

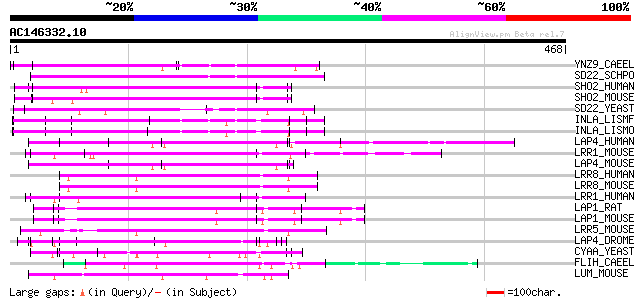
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146332.10 - phase: 0
(468 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
YNZ9_CAEEL (P45969) Hypothetical protein T09A5.9 in chromosome III 97 7e-20
SD22_SCHPO (P22194) Protein phosphatases PP1 regulatory subunit ... 90 1e-17
SHO2_HUMAN (Q9UQ13) Leucine-rich repeat protein SHOC-2 (Ras-bind... 83 2e-15
SHO2_MOUSE (O88520) Leucine-rich repeat protein SHOC-2 (Ras-bind... 82 3e-15
SD22_YEAST (P36047) Protein phosphatases PP1 regulatory subunit ... 78 6e-14
INLA_LISMF (Q723K6) Internalin A precursor 75 3e-13
INLA_LISMO (P25146) Internalin A precursor 75 5e-13
LAP4_HUMAN (Q14160) LAP4 protein (Scribble homolog protein) (hSc... 73 2e-12
LRR1_MOUSE (Q80VQ1) Leucine-rich repeat-containing protein 1 71 5e-12
LAP4_MOUSE (Q80U72) LAP4 protein (Scribble homolog protein) 71 7e-12
LRR8_HUMAN (Q8IWT6) Leucine-rich repeat-containing protein 8 pre... 69 3e-11
LRR8_MOUSE (Q80WG5) Leucine-rich repeat-containing protein 8 pre... 68 6e-11
LRR1_HUMAN (Q9BTT6) Leucine-rich repeat-containing protein 1 (LA... 68 6e-11
LAP1_RAT (P70587) LAP1 protein (Densin-180) 67 8e-11
LAP1_MOUSE (Q80TE7) LAP1 protein (Densin-180) 67 8e-11
LRR5_MOUSE (Q8BGR2) Leucine-rich repeat-containing protein 5 pre... 67 1e-10
LAP4_DROME (Q7KRY7) LAP4 protein (Scribble protein) (Smell-impai... 66 2e-10
CYAA_YEAST (P08678) Adenylate cyclase (EC 4.6.1.1) (ATP pyrophos... 66 2e-10
FLIH_CAEEL (P34268) Flightless-I protein homolog 66 2e-10
LUM_MOUSE (P51885) Lumican precursor (Keratan sulfate proteoglyc... 65 3e-10
>YNZ9_CAEEL (P45969) Hypothetical protein T09A5.9 in chromosome III
Length = 326
Score = 97.4 bits (241), Expect = 7e-20
Identities = 71/248 (28%), Positives = 133/248 (53%), Gaps = 9/248 (3%)
Query: 20 SISSLHLTHKALSDVSCLASFNKLEKLDLKFNNLTSLEGLRACVTLKWLSVVENKLESLE 79
+++SL L L+++S L S L LDL +N + + GL L+ L +V NK+E +E
Sbjct: 82 TLTSLDLYENQLTEISHLESLVNLVSLDLSYNRIRQINGLDKLTKLETLYLVSNKIEKIE 141
Query: 80 GIQGLTKLTVLNAGKNKLKSMDEIGSLSTIRALILNDNEIVSICNLDQMKELNTLVLSKN 139
++ LT+L +L G N++K ++ IG L + L + N+I + ++ +++L+ L L N
Sbjct: 142 NLEALTQLKLLELGDNRIKKIENIGHLVNLDELFIGKNKIRQLEGVETLQKLSVLSLPGN 201
Query: 140 PIRKIGEALKKVKSITKLSLSHCQLEGIDTSLKFCVELTELRLAHNDIKSLPEELMHNSK 199
I KI E ++++ ++ +L LS L+ I ++ L L +A+N+IK+ +
Sbjct: 202 RIVKI-ENVEQLNNLKELYLSDQGLQDIH-GVEPLTNLLLLDVANNEIKTF-SGVERLES 258
Query: 200 LRNLDLGNNVIAKWSDIKVLKSLTKLRNLNLQGNPVATNE--KVIRKIKNALPKLQVFNA 257
L + +N + +S+I+ L L L+ + L+ NP N+ + RK+ L ++ +A
Sbjct: 259 LNDFWANDNKVESFSEIEQLSKLKGLQTVYLERNPFYFNDTNQYRRKVMMTLTQVTQIDA 318
Query: 258 ----KPID 261
KPI+
Sbjct: 319 TTCRKPIE 326
Score = 61.2 bits (147), Expect = 5e-09
Identities = 34/139 (24%), Positives = 73/139 (52%)
Query: 4 LSSEQVLKDNNAVNPSSISSLHLTHKALSDVSCLASFNKLEKLDLKFNNLTSLEGLRACV 63
LS ++ + N + + +L+L + + L + +L+ L+L N + +E + V
Sbjct: 110 LSYNRIRQINGLDKLTKLETLYLVSNKIEKIENLEALTQLKLLELGDNRIKKIENIGHLV 169
Query: 64 TLKWLSVVENKLESLEGIQGLTKLTVLNAGKNKLKSMDEIGSLSTIRALILNDNEIVSIC 123
L L + +NK+ LEG++ L KL+VL+ N++ ++ + L+ ++ L L+D + I
Sbjct: 170 NLDELFIGKNKIRQLEGVETLQKLSVLSLPGNRIVKIENVEQLNNLKELYLSDQGLQDIH 229
Query: 124 NLDQMKELNTLVLSKNPIR 142
++ + L L ++ N I+
Sbjct: 230 GVEPLTNLLLLDVANNEIK 248
Score = 58.5 bits (140), Expect = 3e-08
Identities = 33/143 (23%), Positives = 68/143 (47%), Gaps = 3/143 (2%)
Query: 1 MTRLSSEQVLKDNNAVNPSSISSLHLTHKALSDVSCLASFNKLEKLDLKFNNLTSLEGLR 60
+ L ++ K N + ++ L + + + + + KL L L N + +E +
Sbjct: 151 LLELGDNRIKKIENIGHLVNLDELFIGKNKIRQLEGVETLQKLSVLSLPGNRIVKIENVE 210
Query: 61 ACVTLKWLSVVENKLESLEGIQGLTKLTVLNAGKNKLKSMDEIGSLSTIRALILNDNEIV 120
LK L + + L+ + G++ LT L +L+ N++K+ + L ++ NDN++
Sbjct: 211 QLNNLKELYLSDQGLQDIHGVEPLTNLLLLDVANNEIKTFSGVERLESLNDFWANDNKVE 270
Query: 121 SICNLDQ---MKELNTLVLSKNP 140
S ++Q +K L T+ L +NP
Sbjct: 271 SFSEIEQLSKLKGLQTVYLERNP 293
>SD22_SCHPO (P22194) Protein phosphatases PP1 regulatory subunit
sds22
Length = 332
Score = 90.1 bits (222), Expect = 1e-17
Identities = 68/251 (27%), Positives = 126/251 (50%), Gaps = 5/251 (1%)
Query: 18 PSSISSLHLTHKALSDVSCLASFNKLEKLDLKFNNLTSLEGLRACVTLKWLSVVENKLES 77
P +++ L L + + L + L LDL FNN+ ++ + L+ L V+N++
Sbjct: 81 PETLTELDLYDNLIVRIENLDNVKNLTYLDLSFNNIKTIRNINHLKGLENLFFVQNRIRR 140
Query: 78 LEGIQGLTKLTVLNAGKNKLKSMDEIGSLSTIRALILNDNEIVSICNLDQMKELNTLVLS 137
+E ++GL +LT L G NK++ ++ + +L + L + N+I N +++++L+ L +
Sbjct: 141 IENLEGLDRLTNLELGGNKIRVIENLDTLVNLEKLWVGKNKITKFENFEKLQKLSLLSIQ 200
Query: 138 KNPIRKIGEALKKVKSITKLSLSHCQLEGIDTSLKFCVELTELRLAHNDIKSLPEELMHN 197
N I + + +L +SH L + ++ L L +++N IK L L
Sbjct: 201 SNRITQFENLACLSHCLRELYVSHNGLTSF-SGIEVLENLEILDVSNNMIKHL-SYLAGL 258
Query: 198 SKLRNLDLGNNVIAKWSDIK-VLKSLTKLRNLNLQGNPV-ATNEKVIR-KIKNALPKLQV 254
L L NN ++ + +I+ L L KL + +GNP+ TN V R K++ LP+L+
Sbjct: 259 KNLVELWASNNELSSFQEIEDELSGLKKLETVYFEGNPLQKTNPAVYRNKVRLCLPQLRQ 318
Query: 255 FNAKPIDKDTK 265
+A I K +K
Sbjct: 319 IDATIIPKTSK 329
Score = 36.6 bits (83), Expect = 0.14
Identities = 38/160 (23%), Positives = 59/160 (36%), Gaps = 44/160 (27%)
Query: 12 DNNAVNPSSISSLHLTHKALSDVSCLASFNKLEKLDLKFNNLTSLEGLRACVTLKWLSVV 71
+N A + L+++H N LTS G+ L+ L V
Sbjct: 208 ENLACLSHCLRELYVSH----------------------NGLTSFSGIEVLENLEILDVS 245
Query: 72 ENKLESLEGIQGLTKLTVLNAGKNKLKSMDEIGSLSTIRALILNDNEIVSICNLDQMKEL 131
N ++ L + GL L L A N+L S EI L +K+L
Sbjct: 246 NNMIKHLSYLAGLKNLVELWASNNELSSFQEIED------------------ELSGLKKL 287
Query: 132 NTLVLSKNPIRKIGEALKKVKSITKLSLSHCQLEGIDTSL 171
T+ NP++K A+ + K+ L QL ID ++
Sbjct: 288 ETVYFEGNPLQKTNPAVYR----NKVRLCLPQLRQIDATI 323
>SHO2_HUMAN (Q9UQ13) Leucine-rich repeat protein SHOC-2 (Ras-binding
protein Sur-8)
Length = 582
Score = 82.8 bits (203), Expect = 2e-15
