSSYASFISKGDVALCIVLTSYTTIASVIVTPLLTGLL--IGSVVPVDAVAMSKSI- 252
G S S + GDV L I +T +T+A+ PL + + + S+ V +SK +
Sbjct: 266 GGGGSYLFSLLLGGDVTLAISMTFLSTVAATGFLPLSSAIYSRLLSIHETLHVPISKILG 325
Query: 253 -LQVVLAPVTLGLLLNT-YAKPVVSILRPVMPFVAMICTSLCIGSPLAINRSQILSGEGL 310
L + P+ +G+L+ + K +L+ V PF + L +G R + G+
Sbjct: 326 TLLFIAIPIAVGVLIKSKLPKFSQLLLQVVKPFSFV----LLLGGLFLAYRMGVFILAGI 381
Query: 311 RL 312
RL
Sbjct: 382 RL 383
>YBAS_BACSU (P55190) Hypothetical protein ybaS
Length = 283
Score = 56.6 bits (135), Expect = 1e-07
Identities = 44/162 (27%), Positives = 72/162 (44%), Gaps = 14/162 (8%)
Query: 149 AFKRPLPLSIGFIAQYVLKPVL----GVLIAKAFGLPRMFYAGFVLTACVSGAQLSSYAS 204
A PLP+ + ++ P+ G LI K L G L + S +
Sbjct: 26 ALSHPLPMILALFVLHIFMPLFAWGSGHLIFKGDPLT---ITGLTLAVVIPTGITSLIWA 82
Query: 205 FISKGDVALCIVLTSYTTIASVIVTPLLTGLLIGSVVPVDAVAMSKSILQVVLAPVTLGL 264
+ KG+V L + + T+ S ++ PL LL G+ V +D M K ++ +V+ P LG+
Sbjct: 83 AMYKGNVGLTLSIILVDTVLSPLIVPLSLSLLAGAQVHMDVWGMMKGLIVMVVIPSFLGM 142
Query: 265 LLNTYAKP--VVSILRPVMPFVAMICTSLCIGSPLAINRSQI 304
L N + P + + PF + LC+ + +AIN S I
Sbjct: 143 LFNQMSSPERTAFVSSALSPF-----SKLCLMAVIAINSSAI 179
>YK26_PSEAE (P39879) Hypothetical protein PA2026
Length = 333
Score = 42.7 bits (99), Expect = 0.002
Identities = 46/193 (23%), Positives = 82/193 (41%), Gaps = 11/193 (5%)
Query: 98 ALLPFVVAVTAVAALSQPST-FTWVSKELYAPALGGIMLSIGIGLSMEDFALAFKRPLPL 156
AL+ V+ T + Q + F WV+ +G + G LS +
Sbjct: 14 ALIATVLLATFLPCSGQTAVVFEWVTNI----GIGLLFFLHGAKLSRQAIIAGMTHWRLH 69
Query: 157 SIGFIAQYVLKPVLGVLIAKAFG---LPRMFYAGFVLTACVSGAQLSSYASFISKGDVAL 213
+ F +V+ P+LG+ + A P ++ L A + Q S + +++G+V
Sbjct: 70 LLVFACTFVMFPLLGLALKPALSPMVTPELYLGILFLCALPATVQSSIAFTSLARGNVPA 129
Query: 214 CIVLTSYTTIASVIVTPLLTGLLIGSVVPVDAV--AMSKSILQVVLAPVTLGLLLNTYAK 271
+ S +++ V +TPLL LL+G+ A+ K LQ +L P G +L +
Sbjct: 130 AVCSASVSSLLGVFLTPLLVKLLLGAEGETGNALDAIGKITLQ-LLVPFIAGQVLRRWIG 188
Query: 272 PVVSILRPVMPFV 284
V +PV+ +V
Sbjct: 189 AWVERNKPVLRYV 201
>YFEH_ECOLI (P39836) Hypothetical protein yfeH
Length = 332
Score = 39.3 bits (90), Expect = 0.017
Identities = 43/174 (24%), Positives = 73/174 (41%), Gaps = 36/174 (20%)
Query: 164 YVLKPVLGVLIA--KAFGLPRMFYAGFVLTACVSGAQLSSYASFISKGDVALCIVLTSYT 221
+VL P+LGVL A K + M Y+GF L C+ A + S +F
Sbjct: 76 FVLFPILGVLFAWWKPVNVDPMLYSGF-LYLCILPATVQSAIAF---------------- 118
Query: 222 TIASVIVTPLLTGLLIGSVVPVDAVAMSKSILQVVLAPVTLGLLLNTYA-----KPVVSI 276
T + G+V A + S+L + L+P+ +GL++N + + V I
