

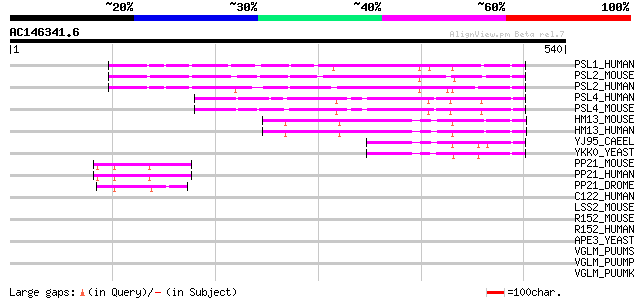
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146341.6 + phase: 0
(540 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
PSL1_HUMAN (Q8TCT7) Signal peptide peptidase-like 2B (EC 3.4.99.... 206 2e-52
PSL2_MOUSE (Q9JJF9) Signal peptide peptidase-like 2A (EC 3.4.99.... 194 5e-49
PSL2_HUMAN (Q8TCT8) Signal peptide peptidase-like 2A (EC 3.4.99.... 188 3e-47
PSL4_HUMAN (Q8TCT6) Signal peptide peptidase-like 3 (EC 3.4.99.-... 110 9e-24
PSL4_MOUSE (Q9CUS9) Signal peptide peptidase-like 3 (EC 3.4.99.-... 106 1e-22
HM13_MOUSE (Q9D8V0) Minor histocompatibility antigen H13 (EC 3.4... 100 7e-21
HM13_HUMAN (Q8TCT9) Minor histocompatibility antigen H13 (EC 3.4... 100 2e-20
YJ95_CAEEL (P49049) Hypothetical protein T05E11.5 in chromosome IV 77 9e-14
YKK0_YEAST (P34248) Hypothetical 67.5 kDa protein in APE1/LAP4-C... 69 2e-11
PP21_MOUSE (Q9D9N8) Protease-associated domain-containing protei... 46 2e-04
PP21_HUMAN (Q9BSG0) Protease-associated domain-containing protei... 46 2e-04
PP21_DROME (Q9W1W9) PAP21-like protein precursor 45 5e-04
C122_HUMAN (Q9BZQ6) Putative alpha-mannosidase C1orf22 (EC 3.2.1.-) 38 0.076
LSS2_MOUSE (Q924Z4) LAG1 longevity assurance homolog 2 (Transloc... 37 0.17
R152_MOUSE (Q8BG47) RING finger protein 152 33 2.5
R152_HUMAN (Q8N8N0) RING finger protein 152 33 2.5
APE3_YEAST (P37302) Aminopeptidase Y precursor (EC 3.4.11.15) 33 2.5
VGLM_PUUMS (P27312) M polyprotein precursor [Contains: Glycoprot... 32 3.2
VGLM_PUUMP (P41266) M polyprotein precursor [Contains: Glycoprot... 32 3.2
VGLM_PUUMK (P41265) M polyprotein precursor [Contains: Glycoprot... 32 3.2
>PSL1_HUMAN (Q8TCT7) Signal peptide peptidase-like 2B (EC 3.4.99.-)
(SPP-like 2B protein) (SPPL2b protein) (Intramembrane
protease 4) (IMP4) (Presenilin-like protein 1)
Length = 564
Score = 206 bits (523), Expect = 2e-52
Identities = 145/419 (34%), Positives = 223/419 (52%), Gaps = 31/419 (7%)
Query: 97 PKNKLTNEIILVHRGKCSFTTKANIADEAGASAILIINYRTELFKMVCEENETDVDIGIP 156
P +N+I LV RG C+F K +A +GA +LI++ R L + + D +IGIP
Sbjct: 54 PARGFSNQIPLVARGNCTFYEKVRLAQGSGARGLLIVS-RERLVPPGGNKTQYD-EIGIP 111
Query: 157 AVMLPQDAGLNLERHIQNNSIVSIQLYSPLRPLVDVAEVFLWLMAVGTILCASYWSAWTA 216
+L L++ V LY+P P++D V +++MAVGT+ YW+ +
Sbjct: 112 VALLSYKDMLDIFTRFGRT--VRAALYAPKEPVLDYNMVIIFIMAVGTVAIGGYWAG--S 167
Query: 217 REAAIEQEKLLKDASDEYVAESVGSRGYVEISTTAAILFVVLASCFLVMLYKLMSFWFLE 276
