

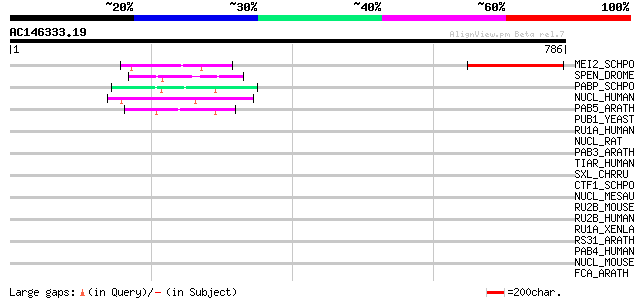
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146333.19 - phase: 0 /pseudo
(786 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
MEI2_SCHPO (P08965) Meiosis protein mei2 138 5e-32
SPEN_DROME (Q8SX83) Split ends protein 49 7e-05
PABP_SCHPO (P31209) Polyadenylate-binding protein (Poly(A)-bindi... 47 2e-04
NUCL_HUMAN (P19338) Nucleolin (Protein C23) 47 2e-04
PAB5_ARATH (Q05196) Polyadenylate-binding protein 5 (Poly(A)-bin... 47 3e-04
PUB1_YEAST (P32588) Nuclear and cytoplasmic polyadenylated RNA-b... 44 0.002
RU1A_HUMAN (P09012) U1 small nuclear ribonucleoprotein A (U1 snR... 44 0.002
NUCL_RAT (P13383) Nucleolin (Protein C23) 44 0.002
PAB3_ARATH (O64380) Polyadenylate-binding protein 3 (Poly(A)-bin... 43 0.003
TIAR_HUMAN (Q01085) Nucleolysin TIAR (TIA-1 related protein) 42 0.005
SXL_CHRRU (O97018) Sex-lethal protein homolog 42 0.005
CTF1_SCHPO (O43040) Cleavage and termination factor 1 (Transcrip... 42 0.005
NUCL_MESAU (P08199) Nucleolin (Protein C23) 42 0.006
RU2B_MOUSE (Q9CQI7) U2 small nuclear ribonucleoprotein B" 42 0.008
RU2B_HUMAN (P08579) U2 small nuclear ribonucleoprotein B" 42 0.008
RU1A_XENLA (P45429) U1 small nuclear ribonucleoprotein A (U1 snR... 42 0.008
RS31_ARATH (P92964) Arginine/serine-rich splicing factor RSP31 42 0.008
PAB4_HUMAN (Q13310) Polyadenylate-binding protein 4 (Poly(A)-bin... 42 0.008
NUCL_MOUSE (P09405) Nucleolin (Protein C23) 42 0.008
FCA_ARATH (O04425) Flowering time control protein FCA 41 0.014
>MEI2_SCHPO (P08965) Meiosis protein mei2
Length = 750
Score = 138 bits (348), Expect = 5e-32
Identities = 69/136 (50%), Positives = 89/136 (64%)
Query: 649 RYELDIDCIKRGEDNRTTLMIKNIPNKYTSKMLLAAIDEHHKGAYDFVYLPIDFRNKCNV 708
R +D I G D RTT+MIKNIPNK+T +ML ID +KG YDF+YL IDF NKCNV
Sbjct: 581 RNSVDYAQIASGIDTRTTVMIKNIPNKFTQQMLRDYIDVTNKGTYDFLYLRIDFVNKCNV 640
Query: 709 GYAFINMTSPSLIVPFYQGFNGKKWEKFNSEKVASLAYARIQGKAALVAHFQNSSLMNED 768
GYAFIN P I+ F + G +W F+SEK+ ++YA IQGK L+ F+NS +M+E+
Sbjct: 641 GYAFINFIEPQSIITFGKARVGTQWNVFHSEKICDISYANIQGKDRLIEKFRNSCVMDEN 700
Query: 769 KRCRPILIDTDGPNAG 784
RP + + GPN G
Sbjct: 701 PAYRPKIFVSHGPNRG 716
Score = 58.2 bits (139), Expect = 8e-08
Identities = 53/177 (29%), Positives = 84/177 (46%), Gaps = 20/177 (11%)
Query: 157 ISGVRNSSIAGENS---YGEHPSRTLFVRNIDSDVKDSVLKALFEQFGDIHTFD-RTCKH 212
++G R+SS+ + H SR LFV N+ V + L LF + GD+ D +
Sbjct: 172 VTGFRSSSLNSNSDDIDIFSHASRYLFVTNLPRIVPYATLLELFSKLGDVKGIDTSSLST 231
Query: 213 QGSAMISYYDIRAAQNAMRALNNRLFGRKKFDIHYPIPKDSPSRNGVNQGTLEVFLYD-- 270
G +++++DIR A A ++L ++ F + +++ + S + +NQG FL D
Sbjct: 232 DGICIVAFFDIRQAIQAAKSLRSQRFFNDRL-LYFQFCQRSSIQKMINQGATIQFLDDNE 290
Query: 271 ----------SSISNTELQHILNVYGGIKEIHENPRSQRHKLI--EFYDFRAADAAL 315
S +S +LQ IL +G + I + RSQ I EFYD R A AL
Sbjct: 291 GQLLLNMQGGSVLSILQLQSILQTFGPLL-IMKPLRSQNVSQIICEFYDTRDASFAL 346
>SPEN_DROME (Q8SX83) Split ends protein
Length = 5560
Score = 48.5 bits (114), Expect = 7e-05
Identities = 42/168 (25%), Positives = 77/168 (45%), Gaps = 18/168 (10%)