Sbjct: 119 -----------TSMAGGNVAAAVCSASASSLLGIFLSPLLVGLVMNVHGAGGSLEQVGKI 167
Query: 277 -LRPVMPFVAMICTSLCIGSPLAINRSQILSGEGLRLVAPVLIFHAAAFTLGYW 329
L+ ++PFV + IG ++ N+ I + ++ V + A G W
Sbjct: 168 MLQLLLPFVLGHLSRPWIGDWVSRNKKWIAKTDQTSILLVVYTAFSEAVVNGIW 221
>YN09_STAEP (Q8CQT3) Hypothetical UPF0324 membrane protein SE2309
Length = 331
Score = 37.4 bits (85), Expect = 0.063
Identities = 54/248 (21%), Positives = 106/248 (41%), Gaps = 52/248 (20%)
Query: 85 GDVGERDWSQILSALLPFVVAVTAVAALSQPSTFTWVSKELYAPALGGIMLSIGIGL--- 141
GD+ + W +L ++ + +++ L+Q +K++ I+L IG G+
Sbjct: 84 GDILGKGWKLLLIDIIVIIFSISLTLLLNQ---IIKGNKDI------SILLGIGTGVCGA 134
Query: 142 ----SMEDFALAFKRPLPLSIGFIAQY-VLKPVLGVLIAKAFGLPRMFYAGF-------- 188
+ + ++ + +S+G IA + ++ I F +P + Y +
Sbjct: 135 AAIAATAPILKSKEKDIAISVGIIALVGTIFALIYTAIEAIFNIPTITYGAWTGISLHEI 194
Query: 189 ---VLTACVSGAQLSSYASFISKGDV----ALCIVLTSYTTIAS--------VIVTPLLT 233
VL A + G++ ++A G V L IVL Y S + + L
Sbjct: 195 AQVVLAAGIGGSEAMTFALLGKLGRVFLLIPLSIVLILYMRYKSHSSQVQQKIDIPYFLI 254
Query: 234 GLLI----GSVVPVDAVAMSK----SILQVVLAPVTLGLLLNTYAKPVVSILRPVMPFVA 285
G +I + VP+ ++ M+ + L +++A V LGL N K V+S + + PF+
Sbjct: 255 GFIIMACINTFVPIPSLLMNIINVITTLCMLMAMVALGL--NIVLKEVIS--KALKPFIV 310
Query: 286 MICTSLCI 293
+ TS+C+
Sbjct: 311 ICITSICL 318
>MLEN_BACSU (P54571) Malate-2H(+)/Na(+)-lactate antiporter
Length = 468
Score = 36.6 bits (83), Expect = 0.11
Identities = 33/123 (26%), Positives = 55/123 (43%), Gaps = 17/123 (13%)
Query: 172 VLIAKAFGLPRMFYAGFVLTACVSGAQLSSY------ASFISKGDVALCIVLTSYTTIAS 225
+ I + G+P AG +L+ G +LS AS +SK DV + Y +I +
Sbjct: 139 IAIGEGLGIPLPLVAGAILSGAYFGDKLSPLSDSTVLASSLSKVDVLAHVRAMLYLSIPA 198
Query: 226 VIVTPLL---TGLLIGSV-VPVDAVAMSKSILQ-------VVLAPVTLGLLLNTYAKPVV 274
++T +L G + G + +D V KS LQ +L P L ++L KP +
Sbjct: 199 YVITAILFTVVGFMYGGKNIDLDKVEFLKSSLQNTFDIHIWMLIPAVLVIVLLAMKKPSM 258
Query: 275 SIL 277
++
Sbjct: 259 PVI 261
>PTFB_HAEIN (P44714) PTS system, fructose-specific IIBC component
(EIIBC-Fru) (Fructose-permease IIBC component)
(Phosphotransferase enzyme II, BC component) (EC
2.7.1.69) (EII-Fru)
Length = 556
Score = 35.8 bits (81), Expect = 0.18
Identities = 34/144 (23%), Positives = 64/144 (43%), Gaps = 14/144 (9%)
Query: 96 LSALLPFVVAVTAVAALSQPSTFTWVSKELYAPALGGIMLSIGIGLSMEDFALAFKRPLP 155
+S +LP VVA + A+S +F + L ++++IG G +AFK +
Sbjct: 232 VSHMLPLVVAGGLLIAISFMFSFNVIENTGVFQDLPNMLINIGSG-------VAFKLMIA 284
Query: 156 LSIGFIAQYVL-KPVLGVLIAKAFGLPRMFYAGFVLTACVSGAQLSSYASFISKGDVALC 214