R+ K +D E + V+++ +FVV+ LV+LY +
Sbjct: 168 RDVKKRYMKHKRDDGPEKQEDEA-----VDVTPVMTCVFVVMCCSMLVLLYYFYDL-LVY 221
Query: 277 VLVVLFCIGGIEGLQTCLTALLSCFRWFQYPAQTYVK--IPFFGAVPYLT-LAVTPFCIV 333
V++ +FC+ GL +CL C R + +P+F P L + FC+
Sbjct: 222 VVIGIFCLASATGLYSCLAP---CVRRLPFGKCRIPNNSLPYFHKRPQARMLLLALFCVA 278
Query: 334 FAVVWAVKR-QASYAWIGQDILGIALIITVLQIVRIPNLKVGTVLLSCAFLYDILWVFVS 392
+VVW V R + +AW+ QD LGIA + +L+ +R+P K T+LL FLYDI +VF++
Sbjct: 279 VSVVWGVFRNEDQWAWVLQDALGIAFCLYMLKTIRLPTFKACTLLLLVLFLYDIFFVFIT 338
Query: 393 KWWFH--ESVMIVVARG--DKSGEDGIPMLLKLPRL-FDPWG----GYSIIGFGDIILPG 443
+ S+M+ VA G D + + +PM+LK+PRL P +S++GFGDI++PG
Sbjct: 339 PFLTKSGSSIMVEVATGPSDSATREKLPMVLKVPRLNSSPLALCDRPFSLLGFGDILVPG 398
Query: 444 LVVAFSLRYDWLAKKNLRAGYFVWAMTAYGLGLLITYVALNLMDGHGQPALLYIVPFTL 502
L+VA+ R+D + + YFV AYG+GLL+T+VAL LM GQPALLY+VP TL
Sbjct: 399 LLVAYCHRFDIQVQSS--RVYFVACTIAYGVGLLVTFVALALMQ-RGQPALLYLVPCTL 454
>PSL2_MOUSE (Q9JJF9) Signal peptide peptidase-like 2A (EC 3.4.99.-)
(SPP-like 2A protein) (SPPL2a protein) (Intramembrane
protease 3) (IMP3) (Presenilin-like protein 2)
Length = 523
Score = 194 bits (493), Expect = 5e-49
Identities = 126/419 (30%), Positives = 221/419 (52%), Gaps = 37/419 (8%)
Query: 97 PKNKLTNEIILVHRGKCSFTTKANIADEAGASAILIINYRTELFKMVCEENETDVDIGIP 156
P + + N+ ++VH G C F KA IA E GA+A+LI N + T ++ +
Sbjct: 82 PPDGIRNKAVVVHWGPCHFLEKARIAQEGGAAALLIAN--NSVLIPSSRNKSTFQNVTVL 139
Query: 157 AVMLPQDAGLNLERHIQNNSIVSIQLYSPLRPLVDVAEVFLWLMAVGTILCASYWSAWTA 216
++ Q +++ + ++ +++++YSP P D V ++++AV T+ YWS
Sbjct: 140 IAVITQKDFKDMKETLGDD--ITVKMYSPSWPNFDYTLVVIFVIAVFTVALGGYWSGLIE 197
Query: 217 REAAIEQEKLLKDASDEYVAESVGSRGYVEISTTAAILFVVLASCFLVMLYKLMSFWFLE 276
+E K ++DA D + Y+ S ++FVV+ +V+LY W +
Sbjct: 198 ----LENMKSVEDAEDRETRKKKDD--YLTFSPLTVVVFVVICCIMIVLLYFFYR-WLVY 250
Query: 277 VLVVLFCIGGIEGLQTCLTALLSCFRWFQYPAQTYVKIPFFGAVPYLTLAVTPFCIVFAV 336
V++ +FCI L CL+AL+ + P + + + ++ CI AV
Sbjct: 251 VMIAIFCIASSMSLYNCLSALIH-----RMPCGQCTILCCGKNIKVSLIFLSGLCISVAV 305
Query: 337 VWAVKRQAS-YAWIGQDILGIALIITVLQIVRIPNLKVGTVLLSCAFLYDILWVFVSKWW 395
VWAV R +AWI QDILGIA + +++ +++PN +LL +YD+ +VF++ +
Sbjct: 306 VWAVFRNEDRWAWILQDILGIAFCLNLIKTMKLPNFMSCVILLGLLLIYDVFFVFITPFI 365
Query: 396 FH--ESVMIVVARGDKSGEDGIPMLLKLPRLFDPWGGY----------SIIGFGDIILPG 443
ES+M+ +A G + +P+++++P+L GY S++GFGDII+PG
Sbjct: 366 TKNGESIMVELAAGPFENAEKLPVVIRVPKLM----GYSVMSVCSVPVSVLGFGDIIVPG 421
