

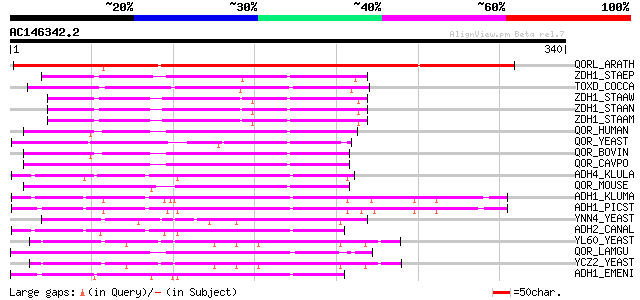
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146342.2 + phase: 0
(340 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
QORL_ARATH (Q9ZUC1) Quinone oxidoreductase-like protein At1g2374... 276 5e-74
ZDH1_STAEP (Q8CRJ7) Zinc-type alcohol dehydrogenase-like protein... 104 4e-22
TOXD_COCCA (P54006) TOXD protein 94 4e-19
ZDH1_STAAW (Q8NVD1) Zinc-type alcohol dehydrogenase-like protein... 90 9e-18
ZDH1_STAAN (P99173) Zinc-type alcohol dehydrogenase-like protein... 89 2e-17
ZDH1_STAAM (P63475) Zinc-type alcohol dehydrogenase-like protein... 89 2e-17
QOR_HUMAN (Q08257) Quinone oxidoreductase (EC 1.6.5.5) (NADPH:qu... 87 5e-17
QOR_YEAST (P38230) Probable quinone oxidoreductase (EC 1.6.5.5) ... 87 6e-17
QOR_BOVIN (O97764) Zeta-crystallin 86 1e-16
QOR_CAVPO (P11415) Quinone oxidoreductase (EC 1.6.5.5) (NADPH:qu... 85 2e-16
ADH4_KLULA (P49385) Alcohol dehydrogenase IV, mitochondrial prec... 84 7e-16
QOR_MOUSE (P47199) Quinone oxidoreductase (EC 1.6.5.5) (NADPH:qu... 83 1e-15
ADH1_KLUMA (Q07288) Alcohol dehydrogenase 1 (EC 1.1.1.1) 82 3e-15
ADH1_PICST (O00097) Alcohol dehydrogenase I (EC 1.1.1.1) (ADH 2) 80 6e-15
YNN4_YEAST (P53912) Hypothetical 41.2 kDa protein in FPR1-TOM22 ... 79 2e-14
ADH2_CANAL (O94038) Alcohol dehydrogenase 2 (EC 1.1.1.1) 78 4e-14
YL60_YEAST (P54007) Hypothetical 41.1 kDa protein ON CDC91-PAU4 ... 77 8e-14
QOR_LAMGU (Q28452) Quinone oxidoreductase (EC 1.6.5.5) (NADPH:qu... 75 2e-13
YCZ2_YEAST (P25608) 40.1 kDa protein in GIT1-PAU3 intergenic region 75 2e-13
ADH1_EMENI (P08843) Alcohol dehydrogenase I (EC 1.1.1.1) (ADH I) 75 2e-13
>QORL_ARATH (Q9ZUC1) Quinone oxidoreductase-like protein At1g23740,
chloroplast precursor (EC 1.-.-.-)
Length = 386
Score = 276 bits (706), Expect = 5e-74
Identities = 144/310 (46%), Positives = 205/310 (65%), Gaps = 5/310 (1%)
Query: 3 KAWFYEEYGPKEVLKL-GDFPIPSPLENQLLVQVYAAALNPIDSKRRMRPIFPSD--FPA 59
KAW Y +YG +VLKL + +P E+Q+L++V AAALNP+D+KRR +D P
Sbjct: 79 KAWVYSDYGGVDVLKLESNIVVPEIKEDQVLIKVVAAALNPVDAKRRQGKFKATDSPLPT 138
Query: 60 VPGCDMAGVVIGKGVNVKKFDIGDEVYGNIQDFNSMEKPKQLGTLAQFIVVEENLVARKP 119
VPG D+AGVV+ G VK GDEVY N+ + ++E PKQ G+LA++ VEE L+A KP
Sbjct: 139 VPGYDVAGVVVKVGSAVKDLKEGDEVYANVSE-KALEGPKQFGSLAEYTAVEEKLLALKP 197
Query: 120 KRLSFEEAASLPLAVQTAIEGFKTGDFKKGETMFVVGGAGGVGTLVLQLAKLMFGASYVV 179
K + F +AA LPLA++TA EG +F G+++ V+ GAGGVG+LV+QLAK ++GAS V
Sbjct: 198 KNIDFAQAAGLPLAIETADEGLVRTEFSAGKSILVLNGAGGVGSLVIQLAKHVYGASKVA 257
Query: 180 SSCSTPKLKFVKQFGADKVVDYTKTKYEDIEEKFDFIYDTVGDCKKSFVVAKKDGAIVDI 239
++ ST KL+ V+ GAD +DYTK ED+ +K+D ++D +G C K+ V K+ G +V +
Sbjct: 258 ATASTEKLELVRSLGADLAIDYTKENIEDLPDKYDVVFDAIGMCDKAVKVIKEGGKVVAL 317
Query: 240 TWPASHERAVYSSLTVCGEILEKLRPYLERGELKAVIDPKGEYDFENVIDAFGYIETGRA 299
T + + +T G++L+KL PY+E G++K V+DPKG + F V DAF Y+ET A