+ G++A + +P L V GL A + G A ++ KG +
Sbjct: 285 VFAGYVAFSIADRPGLAV------GLIAGMLASEAGAGILGGIIAGFLAGYVVKGLNVII 338
Query: 215 IVLTSYTTIASVIVTPLLTGLLIG 238
+ S T++ +++ PLL +++G
Sbjct: 339 RLPASLTSLKPILILPLLGSMIVG 362
>YGJU_HAEIN (P45246) Hypothetical symporter HI1545
Length = 414
Score = 34.3 bits (77), Expect = 0.54
Identities = 43/168 (25%), Positives = 73/168 (42%), Gaps = 13/168 (7%)
Query: 131 GGIMLSIGIGLSMEDFALAFKRPLPLSIGF-IAQYVLKPVLGVLIAKAFGLPRMFYAGFV 189
GG++ I GL + PL +IGF +A+ V VLG + KA F+
Sbjct: 13 GGLVKRIAAGLVLGIVVALISAPLQETIGFNLAEKV--GVLGTIFVKALRAVAPILIFFL 70
Query: 190 LTACVSGAQLSSYASFISKGDVALCIVLTSYTTIASVIVTPLLTGLLIGSVVPVDAVAMS 249
+ A ++ ++ +K ++ IVL T + V ++ G + V + A S
Sbjct: 71 VMAALANRKIG------TKSNMKEIIVLYLLGTFLAAFVA-VIAGFAFPTEVVLAAKEDS 123
Query: 250 KSILQVVLAPVTLGLLLNTYAKPVVSILRPVMPFVAMICTSLCIGSPL 297
S Q V V L L+LN P+ +I + F+ ++ S+ +G L
Sbjct: 124 SSAPQAV-GQVLLTLILNVVDDPLNAIFK--ANFIGVLAWSIGLGLAL 168
>PYRB_PROMA (Q7VDV6) Aspartate carbamoyltransferase (EC 2.1.3.2)
(Aspartate transcarbamylase) (ATCase)
Length = 338
Score = 33.5 bits (75), Expect = 0.92
Identities = 23/66 (34%), Positives = 29/66 (43%), Gaps = 3/66 (4%)
Query: 20 LSFQPLLRTRSSFSLQRVSKGCSISSFACSTSKLHGGRVGFNHREGKTSFLKFGVDESVV 79
L F+P RTRSSF L + SFA S S L G + +++ G VV
Sbjct: 52 LFFEPSTRTRSSFELAAKRLSADVQSFAPSNSSLIKGETPL---DTVMTYVAMGAHVLVV 108
Query: 80 GGGGNG 85
GG G
Sbjct: 109 RHGGTG 114
>GPA1_HUMAN (O43292) Glycosylphosphatidylinositol anchor attachment
1 protein (GPI anchor attachment protein 1) (GAA1
protein homolog) (hGAA1)
Length = 620
Score = 33.1 bits (74), Expect = 1.2
Identities = 53/207 (25%), Positives = 87/207 (41%), Gaps = 37/207 (17%)
Query: 115 PSTFTWVSKELYAPALGGIMLSIGI--------------GLSMEDFALAFKRPLPLSIGF 160
P +VS LY PA+G ++L +G+ GL A PLP S G
Sbjct: 362 PGLSRFVSIGLYMPAVGFLLLVLGLKALELWMQLHEAGMGLEEPGGAPGPSVPLPPSQGV 421
Query: 161 IAQYVLKPVLGVLIAKAFGLPRMFYAGFVLTACVSGAQLSSYASFISKGDVALCIVLTSY 220
++ P +LI++A GL A +VL V G +++ +++ + +VLT
Sbjct: 422 GLASLVAP---LLISQAMGL-----ALYVLP--VLGQHVATQHFPVAEAE---AVVLTLL 468
Query: 221 TTIASVIVTPLLTGLLIGSVVP------VDAVAMSKSILQ---VVLAPVTLGLLLNTYAK 271
A+ + P T ++ + P + VA+ LQ + L +LG LL T
Sbjct: 469 AIYAAGLALPHNTHRVVSTQAPDRGWMALKLVALIYLALQLGCIALTNFSLGFLLATTMV 528
Query: 272 PVVSILRPVMPFVAMICTSLCIGSPLA 298
P ++ +P P + L + SP A
Sbjct: 529 PTAALAKPHGP-RTLYAALLVLTSPAA 554
>PE22_HUMAN (P43116) Prostaglandin E2 receptor, EP2 subtype
(Prostanoid EP2 receptor) (PGE receptor, EP2 subtype)