Identities = 64/238 (26%), Positives = 124/238 (51%), Gaps = 7/238 (2%)
Query: 5 SSEQVLKDNNAVNPSSISSLHLTHKALSDV-SCLASFNKLEKLDLKFNNLTSLEGLRAC- 62
S+ +V+K+ N + L L+ +++ + S + +L +L L N L SL C
Sbjct: 86 SNAEVIKELNKCREENSMRLDLSKRSIHILPSSIKELTQLTELYLYSNKLQSLPAEVGCL 145
Query: 63 VTLKWLSVVENKLESL-EGIQGLTKLTVLNAGKNKLKSMDEIG-SLSTIRALILNDNEIV 120
V L L++ EN L SL + + L KL +L+ NKL+ + + L ++ L L N I
Sbjct: 146 VNLMTLALSENSLTSLPDSLDNLKKLRMLDLRHNKLREIPSVVYRLDSLTTLYLRFNRIT 205
Query: 121 SI-CNLDQMKELNTLVLSKNPIRKIGEALKKVKSITKLSLSHCQLEGIDTSLKFCVELTE 179
++ ++ + +L+ L + +N I+++ + ++ ++ L ++H QLE + + C ++T
Sbjct: 206 TVEKDIKNLSKLSMLSIRENKIKQLPAEIGELCNLITLDVAHNQLEHLPKEIGNCTQITN 265
Query: 180 LRLAHNDIKSLPEELMHNSKLRNLDLGNNVIAKWSDIKVLKSLTKLRNLNLQGNPVAT 237
L L HN++ LP+ + + S L L L N ++ + + L + L LNL+ N ++T
Sbjct: 266 LDLQHNELLDLPDTIGNLSSLSRLGLRYNRLS--AIPRSLAKCSALEELNLENNNIST 321
Score = 70.1 bits (170), Expect = 1e-11
Identities = 55/195 (28%), Positives = 105/195 (53%), Gaps = 6/195 (3%)
Query: 20 SISSLHLTHKALSDV-SCLASFNKLEKLDLKFNNLTSLEGLRACV-TLKWLSVVENKLES 77
++ +L L+ +L+ + L + KL LDL+ N L + + + +L L + N++ +
Sbjct: 147 NLMTLALSENSLTSLPDSLDNLKKLRMLDLRHNKLREIPSVVYRLDSLTTLYLRFNRITT 206
Query: 78 LE-GIQGLTKLTVLNAGKNKLKSMD-EIGSLSTIRALILNDNEIVSICN-LDQMKELNTL 134
+E I+ L+KL++L+ +NK+K + EIG L + L + N++ + + ++ L
Sbjct: 207 VEKDIKNLSKLSMLSIRENKIKQLPAEIGELCNLITLDVAHNQLEHLPKEIGNCTQITNL 266
Query: 135 VLSKNPIRKIGEALKKVKSITKLSLSHCQLEGIDTSLKFCVELTELRLAHNDIKSLPEEL 194
L N + + + + + S+++L L + +L I SL C L EL L +N+I +LPE L
Sbjct: 267 DLQHNELLDLPDTIGNLSSLSRLGLRYNRLSAIPRSLAKCSALEELNLENNNISTLPESL 326
Query: 195 MHN-SKLRNLDLGNN 208
+ + KL +L L N
Sbjct: 327 LSSLVKLNSLTLARN 341
Score = 68.9 bits (167), Expect = 3e-11
Identities = 67/249 (26%), Positives = 121/249 (47%), Gaps = 33/249 (13%)
Query: 17 NPSSISSLHLTHKALSDVS-CLASFNKLEKLDLKFNNLTSL-EGL-------------RA 61
N SS+S L L + LS + LA + LE+L+L+ NN+++L E L R
Sbjct: 282 NLSSLSRLGLRYNRLSAIPRSLAKCSALEELNLENNNISTLPESLLSSLVKLNSLTLARN 341
Query: 62 CV------------TLKWLSVVENKLESLE-GIQGLTK-LTVLNAGKNKLKSMD-EIGSL 106
C T+ L++ N++ + GI K L+ LN N+L S+ + G+
Sbjct: 342 CFQLYPVGGPSQFSTIYSLNMEHNRINKIPFGIFSRAKVLSKLNMKDNQLTSLPLDFGTW 401
Query: 107 STIRALILNDNEIVSIC-NLDQMKELNTLVLSKNPIRKIGEALKKVKSITKLSLSHCQLE 165
+++ L L N++ I ++ + L L+LS N ++K+ L ++ + +L L +LE
Sbjct: 402 TSMVELNLATNQLTKIPEDVSGLVSLEVLILSNNLLKKLPHGLGNLRKLRELDLEENKLE 461
Query: 166 GIDTSLKFCVELTELRLAHNDIKSLPEELMHNSKLRNLDLGNNVIAKWSDIKVLKSLTKL 225
+ + + +L +L L +N + +LP + H + L +L LG N++ + + +L L
Sbjct: 462 SLPNEIAYLKDLQKLVLTNNQLTTLPRGIGHLTNLTHLGLGENLLTHLPE--EIGTLENL 519
Query: 226 RNLNLQGNP 234
L L NP
Sbjct: 520 EELYLNDNP 528
>SHO2_MOUSE (O88520) Leucine-rich repeat protein SHOC-2 (Ras-binding
protein Sur-8)
Length = 582
Score = 82.0 bits (201), Expect = 3e-15
Identities = 64/238 (26%), Positives = 123/238 (50%), Gaps = 7/238 (2%)
Query: 5 SSEQVLKDNNAVNPSSISSLHLTHKALSDVS-CLASFNKLEKLDLKFNNLTSLEGLRAC- 62
S+ +V+K+ N + L L+ +++ + + +L +L L N L SL C
Sbjct: 86 SNAEVIKELNKCREENSMRLDLSKRSIHILPPSVKELTQLTELYLYSNKLQSLPAEVGCL 145
Query: 63 VTLKWLSVVENKLESL-EGIQGLTKLTVLNAGKNKLKSMDEIG-SLSTIRALILNDNEIV 120
V L L++ EN L SL + + L KL +L+ NKL+ + + L ++ L L N I
Sbjct: 146 VNLMTLALSENSLTSLPDSLDNLKKLRMLDLRHNKLREIPSVVYRLDSLTTLYLRFNRIT 205
Query: 121 SI-CNLDQMKELNTLVLSKNPIRKIGEALKKVKSITKLSLSHCQLEGIDTSLKFCVELTE 179
++ ++ + +L+ L + +N I+++ + ++ ++ L ++H QLE + + C ++T
Sbjct: 206 TVEKDIKNLPKLSMLSIRENKIKQLPAEIGELCNLITLDVAHNQLEHLPKEIGNCTQITN 265
Query: 180 LRLAHNDIKSLPEELMHNSKLRNLDLGNNVIAKWSDIKVLKSLTKLRNLNLQGNPVAT 237
L L HND+ LP+ + + S L L L N ++ + + L + L LNL+ N ++T
Sbjct: 266 LDLQHNDLLDLPDTIGNLSSLNRLGLRYNRLS--AIPRSLAKCSALEELNLENNNIST 321
Score = 68.9 bits (167), Expect = 3e-11
Identities = 55/195 (28%), Positives = 103/195 (52%), Gaps = 6/195 (3%)
Query: 20 SISSLHLTHKALSDV-SCLASFNKLEKLDLKFNNLTSLEGLRACV-TLKWLSVVENKLES 77
++ +L L+ +L+ + L + KL LDL+ N L + + + +L L + N++ +
Sbjct: 147 NLMTLALSENSLTSLPDSLDNLKKLRMLDLRHNKLREIPSVVYRLDSLTTLYLRFNRITT 206
Query: 78 LE-GIQGLTKLTVLNAGKNKLKSMD-EIGSLSTIRALILNDNEIVSICN-LDQMKELNTL 134
+E I+ L KL++L+ +NK+K + EIG L + L + N++ + + ++ L
Sbjct: 207 VEKDIKNLPKLSMLSIRENKIKQLPAEIGELCNLITLDVAHNQLEHLPKEIGNCTQITNL 266
Query: 135 VLSKNPIRKIGEALKKVKSITKLSLSHCQLEGIDTSLKFCVELTELRLAHNDIKSLPEEL 194
L N + + + + + S+ +L L + +L I SL C L EL L +N+I +LPE L
Sbjct: 267 DLQHNDLLDLPDTIGNLSSLNRLGLRYNRLSAIPRSLAKCSALEELNLENNNISTLPESL 326
Query: 195 MHN-SKLRNLDLGNN 208
+ + KL +L L N
Sbjct: 327 LSSLVKLNSLTLARN 341
Score = 68.2 bits (165), Expect = 4e-11
Identities = 60/224 (26%), Positives = 112/224 (49%), Gaps = 10/224 (4%)
Query: 19 SSISSLHLTHKALSDV--SCLASFNKLEKLDLKFN--NLTSLEGLRACVTLKWLSVVENK 74
S++ L+L + +S + S L+S KL L L N L + G T+ L++ N+
Sbjct: 307 SALEELNLENNNISTLPESLLSSLVKLNSLTLARNCFQLYPVGGPSQFSTIYSLNMEHNR 366
Query: 75 LESLE-GIQGLTK-LTVLNAGKNKLKSMD-EIGSLSTIRALILNDNEIVSIC-NLDQMKE 130
+ + GI K L+ LN N+L S+ + G+ +++ L L N++ I ++ +
Sbjct: 367 INKIPFGIFSRAKVLSKLNMKDNQLTSLPLDFGTWTSMVELNLATNQLTKIPEDVSGLVS 426
Query: 131 LNTLVLSKNPIRKIGEALKKVKSITKLSLSHCQLEGIDTSLKFCVELTELRLAHNDIKSL 190
L L+LS N ++K+ L ++ + +L L +LE + + + +L +L L +N + +L
Sbjct: 427 LEVLILSNNLLKKLPHGLGNLRKLRELDLEENKLESLPNEIAYLKDLQKLVLTNNQLSTL 486
Query: 191 PEELMHNSKLRNLDLGNNVIAKWSDIKVLKSLTKLRNLNLQGNP 234
P + H + L +L LG N++ + + +L L L L NP
Sbjct: 487 PRGIGHLTNLTHLGLGENLLTHLPE--EIGTLENLEELYLNDNP 528
>SD22_YEAST (P36047) Protein phosphatases PP1 regulatory subunit
SDS22
Length = 338
Score = 77.8 bits (190), Expect = 6e-14
Identities = 70/252 (27%), Positives = 117/252 (45%), Gaps = 32/252 (12%)
Query: 13 NNAVNPSSISSLHLTHKALSDVSCLASFNKLEKLDLKFNNLTSLEGLRACVTLKWLSVVE 72
+N + ++SL L+ + + L + LE L N+++ +E L +LK L +
Sbjct: 107 SNVNKLTKLTSLDLSFNKIKHIKNLENLTDLENLYFVQNSISKIENLSTLKSLKNLELGG 166