Query: 444 LVVAFSLRYDWLAKKNLRAGYFVWAMTAYGLGLLITYVALNLMDGHGQPALLYIVPFTL 502
L++A+ R+D ++ Y++ + AY +G++IT+V L +M GQPALLY+VP TL
Sbjct: 422 LLIAYCRRFDVQTGSSI---YYISSTIAYAVGMIITFVVLMVMK-TGQPALLYLVPCTL 476
>PSL2_HUMAN (Q8TCT8) Signal peptide peptidase-like 2A (EC 3.4.99.-)
(SPP-like 2A protein) (SPPL2a protein) (Intramembrane
protease 3) (IMP3) (Presenilin-like protein 2)
Length = 520
Score = 188 bits (477), Expect = 3e-47
Identities = 128/418 (30%), Positives = 218/418 (51%), Gaps = 35/418 (8%)
Query: 97 PKNKLTNEIILVHRGKCSFTTKANIADEAGASAILIINYRTELFKMVCEENETDVDIGIP 156
P + ++ ++V G C F KA IA + GA A+L++N + LF +E D+ I
Sbjct: 79 PPVGIKSKAVVVPWGSCHFLEKARIAQKGGAEAMLVVN-NSVLFPPSGNRSEFP-DVKIL 136
Query: 157 AVMLPQDAGLNLERHIQNNSIVSIQLYSPLRPLVDVAEVFLWLMAVGTILCASYWSAWTA 216
+ ++ + + +N +++++YSP P D V ++++AV T+ YWS
Sbjct: 137 IAFISYKDFRDMNQTLGDN--ITVKMYSPSWPNFDYTMVVIFVIAVFTVALGGYWSGLVE 194
Query: 217 RE---AAIEQEKLLKDASDEYVAESVGSRGYVEISTTAAILFVVLASCFLVMLYKLMSFW 273
E A +++ ++ +EY+ S ++FVV+ +V+LY W
Sbjct: 195 LENLKAVTTEDREMRKKKEEYLT----------FSPLTVVIFVVICCVMMVLLYFFYK-W 243
Query: 274 FLEVLVVLFCIGGIEGLQTCLTALLSCFRWFQYPAQTYVKIPFFGAVPYLTLAVTPFCIV 333
+ V++ +FCI L CL AL+ + Q K + + ++ CI
Sbjct: 244 LVYVMIAIFCIASAMSLYNCLAALIHKIPYGQCTIACRGK-----NMEVRLIFLSGLCIA 298
Query: 334 FAVVWAVKRQAS-YAWIGQDILGIALIITVLQIVRIPNLKVGTVLLSCAFLYDILWVFVS 392
AVVWAV R +AWI QDILGIA + +++ +++PN K +LL LYD+ +VF++
Sbjct: 299 VAVVWAVFRNEDRWAWILQDILGIAFCLNLIKTLKLPNFKSCVILLGLLLLYDVFFVFIT 358
Query: 393 KWWFH--ESVMIVVARGDKSGEDGIPMLLKLPRL--FDPWG----GYSIIGFGDIILPGL 444
+ ES+M+ +A G + +P+++++P+L F SI+GFGDII+PGL
Sbjct: 359 PFITKNGESIMVELAAGPFGNNEKLPVVIRVPKLIYFSVMSVCLMPVSILGFGDIIVPGL 418
Query: 445 VVAFSLRYDWLAKKNLRAGYFVWAMTAYGLGLLITYVALNLMDGHGQPALLYIVPFTL 502
++A+ R+D + Y+V + AY +G+++T+V L LM GQPALLY+VP TL
Sbjct: 419 LIAYCRRFD--VQTGSSYIYYVSSTVAYAIGMILTFVVLVLMK-KGQPALLYLVPCTL 473
>PSL4_HUMAN (Q8TCT6) Signal peptide peptidase-like 3 (EC 3.4.99.-)
(SPP-like 3 protein) (Intramembrane protease 2) (IMP2)
(Presenilin-like protein 4)
Length = 385
Score = 110 bits (275), Expect = 9e-24
Identities = 101/363 (27%), Positives = 164/363 (44%), Gaps = 55/363 (15%)
Query: 181 QLYSPLRPLVDVAEVFLWLMAVGTILCASYWSAWTAREAAIEQEKLLKDASDEYVAESVG 240
Q YS LVD ++V +L+++ I+ S+ S E ++EK +S + E
Sbjct: 4 QTYSWAYSLVDSSQVSTFLISILLIVYGSFRSLNMDFENQ-DKEKDSNSSSGSFNGEQEP 62
Query: 241 SRGYVEISTTAAILFVVLASCFLVMLYKLMSFWFLEVLVVLFCIGGIEGLQTCLTALLSC 300