Query: 169 NSYGEHPSRTLFVRNIDSDVKDSVLKALFEQFGDIHTFDRTCKHQG---SAMISYYDIRA 225
+ Y +RTLF+ N++ D+ L++ FE FG+I D K QG A Y DI +
Sbjct: 648 DEYHPKSTRTLFIGNLEKDITAGELRSHFEAFGEIIEID--IKKQGLNAYAFCQYSDIVS 705
Query: 226 AQNAMRALNNRLFGRKKFDIHY--PIPKDSPSRNGVNQGTLEVFLYDSSISNTELQHILN 283
AMR ++ G + + + +P + +GV D +S + LQ
Sbjct: 706 VVKAMRKMDGEHLGSNRIKLGFGKSMPTNCVWIDGV----------DEKVSESFLQSQFT 755
Query: 284 VYGGIKEIHENPRSQRHKLIEFYDFRAADAALHGINRNDTTMKRLKVD 331
+G + ++ + R+++ L+ + + A AA+ + K+L+VD
Sbjct: 756 RFGAVTKVSID-RNRQLALVLYDQVQNAQAAVKDMRGTILRRKKLQVD 802
>PABP_SCHPO (P31209) Polyadenylate-binding protein (Poly(A)-binding
protein) (PABP)
Length = 653
Score = 47.0 bits (110), Expect = 2e-04
Identities = 45/216 (20%), Positives = 88/216 (39%), Gaps = 13/216 (6%)
Query: 145 ENPSSIERNSEIISGVRNSSIAGENSYGEHPSRTLFVRNIDSDVKDSVLKALFEQFGDIH 204
E P + SE S N+S S S +L+V +D V +++L LF G +
Sbjct: 48 EEPEAAAEPSESTSTPTNASSVATPSGTAPTSASLYVGELDPSVTEAMLFELFNSIGPVA 107
Query: 205 TFDRTCKHQ------GSAMISYYDIRAAQNAMRALNNRLFGRKKFDIHYPIPKDSPSRNG 258
+ R C+ G A ++++++ + A+ LN L + I + + PS
Sbjct: 108 SI-RVCRDAVTRRSLGYAYVNFHNMEDGEKALDELNYTLIKGRPCRIMW--SQRDPSLRK 164
Query: 259 VNQGTLEVFLYDSSISNTELQHILNVYGGIKE----IHENPRSQRHKLIEFYDFRAADAA 314
+ G + + D +I N L + +G I + E ++ + + F +A+AA
Sbjct: 165 MGTGNVFIKNLDPAIDNKALHDTFSAFGKILSCKVAVDELGNAKGYGFVHFDSVESANAA 224
Query: 315 LHGINRNDTTMKRLKVDQMQSTNSESNIIQPMHPEF 350
+ +N K++ V S + ++ + F
Sbjct: 225 IEHVNGMLLNDKKVYVGHHVSRRERQSKVEALKANF 260
Score = 39.3 bits (90), Expect = 0.041
Identities = 19/68 (27%), Positives = 39/68 (56%), Gaps = 4/68 (5%)
Query: 179 LFVRNIDSDVKDSVLKALFEQFGDIHTF----DRTCKHQGSAMISYYDIRAAQNAMRALN 234
++++N+D+++ + LF QFG+I + D+ K +G ++Y + AQ A+ LN
Sbjct: 263 VYIKNLDTEITEQEFSDLFGQFGEITSLSLVKDQNDKPRGFGFVNYANHECAQKAVDELN 322
Query: 235 NRLFGRKK 242
++ + KK
Sbjct: 323 DKEYKGKK 330
Score = 36.6 bits (83), Expect = 0.26
Identities = 26/95 (27%), Positives = 38/95 (39%), Gaps = 7/95 (7%)
Query: 151 ERNSEIISGVRNSSIAGENSYGEHPSRTLFVRNIDSDVKDSVLKALFEQFGDIHT----F 206
ER E+ + N Y LF++N+ +V D LKA F FG I +
Sbjct: 341 EREEELRKRYEQMKLEKMNKY---QGVNLFIKNLQDEVDDERLKAEFSAFGTITSAKIMT 397
Query: 207 DRTCKHQGSAMISYYDIRAAQNAMRALNNRLFGRK 241
D K +G + Y A A+ +N R+ K
Sbjct: 398 DEQGKSKGFGFVCYTTPEEANKAVTEMNQRMLAGK 432
>NUCL_HUMAN (P19338) Nucleolin (Protein C23)
Length = 706
Score = 47.0 bits (110), Expect = 2e-04
Identities = 50/222 (22%), Positives = 90/222 (40%), Gaps = 15/222 (6%)
Query: 139 FDLGDVENPSSIERNSEII------SGVRNSSIAGENSYGEHPSRTLFVRNIDSDVKDSV 192
F D E+ +E+ E+ + ++ G++S E +RTL +N+ V
Sbjct: 348 FGYVDFESAEDLEKALELTGLKVFGNEIKLEKPKGKDSKKERDARTLLAKNLPYKVTQDE 407
Query: 193 LKALFEQFGDIHTFDRTCKHQGSAMISYYDIRAAQNAMRALNNRLFGRKKFDIHYPIPK- 251
LK +FE +I + K +G A I + A+ + ++Y K
Sbjct: 408 LKEVFEDAAEIRLVSKDGKSKGIAYIEFKTEADAEKTFEEKQGTEIDGRSISLYYTGEKG 467
Query: 252 -DSPSRNGVNQ----GTLEVFLYDSSISNTE--LQHILNVYGGIK-EIHENPRSQRHKLI 303
+ R G N + + L + S S TE LQ + IK ++N +S+ + I
Sbjct: 468 QNQDYRGGKNSTWSGESKTLVLSNLSYSATEETLQEVFEKATFIKVPQNQNGKSKGYAFI 527