Sbjct: 318 TGAVTPPGFRF-VVTSNGDVLKKLNPYIESGKVKPVVDPKGPFPFSRVADAFSYLETNHA 376
Query: 300 WGKVVVTCFP 309
GKVVV P
Sbjct: 377 TGKVVVYPIP 386
>ZDH1_STAEP (Q8CRJ7) Zinc-type alcohol dehydrogenase-like protein
SE1777
Length = 336
Score = 104 bits (259), Expect = 4e-22
Identities = 69/211 (32%), Positives = 111/211 (51%), Gaps = 21/211 (9%)
Query: 20 DFPIPSPLENQLLVQVYAAALNPIDSKRRMRPIFPSDFPAVPGCDMAGVVIGKGVNVKKF 79
+F IP P ++LLV+V + ++NP+D+K+R P+ P V G D GV+ G V F
Sbjct: 22 NFDIPHPSGHELLVKVQSISVNPVDTKQRTMPV--DKVPRVLGFDAVGVIEKIGDQVSMF 79
Query: 80 DIGDEVYGNIQDFNSMEKPKQLGTLAQFIVVEENLVARKPKRLSFEEAASLPLAVQTAIE 139
GD V+ + P Q G+ ++ ++EE LVA+ P L E+AASLPL TA E
Sbjct: 80 QEGDVVFYS-------GSPNQNGSNEEYQLIEEYLVAKAPTNLKSEQAASLPLTGLTAYE 132
Query: 140 GF-------KTGDFKKGETMFVVGGAGGVGTLVLQLAKLMFGASYVVSSCSTPKLKFVKQ 192
K KG+++ ++ GAGGVG++ Q+AK +G + ++ +K+
Sbjct: 133 TLFDVFGISKEPSENKGKSLLIINGAGGVGSIATQIAK-FYGLKVITTASREDTIKWSVN 191
Query: 193 FGADKVVDYTKTKYEDIE----EKFDFIYDT 219
GAD V+++ K + + E D+I+ T
Sbjct: 192 MGADVVLNHKKDLSQQFKDNHIEGVDYIFCT 222
>TOXD_COCCA (P54006) TOXD protein
Length = 297
Score = 94.4 bits (233), Expect = 4e-19
Identities = 71/223 (31%), Positives = 110/223 (48%), Gaps = 21/223 (9%)
Query: 12 PKEVLKLGDFPIPSPLENQLLVQVYAAALNPIDSKRRMRPIFPSDFPAVPGCDMAGVVIG 71
P + D IP ++ +LV+ + ALNP D K +R P + GCD AG+V
Sbjct: 12 PHRARLVSDRLIPKLRDDYILVRTVSVALNPTDWKHILRLSPPG---CLVGCDYAGIVEE 68
Query: 72 KGVNVKK-FDIGDEVYGNIQDFNSMEKPKQLGTLAQFIVVEENLVARKPKRLSFEEAASL 130
G +VKK F GD V G N++ GT A+ I V+ ++ A P+ LSF+EAA+L
Sbjct: 69 VGRSVKKPFKKGDRVCGFAHGGNAVFSDD--GTFAEVITVKGDIQAWIPENLSFQEAATL 126
Query: 131 PLAVQTAIEG--------FKTGDFKKGETMFVVGGAGGVGTLVLQLAKLMFGASYVVSSC 182
+ ++T +G + T + + + GG+ GTL +QLAKL V+++C
Sbjct: 127 GVGIKTVGQGLYQSLKLSWPTTPIEHAVPILIYGGSTATGTLAIQLAKL--SGYRVITTC 184
Query: 183 STPKLKFVKQFGADKVVDYTKTKYED-----IEEKFDFIYDTV 220
S + +K GAD V DY + D + K ++DT+
Sbjct: 185 SPHHFELMKSLGADLVFDYHEITSADHIRRCTQNKLKLVFDTI 227
>ZDH1_STAAW (Q8NVD1) Zinc-type alcohol dehydrogenase-like protein
MW2112
Length = 335
Score = 89.7 bits (221), Expect = 9e-18
Identities = 63/208 (30%), Positives = 109/208 (52%), Gaps = 23/208 (11%)
Query: 24 PSPLENQLLVQVYAAALNPIDSKRRMRPIFPSDFPAVPGCDMAGVVIGKGVNVKKFDIGD 83
P+P + +LV+V + ++NP+D+K+R + + P V G D G V G +V F GD
Sbjct: 26 PTPENDDILVKVNSISVNPVDTKQRQMEV--TQAPRVLGFDAIGTVEAIGPDVTLFSPGD 83
Query: 84 EVYGNIQDFNSMEKPKQLGTLAQFIVVEENLVARKPKRLSFEEAASLPLAVQTAIEGFKT 143
V+ P + G+ A + +V E +VA+ P +S EA SLPL TA E F
Sbjct: 84 VVF-------YAGSPNRQGSNATYQLVSEAIVAKAPHNISANEAVSLPLTGITAYETF-F 135
Query: 144 GDFK--------KGETMFVVGGAGGVGTLVLQLAKLMFGASYVVSSCSTPKLKFVKQFGA 195
FK +G+++ ++ GAGGVG++ Q+AK +G + + ++ ++ ++ GA
Sbjct: 136 DTFKISHNPAENEGKSVLIINGAGGVGSIATQIAK-RYGLTVITTASRQETTEWCEKMGA 194
Query: 196 DKVVDYTKTKYEDIEEK----FDFIYDT 219