G+ + +T A F+ + +C +++ M F+F V VV + LL
Sbjct: 63 IIGFQPMDSTRA-RFLPMGACVSLLV---MFFFFDSVQVVFTICTAVLATIAFAFLLLPM 118
Query: 301 FRWFQYPAQTYVKIPF-----FGAVPYLTLAVTPFCIVFAVVWAVKRQASYAWIGQDILG 355
++ P KI F F A L+ +++ ++ ++W + W+ D L
Sbjct: 119 CQYLTRPCSPQNKISFGCCGRFTAAELLSFSLS---VMLVLIWVLTGH----WLLMDALA 171
Query: 356 IALIITVLQIVRIPNLKVGTVLLSCAFLYDILWVFVSKWWFHESVMIVVAR--------- 406
+ L + ++ VR+P+LKV +LLS +YD+ WVF S + F+ +VM+ VA
Sbjct: 172 MGLCVAMIAFVRLPSLKVSCLLLSGLLIYDVFWVFFSAYIFNSNVMVKVATQPADNPLDV 231
Query: 407 -------GDKSGEDGIPMLLKLPRLFDP---WGGYSIIGFGDIILPGLVVAFSLRYDWLA 456
G G D +P L +L P +S++G GDI++PGL++ F LRYD
Sbjct: 232 LSRKLHLGPNVGRD-VPRLSLPGKLVFPSSTGSHFSMLGIGDIVMPGLLLCFVLRYDNYK 290
Query: 457 K-----------------KNLRAGYFVWAMTAYGLGLLITYVALNLMDGHGQPALLYIVP 499
K + + YF + Y +GLL VA + QPALLY+VP
Sbjct: 291 KQASGDSCGAPGPANISGRMQKVSYFHCTLIGYFVGLLTATVASRIHRA-AQPALLYLVP 349
Query: 500 FTL 502
FTL
Sbjct: 350 FTL 352
>PSL4_MOUSE (Q9CUS9) Signal peptide peptidase-like 3 (EC 3.4.99.-)
(SPP-like 3 protein) (Intramembrane protease 2) (IMP2)
(Presenilin-like protein 4)
Length = 384
Score = 106 bits (265), Expect = 1e-22
Identities = 100/363 (27%), Positives = 162/363 (44%), Gaps = 56/363 (15%)
Query: 181 QLYSPLRPLVDVAEVFLWLMAVGTILCASYWSAWTAREAAIEQEKLLKDASDEYVAESVG 240
Q YS LVD ++V +L+++ I+ S+ S E ++EK +S + S
Sbjct: 4 QTYSWAYSLVDSSQVSTFLISILLIVYGSFRSLNMDFENQ-DKEKDSNSSSGSFNGNSTN 62
Query: 241 SRGYVEISTTAAILFVVLASCFLVMLYKLMSFWFLEVLVVLFCIGGIEGLQTCLTALLSC 300
+ I +T A+ + AS L++++ F+F V VV + LL
Sbjct: 63 N-SIQTIDSTQALFLPIGASVSLLVMF----FFFDSVQVVFTICTAVLATIAFAFLLLPM 117
Query: 301 FRWFQYPAQTYVKIPF-----FGAVPYLTLAVTPFCIVFAVVWAVKRQASYAWIGQDILG 355
++ P KI F F A L+ +++ ++ ++W + W+ D L
Sbjct: 118 CQYLTRPCSPQNKISFGCCGRFTAAELLSFSLS---VMLVLIWVLTGH----WLLMDALA 170
Query: 356 IALIITVLQIVRIPNLKVGTVLLSCAFLYDILWVFVSKWWFHESVMIVVAR--------- 406
+ L + ++ R+P+LKV +LLS +YD+ WVF S + F+ +VM+ VA
Sbjct: 171 MGLCVAMIAFDRLPSLKVSCLLLSGLLIYDVFWVFFSAYIFNSNVMVKVATQPADNPLDV 230
Query: 407 -------GDKSGEDGIPMLLKLPRLFDP---WGGYSIIGFGDIILPGLVVAFSLRYDWLA 456
G G D +P L +L P +S++G GDI++PGL++ F LRYD
Sbjct: 231 LSKKLHLGPNVGRD-VPRLSLPGKLVFPSSTGSHFSMLGIGDIVMPGLLLCFVLRYDNYK 289
Query: 457 K-----------------KNLRAGYFVWAMTAYGLGLLITYVALNLMDGHGQPALLYIVP 499
K + + YF + Y +GLL VA + QPALLY+VP
Sbjct: 290 KQASGDSCGAPGPANISGRMQKVSYFHCTLIGYFVGLLTATVASRIHRA-AQPALLYLVP 348