Query: 304 EFYDFRAADAALHGINRNDTTMKRLKVDQMQSTNSESNIIQP 345
EF F A AL+ N+ + + ++++ S + QP
Sbjct: 528 EFASFEDAKEALNSCNKREIEGRAIRLELQGPRGSPNARSQP 569
Score = 35.8 bits (81), Expect = 0.45
Identities = 36/167 (21%), Positives = 70/167 (41%), Gaps = 4/167 (2%)
Query: 167 GENSYGEHPSRTLFVRNIDSDVKDSVLKALFEQFGDIHT-FDRTCKHQGSAMISYYDIRA 225
G+NS S+TL + N+ + L+ +FE+ I ++ K +G A I +
Sbjct: 475 GKNSTWSGESKTLVLSNLSYSATEETLQEVFEKATFIKVPQNQNGKSKGYAFIEFASFED 534
Query: 226 AQNAMRALNNRLFGRKKFDIHYPIPKDSPSRNGVNQGTLEVFLYDSSISNTELQHILNVY 285
A+ A+ + N R + + P+ SP N +Q + +F+ S TE +
Sbjct: 535 AKEALNSCNKREIEGRAIRLELQGPRGSP--NARSQPSKTLFVKGLSEDTTEETLKESFD 592
Query: 286 GGIK-EIHENPRSQRHKLIEFYDFRAADAALHGINRNDTTMKRLKVD 331
G ++ I + + K F DF + + A + + ++ +D
Sbjct: 593 GSVRARIVTDRETGSSKGFGFVDFNSEEDAKEAMEDGEIDGNKVTLD 639
Score = 33.1 bits (74), Expect = 2.9
Identities = 23/97 (23%), Positives = 40/97 (40%), Gaps = 3/97 (3%)
Query: 167 GENSYGEHPSRTLFVRNIDSDVKDSVLKALFEQFGDIHTFDRTCKHQGSAM-ISYYDIRA 225
G + PS+TLFV+ + D + LK F+ G + T + GS+ + D +
Sbjct: 561 GSPNARSQPSKTLFVKGLSEDTTEETLKESFD--GSVRARIVTDRETGSSKGFGFVDFNS 618
Query: 226 AQNAMRALNNRLFGRKKFDIHYPIPKDSPSRNGVNQG 262
++A A+ + K + + PK G G
Sbjct: 619 EEDAKEAMEDGEIDGNKVTLDWAKPKGEGGFGGRGGG 655
>PAB5_ARATH (Q05196) Polyadenylate-binding protein 5
(Poly(A)-binding protein 5) (PABP 5)
Length = 668
Score = 46.6 bits (109), Expect = 3e-04
Identities = 40/165 (24%), Positives = 71/165 (42%), Gaps = 10/165 (6%)
Query: 163 SSIAGENSYGEHPSRTLFVRNIDSDVKDSVLKALFEQFGDIHTF----DRTCKHQGSAMI 218
++ A + HP+ +L+V ++D V +S L LF Q +H D T + G A +
Sbjct: 31 AAAAAAEALQTHPNSSLYVGDLDPSVNESHLLDLFNQVAPVHNLRVCRDLTHRSLGYAYV 90
Query: 219 SYYDIRAAQNAMRALNNRLFGRKKFDIHYPIPKDSPSRNGVNQGTLEVFLYDSSISNTEL 278
++ + A AM +LN + I + PS +G + + D+SI N L
Sbjct: 91 NFANPEDASRAMESLNYAPI--RDRPIRIMLSNRDPSTRLSGKGNVFIKNLDASIDNKAL 148
Query: 279 QHILNVYGGIKE----IHENPRSQRHKLIEFYDFRAADAALHGIN 319
+ +G I + RS+ + ++F A AA+ +N
Sbjct: 149 YETFSSFGTILSCKVAMDVVGRSKGYGFVQFEKEETAQAAIDKLN 193
Score = 39.7 bits (91), Expect = 0.031
Identities = 19/67 (28%), Positives = 37/67 (54%), Gaps = 4/67 (5%)
Query: 179 LFVRNIDSDVKDSVLKALFEQFGDIHTFDRTCKHQGSA----MISYYDIRAAQNAMRALN 234
L+++N+D V D LK +F ++G++ + QG + ++Y + A AM+ +N
Sbjct: 330 LYLKNLDDSVNDEKLKEMFSEYGNVTSCKVMMNSQGLSRGFGFVAYSNPEEALLAMKEMN 389
Query: 235 NRLFGRK 241
++ GRK
Sbjct: 390 GKMIGRK 396
Score = 32.7 bits (73), Expect = 3.8
Identities = 32/136 (23%), Positives = 63/136 (45%), Gaps = 19/136 (13%)
Query: 172 GEHPSRT-LFVRNIDSDVKDSVLKALFEQFGDIHTF----DRTCKHQGSAMISYYDIRAA 226
G PS T ++V+N+ ++ D LK F ++GDI + D++ + +++ AA
Sbjct: 219 GAVPSFTNVYVKNLPKEITDDELKKTFGKYGDISSAVVMKDQSGNSRSFGFVNFVSPEAA 278
Query: 227 QNAMRALNNRLFG---------RKKFD----IHYPIPKDSPSRNGVNQGT-LEVFLYDSS 272
A+ +N G +KK D + ++ SR QG+ L + D S
Sbjct: 279 AVAVEKMNGISLGEDVLYVGRAQKKSDREEELRRKFEQERISRFEKLQGSNLYLKNLDDS 338
Query: 273 ISNTELQHILNVYGGI 288
+++ +L+ + + YG +
Sbjct: 339 VNDEKLKEMFSEYGNV 354
>PUB1_YEAST (P32588) Nuclear and cytoplasmic polyadenylated
RNA-binding protein PUB1 (ARS consensus binding protein