Query: 73 NKLESLE--GIQGLTKLTVLNAGKNKLKSMDEIGSLSTIRALILNDNEIVSICNLDQMKE 130
NK+ S+E +GL+ L + GKN + + + L ++ L + N++ I NL+++
Sbjct: 167 NKVHSIEPDSFEGLSNLEEIWLGKNSIPRLINLHPLKNLKILSIQSNKLKKIENLEELTN 226
Query: 131 LNTLVLSKNPIRKIGEALKKVKSITKLSLSHCQLEGIDTSLKFCVELTELRLAHNDIKSL 190
L L LS N I KI EG++ +LK LT L + N I SL
Sbjct: 227 LEELYLSHNFITKI--------------------EGLEKNLK----LTTLDVTSNKITSL 262
Query: 191 PEELMHNSKLRNLDLGNNVIAKWSDI--KVLKSLTKLRNLNLQGNPVATNEKVIRKIK-- 246
E L H S L ++ N I + + + L +L++L + L+GNP+ K + K
Sbjct: 263 -ENLNHLSNLTDIWASFNKIDQSFESLGENLSALSRLETIYLEGNPIQLENKTSYRRKLT 321
Query: 247 -NALPKLQVFNA 257
N P LQ +A
Sbjct: 322 MNLPPSLQKIDA 333
Score = 49.7 bits (117), Expect = 2e-05
Identities = 43/188 (22%), Positives = 83/188 (43%), Gaps = 27/188 (14%)
Query: 4 LSSEQVLKDNNAVNPSSISSLHLTHKALSDVSCLASFNKLEKLDLKFNNLTSLE------ 57
LS ++ N N + + +L+ ++S + L++ L+ L+L N + S+E
Sbjct: 120 LSFNKIKHIKNLENLTDLENLYFVQNSISKIENLSTLKSLKNLELGGNKVHSIEPDSFEG 179
Query: 58 ------------------GLRACVTLKWLSVVENKLESLEGIQGLTKLTVLNAGKNKLKS 99
L LK LS+ NKL+ +E ++ LT L L N +
Sbjct: 180 LSNLEEIWLGKNSIPRLINLHPLKNLKILSIQSNKLKKIENLEELTNLEELYLSHNFITK 239
Query: 100 MDEIGSLSTIRALILNDNEIVSICNLDQMKELNTLVLSKNPIRKIGEAL-KKVKSITKLS 158
++ + + L + N+I S+ NL+ + L + S N I + E+L + + ++++L
Sbjct: 240 IEGLEKNLKLTTLDVTSNKITSLENLNHLSNLTDIWASFNKIDQSFESLGENLSALSRLE 299
Query: 159 LSHCQLEG 166
+ LEG
Sbjct: 300 TIY--LEG 305
>INLA_LISMF (Q723K6) Internalin A precursor
Length = 800
Score = 75.5 bits (184), Expect = 3e-13
Identities = 65/247 (26%), Positives = 112/247 (45%), Gaps = 29/247 (11%)
Query: 4 LSSEQVLKDNNAVNPSSISSLHLTHKALSDVSCLASFNKLEKLDLKFNNLTSLEGLRACV 63
+SS +V + +++ SL T+ +SD++ L L++L L N L + L +
Sbjct: 214 ISSNKVSDISVLAKLTNLESLIATNNQISDITPLGILTNLDELSLNGNQLKDIGTLASLT 273
Query: 64 TLKWLSVVENKLESLEGIQGLTKLTVLNAGKNKLKSMDEIGSLSTIRALILNDNEIVSIC 123
L L + N++ +L + GLTKLT L G N++ ++ + L+ + L LN+N++ I
Sbjct: 274 NLTDLDLANNQISNLAPLSGLTKLTELKLGANQISNISPLAGLTALTNLELNENQLEDIS 333
Query: 124 NLDQMKELNTLVLSKNPIRKIGEALKKVKSITKLSLSHCQLEGIDTSLKFCVELTELRLA 183
+ +K L L L N I I V S+TKL +
Sbjct: 334 PISNLKNLTYLTLYFNNISDI----SPVSSLTKL---------------------QRLFF 368
Query: 184 HNDIKSLPEELMHNSKLRNLDLGNNVIAKWSDIKVLKSLTKLRNLNLQGNPVATNEKVIR 243
+N+ S L + + + L G+N I SD+ L +LT++ L L + TN V
Sbjct: 369 YNNKVSDVSSLANLTNINWLSAGHNQI---SDLTPLANLTRITQLGL-NDQEWTNPPVNY 424
Query: 244 KIKNALP 250
K+ ++P
Sbjct: 425 KVNVSIP 431
Score = 72.8 bits (177), Expect = 2e-12
Identities = 59/206 (28%), Positives = 102/206 (48%), Gaps = 6/206 (2%)
Query: 31 LSDVSCLASFNKLEKLDLKFNNLTSLEGLRACVTLKWLSVVENKLESLEGIQGLTKLTVL 90
++D+ LA+ LE+LD+ N ++ + L L+ L N++ + + LT L L
Sbjct: 197 VTDLKPLANLTTLERLDISSNKVSDISVLAKLTNLESLIATNNQISDITPLGILTNLDEL 256
Query: 91 NAGKNKLKSMDEIGSLSTIRALILNDNEIVSICNLDQMKELNTLVLSKNPIRKIGEALKK 150
+ N+LK + + SL+ + L L +N+I ++ L + +L L L N I I L
Sbjct: 257 SLNGNQLKDIGTLASLTNLTDLDLANNQISNLAPLSGLTKLTELKLGANQISNI-SPLAG 315
Query: 151 VKSITKLSLSHCQLEGIDTSLKFCVELTELRLAHNDIKSLPEELMHNSKLRNLDLGNNVI 210
+ ++T L L+ QLE I + + LT L L N+I + + +KL+ L NN
Sbjct: 316 LTALTNLELNENQLEDI-SPISNLKNLTYLTLYFNNISDI-SPVSSLTKLQRLFFYNN-- 371
Query: 211 AKWSDIKVLKSLTKLRNLNLQGNPVA 236
K SD+ L +LT + L+ N ++
Sbjct: 372 -KVSDVSSLANLTNINWLSAGHNQIS 396
Score = 62.0 bits (149), Expect = 3e-09
Identities = 63/219 (28%), Positives = 97/219 (43%), Gaps = 16/219 (7%)
Query: 53 LTSLEGLRACVTLKWLSVVENKLESLEGIQGLTKLTVLNAGKNKLKSMDEIGSLSTIRAL 112
+ S++GL L ++ N+L + ++ LTKL + N++ + + +LS + L
Sbjct: 88 IKSIDGLEYLNNLTQINFSNNQLTDITPLKDLTKLVDILMNNNQIADITPLANLSNLTGL 147
Query: 113 ILNDNEIVSICNLDQMKELNTLVLSKNPIRKIGEALKKVKSITKLSLSHCQLEGIDTSLK 172
L +N+I I L + LN L LS N I I AL + S+ +LS + T LK
Sbjct: 148 TLFNNQITDIDPLKNLTNLNRLELSSNTISDI-SALSGLTSLQQLSFGNQV-----TDLK 201
Query: 173 FCVELTELR---LAHNDIKSLPEELMHNSKLRNLDLGNNVIAKWSDIKVLKSLTKLRNLN 229
LT L ++ N + + L + L +L NN I SDI L LT L L+
Sbjct: 202 PLANLTTLERLDISSNKVSDI-SVLAKLTNLESLIATNNQI---SDITPLGILTNLDELS 257
Query: 230 LQGN---PVATNEKVIRKIKNALPKLQVFNAKPIDKDTK 265
L GN + T + L Q+ N P+ TK
Sbjct: 258 LNGNQLKDIGTLASLTNLTDLDLANNQISNLAPLSGLTK 296
Score = 58.2 bits (139), Expect = 5e-08
Identities = 30/116 (25%), Positives = 60/116 (50%)
Query: 3 RLSSEQVLKDNNAVNPSSISSLHLTHKALSDVSCLASFNKLEKLDLKFNNLTSLEGLRAC 62
+L + Q+ + +++++L L L D+S +++ L L L FNN++ + + +
Sbjct: 301 KLGANQISNISPLAGLTALTNLELNENQLEDISPISNLKNLTYLTLYFNNISDISPVSSL 360
Query: 63 VTLKWLSVVENKLESLEGIQGLTKLTVLNAGKNKLKSMDEIGSLSTIRALILNDNE 118
L+ L NK+ + + LT + L+AG N++ + + +L+ I L LND E
Sbjct: 361 TKLQRLFFYNNKVSDVSSLANLTNINWLSAGHNQISDLTPLANLTRITQLGLNDQE 416
>INLA_LISMO (P25146) Internalin A precursor
Length = 800
Score = 74.7 bits (182), Expect = 5e-13
Identities = 65/247 (26%), Positives = 111/247 (44%), Gaps = 29/247 (11%)
Query: 4 LSSEQVLKDNNAVNPSSISSLHLTHKALSDVSCLASFNKLEKLDLKFNNLTSLEGLRACV 63
+SS +V + +++ SL T+ +SD++ L L++L L N L + L +
Sbjct: 214 ISSNKVSDISVLAKLTNLESLIATNNQISDITPLGILTNLDELSLNGNQLKDIGTLASLT 273
Query: 64 TLKWLSVVENKLESLEGIQGLTKLTVLNAGKNKLKSMDEIGSLSTIRALILNDNEIVSIC 123
L L + N++ +L + GLTKLT L G N++ ++ + L+ + L LN+N++ I
Sbjct: 274 NLTDLDLANNQISNLAPLSGLTKLTELKLGANQISNISPLAGLTALTNLELNENQLEDIS 333
Query: 124 NLDQMKELNTLVLSKNPIRKIGEALKKVKSITKLSLSHCQLEGIDTSLKFCVELTELRLA 183
+ +K L L L N I I V S+TKL +
Sbjct: 334 PISNLKNLTYLTLYFNNISDI----SPVSSLTKL---------------------QRLFF 368
Query: 184 HNDIKSLPEELMHNSKLRNLDLGNNVIAKWSDIKVLKSLTKLRNLNLQGNPVATNEKVIR 243
+N+ S L + + + L G+N I SD+ L +LT++ L L + TN V
Sbjct: 369 YNNKVSDVSSLANLTNINWLSAGHNQI---SDLTPLANLTRITQLGL-NDQAWTNAPVNY 424
Query: 244 KIKNALP 250
K ++P
Sbjct: 425 KANVSIP 431
Score = 72.8 bits (177), Expect = 2e-12
Identities = 59/206 (28%), Positives = 102/206 (48%), Gaps = 6/206 (2%)