Query: 500 FTL 502
FTL
Sbjct: 349 FTL 351
>HM13_MOUSE (Q9D8V0) Minor histocompatibility antigen H13 (EC
3.4.99.-) (Signal peptide peptidase) (Presenilin-like
protein 3)
Length = 378
Score = 100 bits (250), Expect = 7e-21
Identities = 77/272 (28%), Positives = 136/272 (49%), Gaps = 28/272 (10%)
Query: 247 ISTTAAILFVVLASCFLVMLY---KLMSFWFLEVLVVL-FCIGGIEGLQTCLTALLSCFR 302
I++ A F ++ASC L+ LY K+ S ++ +L+ + F + GI L ++ ++ F
Sbjct: 70 ITSRDAARFPIIASCTLLGLYLFFKIFSQEYINLLLSMYFFVLGILALSHTISPFMNKFF 129
Query: 303 WFQYPAQTYVKIPFFGA-------VPYLTLAVTPFCIVFAVVWAVKRQASYAWIGQDILG 355
+P + Y + G+ + Y C+ + V V WI ++ G
Sbjct: 130 PANFPNRQYQLLFTQGSGENKEEIINYEFDTKDLVCLGLSSVVGVWYLLRKHWIANNLFG 189
Query: 356 IALIITVLQIVRIPNLKVGTVLLSCAFLYDILWVFVSKWWFHESVMIVVARGDKSGEDGI 415
+A + ++++ + N+ G +LL F+YDI WVF + +VM+ VA+ ++
Sbjct: 190 LAFSLNGVELLHLNNVSTGCILLGGLFIYDIFWVFGT------NVMVTVAKSFEA----- 238
Query: 416 PMLLKLPRLFDPWG----GYSIIGFGDIILPGLVVAFSLRYDWLAKKNLRAGYFVWAMTA 471
P+ L P+ G ++++G GDI++PG+ +A LR+D KKN YF + A
Sbjct: 239 PIKLVFPQDLLEKGLEADNFAMLGLGDIVIPGIFIALLLRFDISLKKNTHT-YFYTSFAA 297
Query: 472 YGLGLLITYVALNLMDGHGQPALLYIVPFTLG 503
Y GL +T +++ H QPALLY+VP +G
Sbjct: 298 YIFGLGLTIFIMHIFK-HAQPALLYLVPACIG 328
>HM13_HUMAN (Q8TCT9) Minor histocompatibility antigen H13 (EC
3.4.99.-) (Signal peptide peptidase) (Presenilin-like
protein 3)
Length = 377
Score = 99.8 bits (247), Expect = 2e-20
Identities = 74/272 (27%), Positives = 136/272 (49%), Gaps = 28/272 (10%)
Query: 247 ISTTAAILFVVLASCFLVMLY---KLMSFWFLEVLVVL-FCIGGIEGLQTCLTALLSCFR 302
I++ A F ++ASC L+ LY K+ S ++ +L+ + F + GI L ++ ++ F
Sbjct: 70 ITSRDAARFPIIASCTLLGLYLFFKIFSQEYINLLLSMYFFVLGILALSHTISPFMNKFF 129
Query: 303 WFQYPAQTYVKIPFFGA-------VPYLTLAVTPFCIVFAVVWAVKRQASYAWIGQDILG 355
+P + Y + G+ + Y C+ + + V WI ++ G
Sbjct: 130 PASFPNRQYQLLFTQGSGENKEEIINYEFDTKDLVCLGLSSIVGVWYLLRKHWIANNLFG 189
Query: 356 IALIITVLQIVRIPNLKVGTVLLSCAFLYDILWVFVSKWWFHESVMIVVARGDKSGEDGI 415
+A + ++++ + N+ G +LL F+YD+ WVF + +VM+ VA+ ++
Sbjct: 190 LAFSLNGVELLHLNNVSTGCILLGGLFIYDVFWVFGT------NVMVTVAKSFEA----- 238
Query: 416 PMLLKLPRLFDPWG----GYSIIGFGDIILPGLVVAFSLRYDWLAKKNLRAGYFVWAMTA 471
P+ L P+ G ++++G GD+++PG+ +A LR+D KKN YF + A
Sbjct: 239 PIKLVFPQDLLEKGLEANNFAMLGLGDVVIPGIFIALLLRFDISLKKNTHT-YFYTSFAA 297
Query: 472 YGLGLLITYVALNLMDGHGQPALLYIVPFTLG 503
Y GL +T +++ H QPALLY+VP +G
Sbjct: 298 YIFGLGLTIFIMHIFK-HAQPALLYLVPACIG 328