ACBP-60) (Poly(U)-binding protein) (Poly
uridylate-binding protein)
Length = 452
Score = 43.9 bits (102), Expect = 0.002
Identities = 46/221 (20%), Positives = 88/221 (39%), Gaps = 23/221 (10%)
Query: 144 VENPSSIERNSEIISGVRNSSIAGENSYGEH-PS---------------RTLFVRNIDSD 187
VE P+S + +SE V +S E++ GE+ PS R L+V N+D
Sbjct: 25 VEAPASADPSSEQSVAVEGNSEQAEDNQGENDPSVVPANAITGGRETSDRVLYVGNLDKA 84
Query: 188 VKDSVLKALFEQFGDIHT----FDRTCKHQGSAMISYYDIRAAQNAMRALNNRLFGRKKF 243
+ + +LK F+ G I D+ K+ A + Y+ A A++ LN +
Sbjct: 85 ITEDILKQYFQVGGPIANIKIMIDKNNKNVNYAFVEYHQSHDANIALQTLNGKQIENNIV 144
Query: 244 DIHYPIPKDSPSRN---GVNQGTLEVFLYDSSISNTELQHILNVYGGIKEIHENPRSQRH 300
I++ S + + G L V + D ++ N + G + + S+ +
Sbjct: 145 KINWAFQSQQSSSDDTFNLFVGDLNVNVDDETLRNAFKDFPSYLSGHVMWDMQTGSSRGY 204
Query: 301 KLIEFYDFRAADAALHGINRNDTTMKRLKVDQMQSTNSESN 341
+ F A A+ + D + L+++ ++ +N
Sbjct: 205 GFVSFTSQDDAQNAMDSMQGQDLNGRPLRINWAAKRDNNNN 245
>RU1A_HUMAN (P09012) U1 small nuclear ribonucleoprotein A (U1 snRNP
protein A) (U1A protein) (U1-A)
Length = 281
Score = 43.5 bits (101), Expect = 0.002
Identities = 24/79 (30%), Positives = 42/79 (52%), Gaps = 6/79 (7%)
Query: 175 PSRTLFVRNIDSDVKDSVLK----ALFEQFGDIHTF--DRTCKHQGSAMISYYDIRAAQN 228
P+ T+++ N++ +K LK A+F QFG I R+ K +G A + + ++ +A N
Sbjct: 7 PNHTIYINNLNEKIKKDELKKSLYAIFSQFGQILDILVSRSLKMRGQAFVIFKEVSSATN 66
Query: 229 AMRALNNRLFGRKKFDIHY 247
A+R++ F K I Y
Sbjct: 67 ALRSMQGFPFYDKPMRIQY 85
>NUCL_RAT (P13383) Nucleolin (Protein C23)
Length = 712
Score = 43.5 bits (101), Expect = 0.002
Identities = 49/226 (21%), Positives = 93/226 (40%), Gaps = 16/226 (7%)
Query: 135 SNGGFDLGDVENPSSIERNSEII------SGVRNSSIAGENSYGEHPSRTLFVRNIDSDV 188
+N F D E+ +E+ E+ + ++ G +S +RTL +N+ ++
Sbjct: 348 TNRKFGYVDFESAEDLEKALELTGLKVFGNEIKLEKPKGRDSKKVRAARTLLAKNLSFNI 407
Query: 189 KDSVLKALFEQFGDIHTFDRTCKHQGSAMISYYDIRAAQNAMRALNNRLFGRKKFDIHYP 248
+ LK +FE +I + + +G A I + A+ + + ++Y
Sbjct: 408 TEDELKEVFEDAVEIRLVSQDGRSKGIAYIEFKSEADAEKNLEEKQGAEIDGRSVSLYYT 467
Query: 249 IPK-DSPSRNGVNQ----GTLEVFLYDSSISNTE--LQHILNVYGGIKEIHENP--RSQR 299
K R G N + + L + S S TE LQ + IK + +NP +S+
Sbjct: 468 GEKGQRQERTGKNSTWSGESKTLVLSNLSYSATEETLQEVFEKATFIK-VPQNPHGKSKG 526
Query: 300 HKLIEFYDFRAADAALHGINRNDTTMKRLKVDQMQSTNSESNIIQP 345
+ IEF F A AL+ N+ + + ++++ S + QP
Sbjct: 527 YAFIEFASFEDAKEALNSCNKMEIEGRTIRLELQGPRGSPNARSQP 572
>PAB3_ARATH (O64380) Polyadenylate-binding protein 3
(Poly(A)-binding protein 3) (PABP 3)
Length = 660
Score = 43.1 bits (100), Expect = 0.003
Identities = 41/156 (26%), Positives = 71/156 (45%), Gaps = 14/156 (8%)
Query: 174 HPSRTLFVRNIDSDVKDSVLKALFEQFGDIHTFDRTCKHQ-----GSAMISYYDIRAAQN 228
HP+ +L+ ++D V ++ L LF+ ++ + R C+ Q G A I++ + A
Sbjct: 46 HPNSSLYAGDLDPKVTEAHLFDLFKHVANVVSV-RVCRDQNRRSLGYAYINFSNPNDAYR 104
Query: 229 AMRALN-NRLFGRKKFDIHYPIPKDSPSRNGVNQGTLEVFLYDSSISNTELQHILNVYGG 287
AM ALN LF R I + PS +G + + D+SI N L + +G
Sbjct: 105 AMEALNYTPLFDR---PIRIMLSNRDPSTRLSGKGNIFIKNLDASIDNKALFETFSSFGT 161
Query: 288 IKE----IHENPRSQRHKLIEFYDFRAADAALHGIN 319
I + RS+ + ++F +A AA+ +N