D V+++ + +EK D+I+ T
Sbjct: 195 DIVLNHKEDLVRQFKEKEIPLVDYIFCT 222
>ZDH1_STAAN (P99173) Zinc-type alcohol dehydrogenase-like protein
SA1988
Length = 335
Score = 88.6 bits (218), Expect = 2e-17
Identities = 63/208 (30%), Positives = 108/208 (51%), Gaps = 23/208 (11%)
Query: 24 PSPLENQLLVQVYAAALNPIDSKRRMRPIFPSDFPAVPGCDMAGVVIGKGVNVKKFDIGD 83
P+P + +LV+V + ++NP+D+K+R + + P V G D G V G +V F GD
Sbjct: 26 PTPENDDILVKVNSISVNPVDTKQRQMKV--TQAPRVLGFDAIGTVEAIGPDVTLFSPGD 83
Query: 84 EVYGNIQDFNSMEKPKQLGTLAQFIVVEENLVARKPKRLSFEEAASLPLAVQTAIEGFKT 143
V+ P + G+ A + +V E +VA+ P +S EA SLPL TA E F
Sbjct: 84 VVF-------YAGSPNRQGSNATYQLVSEAIVAKAPHNISANEAVSLPLTGITAYETF-F 135
Query: 144 GDFK--------KGETMFVVGGAGGVGTLVLQLAKLMFGASYVVSSCSTPKLKFVKQFGA 195
FK G+++ ++ GAGGVG++ Q+AK +G + + ++ ++ ++ GA
Sbjct: 136 DTFKISHNPSENVGKSVLIINGAGGVGSIATQIAK-RYGLTVITTASRQETTEWCEKMGA 194
Query: 196 DKVVDYTKTKYEDIEEK----FDFIYDT 219
D V+++ + +EK D+I+ T
Sbjct: 195 DIVLNHKEDLVRQFKEKEIPLVDYIFCT 222
>ZDH1_STAAM (P63475) Zinc-type alcohol dehydrogenase-like protein
SAV2186
Length = 335
Score = 88.6 bits (218), Expect = 2e-17
Identities = 63/208 (30%), Positives = 108/208 (51%), Gaps = 23/208 (11%)
Query: 24 PSPLENQLLVQVYAAALNPIDSKRRMRPIFPSDFPAVPGCDMAGVVIGKGVNVKKFDIGD 83
P+P + +LV+V + ++NP+D+K+R + + P V G D G V G +V F GD
Sbjct: 26 PTPENDDILVKVNSISVNPVDTKQRQMKV--TQAPRVLGFDAIGTVEAIGPDVTLFSPGD 83
Query: 84 EVYGNIQDFNSMEKPKQLGTLAQFIVVEENLVARKPKRLSFEEAASLPLAVQTAIEGFKT 143
V+ P + G+ A + +V E +VA+ P +S EA SLPL TA E F
Sbjct: 84 VVF-------YAGSPNRQGSNATYQLVSEAIVAKAPHNISANEAVSLPLTGITAYETF-F 135
Query: 144 GDFK--------KGETMFVVGGAGGVGTLVLQLAKLMFGASYVVSSCSTPKLKFVKQFGA 195
FK G+++ ++ GAGGVG++ Q+AK +G + + ++ ++ ++ GA
Sbjct: 136 DTFKISHNPSENVGKSVLIINGAGGVGSIATQIAK-RYGLTVITTASRQETTEWCEKMGA 194
Query: 196 DKVVDYTKTKYEDIEEK----FDFIYDT 219
D V+++ + +EK D+I+ T
Sbjct: 195 DIVLNHKEDLVRQFKEKEIPLVDYIFCT 222
>QOR_HUMAN (Q08257) Quinone oxidoreductase (EC 1.6.5.5)
(NADPH:quinone reductase) (Zeta-crystallin)
Length = 329
Score = 87.4 bits (215), Expect = 5e-17
Identities = 61/212 (28%), Positives = 108/212 (50%), Gaps = 21/212 (9%)
Query: 9 EYGPKEVLKL-GDFPIPSPLENQLLVQVYAAALNPIDSKRR-----MRPIFPSDFPAVPG 62
E+G EVLKL D +P P ++Q+L++V+A +NP+++ R +P+ P PG
Sbjct: 15 EFGGPEVLKLRSDIAVPIPKDHQVLIKVHACGVNPVETYIRSGTYSRKPL----LPYTPG 70
Query: 63 CDMAGVVIGKGVNVKKFDIGDEVYGNIQDFNSMEKPKQLGTLAQFIVVEENLVARKPKRL 122
D+AGV+ G N F GD V+ G A++ + ++ V + P++L
Sbjct: 71 SDVAGVIEAVGDNASAFKKGDRVF---------TSSTISGGYAEYALAADHTVYKLPEKL 121
Query: 123 SFEEAASLPLAVQTAIEG-FKTGDFKKGETMFVVGGAGGVGTLVLQLAKLMFGASYVVSS 181
F++ A++ + TA + K GE++ V G +GGVG Q+A+ +G + ++
Sbjct: 122 DFKQGAAIGIPYFTAYRALIHSACVKAGESVLVHGASGGVGLAACQIAR-AYGLKILGTA 180
Query: 182 CSTPKLKFVKQFGADKVVDYTKTKYEDIEEKF 213
+ K V Q GA +V ++ + Y D +K+
Sbjct: 181 GTEEGQKIVLQNGAHEVFNHREVNYIDKIKKY 212