>YJ95_CAEEL (P49049) Hypothetical protein T05E11.5 in chromosome IV
Length = 468
Score = 77.4 bits (189), Expect = 9e-14
Identities = 56/171 (32%), Positives = 87/171 (50%), Gaps = 28/171 (16%)
Query: 348 WIGQDILGIALIITVLQIVRIPNLKVGTVLLSCAFLYDILWVFVSKWWFHESVMIVVARG 407
WI +I+G++ I ++ + + + K G++LL F YDI WVF + VM VA+G
Sbjct: 269 WITNNIIGVSFSILGIERLHLASFKAGSLLLVGLFFYDIFWVFGT------DVMTSVAKG 322
Query: 408 DKSGEDGIPMLLKLPRLFDPWG-----GYSIIGFGDIILPGLVVAFSLRYDW-----LAK 457
+ P+LL+ P+ G +S++G GDI++PG+ +A R+D+ A+
Sbjct: 323 IDA-----PILLQFPQDIYRNGIMEASKHSMLGLGDIVIPGIFIALLRRFDYRVVQTTAE 377
Query: 458 KNLRAG------YFVWAMTAYGLGLLITYVALNLMDGHGQPALLYIVPFTL 502
G YFV + AY GL IT ++ QPALLY+VP L
Sbjct: 378 SKAPQGSLKGRYYFVVTVVAYMAGLFITMAVMHHFKA-AQPALLYLVPCCL 427
>YKK0_YEAST (P34248) Hypothetical 67.5 kDa protein in APE1/LAP4-CWP1
intergenic region
Length = 587
Score = 69.3 bits (168), Expect = 2e-11
Identities = 54/171 (31%), Positives = 83/171 (47%), Gaps = 28/171 (16%)
Query: 348 WIGQDILGIALIITVLQIVRIPNLKVGTVLLSCAFLYDILWVFVSKWWFHESVMIVVARG 407
W+ + + + + I + +++ NLK G ++L F YDI +VF + VM+ VA
Sbjct: 329 WLISNAVSMNMAIWSIAQLKLKNLKSGALILIALFFYDICFVFGT------DVMVTVATN 382
Query: 408 DKSGEDGIPMLLKLPRLFDPWGG---YSIIGFGDIILPGLVVAFSLRYD-W--------- 454
IP+ L LP F+ +SI+G GDI LPG+ +A +YD W
Sbjct: 383 LD-----IPVKLSLPVKFNTAQNNFNFSILGLGDIALPGMFIAMCYKYDIWKWHLDHDDT 437
Query: 455 ---LAKKNLRAGYFVWAMTAYGLGLLITYVALNLMDGHGQPALLYIVPFTL 502
+ YF+ AM +Y L+ V+L++ + QPALLYIVP L
Sbjct: 438 EFHFLNWSYVGKYFITAMVSYVASLVSAMVSLSIFN-TAQPALLYIVPSLL 487
>PP21_MOUSE (Q9D9N8) Protease-associated domain-containing protein
of 21 kDa precursor
Length = 188
Score = 46.2 bits (108), Expect = 2e-04
Identities = 30/105 (28%), Positives = 52/105 (48%), Gaps = 9/105 (8%)
Query: 82 HTR-----VVMADPPDCCSKPKNK--LTNEIILVHRGKCSFTTKANIADEAGASAILIIN 134
HTR +V A+PP+ C + N + ++I LV RG CSF +K + E G A++I +
Sbjct: 57 HTRYEQIHLVPAEPPEACGELSNGFFIQDQIALVERGGCSFLSKTRVVQEHGGRAVIISD 116
Query: 135 --YRTELFKMVCEENETDVDIGIPAVMLPQDAGLNLERHIQNNSI 177
+ F + ++ T IPA+ L G + R ++ + +
Sbjct: 117 NAVDNDSFYVEMIQDSTQRTADIPALFLLGRDGYMIRRSLEQHGL 161
>PP21_HUMAN (Q9BSG0) Protease-associated domain-containing protein
of 21 kDa precursor (hPAP21) (UNQ833/PRO1760)
Length = 188
Score = 46.2 bits (108), Expect = 2e-04
Identities = 30/105 (28%), Positives = 52/105 (48%), Gaps = 9/105 (8%)
Query: 82 HTR-----VVMADPPDCCSKPKNK--LTNEIILVHRGKCSFTTKANIADEAGASAILIIN 134