Sbjct: 162 ILSCKVAMDVTGRSKGYGFVQFEKEESAQAAIDKLN 197
Score = 39.7 bits (91), Expect = 0.031
Identities = 19/77 (24%), Positives = 39/77 (49%), Gaps = 4/77 (5%)
Query: 169 NSYGEHPSRTLFVRNIDSDVKDSVLKALFEQFGDIHTFDRTCKHQGSA----MISYYDIR 224
N + + L+++N+D V D LK +F ++G++ + QG + ++Y +
Sbjct: 324 NRFEKSQGANLYLKNLDDSVDDEKLKEMFSEYGNVTSSKVMLNPQGMSRGFGFVAYSNPE 383
Query: 225 AAQNAMRALNNRLFGRK 241
A A+ +N ++ GRK
Sbjct: 384 EALRALSEMNGKMIGRK 400
Score = 36.6 bits (83), Expect = 0.26
Identities = 37/184 (20%), Positives = 75/184 (40%), Gaps = 9/184 (4%)
Query: 179 LFVRNIDSDVKDSVLKALFEQFGDIHT----FDRTCKHQGSAMISYYDIRAAQNAMRALN 234
+F++N+D+ + + L F FG I + D T + +G + + +AQ A+ LN
Sbjct: 138 IFIKNLDASIDNKALFETFSSFGTILSCKVAMDVTGRSKGYGFVQFEKEESAQAAIDKLN 197
Query: 235 NRLFGRKKFDIHYPIPKDSPSR--NGVNQGTLEVFLYD--SSISNTELQHILNVYGGIKE 290
L K+ + + I + +R N V++ + I EL+ +G I
Sbjct: 198 GMLMNDKQVFVGHFIRRQERARDENTPTPRFTNVYVKNLPKEIGEDELRKTFGKFGVISS 257
Query: 291 -IHENPRSQRHKLIEFYDFRAADAALHGINRNDTTMKRLKVDQMQSTNSESNIIQPMHPE 349
+ +S + F +F +AA + + + V + +S + + +
Sbjct: 258 AVVMRDQSGNSRCFGFVNFECTEAAASAVEKMNGISLGDDVLYVGRAQKKSEREEELRRK 317
Query: 350 FKQE 353
F+QE
Sbjct: 318 FEQE 321
>TIAR_HUMAN (Q01085) Nucleolysin TIAR (TIA-1 related protein)
Length = 375
Score = 42.4 bits (98), Expect = 0.005
Identities = 34/149 (22%), Positives = 67/149 (44%), Gaps = 12/149 (8%)
Query: 177 RTLFVRNIDSDVKDSVLKALFEQFGDIHTFDRTCKHQGS---AMISYYDIRAAQNAMRAL 233
RTL+V N+ DV + ++ LF Q G + +H + + +Y+ R A A+ A+
Sbjct: 9 RTLYVGNLSRDVTEVLILQLFSQIGPCKSCKMITEHTSNDPYCFVEFYEHRDAAAALAAM 68
Query: 234 NNRLFGRKKFDIHYPIPKDSPSRNGVNQGTLEVFLYDSS--ISNTELQHILNVYGGIKEI 291
N R K+ +++ S ++ N VF+ D S I+ +++ +G I +
Sbjct: 69 NGRKILGKEVKVNWATTPSSQKKDTSNH--FHVFVGDLSPEITTEDIKSAFAPFGKISDA 126
Query: 292 H-----ENPRSQRHKLIEFYDFRAADAAL 315
+S+ + + FY+ A+ A+
Sbjct: 127 RVVKDMATGKSKGYGFVSFYNKLDAENAI 155
Score = 32.7 bits (73), Expect = 3.8
Identities = 39/172 (22%), Positives = 72/172 (41%), Gaps = 28/172 (16%)
Query: 179 LFVRNIDSDVKDSVLKALFEQFGDIHTFD-----RTCKHQGSAMISYYDIRAAQNAMRAL 233
+FV ++ ++ +K+ F FG I T K +G +S+Y+ A+NA+ +
Sbjct: 99 VFVGDLSPEITTEDIKSAFAPFGKISDARVVKDMATGKSKGYGFVSFYNKLDAENAIVHM 158
Query: 234 NNRLFGRKKFDIHY-----PIPKDSPSRNG--------VNQG-----TLEVFLYDSSISN 275
+ G ++ ++ P PK + N VNQ T+ S +++
Sbjct: 159 GGQWLGGRQIRTNWATRKPPAPKSTQENNTKQLRFEDVVNQSSPKNCTVYCGGIASGLTD 218
Query: 276 TELQHILNVYGGIKEIHENPRSQRHKLIEFYDFRAADAALHGI-NRNDTTMK 326
++ + +G I EI P K F F ++A H I + N TT++
Sbjct: 219 QLMRQTFSPFGQIMEIRVFP----EKGYSFVRFSTHESAAHAIVSVNGTTIE 266
>SXL_CHRRU (O97018) Sex-lethal protein homolog
Length = 307
Score = 42.4 bits (98), Expect = 0.005
Identities = 47/192 (24%), Positives = 76/192 (39%), Gaps = 16/192 (8%)
Query: 135 SNGGFDLGDVENPSSIERNSEIISGVRNSSIAGENSYGEHPSRTLFVRNIDSDVKDSVLK 194
S GG +G + N + + + SG G+N L V + D+ D L
Sbjct: 48 SGGGSAMGSMCNMAPAISTNSVNSG---GGDCGDNQGCN--GTNLIVNYLPQDMTDRELY 102
Query: 195 ALFEQFGDIHTFD-----RTCKHQGSAMISYYDIRAAQNAMRALNNRLFGRKKFDIHYPI 249
ALF G I+T +T G A + + AQNA+++LN K+ + Y
Sbjct: 103 ALFRTCGPINTCRIMKDYKTGYSFGYAFVDFASEIDAQNAIKSLNGVTVRNKRLKVSYA- 161