Length = 358
Score = 32.3 bits (72), Expect = 2.0
Identities = 34/133 (25%), Positives = 57/133 (42%), Gaps = 10/133 (7%)
Query: 208 KGDVALCIVLTSYTTIASVIVTPLLTGLLIGS--VVPVDAVAMSKSILQVVLAPVTLGLL 265
+GDV S ++ V+VT L+ L+G+ + PV + +++ V LAP +
Sbjct: 51 RGDVGCSAGRRSSLSLFHVLVTELVFTDLLGTCLISPVVLASYARNQTLVALAPESRACT 110
Query: 266 LNTYAKPVVSILRPVMPFVAMICTSLCIGSPLAINRSQILSGEGLRLVAPVLIFHAAAFT 325
+A S+ +M F + L IG P R +S G V PV+ +
Sbjct: 111 YFAFAMTFFSLATMLMLFAMALERYLSIGHPYFYQRR--VSRSGGLAVLPVI------YA 162
Query: 326 LGYWFSNLPSLRF 338
+ F +LP L +
Sbjct: 163 VSLLFCSLPLLDY 175
>DCTA_AGRT5 (P58734) C4-dicarboxylate transport protein
Length = 448
Score = 32.3 bits (72), Expect = 2.0
Identities = 64/261 (24%), Positives = 97/261 (36%), Gaps = 60/261 (22%)
Query: 107 TAVAALSQPSTFTWVSKELYAPALGGIMLSIGIGLSMEDFALAFKRPLPLSIGFI--AQY 164
TAVAA F K LY L I+ I +G F K PL GFI +
Sbjct: 5 TAVAAGHAKQPFY---KHLYFQVLVAIIAGIALGHFYPTFGEQLK---PLGDGFIRLVKM 58
Query: 165 VLKPVLGVLIAKAF-GLPRMFYAGFVLTACVSGAQLSSYASFISKGDVALCIVLTSYTTI 223
++ PV+ + +A G+ M G V +G + + F + + IV +
Sbjct: 59 IIAPVIFLTVATGIAGMNDMKKVGRV-----AGKAMIYFLVFSTLALIVGLIVANTVQPG 113
Query: 224 ASVIVTPL------------------LTGLLIGSVVPVDAVAM--SKSILQVVLAPVTLG 263
A + + P +TG L+ +++P V S ILQV+ V G
Sbjct: 114 AGMNIDPATLDAKAVATYADKAHEQTITGFLM-NIIPTTIVGAFASGDILQVLFFSVLFG 172
Query: 264 LLLNTYA---KPVVSILRPVM-PFVAMICTSLCIGSPLAINRSQILSGEGLRLVAPVLIF 319
+ L KPV + +M P ++ L AP+ F
Sbjct: 173 IALGIVGEKGKPVTDFMHAMMYPIFKLVAI--------------------LMKAAPIGAF 212
Query: 320 HAAAFTLG-YWFSNLPSLRFL 339
A AFT+G Y S++ +L L
Sbjct: 213 GAMAFTIGKYGISSVTNLAML 233
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.324 0.138 0.409
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 41,668,435
Number of Sequences: 164201
Number of extensions: 1689041
Number of successful extensions: 5545
Number of sequences better than 10.0: 40
Number of HSP's better than 10.0 without gapping: 15
Number of HSP's successfully gapped in prelim test: 25
Number of HSP's that attempted gapping in prelim test: 5517
Number of HSP's gapped (non-prelim): 49
length of query: 379
length of database: 59,974,054
effective HSP length: 112
effective length of query: 267
effective length of database: 41,583,542
effective search space: 11102805714
effective search space used: 11102805714
T: 11
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 67 (30.4 bits)
Medicago: description of AC146343.8