>QOR_YEAST (P38230) Probable quinone oxidoreductase (EC 1.6.5.5)
(NADPH:quinone reductase)
Length = 334
Score = 87.0 bits (214), Expect = 6e-17
Identities = 71/214 (33%), Positives = 110/214 (51%), Gaps = 23/214 (10%)
Query: 2 QKAWFYEEYGPKEVLKLGDFPIPSPLENQLLVQVYAAALNPIDSKRRMRPIFPSDFPAVP 61
QK +E G +V+K D+P+PS E +LL++ +N I+S R + I+P + P V
Sbjct: 9 QKVILIDEIGGYDVIKYEDYPVPSISEEELLIKNKYTGVNYIESYFR-KGIYPCEKPYVL 67
Query: 62 GCDMAGVVIGKGVNVKKFDIGDEV-YGNIQDFNSMEKPKQLGTLAQFIVVEENLVARKPK 120
G + +G V+ KG V F++GD+V Y + F K + + V + PK
Sbjct: 68 GREASGTVVAKGKGVTNFEVGDQVAYISNSTFAQYSK-----------ISSQGPVMKLPK 116
Query: 121 RLSFEE----AASLPLAVQTAIEGFKTG-DFKKGETMFVVGGAGGVGTLVLQLAKLMFGA 175
S EE AA L L V TA+ KKG+ + + AGGVG ++ QL K M GA
Sbjct: 117 GTSDEELKLYAAGL-LQVLTALSFTNEAYHVKKGDYVLLFAAAGGVGLILNQLLK-MKGA 174
Query: 176 SYVVSSCSTPKLKFVKQFGADKVVDYTKTKYEDI 209
+ + + KLK K++GA+ +++ +K EDI
Sbjct: 175 HTIAVASTDEKLKIAKEYGAEYLINASK---EDI 205
>QOR_BOVIN (O97764) Zeta-crystallin
Length = 330
Score = 85.9 bits (211), Expect = 1e-16
Identities = 60/207 (28%), Positives = 105/207 (49%), Gaps = 21/207 (10%)
Query: 9 EYGPKEVLKL-GDFPIPSPLENQLLVQVYAAALNPIDSKRR-----MRPIFPSDFPAVPG 62
E+G EVLKL D +P P ++Q+L++V A +NP+D+ R ++P+ P PG
Sbjct: 15 EFGGPEVLKLQSDVAVPIPKDHQVLIKVQACGVNPVDTYIRSGTHNIKPL----LPYTPG 70
Query: 63 CDMAGVVIGKGVNVKKFDIGDEVYGNIQDFNSMEKPKQLGTLAQFIVVEENLVARKPKRL 122
D+AG++ G +V F GD V+ G A++ + ++ V P++L
Sbjct: 71 FDVAGIIEAVGESVSAFKKGDRVF---------TTRTISGGYAEYALAADHTVYTLPEKL 121
Query: 123 SFEEAASLPLAVQTAIEG-FKTGDFKKGETMFVVGGAGGVGTLVLQLAKLMFGASYVVSS 181
F++ A++ + TA + K GE++ V G +GGVG Q+A+ +G + ++
Sbjct: 122 DFKQGAAIGIPYFTAYRALLYSAPVKPGESVLVHGASGGVGIAACQIAR-AYGLKVLGTA 180
Query: 182 CSTPKLKFVKQFGADKVVDYTKTKYED 208
+ K V + GA KV ++ + Y D
Sbjct: 181 STEEGQKIVLENGAHKVFNHKEANYID 207
>QOR_CAVPO (P11415) Quinone oxidoreductase (EC 1.6.5.5)
(NADPH:quinone reductase) (Zeta-crystallin)
Length = 329
Score = 85.1 bits (209), Expect = 2e-16
Identities = 61/203 (30%), Positives = 102/203 (50%), Gaps = 13/203 (6%)
Query: 9 EYGPKEVLKL-GDFPIPSPLENQLLVQVYAAALNPIDSKRRMRPIFPSD-FPAVPGCDMA 66
E+G EVLK+ D +P P ++Q+L++V+A +NP+++ R P PG D+A
Sbjct: 15 EFGGPEVLKVQSDVAVPIPKDHQVLIKVHACGINPVETYIRSGTYTRIPLLPYTPGTDVA 74
Query: 67 GVVIGKGVNVKKFDIGDEVYGNIQDFNSMEKPKQLGTLAQFIVVEENLVARKPKRLSFEE 126
GVV G +V F GD V+ G A++ + ++ V R P++L F +
Sbjct: 75 GVVESIGNDVSAFKKGDRVF---------TTSTISGGYAEYALASDHTVYRLPEKLDFRQ 125
Query: 127 AASLPLAVQTAIEG-FKTGDFKKGETMFVVGGAGGVGTLVLQLAKLMFGASYVVSSCSTP 185
A++ + TA F + K GE++ V G +GGVG Q+A+ +G + ++ +
Sbjct: 126 GAAIGIPYFTACRALFHSARAKAGESVLVHGASGGVGLAACQIAR-AYGLKVLGTAGTEE 184
Query: 186 KLKFVKQFGADKVVDYTKTKYED 208
K V Q GA +V ++ Y D
Sbjct: 185 GQKVVLQNGAHEVFNHRDAHYID 207
>ADH4_KLULA (P49385) Alcohol dehydrogenase IV, mitochondrial
precursor (EC 1.1.1.1)