Query: 31 LSDVSCLASFNKLEKLDLKFNNLTSLEGLRACVTLKWLSVVENKLESLEGIQGLTKLTVL 90
++D+ LA+ LE+LD+ N ++ + L L+ L N++ + + LT L L
Sbjct: 197 VTDLKPLANLTTLERLDISSNKVSDISVLAKLTNLESLIATNNQISDITPLGILTNLDEL 256
Query: 91 NAGKNKLKSMDEIGSLSTIRALILNDNEIVSICNLDQMKELNTLVLSKNPIRKIGEALKK 150
+ N+LK + + SL+ + L L +N+I ++ L + +L L L N I I L
Sbjct: 257 SLNGNQLKDIGTLASLTNLTDLDLANNQISNLAPLSGLTKLTELKLGANQISNI-SPLAG 315
Query: 151 VKSITKLSLSHCQLEGIDTSLKFCVELTELRLAHNDIKSLPEELMHNSKLRNLDLGNNVI 210
+ ++T L L+ QLE I + + LT L L N+I + + +KL+ L NN
Sbjct: 316 LTALTNLELNENQLEDI-SPISNLKNLTYLTLYFNNISDI-SPVSSLTKLQRLFFYNN-- 371
Query: 211 AKWSDIKVLKSLTKLRNLNLQGNPVA 236
K SD+ L +LT + L+ N ++
Sbjct: 372 -KVSDVSSLANLTNINWLSAGHNQIS 396
Score = 60.1 bits (144), Expect = 1e-08
Identities = 61/219 (27%), Positives = 97/219 (43%), Gaps = 16/219 (7%)
Query: 53 LTSLEGLRACVTLKWLSVVENKLESLEGIQGLTKLTVLNAGKNKLKSMDEIGSLSTIRAL 112
+ S++G+ L ++ N+L + ++ LTKL + N++ + + +L+ + L
Sbjct: 88 IKSIDGVEYLNNLTQINFSNNQLTDITPLKNLTKLVDILMNNNQIADITPLANLTNLTGL 147
Query: 113 ILNDNEIVSICNLDQMKELNTLVLSKNPIRKIGEALKKVKSITKLSLSHCQLEGIDTSLK 172
L +N+I I L + LN L LS N I I AL + S+ +LS + T LK
Sbjct: 148 TLFNNQITDIDPLKNLTNLNRLELSSNTISDI-SALSGLTSLQQLSFGNQV-----TDLK 201
Query: 173 FCVELTELR---LAHNDIKSLPEELMHNSKLRNLDLGNNVIAKWSDIKVLKSLTKLRNLN 229
LT L ++ N + + L + L +L NN I SDI L LT L L+
Sbjct: 202 PLANLTTLERLDISSNKVSDI-SVLAKLTNLESLIATNNQI---SDITPLGILTNLDELS 257
Query: 230 LQGN---PVATNEKVIRKIKNALPKLQVFNAKPIDKDTK 265
L GN + T + L Q+ N P+ TK
Sbjct: 258 LNGNQLKDIGTLASLTNLTDLDLANNQISNLAPLSGLTK 296
Score = 56.2 bits (134), Expect = 2e-07
Identities = 29/114 (25%), Positives = 59/114 (51%)
Query: 3 RLSSEQVLKDNNAVNPSSISSLHLTHKALSDVSCLASFNKLEKLDLKFNNLTSLEGLRAC 62
+L + Q+ + +++++L L L D+S +++ L L L FNN++ + + +
Sbjct: 301 KLGANQISNISPLAGLTALTNLELNENQLEDISPISNLKNLTYLTLYFNNISDISPVSSL 360
Query: 63 VTLKWLSVVENKLESLEGIQGLTKLTVLNAGKNKLKSMDEIGSLSTIRALILND 116
L+ L NK+ + + LT + L+AG N++ + + +L+ I L LND
Sbjct: 361 TKLQRLFFYNNKVSDVSSLANLTNINWLSAGHNQISDLTPLANLTRITQLGLND 414
>LAP4_HUMAN (Q14160) LAP4 protein (Scribble homolog protein)
(hScrib)
Length = 1630
Score = 72.8 bits (177), Expect = 2e-12
Identities = 99/447 (22%), Positives = 187/447 (41%), Gaps = 43/447 (9%)
Query: 17 NPSSISSLHLTHKALSDVSCLASFN-KLEKLDLKFNNLTSL-EGLRACVTLKWLSVVENK 74
N +++ +L L L + SF KLE+LDL N+L L + L A L+ L + N+
Sbjct: 149 NLANLVTLELRENLLKSLPASLSFLVKLEQLDLGGNDLEVLPDTLGALPNLRELWLDRNQ 208
Query: 75 LESLEGIQG-LTKLTVLNAGKNKLKSMD-EIGSLSTIRALILNDNEI------------V 120
L +L G L +L L+ +N+L+ + E+G L + L+L+ N + +
Sbjct: 209 LSALPPELGNLRRLVCLDVSENRLEELPAELGGLVLLTDLLLSQNLLRRLPDGIGQLKQL 268
Query: 121 SICNLDQMK------------ELNTLVLSKNPIRKIGEALKKVKSITKLSLSHCQLEGID 168
SI +DQ + L+ L+L++N + + +L K+ +T L++ LE +
Sbjct: 269 SILKVDQNRLCEVTEAIGDCENLSELILTENLLMALPRSLGKLTKLTNLNVDRNHLEALP 328
Query: 169 TSLKFCVELTELRLAHNDIKSLPEELMHNSKLRNLDLGNNVIAKWSDIKVLKSLTKLRNL 228
+ CV L+ L L N + LP EL H ++L LD+ N + +L L
Sbjct: 329 PEIGGCVALSVLSLRDNRLAVLPPELAHTTELHVLDVAGNRLQSLPFALTHLNLKALWLA 388
Query: 229 NLQGNPVA--TNEKVIRKIKNALPKLQVFNAKPIDKDTKNEKGHMTDDAHD--------F 278
Q P+ E R + L + P+ + ++G +++ D
Sbjct: 389 ENQAQPMLRFQTEDDARTGEKVLTCYLLPQQPPLSLEDAGQQGSLSETWSDAPPSRVSVI 448
Query: 279 SFDHVDQNEDDHLEAADKRKSNKKRKETADASEKEAGVYDKENTGHNKDNGNKKKDKLTG 338
F ++D EAA +++ ++R A E V + G + + D +
Sbjct: 449 QFLEAPIGDEDAEEAAAEKRGLQRR---ATPHPSELKVMKRSIEGRRSEACPCQPDSGSP 505
Query: 339 TVDPDTKNKSTKKKLKKDDNKPSEKALALEENVNRTEKKKKNRKNKEQSEFDIIDDAEAS 398
+ K S + L +D++PS ++ E + + +E + +DAE
Sbjct: 506 LPAEEEKRLSAESGL-SEDSRPSASTVS-EAEPEGPSAEAQGGSQQEATTAGGEEDAEED 563
Query: 399 FAEIFNIKDQENLNHGGEMKLQDQVPK 425
+ E ++ L G + ++++ P+
Sbjct: 564 YQEPTVHFAEDALLPGDDREIEEGQPE 590
Score = 51.2 bits (121), Expect = 6e-06
Identities = 42/155 (27%), Positives = 72/155 (46%), Gaps = 4/155 (2%)
Query: 84 LTKLTVLNAGKNKLKSMD-EIGSLSTIRALILNDNEIVSIC-NLDQMKELNTLVLSKNPI 141
L L L N+++ + E+ + + L ++ N+I I ++ K L S NP+
Sbjct: 58 LLNLRKLGLSDNEIQRLPPEVANFMQLVELDVSRNDIPEIPESIKFCKALEIADFSGNPL 117
Query: 142 RKIGEALKKVKSITKLSLSHCQLEGIDTSLKFCVELTELRLAHNDIKSLPEELMHNSKLR 201
++ + +++S+ L+L+ L+ + + L L L N +KSLP L KL
Sbjct: 118 SRLPDGFTQLRSLAHLALNDVSLQALPGDVGNLANLVTLELRENLLKSLPASLSFLVKLE 177
Query: 202 NLDLGNNVIAKWSDIKVLKSLTKLRNLNLQGNPVA 236
LDLG N + D L +L LR L L N ++
Sbjct: 178 QLDLGGNDLEVLPD--TLGALPNLRELWLDRNQLS 210
Score = 44.7 bits (104), Expect = 5e-04
Identities = 58/241 (24%), Positives = 105/241 (43%), Gaps = 13/241 (5%)
Query: 43 LEKLDLKFNNLTSLEGLRA-CVTLKWLSVVENKLESL-EGIQGLTKLTVLNAGKNKLKSM 100
L KL L N + L A + L L V N + + E I+ L + + N L +
Sbjct: 61 LRKLGLSDNEIQRLPPEVANFMQLVELDVSRNDIPEIPESIKFCKALEIADFSGNPLSRL 120
Query: 101 -DEIGSLSTIRALILNDNEIVSIC-NLDQMKELNTLVLSKNPIRKIGEALKKVKSITKLS 158
D L ++ L LND + ++ ++ + L TL L +N ++ + +L + + +L
Sbjct: 121 PDGFTQLRSLAHLALNDVSLQALPGDVGNLANLVTLELRENLLKSLPASLSFLVKLEQLD 180
Query: 159 LSHCQLEGIDTSLKFCVELTELRLAHNDIKSLPEELMHNSKLRNLDLGNNVIAKWSDIKV 218
L LE + +L L EL L N + +LP EL + +L LD+ N + +
Sbjct: 181 LGGNDLEVLPDTLGALPNLRELWLDRNQLSALPPELGNLRRLVCLDVSENRLEELP--AE 238
Query: 219 LKSLTKLRNLNLQGNPVATNEKVIRKIKNALPKLQVFNAKPIDKDTKNEKGHMTDDAHDF 278
L L L +L L N ++R++ + + +L+ + +D++ E D +
Sbjct: 239 LGGLVLLTDLLLSQN-------LLRRLPDGIGQLKQLSILKVDQNRLCEVTEAIGDCENL 291
Query: 279 S 279
S
Sbjct: 292 S 292
Score = 43.9 bits (102), Expect = 9e-04
Identities = 28/109 (25%), Positives = 56/109 (50%), Gaps = 3/109 (2%)
Query: 129 KELNTLVLSKNPIRKIGEALKKVKSITKLSLSHCQLEGIDTSLKFCVELTELRLAHNDIK 188
+ L L+L N +R++ + ++ ++ KL LS +++ + + ++L EL ++ NDI
Sbjct: 36 RSLEELLLDANQLRELPKPFFRLLNLRKLGLSDNEIQRLPPEVANFMQLVELDVSRNDIP 95
Query: 189 SLPEELMHNSKLRNLDLGNNVIAKWSD-IKVLKSLT--KLRNLNLQGNP 234
+PE + L D N +++ D L+SL L +++LQ P