Query: 250 PKDSPSRNGVNQGTLEVFLYDSSISNTELQHILNVYGGI--KEIHENPRSQRHKLIEFYD 307
P + L V +I++ EL+ I YG I K I + + + + + F
Sbjct: 162 ---RPGGESIKDTNLYVTNLPRTITDDELEKIFGKYGNIVQKNILRDKLTGKPRGVAFVR 218
Query: 308 FRAADAALHGIN 319
F + A I+
Sbjct: 219 FNKREEAQEAIS 230
>CTF1_SCHPO (O43040) Cleavage and termination factor 1
(Transcription termination factor ctf1)
Length = 363
Score = 42.4 bits (98), Expect = 0.005
Identities = 25/84 (29%), Positives = 38/84 (44%), Gaps = 7/84 (8%)
Query: 179 LFVRNIDSDVKDSVLKALFEQFGDIHTF-----DRTCKHQGSAMISYYDIRAAQNAMRAL 233
+FV NI DV + + +F Q G + TF T +G ++D A+R L
Sbjct: 9 VFVGNIPYDVSEQQMTEIFNQVGPVKTFKLVLDPETGSGKGYGFCEFFDSETTAMAVRKL 68
Query: 234 NNRLFGRKKFDIHYPIPKDSPSRN 257
NN G +K + + P + P RN
Sbjct: 69 NNSELGPRKIRVEF--PSNDPRRN 90
>NUCL_MESAU (P08199) Nucleolin (Protein C23)
Length = 713
Score = 42.0 bits (97), Expect = 0.006
Identities = 48/225 (21%), Positives = 91/225 (40%), Gaps = 14/225 (6%)
Query: 135 SNGGFDLGDVENPSSIERNSEII------SGVRNSSIAGENSYGEHPSRTLFVRNIDSDV 188
+N F D E+ +E+ E+ + ++ G +S +RTL +N+ ++
Sbjct: 345 TNRKFGYVDFESAEDLEKALELTGLKVFGNEIKLEKPKGRDSKKVRAARTLLAKNLSFNI 404
Query: 189 KDSVLKALFEQFGDIHTFDRTCKHQGSAMISYYDIRAAQNAMRALNNRLFGRKKFDIHYP 248
+ LK +FE +I + K +G A I + A+ + + ++Y
Sbjct: 405 TEDELKEVFEDALEIRLVSQDGKSKGIAYIEFKSEADAEKNLEEKQGAEIDGRSVSLYYT 464
Query: 249 IPK-DSPSRNGVNQ----GTLEVFLYDSSISNTE--LQHILNVYGGIK-EIHENPRSQRH 300
K R G N + + L + S S TE LQ + IK ++ +S+ +
Sbjct: 465 GEKGQRQERTGKNSTWSGESKTLVLSNLSYSATEETLQEVFEKATFIKVPQNQQGKSKGY 524
Query: 301 KLIEFYDFRAADAALHGINRNDTTMKRLKVDQMQSTNSESNIIQP 345
IEF F A AL+ N+ + + ++++ S + QP
Sbjct: 525 AFIEFASFEDAKEALNSCNKMEIEGRTIRLELQGPRGSPNARSQP 569
>RU2B_MOUSE (Q9CQI7) U2 small nuclear ribonucleoprotein B"
Length = 225
Score = 41.6 bits (96), Expect = 0.008
Identities = 26/81 (32%), Positives = 42/81 (51%), Gaps = 10/81 (12%)
Query: 175 PSRTLFVRNIDSDVKDSVLK----ALFEQFGDIHTFD----RTCKHQGSAMISYYDIRAA 226
P+ T+++ N++ +K LK ALF QFG H D +T K +G A + + ++ ++
Sbjct: 5 PNHTIYINNMNDKIKKEELKRSLYALFSQFG--HVVDIVALKTMKMRGQAFVIFKELGSS 62
Query: 227 QNAMRALNNRLFGRKKFDIHY 247
NA+R L F K I Y
Sbjct: 63 TNALRQLQGFPFYGKPMRIQY 83
>RU2B_HUMAN (P08579) U2 small nuclear ribonucleoprotein B"
Length = 225
Score = 41.6 bits (96), Expect = 0.008
Identities = 26/81 (32%), Positives = 42/81 (51%), Gaps = 10/81 (12%)
Query: 175 PSRTLFVRNIDSDVKDSVLK----ALFEQFGDIHTFD----RTCKHQGSAMISYYDIRAA 226
P+ T+++ N++ +K LK ALF QFG H D +T K +G A + + ++ ++
Sbjct: 5 PNHTIYINNMNDKIKKEELKRSLYALFSQFG--HVVDIVALKTMKMRGQAFVIFKELGSS 62
Query: 227 QNAMRALNNRLFGRKKFDIHY 247
NA+R L F K I Y
Sbjct: 63 TNALRQLQGFPFYGKPMRIQY 83
>RU1A_XENLA (P45429) U1 small nuclear ribonucleoprotein A (U1 snRNP
protein A) (U1A protein) (U1-A)
Length = 282
Score = 41.6 bits (96), Expect = 0.008
Identities = 24/79 (30%), Positives = 40/79 (50%), Gaps = 6/79 (7%)
Query: 175 PSRTLFVRNIDSDVKDSVLK----ALFEQFGDIHTF--DRTCKHQGSAMISYYDIRAAQN 228
P+ T+++ N++ +K LK A+F QFG I R K +G A + + + +A N