Length = 375
Score = 83.6 bits (205), Expect = 7e-16
Identities = 70/234 (29%), Positives = 106/234 (44%), Gaps = 30/234 (12%)
Query: 2 QKAWFYEEYGPKEVLKLGDFPIPSPLENQLLVQVYAAALNPID--SKRRMRPIFPSDFPA 59
QK + E G K L+ D P+P P N++L+ V + + D + + P+ P P
Sbjct: 34 QKGVIFYENGGK--LEYKDLPVPKPKANEILINVKYSGVCHTDLHAWKGDWPL-PVKLPL 90
Query: 60 VPGCDMAGVVIGKGVNVKKFDIGDEVYGNIQDFNSMEKPKQL------------------ 101
V G + AG+V+ KG NVK F+IGD Y I+ N +L
Sbjct: 91 VGGHEGAGIVVAKGENVKNFEIGD--YAGIKWLNGSCMSCELCEQGYESNCLQADLSGYT 148
Query: 102 --GTLAQFIVVEENLVARKPKRLSFEEAASLPLAVQTAIEGFKTGDFKKGETMFVVGGAG 159
G+ Q+ + A+ PK E A + A T + KT D K G+ + + G AG
Sbjct: 149 HDGSFQQYATADAVQAAQIPKGTDLAEIAPILCAGVTVYKALKTADLKPGQWVAISGAAG 208
Query: 160 GVGTLVLQLAKLMFGASYVVSSCSTPKLKFVKQFGADKVVDYTKTK--YEDIEE 211
G+G+L +Q AK M G + K + KQ G + +D+ K+K DI+E
Sbjct: 209 GLGSLAVQYAKAM-GLRVLGIDGGDGKEELFKQCGGEVFIDFRKSKDMVADIQE 261
>QOR_MOUSE (P47199) Quinone oxidoreductase (EC 1.6.5.5)
(NADPH:quinone reductase) (Zeta-crystallin)
Length = 331
Score = 82.8 bits (203), Expect = 1e-15
Identities = 61/205 (29%), Positives = 100/205 (48%), Gaps = 17/205 (8%)
Query: 9 EYGPKEVLKL-GDFPIPSPLENQLLVQVYAAALNPIDSKRRMRPIFPSD-FPAVPGCDMA 66
E+G EVLKL D +P P +Q+L++V+A +NP+++ R P PG D+A
Sbjct: 15 EFGGPEVLKLQSDVVVPVPQSHQVLIKVHACGVNPVETYIRSGAYSRKPALPYTPGSDVA 74
Query: 67 GVVIGKGVNVKKFDIGDEV--YGNIQDFNSMEKPKQLGTLAQFIVVEENLVARKPKRLSF 124
G++ G V F GD V Y + G A+F + ++ + P+ L+F
Sbjct: 75 GIIESVGDKVSAFKKGDRVFCYSTVS-----------GGYAEFALAADDTIYPLPETLNF 123
Query: 125 EEAASLPLAVQTAIEG-FKTGDFKKGETMFVVGGAGGVGTLVLQLAKLMFGASYVVSSCS 183
+ A+L + TA F + + GE++ V G +GGVG Q+A+ G + ++ S
Sbjct: 124 RQGAALGIPYFTACRALFHSARARAGESVLVHGASGGVGLATCQIAR-AHGLKVLGTAGS 182
Query: 184 TPKLKFVKQFGADKVVDYTKTKYED 208
K V Q GA +V ++ + Y D
Sbjct: 183 EEGKKLVLQNGAHEVFNHKEANYID 207
>ADH1_KLUMA (Q07288) Alcohol dehydrogenase 1 (EC 1.1.1.1)
Length = 348
Score = 81.6 bits (200), Expect = 3e-15
Identities = 88/347 (25%), Positives = 147/347 (42%), Gaps = 52/347 (14%)
Query: 2 QKAWFYEEYGPKEVLKLGDFPIPSPLENQLLVQVYAAALNPIDSKRRMRPIFPSD--FPA 59
QK + E+G + L+ D P+P P N+LL+ V + + D + +P D P
Sbjct: 7 QKGVIFYEHGGE--LQYKDIPVPKPKPNELLINVKYSGVCHTDL-HAWQGDWPLDTKLPL 63
Query: 60 VPGCDMAGVVIGKGVNVKKFDIGDEVYGNIQDFN----SMEK---------PK------- 99
V G + AG+V+ G NV ++IGD Y I+ N S E+ PK
Sbjct: 64 VGGHEGAGIVVAMGENVTGWEIGD--YAGIKWLNGSCMSCEECELSNEPNCPKADLSGYT 121
Query: 100 QLGTLAQFIVVEENLVARKPKRLSFEEAASLPLAVQTAIEGFKTGDFKKGETMFVVGGAG 159
G+ Q+ + AR PK + E A + A T + K+ K G+ + + G G
Sbjct: 122 HDGSFQQYATADAVQAARIPKNVDLAEVAPILCAGVTVYKALKSAHIKAGDWVAISGACG 181
Query: 160 GVGTLVLQLAKLMFGASYVVSSCSTPKLKFVKQFGADKVVDYTKTK--YEDIEEKFDFIY 217
G+G+L +Q AK M G + K K K+ G + +D+TKTK ++ E + +
Sbjct: 182 GLGSLAIQYAKAM-GYRVLGIDAGDEKAKLFKELGGEYFIDFTKTKDMVAEVIEATNGVA 240