Sbjct: 96 EIPESIKFCKALEIADFSGNPLSRLPDGFTQLRSLAHLALNDVSLQALP 144
Score = 38.1 bits (87), Expect = 0.049
Identities = 44/170 (25%), Positives = 78/170 (45%), Gaps = 30/170 (17%)
Query: 108 TIRALILNDNEIVSICN-LDQMKELNTLVLSKNPIRKIGEALKKVKSITKLSLSHCQLEG 166
++ L+L+ N++ + ++ L L LS N I+++ + + +L +S +
Sbjct: 37 SLEELLLDANQLRELPKPFFRLLNLRKLGLSDNEIQRLPPEVANFMQLVELDVSRNDIPE 96
Query: 167 IDTSLKFCVEL-----------------TELR----LAHNDI--KSLPEELMHNSKLRNL 203
I S+KFC L T+LR LA ND+ ++LP ++ + + L L
Sbjct: 97 IPESIKFCKALEIADFSGNPLSRLPDGFTQLRSLAHLALNDVSLQALPGDVGNLANLVTL 156
Query: 204 DLGNNVIAKWSDIKVLKSLTKLRNLNLQGNPVATNEKVIRKIKNALPKLQ 253
+L N++ S L L KL L+L GN + +V+ ALP L+
Sbjct: 157 ELRENLLK--SLPASLSFLVKLEQLDLGGNDL----EVLPDTLGALPNLR 200
Score = 32.0 bits (71), Expect = 3.5
Identities = 24/91 (26%), Positives = 44/91 (47%), Gaps = 2/91 (2%)
Query: 146 EALKKVKSITKLSLSHCQLEGIDTSLKFCVELTELRLAHNDIKSLPEELMHNSKLRNLDL 205
E + +S+ +L L QL + + L +L L+ N+I+ LP E+ + +L LD+
Sbjct: 30 EIYRYSRSLEELLLDANQLRELPKPFFRLLNLRKLGLSDNEIQRLPPEVANFMQLVELDV 89
Query: 206 GNNVIAKWSDIKVLKSLTKLRNLNLQGNPVA 236
N I + + +K L + GNP++
Sbjct: 90 SRNDIPEIPE--SIKFCKALEIADFSGNPLS 118
>LRR1_MOUSE (Q80VQ1) Leucine-rich repeat-containing protein 1
Length = 524
Score = 71.2 bits (173), Expect = 5e-12
Identities = 96/391 (24%), Positives = 167/391 (42%), Gaps = 59/391 (15%)
Query: 14 NAVNPSSISSLHLTHKALSDV-SCLASFNKLEKLDLKFNNLTSL-EGLRACVTLK--WLS 69
N N +++SL L L+ + L +LE+LDL N + +L E + A + LK WL
Sbjct: 146 NIGNLYNLASLELRENLLTYLPDSLTQLRRLEELDLGNNEIYNLPESIGALLHLKDLWLD 205
Query: 70 ---------------------VVENKLESL-EGIQGLTKLTVLNAGKNKLKSMDE-IGSL 106
V EN+LE L E I GLT LT L +N L+++ E IG L
Sbjct: 206 GNQLSELPQEIGNLKNLLCLDVSENRLERLPEEISGLTSLTYLVISQNLLETIPEGIGKL 265
Query: 107 STIRALILNDNEIVSICN-LDQMKELNTLVLSKNPIRKIGEALKKVKSITKLSLSHCQLE 165
+ L L+ N + + + + L LVL++N + + +++ K+K ++ L+ +L
Sbjct: 266 KKLSILKLDQNRLTQLPEAIGDCENLTELVLTENRLLTLPKSIGKLKKLSNLNADRNKLV 325
Query: 166 GIDTSLKFCVELTELRLAHNDIKSLPEELMHNSKLRNLDLGNNVIAKWSDIKVLKSLTKL 225
+ + C LT + N + LP E+ +L LD+ N + + + + KL
Sbjct: 326 SLPKEIGGCCSLTMFCIRDNRLTRLPAEVSQAVELHVLDVAGN---RLHHLPLSLTTLKL 382
Query: 226 RNLNLQGNPV------------ATNEKVIRKIKNALPKLQVFNAKPIDKDTKNEKGHMTD 273
+ L L N AT EK++ + LP++ ++PI +++ G +
Sbjct: 383 KALWLSDNQSQPLLTFQTDIDRATGEKILTCV--LLPQMP---SEPICQESLPRCGALES 437
Query: 274 DAHDFSFDHVDQNEDDHLEAADKRKSNKKRKETADASEKEAGVYDKENTGHNKDNGNKKK 333
D S + + H +A + ++K D E E + T H + N KK
Sbjct: 438 LVTDMS-EEAWNDRSVHRVSAIRFLEDEK-----DEDENETRTLQRRATPHPGELKNMKK 491
Query: 334 DKLTGTVDPDTKNKSTKKKLKKDDNKPSEKA 364
TV+ + + K L + N+ + A
Sbjct: 492 -----TVENLRNDMNAAKGLDSNKNEVNHAA 517
Score = 63.2 bits (152), Expect = 1e-09
Identities = 58/202 (28%), Positives = 100/202 (48%), Gaps = 11/202 (5%)
Query: 18 PSSISSL-HLTHKALSDVSC------LASFNKLEKLDLKFNNLTSL-EGLRACVTLKWLS 69
P S L +LT +++D+S + + L L+L+ N LT L + L L+ L
Sbjct: 121 PESFPELQNLTCLSVNDISLQSLPENIGNLYNLASLELRENLLTYLPDSLTQLRRLEELD 180
Query: 70 VVENKLESL-EGIQGLTKLTVLNAGKNKLKSM-DEIGSLSTIRALILNDNEIVSICN-LD 126
+ N++ +L E I L L L N+L + EIG+L + L +++N + + +
Sbjct: 181 LGNNEIYNLPESIGALLHLKDLWLDGNQLSELPQEIGNLKNLLCLDVSENRLERLPEEIS 240
Query: 127 QMKELNTLVLSKNPIRKIGEALKKVKSITKLSLSHCQLEGIDTSLKFCVELTELRLAHND 186
+ L LV+S+N + I E + K+K ++ L L +L + ++ C LTEL L N
Sbjct: 241 GLTSLTYLVISQNLLETIPEGIGKLKKLSILKLDQNRLTQLPEAIGDCENLTELVLTENR 300
Query: 187 IKSLPEELMHNSKLRNLDLGNN 208
+ +LP+ + KL NL+ N
Sbjct: 301 LLTLPKSIGKLKKLSNLNADRN 322
Score = 60.8 bits (146), Expect = 7e-09
Identities = 52/189 (27%), Positives = 96/189 (50%), Gaps = 5/189 (2%)
Query: 64 TLKWLSVVENKLESL-EGIQGLTKLTVLNAGKNKLKSMD-EIGSLSTIRALILNDNEIVS 121
+L+ L + N+L L E L KL L N+++ + EI + + L ++ N+I
Sbjct: 37 SLEELLLDANQLRELPEQFFQLVKLRKLGLSDNEIQRLPPEIANFMQLVELDVSRNDIPE 96
Query: 122 IC-NLDQMKELNTLVLSKNPIRKIGEALKKVKSITKLSLSHCQLEGIDTSLKFCVELTEL 180
I ++ K L S NP+ ++ E+ +++++T LS++ L+ + ++ L L
Sbjct: 97 IPESIAFCKALQVADFSGNPLTRLPESFPELQNLTCLSVNDISLQSLPENIGNLYNLASL 156
Query: 181 RLAHNDIKSLPEELMHNSKLRNLDLGNNVIAKWSDIKVLKSLTKLRNLNLQGNPVATNEK 240
L N + LP+ L +L LDLGNN I ++ + + +L L++L L GN ++ +
Sbjct: 157 ELRENLLTYLPDSLTQLRRLEELDLGNNEI--YNLPESIGALLHLKDLWLDGNQLSELPQ 214
Query: 241 VIRKIKNAL 249
I +KN L
Sbjct: 215 EIGNLKNLL 223
Score = 43.5 bits (101), Expect = 0.001
Identities = 27/109 (24%), Positives = 56/109 (50%), Gaps = 3/109 (2%)
Query: 129 KELNTLVLSKNPIRKIGEALKKVKSITKLSLSHCQLEGIDTSLKFCVELTELRLAHNDIK 188
+ L L+L N +R++ E ++ + KL LS +++ + + ++L EL ++ NDI
Sbjct: 36 RSLEELLLDANQLRELPEQFFQLVKLRKLGLSDNEIQRLPPEIANFMQLVELDVSRNDIP 95
Query: 189 SLPEELMHNSKLRNLDLGNNVIAKWSD-IKVLKSLT--KLRNLNLQGNP 234
+PE + L+ D N + + + L++LT + +++LQ P
Sbjct: 96 EIPESIAFCKALQVADFSGNPLTRLPESFPELQNLTCLSVNDISLQSLP 144
Score = 33.9 bits (76), Expect = 0.92
Identities = 27/108 (25%), Positives = 51/108 (47%), Gaps = 9/108 (8%)
Query: 146 EALKKVKSITKLSLSHCQLEGIDTSLKFCVELTELRLAHNDIKSLPEELMHNSKLRNLDL 205
E + +S+ +L L QL + V+L +L L+ N+I+ LP E+ + +L LD+
Sbjct: 30 EIYRYARSLEELLLDANQLRELPEQFFQLVKLRKLGLSDNEIQRLPPEIANFMQLVELDV 89
Query: 206 GNNVIAKWSDIKVLKSLTKLRNLNLQGNPVATNEKVIRKIKNALPKLQ 253
N I + + + L+ + GNP + ++ + P+LQ
Sbjct: 90 SRNDIPEIPE--SIAFCKALQVADFSGNP-------LTRLPESFPELQ 128
>LAP4_MOUSE (Q80U72) LAP4 protein (Scribble homolog protein)
Length = 1612
Score = 70.9 bits (172), Expect = 7e-12
Identities = 72/252 (28%), Positives = 122/252 (47%), Gaps = 38/252 (15%)
Query: 17 NPSSISSLHLTHKALSDVSCLASFN-KLEKLDLKFNNLTSL-EGLRACVTLKWLSVVENK 74
N +++ +L L L + SF KLE+LDL N+L L + L A L+ L + N+
Sbjct: 149 NLANLVTLELRENLLKSLPASLSFLVKLEQLDLGGNDLEVLPDTLGALPNLRELWLDRNQ 208
Query: 75 LESLEGIQG-LTKLTVLNAGKNKLKSMD-EIGSLSTIRALILNDNEI------------V 120
L +L G L +L L+ +N+L+ + E+G L+ + L+L+ N + +