Sbjct: 8 PNNTIYINNLNEKIKKDELKKSLYAIFSQFGQILDILVSRNLKMRGQAFVIFKETSSATN 67
Query: 229 AMRALNNRLFGRKKFDIHY 247
A+R++ F K I Y
Sbjct: 68 ALRSMQGFPFYDKPMRIQY 86
>RS31_ARATH (P92964) Arginine/serine-rich splicing factor RSP31
Length = 264
Score = 41.6 bits (96), Expect = 0.008
Identities = 23/66 (34%), Positives = 39/66 (58%), Gaps = 3/66 (4%)
Query: 177 RTLFVRNIDSDVKDSVLKALFEQFGDIHTFDRTCKHQGSAMISYYDIRAAQNAMRALNNR 236
R +FV N + + + S L+ LF+++G + DR G A + + D R A++A+R L+N
Sbjct: 2 RPVFVGNFEYETRQSDLERLFDKYGRV---DRVDMKSGYAFVYFEDERDAEDAIRKLDNF 58
Query: 237 LFGRKK 242
FG +K
Sbjct: 59 PFGYEK 64
>PAB4_HUMAN (Q13310) Polyadenylate-binding protein 4
(Poly(A)-binding protein 4) (PABP 4) (Inducible
poly(A)-binding protein) (iPABP) (Activated-platelet
protein-1) (APP-1)
Length = 644
Score = 41.6 bits (96), Expect = 0.008
Identities = 37/155 (23%), Positives = 69/155 (43%), Gaps = 12/155 (7%)
Query: 174 HPSRTLFVRNIDSDVKDSVLKALFEQFGDIHTFDRTCKHQ------GSAMISYYDIRAAQ 227
+P +L+V ++ SDV +++L F G + + R C+ G A +++ A+
Sbjct: 8 YPMASLYVGDLHSDVTEAMLYEKFSPAGPVLSI-RVCRDMITRRSLGYAYVNFQQPADAE 66
Query: 228 NAMRALNNRLFGRKKFDIHYPIPKDSPSRNGVNQGTLEVFLYDSSISNTELQHILNVYGG 287
A+ +N + K I + S ++GV G + + D SI N L + +G
Sbjct: 67 RALDTMNFDVIKGKPIRIMWSQRDPSLRKSGV--GNVFIKNLDKSIDNKALYDTFSAFGN 124
Query: 288 I---KEIHENPRSQRHKLIEFYDFRAADAALHGIN 319
I K + + S+ + + F AAD A+ +N
Sbjct: 125 ILSCKVVCDENGSKGYAFVHFETQEAADKAIEKMN 159
Score = 40.0 bits (92), Expect = 0.024
Identities = 46/234 (19%), Positives = 97/234 (40%), Gaps = 18/234 (7%)
Query: 138 GFDLGDVENPSSIER-----NSEIISG--VRNSSIAGENSYGEHPSRTLFVRNIDSDVKD 190
G+ + + P+ ER N ++I G +R + S + +F++N+D + +
Sbjct: 53 GYAYVNFQQPADAERALDTMNFDVIKGKPIRIMWSQRDPSLRKSGVGNVFIKNLDKSIDN 112
Query: 191 SVLKALFEQFGDIHTFDRTCKHQGS---AMISYYDIRAAQNAMRALNNRLFG-RKKFDIH 246
L F FG+I + C GS A + + AA A+ +N L RK F
Sbjct: 113 KALYDTFSAFGNILSCKVVCDENGSKGYAFVHFETQEAADKAIEKMNGMLLNDRKVFVGR 172
Query: 247 YPIPKDSPSRNGVN-QGTLEVFL--YDSSISNTELQHILNVYG---GIKEIHE-NPRSQR 299
+ K+ + G + V++ + + + L+ + + +G +K + + N +S+
Sbjct: 173 FKSRKEREAELGAKAKEFTNVYIKNFGEEVDDESLKELFSQFGKTLSVKVMRDPNGKSKG 232
Query: 300 HKLIEFYDFRAADAALHGINRNDTTMKRLKVDQMQSTNSESNIIQPMHPEFKQE 353
+ + A+ A+ +N + + K + V + Q ++ + KQE
Sbjct: 233 FGFVSYEKHEDANKAVEEMNGKEISGKIIFVGRAQKKVERQAELKRKFEQLKQE 286
Score = 36.6 bits (83), Expect = 0.26
Identities = 46/216 (21%), Positives = 85/216 (39%), Gaps = 20/216 (9%)
Query: 57 SKKLRLSANNAFYGHSVDTIASNYEEEKLSDSLEELEAQIIGNLLPDEDDLLSGVTDGNN 116
S+K R + A + N+ EE +SL+EL +Q L +G + G
Sbjct: 175 SRKEREAELGAKAKEFTNVYIKNFGEEVDDESLKELFSQFGKTLSVKVMRDPNGKSKGFG 234
Query: 117 YIICDSNGDD---IDELDLFSSNG-----GFDLGDVENPSSIERNSEIISGVRNSSIAGE 168
++ + + D ++E++ +G G VE + ++R E + R S G
Sbjct: 235 FVSYEKHEDANKAVEEMNGKEISGKIIFVGRAQKKVERQAELKRKFEQLKQERISRYQGV 294
Query: 169 NSYGEHPSRTLFVRNIDSDVKDSVLKALFEQFGDIHTFD---RTCKHQGSAMISYYDIRA 225
N L+++N+D + D L+ F FG I + + +G + +
Sbjct: 295 N---------LYIKNLDDTIDDEKLRKEFSPFGSITSAKVMLEDGRSKGFGFVCFSSPEE 345
Query: 226 AQNAMRALNNRLFGRKKFDIHYPIPKDSPSRNGVNQ 261
A A+ +N R+ G K + K+ + NQ