Query: 218 DTV-------GDCKKSFVVAKKDGAIVDITWPASHE------RAVYSSLTVCGEIL---- 260
V S + + +G +V + P + V S+++ G +
Sbjct: 241 HAVINVSVSEAAISTSVLYTRSNGTVVLVGLPRDAQCKSDVFNQVVKSISIVGSYVGNRA 300
Query: 261 --EKLRPYLERGELKAVIDPKGEYDFENVIDAFGYIETGRAWGKVVV 305
+ + RG +KA I G + +V D + G+ G++VV
Sbjct: 301 DTREALDFFSRGLVKAPIKILGLSELASVYDK---MVKGQIVGRIVV 344
>ADH1_PICST (O00097) Alcohol dehydrogenase I (EC 1.1.1.1) (ADH 2)
Length = 348
Score = 80.5 bits (197), Expect = 6e-15
Identities = 91/347 (26%), Positives = 148/347 (42%), Gaps = 52/347 (14%)
Query: 2 QKAWFYEEYGPKEVLKLGDFPIPSPLENQLLVQVYAAALNPIDSKRRMRPIFPSD--FPA 59
QKA +E G + K D P+P+P N++L+ V + + D + +P D P
Sbjct: 7 QKAVVFESNGGPLLYK--DIPVPTPKPNEILINVKYSGVCHTDL-HAWKGDWPLDTKLPL 63
Query: 60 VPGCDMAGVVIGKGVNVKKFDIGDEVYGNIQDFNSM----------EKPKQL-------- 101
V G + AGVV+G G NV +++GD Y I+ N ++P
Sbjct: 64 VGGHEGAGVVVGIGSNVTGWELGD--YAGIKWLNGSCLNCEFCQHSDEPNCAKADLSGYT 121
Query: 102 --GTLAQFIVVEENLVARKPKRLSFEEAASLPLAVQTAIEGFKTGDFKKGETMFVVGGAG 159
G+ Q+ + AR PK +AA + A T + KT + G + + G G
Sbjct: 122 HDGSFQQYATADAVQAARLPKGTDLAQAAPILCAGITVYKALKTAQIQPGNWVCISGAGG 181
Query: 160 GVGTLVLQLAKLMFGASYVVSSCSTPKLKFVKQFGADKVVDYTKTK--YEDIEEKFD--- 214
G+G+L +Q AK M G + K +FVK GA+ VD+T +K +DI+ D
Sbjct: 182 GLGSLAIQYAKAM-GFRVIAIDGGEEKGEFVKSLGAEAYVDFTVSKDIVKDIQTATDGGP 240
Query: 215 --FIYDTVGD--CKKSFVVAKKDGAIVDITWPASHE------RAVYSSLTVCGEIL---- 260
I +V + +S + G +V + PA + AV S+++ G +
Sbjct: 241 HAAINVSVSEKAIAQSCQYVRSTGTVVLVGLPAGAKVVAPVFDAVVKSISIRGSYVGNRA 300
Query: 261 --EKLRPYLERGELKAVIDPKGEYDFENVIDAFGYIETGRAWGKVVV 305
+ + RG +K I G + V + +E G+ G+ VV
Sbjct: 301 DSAEAIDFFTRGLIKCPIKVVGLSELPKV---YELMEAGKVIGRYVV 344
>YNN4_YEAST (P53912) Hypothetical 41.2 kDa protein in FPR1-TOM22
intergenic region
Length = 376
Score = 79.0 bits (193), Expect = 2e-14
Identities = 72/235 (30%), Positives = 106/235 (44%), Gaps = 40/235 (17%)
Query: 20 DFPIPSPLENQLLVQVYAAALNPIDSKRRMRPIFPSDFPAVPGCDMAGVVIGKGVNVK-- 77
D PIP E +L++ A A NP D K I P A+ GCD AG ++ G NV
Sbjct: 24 DIPIPELEEGFVLIKTVAVAGNPTDWKHIDFKIGPQG--ALLGCDAAGQIVKLGPNVDAA 81
Query: 78 KFDIGDEVYGNIQDFNSMEKPKQLGTLAQFIVVEENLVARKPKR---------------L 122
+F IGD +YG I S+ P G A++ + A KP R
Sbjct: 82 RFAIGDYIYGVIHGA-SVRFPSN-GAFAEYSAISSE-TAYKPAREFRLCGKDKLPEGPVK 138
Query: 123 SFEEAASLPLAVQTA----------IEGFKTGDFKKGETMFVVGGAGGVGTLVLQLAKLM 172
S E A SLP+++ TA +K ++ + + GGA VG +++QLAK +
Sbjct: 139 SLEGAVSLPVSLTTAGMILTHSFGLDMTWKPSKAQRDQPILFWGGATAVGQMLIQLAKKL 198
Query: 173 FGASYVVSSCSTPKLKFVKQFGADKVVDY--------TKTKYEDIEEKFDFIYDT 219
G S ++ S K +K++GAD++ DY K KY +I D + +T
Sbjct: 199 NGFSKIIVVASRKHEKLLKEYGADELFDYHDADVIEQIKKKYNNIPYLVDCVSNT 253
>ADH2_CANAL (O94038) Alcohol dehydrogenase 2 (EC 1.1.1.1)
Length = 348
Score = 77.8 bits (190), Expect = 4e-14
Identities = 66/226 (29%), Positives = 101/226 (44%), Gaps = 28/226 (12%)