Sbjct: 209 LSALPPELGNLRRLVCLDVSENRLEELPVELGGLALLTDLLLSQNLLQRLPEGIGQLKQL 268
Query: 121 SICNLDQMK------------ELNTLVLSKNPIRKIGEALKKVKSITKLSLSHCQLEGID 168
SI +DQ + L+ L+L++N + + +L K+ +T L++ LE +
Sbjct: 269 SILKVDQNRLCEVTEAIGDCENLSELILTENLLTALPHSLGKLTKLTNLNVDRNHLEVLP 328
Query: 169 TSLKFCVELTELRLAHNDIKSLPEELMHNSKLRNLDLGNNVIAKWSDIKVLKSLT-KLRN 227
+ CV L+ L L N + LP EL H ++L LD+ N L+SL L +
Sbjct: 329 PEIGGCVALSVLSLRDNRLAVLPPELAHTAELHVLDVAGN---------RLRSLPFALTH 379
Query: 228 LNLQGNPVATNE 239
LNL+ +A N+
Sbjct: 380 LNLKALWLAENQ 391
Score = 51.2 bits (121), Expect = 6e-06
Identities = 42/155 (27%), Positives = 72/155 (46%), Gaps = 4/155 (2%)
Query: 84 LTKLTVLNAGKNKLKSMD-EIGSLSTIRALILNDNEIVSIC-NLDQMKELNTLVLSKNPI 141
L L L N+++ + E+ + + L ++ N+I I ++ K L S NP+
Sbjct: 58 LLNLRKLGLSDNEIQRLPPEVANFMQLVELDVSRNDIPEIPESIKFCKALEIADFSGNPL 117
Query: 142 RKIGEALKKVKSITKLSLSHCQLEGIDTSLKFCVELTELRLAHNDIKSLPEELMHNSKLR 201
++ + +++S+ L+L+ L+ + + L L L N +KSLP L KL
Sbjct: 118 SRLPDGFTQLRSLAHLALNDVSLQALPGDVGNLANLVTLELRENLLKSLPASLSFLVKLE 177
Query: 202 NLDLGNNVIAKWSDIKVLKSLTKLRNLNLQGNPVA 236
LDLG N + D L +L LR L L N ++
Sbjct: 178 QLDLGGNDLEVLPD--TLGALPNLRELWLDRNQLS 210
Score = 43.9 bits (102), Expect = 9e-04
Identities = 28/109 (25%), Positives = 56/109 (50%), Gaps = 3/109 (2%)
Query: 129 KELNTLVLSKNPIRKIGEALKKVKSITKLSLSHCQLEGIDTSLKFCVELTELRLAHNDIK 188
+ L L+L N +R++ + ++ ++ KL LS +++ + + ++L EL ++ NDI
Sbjct: 36 RSLEELLLDANQLRELPKPFFRLLNLRKLGLSDNEIQRLPPEVANFMQLVELDVSRNDIP 95
Query: 189 SLPEELMHNSKLRNLDLGNNVIAKWSD-IKVLKSLT--KLRNLNLQGNP 234
+PE + L D N +++ D L+SL L +++LQ P
Sbjct: 96 EIPESIKFCKALEIADFSGNPLSRLPDGFTQLRSLAHLALNDVSLQALP 144
Score = 43.1 bits (100), Expect = 0.002
Identities = 58/242 (23%), Positives = 106/242 (42%), Gaps = 15/242 (6%)
Query: 43 LEKLDLKFNNLTSLEGLRA-CVTLKWLSVVENKLESL-EGIQGLTKLTVLNAGKNKLKSM 100
L KL L N + L A + L L V N + + E I+ L + + N L +
Sbjct: 61 LRKLGLSDNEIQRLPPEVANFMQLVELDVSRNDIPEIPESIKFCKALEIADFSGNPLSRL 120
Query: 101 -DEIGSLSTIRALILNDNEIVSIC-NLDQMKELNTLVLSKNPIRKIGEALKKVKSITKLS 158
D L ++ L LND + ++ ++ + L TL L +N ++ + +L + + +L
Sbjct: 121 PDGFTQLRSLAHLALNDVSLQALPGDVGNLANLVTLELRENLLKSLPASLSFLVKLEQLD 180
Query: 159 LSHCQLEGIDTSLKFCVELTELRLAHNDIKSLPEELMHNSKLRNLDLGNNVIAKWSDIKV 218
L LE + +L L EL L N + +LP EL + +L LD+ N + ++ V
Sbjct: 181 LGGNDLEVLPDTLGALPNLRELWLDRNQLSALPPELGNLRRLVCLDVSEN---RLEELPV 237
Query: 219 -LKSLTKLRNLNLQGNPVATNEKVIRKIKNALPKLQVFNAKPIDKDTKNEKGHMTDDAHD 277
L L L +L L N +++++ + +L+ + +D++ E D +
Sbjct: 238 ELGGLALLTDLLLSQN-------LLQRLPEGIGQLKQLSILKVDQNRLCEVTEAIGDCEN 290
Query: 278 FS 279
S
Sbjct: 291 LS 292
Score = 38.1 bits (87), Expect = 0.049
Identities = 44/170 (25%), Positives = 78/170 (45%), Gaps = 30/170 (17%)
Query: 108 TIRALILNDNEIVSICN-LDQMKELNTLVLSKNPIRKIGEALKKVKSITKLSLSHCQLEG 166
++ L+L+ N++ + ++ L L LS N I+++ + + +L +S +
Sbjct: 37 SLEELLLDANQLRELPKPFFRLLNLRKLGLSDNEIQRLPPEVANFMQLVELDVSRNDIPE 96
Query: 167 IDTSLKFCVEL-----------------TELR----LAHNDI--KSLPEELMHNSKLRNL 203
I S+KFC L T+LR LA ND+ ++LP ++ + + L L
Sbjct: 97 IPESIKFCKALEIADFSGNPLSRLPDGFTQLRSLAHLALNDVSLQALPGDVGNLANLVTL 156
Query: 204 DLGNNVIAKWSDIKVLKSLTKLRNLNLQGNPVATNEKVIRKIKNALPKLQ 253
+L N++ S L L KL L+L GN + +V+ ALP L+
Sbjct: 157 ELRENLLK--SLPASLSFLVKLEQLDLGGNDL----EVLPDTLGALPNLR 200
Score = 32.0 bits (71), Expect = 3.5
Identities = 24/91 (26%), Positives = 44/91 (47%), Gaps = 2/91 (2%)
Query: 146 EALKKVKSITKLSLSHCQLEGIDTSLKFCVELTELRLAHNDIKSLPEELMHNSKLRNLDL 205
E + +S+ +L L QL + + L +L L+ N+I+ LP E+ + +L LD+
Sbjct: 30 EIYRYSRSLEELLLDANQLRELPKPFFRLLNLRKLGLSDNEIQRLPPEVANFMQLVELDV 89
Query: 206 GNNVIAKWSDIKVLKSLTKLRNLNLQGNPVA 236
N I + + +K L + GNP++
Sbjct: 90 SRNDIPEIPE--SIKFCKALEIADFSGNPLS 118
>LRR8_HUMAN (Q8IWT6) Leucine-rich repeat-containing protein 8
precursor
Length = 810
Score = 68.6 bits (166), Expect = 3e-11
Identities = 69/229 (30%), Positives = 101/229 (43%), Gaps = 14/229 (6%)
Query: 43 LEKLDL--KFNNLTSLEGLRACVTLKWLSVVENKLESL-EGIQGLTKLTVLNAGKNKLKS 99
L+KL + + L L L+ L L ++ LE + I L L ++ N LK+
Sbjct: 569 LQKLSINNEGTKLIVLNSLKKMANLTELELIRCDLERIPHSIFSLHNLQEIDLKDNNLKT 628
Query: 100 MDEIGS---LSTIRALILNDNEIVSI-CNLDQMKELNTLVLSKNPIRKIGEALKKVKSIT 155
++EI S L + L L N I I + + L L L++N I KI L + +
Sbjct: 629 IEEIISFQHLHRLTCLKLWYNHIAYIPIQIGNLTNLERLYLNRNKIEKIPTQLFYCRKLR 688
Query: 156 KLSLSHCQLEGIDTSLKFCVELTELRLAHNDIKSLPEELMHNSKLRNLDLGNNVIAKWSD 215
L LSH L + + L L + N I++LP EL KLR L LGNNV+ S
Sbjct: 689 YLDLSHNNLTFLPADIGLLQNLQNLAITANRIETLPPELFQCRKLRALHLGNNVLQ--SL 746
Query: 216 IKVLKSLTKLRNLNLQGN-----PVATNEKVIRKIKNALPKLQVFNAKP 259
+ LT L + L+GN PV E + K + + +FN P
Sbjct: 747 PSRVGELTNLTQIELRGNRLECLPVELGECPLLKRSGLVVEEDLFNTLP 795
>LRR8_MOUSE (Q80WG5) Leucine-rich repeat-containing protein 8
precursor
Length = 810
Score = 67.8 bits (164), Expect = 6e-11
Identities = 69/229 (30%), Positives = 102/229 (44%), Gaps = 14/229 (6%)
Query: 43 LEKLDL--KFNNLTSLEGLRACVTLKWLSVVENKLESL-EGIQGLTKLTVLNAGKNKLKS 99
L+KL + + L L L+ V L L ++ LE + I L L ++ N LK+
Sbjct: 569 LQKLSINNEGTKLIVLNSLKKMVNLTELELIRCDLERIPHSIFSLHNLQEIDLKDNNLKT 628
Query: 100 MDEIGS---LSTIRALILNDNEIVSI-CNLDQMKELNTLVLSKNPIRKIGEALKKVKSIT 155
++EI S L + L L N I I + + L L L++N I KI L + +
Sbjct: 629 IEEIISFQHLHRLTCLKLWYNHIAYIPIQIGNLTNLERLYLNRNKIEKIPTQLFYCRKLR 688
Query: 156 KLSLSHCQLEGIDTSLKFCVELTELRLAHNDIKSLPEELMHNSKLRNLDLGNNVIAKWSD 215
L LSH L + + L L + N I++LP EL KLR L LGNNV+ S
Sbjct: 689 YLDLSHNNLTFLPADIGLLQNLQNLAVTANRIEALPPELFQCRKLRALHLGNNVLQ--SL 746
Query: 216 IKVLKSLTKLRNLNLQGN-----PVATNEKVIRKIKNALPKLQVFNAKP 259
+ LT L + L+GN PV E + K + + +F+ P
Sbjct: 747 PSRVGELTNLTQIELRGNRLECLPVELGECPLLKRSGLVVEEDLFSTLP 795
>LRR1_HUMAN (Q9BTT6) Leucine-rich repeat-containing protein 1 (LAP
and no PDZ protein) (LANO adapter protein)