Sbjct: 346 ATKAVTEMNGRIVGSKPLYVALAQRKEERKAHLTNQ 381
>NUCL_MOUSE (P09405) Nucleolin (Protein C23)
Length = 706
Score = 41.6 bits (96), Expect = 0.008
Identities = 46/221 (20%), Positives = 90/221 (39%), Gaps = 17/221 (7%)
Query: 135 SNGGFDLGDVENPSSIERNSEII------SGVRNSSIAGENSYGEHPSRTLFVRNIDSDV 188
+N F D E+ +E+ E+ + ++ G +S +RTL +N+ ++
Sbjct: 346 TNRKFGYVDFESAEDLEKALELTGLKVFGNEIKLEKPKGRDSKKVRAARTLLAKNLSFNI 405
Query: 189 KDSVLKALFEQFGDIHTFDRTCKHQGSAMISYYDIRAAQNAMRALNNRLFGRKKFDIHYP 248
+ LK +FE +I + K +G A I + A+ + + ++Y
Sbjct: 406 TEDELKEVFEDAMEIRLVSQDGKSKGIAYIEFKSEADAEKNLEEKQGAEIDGRSVSLYYT 465
Query: 249 IPK-DSPSRNGV------NQGTLEVFLYDSSISNTELQHILNVYGGIKEIHENP--RSQR 299
K R G TL + S + L+ + IK + +NP + +
Sbjct: 466 GEKGQRQERTGKTSTWSGESKTLVLSNLSYSATKETLEEVFEKATFIK-VPQNPHGKPKG 524
Query: 300 HKLIEFYDFRAADAALHGINRNDTTMKRLKVDQMQSTNSES 340
+ IEF F A AL+ N+ + + +++ ++Q +NS S
Sbjct: 525 YAFIEFASFEDAKEALNSCNKMEIEGRTIRL-ELQGSNSRS 564
Score = 32.3 bits (72), Expect = 5.0
Identities = 26/100 (26%), Positives = 40/100 (40%), Gaps = 3/100 (3%)
Query: 165 IAGENSYGEHPSRTLFVRNIDSDVKDSVLKALFEQF--GDIHTFDRTCKHQGSAMISYYD 222
+ G NS + PS+TLFV+ + D + LK FE I T T +G + +
Sbjct: 557 LQGSNSRSQ-PSKTLFVKGLSEDTTEETLKESFEGSVRARIVTDRETGSSKGFGFVDFNS 615
Query: 223 IRAAQNAMRALNNRLFGRKKFDIHYPIPKDSPSRNGVNQG 262
A+ A A+ + K + + PK G G
Sbjct: 616 EEDAKAAKEAMEDGEIDGNKVTLDWAKPKGEGGFGGRGGG 655
>FCA_ARATH (O04425) Flowering time control protein FCA
Length = 747
Score = 40.8 bits (94), Expect = 0.014
Identities = 44/192 (22%), Positives = 84/192 (42%), Gaps = 24/192 (12%)
Query: 144 VENPSSIERNSEIISGVRNSSIAGENSYGEHPSRTLFVRNIDSDVKDSVLKALFEQFGDI 203
V+ P S ++ IS + S G + + LFV ++ + ++ FEQ G++
Sbjct: 89 VQQPLSGQKRGYPISD--HGSFTGTDVSDRSSTVKLFVGSVPRTATEEEIRPYFEQHGNV 146
Query: 204 HTF-----DRTCKHQGSAMISYYDIRAAQNAMRALNNRLF---GRKKFDIHYPIPKDSPS 255
RT + QG + Y + A A+RAL+N++ G + Y
Sbjct: 147 LEVALIKDKRTGQQQGCCFVKYATSKDADRAIRALHNQITLPGGTGPVQVRYA----DGE 202
Query: 256 RNGVNQGTLEVFLYDSSI----SNTELQHILNVYGGIKEIH----ENPRSQRHKLIEFYD 307
R + GTLE L+ S+ + E++ I +G +++++ E +S+ +++
Sbjct: 203 RERI--GTLEFKLFVGSLNKQATEKEVEEIFLQFGHVEDVYLMRDEYRQSRGCGFVKYSS 260
Query: 308 FRAADAALHGIN 319
A AA+ G+N
Sbjct: 261 KETAMAAIDGLN 272
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.324 0.139 0.419
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 92,968,325
Number of Sequences: 164201
Number of extensions: 4014337
Number of successful extensions: 12410
Number of sequences better than 10.0: 141
Number of HSP's better than 10.0 without gapping: 16
Number of HSP's successfully gapped in prelim test: 125
Number of HSP's that attempted gapping in prelim test: 12235
Number of HSP's gapped (non-prelim): 267
length of query: 786
length of database: 59,974,054
effective HSP length: 118
effective length of query: 668
effective length of database: 40,598,336
effective search space: 27119688448
effective search space used: 27119688448
T: 11
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 70 (31.6 bits)
Medicago: description of AC146333.19