Query: 2 QKAWFYEEYGPKEVLKLGDFPIPSPLENQLLVQVYAAALNPIDSKRRMRPIFP--SDFPA 59
QKA +E G K L+ D P+P P N+LL+ V + + D + +P + P
Sbjct: 7 QKAVIFETNGGK--LEYKDIPVPKPKANELLINVKYSGVCHTDL-HAWKGDWPLATKLPL 63
Query: 60 VPGCDMAGVVIGKGVNVKKFDIGDEVYGNIQDFN----------SMEKPKQL-------- 101
V G + AGVV+ G NVK + +GD Y ++ N S +P
Sbjct: 64 VGGHEGAGVVVALGENVKGWKVGD--YAGVKWLNGSCLNCEYCQSGAEPNCAEADLSGYT 121
Query: 102 --GTLAQFIVVEENLVARKPKRLSFEEAASLPLAVQTAIEGFKTGDFKKGETMFVVGGAG 159
G+ Q+ + AR P A + A T + KT + + G+ + + G AG
Sbjct: 122 HDGSFQQYATADAVQAARIPAGTDLANVAPILCAGVTVYKALKTAELEAGQWVAISGAAG 181
Query: 160 GVGTLVLQLAKLMFGASYVVSSCSTPKLKFVKQFGADKVVDYTKTK 205
G+G+L +Q AK M G + K +FVK GA+ +D+TK K
Sbjct: 182 GLGSLAVQYAKAM-GYRVLAIDGGEDKGEFVKSLGAETFIDFTKEK 226
>YL60_YEAST (P54007) Hypothetical 41.1 kDa protein ON CDC91-PAU4
intergenic region
Length = 376
Score = 76.6 bits (187), Expect = 8e-14
Identities = 76/263 (28%), Positives = 118/263 (43%), Gaps = 42/263 (15%)
Query: 13 KEVLKLGDFPIPSPLENQLLVQVYAAALNPIDSKRRMRPIFPSDFPAVPGCDMAGVVI-- 70
K V+K G PIP E +L++ A A NP D I P ++ GCD AG ++
Sbjct: 18 KAVVKEG-IPIPELEEGFVLIKTLAVAGNPTDWAHIDYKIGPQG--SILGCDAAGQIVKL 74
Query: 71 GKGVNVKKFDIGDEVYGNIQDFNSMEKPKQLGTLAQFIVVEENLVARKPKRLSF------ 124
G VN K F IGD +YG I +S+ P G A++ + + + P L F
Sbjct: 75 GPAVNPKDFSIGDYIYGFIHG-SSVRFPSN-GAFAEYSAISTVVAYKSPNELKFLGEDVL 132
Query: 125 --------EEAASLPLAVQTA--IEGFKTGDFKKGET--------MFVVGGAGGVGTLVL 166
E A++P+++ TA + + G K E + + GGA VG ++
Sbjct: 133 PAGPVRSLEGVATIPVSLTTAGLVLTYNLGLDLKWEPSTPQRKGPILLWGGATAVGQSLI 192
Query: 167 QLAKLMFGASYVVSSCSTPKLKFVKQFGADKVVDY--------TKTKYEDIEEKFDFI-- 216
QLA + G + ++ S K +K++GAD++ DY K KY +I D +
Sbjct: 193 QLANKLNGFTKIIVVASRKHEKLLKEYGADELFDYHDIDVVEQIKHKYNNISYLVDCVAN 252
Query: 217 YDTVGDCKKSFVVAKKDGAIVDI 239
DT+ K K+D IV++
Sbjct: 253 QDTLQQVYKC-AADKQDATIVEL 274
>QOR_LAMGU (Q28452) Quinone oxidoreductase (EC 1.6.5.5)
(NADPH:quinone reductase) (Zeta-crystallin)
Length = 330
Score = 75.5 bits (184), Expect = 2e-13
Identities = 66/226 (29%), Positives = 107/226 (47%), Gaps = 21/226 (9%)
Query: 1 MQKAWFYEEYGPKEVLKL-GDFPIPSPLENQLLVQVYAAALNPIDSKRRMRPIFPSD-FP 58
+ +A E+G EVLKL D +P P E+Q+L++V A +NP+D+ R P
Sbjct: 7 LMRAIRVSEFGGPEVLKLQSDVAVPIPEEHQVLIKVQACGVNPVDTYIRSGTYSRKPRLP 66
Query: 59 AVPGCDMAGVVIGKGVNVKKFDIGDEVYGNIQDFNSMEKPKQLGTLAQFIVVEENLVARK 118
PG D+AG++ G V F GD V+ G A++ + ++ V +
Sbjct: 67 YTPGLDVAGLIEAVGERVSAFKKGDRVF---------TTSTVSGGYAEYALAADHTVYKL 117
Query: 119 PKRLSFEEAASLPLAVQTAIEG-FKTGDFKKGETMFVVGGAGGVGTLVLQLAKLMFGASY 177
P L F++ A++ + TA + K GE++ V G +GGVG Q+A+
Sbjct: 118 PGELDFQKGAAIGVPYFTAYRALLHSACAKAGESVLVHGASGGVGLAACQIARAC--CFK 175
Query: 178 VVSSCSTPK-LKFVKQFGADKVVDYTKTKYEDIEEKFDFIYDTVGD 222
V+ + T + + V Q GA +V ++ EDI D I +VG+
Sbjct: 176 VLGTAGTEEGQRVVLQNGAHEVFNHR----EDI--NIDKIKKSVGE 215