HTR +V A+PP+ C + N + ++I LV RG CSF +K + E G A++I +
Sbjct: 57 HTRYEQIHLVPAEPPEACGELSNGFFIQDQIALVERGGCSFLSKTRVVQEHGGRAVIISD 116
Query: 135 --YRTELFKMVCEENETDVDIGIPAVMLPQDAGLNLERHIQNNSI 177
+ F + ++ T IPA+ L G + R ++ + +
Sbjct: 117 NAVDNDSFYVEMIQDSTQRTADIPALFLLGRDGYMIRRSLEQHGL 161
>PP21_DROME (Q9W1W9) PAP21-like protein precursor
Length = 196
Score = 45.1 bits (105), Expect = 5e-04
Identities = 30/97 (30%), Positives = 48/97 (48%), Gaps = 10/97 (10%)
Query: 85 VVMADPPDCCSKPKNK--LTNEIILVHRGKCSFTTKANIADEAGASAILIINYR------ 136
+V+ DPP C + +N L + L+ RG+CSF TK A+ AGA A +I Y
Sbjct: 71 LVITDPPGACQEIRNARDLNGGVALIDRGECSFLTKTLRAEAAGALAAIITEYNPSSPEF 130
Query: 137 TELFKMVCEENETDVDIGIPAVMLPQDAGLNLERHIQ 173
+M+ + ++ D + IPA L G+ + +Q
Sbjct: 131 EHYIEMIHDNSQQDAN--IPAGFLLGKNGVIIRSTLQ 165
>C122_HUMAN (Q9BZQ6) Putative alpha-mannosidase C1orf22 (EC 3.2.1.-)
Length = 889
Score = 37.7 bits (86), Expect = 0.076
Identities = 34/103 (33%), Positives = 49/103 (47%), Gaps = 12/103 (11%)
Query: 67 ARFGPTLESKEKHANHTRVVMADPPDCCSKPKNK--LTNEIILVHRGKCSFTTKANIADE 124
A+FG L SK K V + P + CS+ N + +I L+ RG+C F KA
Sbjct: 625 AQFGLDL-SKHKETRGF-VASSKPSNGCSELTNPEAVMGKIALIQRGQCMFAEKARNIQN 682
Query: 125 AGASAILIINYR-------TELFKMVCEENETDVDIGIPAVML 160
AGA ++I+ LF+M + +TD DI IP + L
Sbjct: 683 AGAIGGIVIDDNEGSSSDTAPLFQMAGDGKDTD-DIKIPMLFL 724
>LSS2_MOUSE (Q924Z4) LAG1 longevity assurance homolog 2
(Translocating chain-associating membrane protein
homolog 3) (TRAM homolog 3)
Length = 380
Score = 36.6 bits (83), Expect = 0.17
Identities = 26/90 (28%), Positives = 40/90 (43%), Gaps = 4/90 (4%)
Query: 191 DVAEVFLWLMAVGTILCASYWSAWTAREAAIEQEKLLKDASDEYVAESVGSRGYVEISTT 250
D E + +A +LC S+++ + I L DASD Y+ ES Y T
Sbjct: 205 DFKEQIIHHVATIILLCFSWFANYVRAGTLI---MALHDASD-YLLESAKMFNYAGWKNT 260
Query: 251 AAILFVVLASCFLVMLYKLMSFWFLEVLVV 280
LF+V A F++ +M FW L ++
Sbjct: 261 CNNLFIVFAIVFIITRLVIMPFWILHCTMI 290
>R152_MOUSE (Q8BG47) RING finger protein 152
Length = 203
Score = 32.7 bits (73), Expect = 2.5
Identities = 21/62 (33%), Positives = 29/62 (45%)
Query: 205 ILCASYWSAWTAREAAIEQEKLLKDASDEYVAESVGSRGYVEISTTAAILFVVLASCFLV 264
+L S + T EQ+ L A E V E RG V+ ST + + V+L +C LV
Sbjct: 121 LLPGSQQKSLTVVTIPAEQQPLQGGAPPEAVEEEPDRRGVVKSSTWSGVCTVILVACVLV 180
Query: 265 ML 266
L
Sbjct: 181 FL 182
>R152_HUMAN (Q8N8N0) RING finger protein 152
Length = 203
Score = 32.7 bits (73), Expect = 2.5