Length = 524
Score = 67.8 bits (164), Expect = 6e-11
Identities = 54/186 (29%), Positives = 106/186 (56%), Gaps = 5/186 (2%)
Query: 14 NAVNPSSISSLHLTHKALSDV-SCLASFNKLEKLDLKFNNLTSL-EGLRACVTLKWLSVV 71
N N +++SL L L+ + L +LE+LDL N + +L E + A + LK L +
Sbjct: 146 NIGNLYNLASLELRENLLTYLPDSLTQLRRLEELDLGNNEIYNLPESIGALLHLKDLWLD 205
Query: 72 ENKLESL-EGIQGLTKLTVLNAGKNKLKSM-DEIGSLSTIRALILNDNEIVSICN-LDQM 128
N+L L + I L L L+ +N+L+ + +EI L+++ L+++ N + +I + + ++
Sbjct: 206 GNQLSELPQEIGNLKNLLCLDVSENRLERLPEEISGLTSLTDLVISQNLLETIPDGIGKL 265
Query: 129 KELNTLVLSKNPIRKIGEALKKVKSITKLSLSHCQLEGIDTSLKFCVELTELRLAHNDIK 188
K+L+ L + +N + ++ EA+ + +S+T+L L+ QL + S+ +L+ L N +
Sbjct: 266 KKLSILKVDQNRLTQLPEAVGECESLTELVLTENQLLTLPKSIGKLKKLSNLNADRNKLV 325
Query: 189 SLPEEL 194
SLP+E+
Sbjct: 326 SLPKEI 331
Score = 63.2 bits (152), Expect = 1e-09
Identities = 58/212 (27%), Positives = 106/212 (49%), Gaps = 7/212 (3%)
Query: 43 LEKLDLKFNNLTSL--EGLRACVTLKWLSVVENKLESL-EGIQGLTKLTVLNAGKNKLKS 99
+E +D + +L + E R +L+ L + N+L L E L KL L N+++
Sbjct: 14 VESIDKRHCSLVYVPEEIYRYARSLEELLLDANQLRELPEQFFQLVKLRKLGLSDNEIQR 73
Query: 100 MD-EIGSLSTIRALILNDNEIVSIC-NLDQMKELNTLVLSKNPIRKIGEALKKVKSITKL 157
+ EI + + L ++ NEI I ++ K L S NP+ ++ E+ +++++T L
Sbjct: 74 LPPEIANFMQLVELDVSRNEIPEIPESISFCKALQVADFSGNPLTRLPESFPELQNLTCL 133
Query: 158 SLSHCQLEGIDTSLKFCVELTELRLAHNDIKSLPEELMHNSKLRNLDLGNNVIAKWSDIK 217
S++ L+ + ++ L L L N + LP+ L +L LDLGNN I ++ +
Sbjct: 134 SVNDISLQSLPENIGNLYNLASLELRENLLTYLPDSLTQLRRLEELDLGNNEI--YNLPE 191
Query: 218 VLKSLTKLRNLNLQGNPVATNEKVIRKIKNAL 249
+ +L L++L L GN ++ + I +KN L
Sbjct: 192 SIGALLHLKDLWLDGNQLSELPQEIGNLKNLL 223
Score = 61.6 bits (148), Expect = 4e-09
Identities = 56/202 (27%), Positives = 100/202 (48%), Gaps = 11/202 (5%)
Query: 18 PSSISSL-HLTHKALSDVSC------LASFNKLEKLDLKFNNLTSL-EGLRACVTLKWLS 69
P S L +LT +++D+S + + L L+L+ N LT L + L L+ L
Sbjct: 121 PESFPELQNLTCLSVNDISLQSLPENIGNLYNLASLELRENLLTYLPDSLTQLRRLEELD 180
Query: 70 VVENKLESL-EGIQGLTKLTVLNAGKNKLKSM-DEIGSLSTIRALILNDNEIVSICN-LD 126
+ N++ +L E I L L L N+L + EIG+L + L +++N + + +
Sbjct: 181 LGNNEIYNLPESIGALLHLKDLWLDGNQLSELPQEIGNLKNLLCLDVSENRLERLPEEIS 240
Query: 127 QMKELNTLVLSKNPIRKIGEALKKVKSITKLSLSHCQLEGIDTSLKFCVELTELRLAHND 186
+ L LV+S+N + I + + K+K ++ L + +L + ++ C LTEL L N
Sbjct: 241 GLTSLTDLVISQNLLETIPDGIGKLKKLSILKVDQNRLTQLPEAVGECESLTELVLTENQ 300
Query: 187 IKSLPEELMHNSKLRNLDLGNN 208
+ +LP+ + KL NL+ N
Sbjct: 301 LLTLPKSIGKLKKLSNLNADRN 322
>LAP1_RAT (P70587) LAP1 protein (Densin-180)
Length = 1495
Score = 67.4 bits (163), Expect = 8e-11
Identities = 66/230 (28%), Positives = 106/230 (45%), Gaps = 27/230 (11%)
Query: 21 ISSLHLTHKALSDVSC-LASFNK-LEKLDLKFNNLTSL-EGLRACVTLKWLSVVENKLES 77
IS L +H +L V + +F + LE+L L N + L + L C L+ LS+ +N L S
Sbjct: 29 ISVLDYSHCSLQQVPKEVFNFERTLEELYLDANQIEELPKQLFNCQALRKLSIPDNDLSS 88
Query: 78 LE-GIQGLTKLTVLNAGKNKLKSMDEIGSLSTIRALILNDNEIVSICNLDQMKELNTLVL 136
L I L L L+ KN ++ E N+ K L +
Sbjct: 89 LPTSIASLVNLKELDISKNGVQEFPE---------------------NIKCCKCLTIIEA 127
Query: 137 SKNPIRKIGEALKKVKSITKLSLSHCQLEGIDTSLKFCVELTELRLAHNDIKSLPEELMH 196
S NPI K+ + ++ ++T+L L+ LE + + V+L L L N +K+LP+ +
Sbjct: 128 SVNPISKLPDGFTQLLNLTQLYLNDAFLEFLPANFGRLVKLRILELRENHLKTLPKSMHK 187
Query: 197 NSKLRNLDLGNNVIAKWSDIKVLKSLTKLRNLNLQGNPVATNEKVIRKIK 246
++L LDLGNN ++ + VL + LR L + N + I K+K
Sbjct: 188 LAQLERLDLGNNEFSELPE--VLDQIQNLRELWMDNNALQVLPGSIGKLK 235
Score = 57.0 bits (136), Expect = 1e-07
Identities = 47/174 (27%), Positives = 85/174 (48%), Gaps = 12/174 (6%)
Query: 38 ASFNKLEKLDLKFNNLTSLEGLRACVTLKWLSVVENKLESLEGIQG-LTKLTVLNAGKNK 96
AS N + KL +G + L L + + LE L G L KL +L +N
Sbjct: 127 ASVNPISKLP---------DGFTQLLNLTQLYLNDAFLEFLPANFGRLVKLRILELRENH 177
Query: 97 LKSMDE-IGSLSTIRALILNDNEIVSICN-LDQMKELNTLVLSKNPIRKIGEALKKVKSI 154
LK++ + + L+ + L L +NE + LDQ++ L L + N ++ + ++ K+K +
Sbjct: 178 LKTLPKSMHKLAQLERLDLGNNEFSELPEVLDQIQNLRELWMDNNALQVLPGSIGKLKML 237
Query: 155 TKLSLSHCQLEGIDTSLKFCVELTELRLAHNDIKSLPEELMHNSKLRNLDLGNN 208
L +S ++E +D + C L +L L+ N ++ LP+ + KL L + +N
Sbjct: 238 VYLDMSKNRIETVDMDISGCEALEDLLLSSNMLQQLPDSIGLLKKLTTLKVDDN 291
Score = 53.5 bits (127), Expect = 1e-06
Identities = 67/296 (22%), Positives = 130/296 (43%), Gaps = 40/296 (13%)
Query: 42 KLEKLDLKFNNLTSL-EGLRACVTLKWLSVVENKLESLEGIQGLTKLTV-LNAGKNKLKS 99
+LE+LDL N + L E L L+ L + N L+ L G G K+ V L+ KN++++
Sbjct: 190 QLERLDLGNNEFSELPEVLDQIQNLRELWMDNNALQVLPGSIGKLKMLVYLDMSKNRIET 249
Query: 100 MD-EIGSLSTIRALILNDNEIVSICN-LDQMKELNTLVLSKNPIRKIGEALKKVKSITKL 157
+D +I + L+L+ N + + + + +K+L TL + N + + + + + +
Sbjct: 250 VDMDISGCEALEDLLLSSNMLQQLPDSIGLLKKLTTLKVDDNQLTMLPNTIGNLSLLEEF 309
Query: 158 SLSHCQLEGIDTSLKF-----------------------CVELTELRLAHNDIKSLPEEL 194
S +LE + ++ + C +T + L N ++ LPEE+
Sbjct: 310 DCSCNELESLPPTIGYLHSLRTLAVDENFLPELPREIGSCKNVTVMSLRSNKLEFLPEEI 369
Query: 195 MHNSKLRNLDLGNNVIA----KWSDIKVLKSLTKLRNLNLQGNPVATN---EKVIRKIKN 247
+LR L+L +N + ++ +K L +L N + P+ T E R + N
Sbjct: 370 GQMQRLRVLNLSDNRLKNLPFSFTKLKELAALWLSDNQSKALIPLQTEAHPETKQRVLTN 429
Query: 248 ALPKLQVFNAKPIDKDTKNEKGHMTDDAHD----FSFDHVDQNEDDHLEAADKRKS 299
+ Q + D+ + + ++ +F+ D+ EDD E+A K K+
Sbjct: 430 YMFPQQPRGDEDFQSDSDSFNPTLWEEQRQQRMTVAFEFEDKKEDD--ESAGKVKA 483
>LAP1_MOUSE (Q80TE7) LAP1 protein (Densin-180)
Length = 1490
Score = 67.4 bits (163), Expect = 8e-11
Identities = 66/230 (28%), Positives = 106/230 (45%), Gaps = 27/230 (11%)
Query: 21 ISSLHLTHKALSDVSC-LASFNK-LEKLDLKFNNLTSL-EGLRACVTLKWLSVVENKLES 77
IS L +H +L V + +F + LE+L L N + L + L C L+ LS+ +N L S
Sbjct: 24 ISVLDYSHCSLQQVPKEVFNFERTLEELYLDANQIEELPKQLFNCQALRKLSIPDNDLSS 83
Query: 78 LE-GIQGLTKLTVLNAGKNKLKSMDEIGSLSTIRALILNDNEIVSICNLDQMKELNTLVL 136
L I L L L+ KN ++ E N+ K L +
Sbjct: 84 LPTSIASLVNLKELDISKNGVQEFPE---------------------NIKCCKCLTIIEA 122