>YCZ2_YEAST (P25608) 40.1 kDa protein in GIT1-PAU3 intergenic region
Length = 368
Score = 75.1 bits (183), Expect = 2e-13
Identities = 74/264 (28%), Positives = 120/264 (45%), Gaps = 42/264 (15%)
Query: 13 KEVLKLGDFPIPSPLENQLLVQVYAAALNPIDSKRRMRPIFPSDFPAVPGCDMAGVVI-- 70
K V+K G PIP E +L++ A A NP D + P ++ GCD AG ++
Sbjct: 10 KAVVKEG-VPIPELEEGFVLIKTLAVAGNPTDWAHIDYKVGPQG--SILGCDAAGQIVKL 66
Query: 71 GKGVNVKKFDIGDEVYGNIQDFNSMEKPKQLGTLAQFIVVEENLVARKPKRLSF------ 124
G V+ K F IGD +YG I +S+ P G A++ + + + P L F
Sbjct: 67 GPAVDPKDFSIGDYIYGFIHG-SSVRFPSN-GAFAEYSAISTVVAYKSPNELKFLGEDVL 124
Query: 125 --------EEAASLPLAVQTA--IEGFKTGDFKKGET--------MFVVGGAGGVGTLVL 166
E AA++P+++ TA + + G K E + + GGA VG ++
Sbjct: 125 PAGPVRSLEGAATIPVSLTTAGLVLTYNLGLNLKWEPSTPQRNGPILLWGGATAVGQSLI 184
Query: 167 QLAKLMFGASYVVSSCSTPKLKFVKQFGADKVVDY--------TKTKYEDIEEKFDFI-- 216
QLA + G + ++ S K +K++GAD++ DY K KY +I D +
Sbjct: 185 QLANKLNGFTKIIVVASRKHEKLLKEYGADQLFDYHDIDVVEQIKHKYNNISYLVDCVAN 244
Query: 217 YDTVGDCKKSFVVAKKDGAIVDIT 240
+T+ K K+D +V++T
Sbjct: 245 QNTLQQVYKC-AADKQDATVVELT 267
>ADH1_EMENI (P08843) Alcohol dehydrogenase I (EC 1.1.1.1) (ADH I)
Length = 349
Score = 75.1 bits (183), Expect = 2e-13
Identities = 66/225 (29%), Positives = 102/225 (45%), Gaps = 24/225 (10%)
Query: 1 MQKAWFYEEYGPKEVLKLGDFPIPSPLENQLLVQVYAAALNPIDSKRRMR--PIFPSDFP 58
MQ A E+ G V K P+P P +Q+LV++ + + D M PI P P
Sbjct: 6 MQWAQVAEKVGGPLVYK--QIPVPKPGPDQILVKIRYSGVCHTDLHAMMGHWPI-PVKMP 62
Query: 59 AVPGCDMAGVVIGKGVNVKKFDIGDEV--------YGNIQDFNSMEKP----KQL----- 101
V G + AG+V+ KG V +F+IGD+ G + + P QL
Sbjct: 63 LVGGHEGAGIVVAKGELVHEFEIGDQAGIKWLNGSCGECEFCRQSDDPLCARAQLSGYTV 122
Query: 102 -GTLAQFIVVEENLVARKPKRLSFEEAASLPLAVQTAIEGFKTGDFKKGETMFVVGGAGG 160
GT Q+ + + + ++ P + + AA + A T +G K + G+T+ +VG GG
Sbjct: 123 DGTFQQYALGKASHASKIPAGVPVDAAAPVLCAGITVYKGLKEAGVRPGQTVAIVGAGGG 182
Query: 161 VGTLVLQLAKLMFGASYVVSSCSTPKLKFVKQFGADKVVDYTKTK 205
+G+L Q AK M G V K + G + VD+TK+K
Sbjct: 183 LGSLAQQYAKAM-GIRVVAVDGGDEKRAMCESLGTETYVDFTKSK 226
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.319 0.139 0.411
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 41,999,263
Number of Sequences: 164201
Number of extensions: 1858088
Number of successful extensions: 5083
Number of sequences better than 10.0: 212
Number of HSP's better than 10.0 without gapping: 104
Number of HSP's successfully gapped in prelim test: 108
Number of HSP's that attempted gapping in prelim test: 4773
Number of HSP's gapped (non-prelim): 285
length of query: 340
length of database: 59,974,054
effective HSP length: 111
effective length of query: 229
effective length of database: 41,747,743
effective search space: 9560233147
effective search space used: 9560233147
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 66 (30.0 bits)
Medicago: description of AC146342.2