Identities = 18/45 (40%), Positives = 24/45 (53%)
Query: 222 EQEKLLKDASDEYVAESVGSRGYVEISTTAAILFVVLASCFLVML 266
EQ+ L A E V E RG V+ ST + + V+L +C LV L
Sbjct: 138 EQQPLQGGAPQEAVEEEQDRRGVVKSSTWSGVCTVILVACVLVFL 182
>APE3_YEAST (P37302) Aminopeptidase Y precursor (EC 3.4.11.15)
Length = 537
Score = 32.7 bits (73), Expect = 2.5
Identities = 14/36 (38%), Positives = 22/36 (60%)
Query: 97 PKNKLTNEIILVHRGKCSFTTKANIADEAGASAILI 132
P +I L+ RGKC F K+N+A + G +A++I
Sbjct: 197 PPRHNEKQIALIERGKCPFGDKSNLAGKFGFTAVVI 232
>VGLM_PUUMS (P27312) M polyprotein precursor [Contains: Glycoprotein
G1; Glycoprotein G2]
Length = 1148
Score = 32.3 bits (72), Expect = 3.2
Identities = 16/46 (34%), Positives = 20/46 (42%)
Query: 278 LVVLFCIGGIEGLQTCLTALLSCFRWFQYPAQTYVKIPFFGAVPYL 323
L V C+ G+ G T L L CF W P T + + A YL
Sbjct: 480 LAVELCVPGLHGWATVLLLLTFCFGWVLIPTITMILLKILIAFAYL 525
>VGLM_PUUMP (P41266) M polyprotein precursor [Contains: Glycoprotein
G1; Glycoprotein G2]
Length = 1148
Score = 32.3 bits (72), Expect = 3.2
Identities = 16/46 (34%), Positives = 20/46 (42%)
Query: 278 LVVLFCIGGIEGLQTCLTALLSCFRWFQYPAQTYVKIPFFGAVPYL 323
L V C+ G+ G T L L CF W P T + + A YL
Sbjct: 480 LAVELCVPGLHGWATMLLLLTFCFGWVLIPTITMILLKILIAFAYL 525
>VGLM_PUUMK (P41265) M polyprotein precursor [Contains: Glycoprotein
G1; Glycoprotein G2]
Length = 1148
Score = 32.3 bits (72), Expect = 3.2
Identities = 16/46 (34%), Positives = 20/46 (42%)
Query: 278 LVVLFCIGGIEGLQTCLTALLSCFRWFQYPAQTYVKIPFFGAVPYL 323
L V C+ G+ G T L L CF W P T + + A YL
Sbjct: 480 LAVELCVPGLHGWATMLLLLTLCFGWVLIPTITMILLKILIAFAYL 525
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.327 0.142 0.447
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 62,864,696
Number of Sequences: 164201
Number of extensions: 2641633
Number of successful extensions: 7304
Number of sequences better than 10.0: 29
Number of HSP's better than 10.0 without gapping: 21
Number of HSP's successfully gapped in prelim test: 8
Number of HSP's that attempted gapping in prelim test: 7234
Number of HSP's gapped (non-prelim): 35
length of query: 540
length of database: 59,974,054
effective HSP length: 115
effective length of query: 425
effective length of database: 41,090,939
effective search space: 17463649075
effective search space used: 17463649075
T: 11
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.7 bits)
S2: 68 (30.8 bits)
Medicago: description of AC146341.6