Query: 137 SKNPIRKIGEALKKVKSITKLSLSHCQLEGIDTSLKFCVELTELRLAHNDIKSLPEELMH 196
S NPI K+ + ++ ++T+L L+ LE + + V+L L L N +K+LP+ +
Sbjct: 123 SVNPISKLPDGFTQLLNLTQLYLNDAFLEFLPANFGRLVKLRILELRENHLKTLPKSMHK 182
Query: 197 NSKLRNLDLGNNVIAKWSDIKVLKSLTKLRNLNLQGNPVATNEKVIRKIK 246
++L LDLGNN ++ + VL + LR L + N + I K+K
Sbjct: 183 LAQLERLDLGNNEFSELPE--VLDQIQNLRELWMDNNALQVLPGSIGKLK 230
Score = 57.0 bits (136), Expect = 1e-07
Identities = 47/174 (27%), Positives = 85/174 (48%), Gaps = 12/174 (6%)
Query: 38 ASFNKLEKLDLKFNNLTSLEGLRACVTLKWLSVVENKLESLEGIQG-LTKLTVLNAGKNK 96
AS N + KL +G + L L + + LE L G L KL +L +N
Sbjct: 122 ASVNPISKLP---------DGFTQLLNLTQLYLNDAFLEFLPANFGRLVKLRILELRENH 172
Query: 97 LKSMDE-IGSLSTIRALILNDNEIVSICN-LDQMKELNTLVLSKNPIRKIGEALKKVKSI 154
LK++ + + L+ + L L +NE + LDQ++ L L + N ++ + ++ K+K +
Sbjct: 173 LKTLPKSMHKLAQLERLDLGNNEFSELPEVLDQIQNLRELWMDNNALQVLPGSIGKLKML 232
Query: 155 TKLSLSHCQLEGIDTSLKFCVELTELRLAHNDIKSLPEELMHNSKLRNLDLGNN 208
L +S ++E +D + C L +L L+ N ++ LP+ + KL L + +N
Sbjct: 233 VYLDMSKNRIETVDMDISGCEALEDLLLSSNMLQQLPDSIGLLKKLTTLKVDDN 286
Score = 53.5 bits (127), Expect = 1e-06
Identities = 67/296 (22%), Positives = 130/296 (43%), Gaps = 40/296 (13%)
Query: 42 KLEKLDLKFNNLTSL-EGLRACVTLKWLSVVENKLESLEGIQGLTKLTV-LNAGKNKLKS 99
+LE+LDL N + L E L L+ L + N L+ L G G K+ V L+ KN++++
Sbjct: 185 QLERLDLGNNEFSELPEVLDQIQNLRELWMDNNALQVLPGSIGKLKMLVYLDMSKNRIET 244
Query: 100 MD-EIGSLSTIRALILNDNEIVSICN-LDQMKELNTLVLSKNPIRKIGEALKKVKSITKL 157
+D +I + L+L+ N + + + + +K+L TL + N + + + + + +
Sbjct: 245 VDMDISGCEALEDLLLSSNMLQQLPDSIGLLKKLTTLKVDDNQLTMLPNTIGNLSLLEEF 304
Query: 158 SLSHCQLEGIDTSLKF-----------------------CVELTELRLAHNDIKSLPEEL 194
S +LE + ++ + C +T + L N ++ LPEE+
Sbjct: 305 DCSCNELESLPPTIGYLHSLRTLAVDENFLPELPREIGSCKNVTVMSLRSNKLEFLPEEI 364
Query: 195 MHNSKLRNLDLGNNVIA----KWSDIKVLKSLTKLRNLNLQGNPVATN---EKVIRKIKN 247
+LR L+L +N + ++ +K L +L N + P+ T E R + N
Sbjct: 365 GQMQRLRVLNLSDNRLKNLPFSFTKLKELAALWLSDNQSKALIPLQTEAHPETKQRVLTN 424
Query: 248 ALPKLQVFNAKPIDKDTKNEKGHMTDDAHD----FSFDHVDQNEDDHLEAADKRKS 299
+ Q + D+ + + ++ +F+ D+ EDD E+A K K+
Sbjct: 425 YMFPQQPRGDEDFQSDSDSFNPTLWEEQRQQRMTVAFEFEDKKEDD--ESAGKVKA 478
>LRR5_MOUSE (Q8BGR2) Leucine-rich repeat-containing protein 5
precursor
Length = 859
Score = 67.0 bits (162), Expect = 1e-10
Identities = 73/272 (26%), Positives = 129/272 (46%), Gaps = 32/272 (11%)
Query: 10 LKDNNAVNPSSISSL--HLTHKAL-SDVSCLASFNKLEKLDLKFNNLTSLEGLRACVTLK 66
+K N PS+I+ + HLT + +D + L N L+K+ N+ LE L+ C
Sbjct: 596 VKSNLTKVPSNITDVAPHLTKLVIHNDGTKLLVLNSLKKM----MNVAELE-LQNC---- 646
Query: 67 WLSVVENKLESL-EGIQGLTKLTVLNAGKNKLKSMDEIGS---LSTIRALILNDNEIVSI 122
+LE + I L+ L L+ N +++++EI S L + L L N+IV+I
Sbjct: 647 -------ELERIPHAIFSLSNLQELDLKSNNIRTIEEIISFQHLKRLTCLKLWHNKIVAI 699
Query: 123 C-NLDQMKELNTLVLSKNPIRKIGEALKKVKSITKLSLSHCQLEGIDTSLKFCVELTELR 181
++ +K L +L S N + + A+ ++ + L +S+ + I + L L
Sbjct: 700 PPSITHVKNLESLYFSNNKLESLPTAVFSLQKLRCLDVSYNNISTIPIEIGLLQNLQHLH 759
Query: 182 LAHNDIKSLPEELMHNSKLRNLDLGNNVIAKWSDIKVLKSLTKLRNLNLQGN-----PVA 236
+ N + LP++L KLR L+LG N IA + + LT+L L L+GN P
Sbjct: 760 ITGNKVDILPKQLFKCVKLRTLNLGQNCIASLPE--KISQLTQLTQLELKGNCLDRLPAQ 817
Query: 237 TNEKVIRKIKNALPKLQVFNAKPID-KDTKNE 267
+ + K + + Q+F+ P++ K+ N+
Sbjct: 818 LGQCRMLKKSGLVVEDQLFDTLPLEVKEALNQ 849
>LAP4_DROME (Q7KRY7) LAP4 protein (Scribble protein) (Smell-impaired
protein)
Length = 1851
Score = 66.2 bits (160), Expect = 2e-10
Identities = 58/204 (28%), Positives = 102/204 (49%), Gaps = 11/204 (5%)
Query: 18 PSSISSL-HLTHKALSDVSC------LASFNKLEKLDLKFNNLTSL-EGLRACVTLKWLS 69
PS S L +LT L+D+S S +LE L+L+ N L L E + LK L
Sbjct: 122 PSGFSQLKNLTVLGLNDMSLTTLPADFGSLTQLESLELRENLLKHLPETISQLTKLKRLD 181
Query: 70 VVENKLESLEGIQG-LTKLTVLNAGKNKLKSMD-EIGSLSTIRALILNDNEIVSICN-LD 126
+ +N++E L G L L L N+L+ + E+G L+ + L +++N + + N +
Sbjct: 182 LGDNEIEDLPPYLGYLPGLHELWLDHNQLQRLPPELGLLTKLTYLDVSENRLEELPNEIS 241
Query: 127 QMKELNTLVLSKNPIRKIGEALKKVKSITKLSLSHCQLEGIDTSLKFCVELTELRLAHND 186
+ L L L++N + + + + K+ +T L L +L+ ++ +L C + EL L N
Sbjct: 242 GLVSLTDLDLAQNLLEALPDGIAKLSRLTILKLDQNRLQRLNDTLGNCENMQELILTENF 301
Query: 187 IKSLPEELMHNSKLRNLDLGNNVI 210
+ LP + +KL NL++ N +
Sbjct: 302 LSELPASIGQMTKLNNLNVDRNAL 325
Score = 63.5 bits (153), Expect = 1e-09
Identities = 65/244 (26%), Positives = 112/244 (45%), Gaps = 21/244 (8%)
Query: 7 EQVLKDNNAVN--PSSISSLHLTHK-ALSDVSC------LASFNKLEKLDLKFNNLTSL- 56
E++ D N + P + LH K LSD + +F L +LD+ N++ +
Sbjct: 40 EELFLDANHIRDLPKNFFRLHRLRKLGLSDNEIGRLPPDIQNFENLVELDVSRNDIPDIP 99
Query: 57 EGLRACVTLKWLSVVENKLESL-EGIQGLTKLTVLNAGKNKLKSMD-EIGSLSTIRALIL 114
+ ++ +L+ N + L G L LTVL L ++ + GSL+ + +L L
Sbjct: 100 DDIKHLQSLQVADFSSNPIPKLPSGFSQLKNLTVLGLNDMSLTTLPADFGSLTQLESLEL 159
Query: 115 NDNEIVSICN-LDQMKELNTLVLSKNPIRKIGEALKKVKSITKLSLSHCQLEGIDTSLKF 173
+N + + + Q+ +L L L N I + L + + +L L H QL+ + L
Sbjct: 160 RENLLKHLPETISQLTKLKRLDLGDNEIEDLPPYLGYLPGLHELWLDHNQLQRLPPELGL 219
Query: 174 CVELTELRLAHNDIKSLPEELMHNSKLRNLDLGNNV-------IAKWSDIKVLK-SLTKL 225
+LT L ++ N ++ LP E+ L +LDL N+ IAK S + +LK +L
Sbjct: 220 LTKLTYLDVSENRLEELPNEISGLVSLTDLDLAQNLLEALPDGIAKLSRLTILKLDQNRL 279
Query: 226 RNLN 229
+ LN
Sbjct: 280 QRLN 283
Score = 63.5 bits (153), Expect = 1e-09
Identities = 51/170 (30%), Positives = 86/170 (50%), Gaps = 4/170 (2%)
Query: 43 LEKLDLKFNNLTSLEGLRACVT-LKWLSVVENKLESLEG-IQGLTKLTVLNAGKNKLKSM 100
L +L L N L L +T L +L V EN+LE L I GL LT L+ +N L+++
Sbjct: 200 LHELWLDHNQLQRLPPELGLLTKLTYLDVSENRLEELPNEISGLVSLTDLDLAQNLLEAL 259
Query: 101 -DEIGSLSTIRALILNDNEIVSICN-LDQMKELNTLVLSKNPIRKIGEALKKVKSITKLS 158
D I LS + L L+ N + + + L + + L+L++N + ++ ++ ++ + L+
Sbjct: 260 PDGIAKLSRLTILKLDQNRLQRLNDTLGNCENMQELILTENFLSELPASIGQMTKLNNLN 319
Query: 159 LSHCQLEGIDTSLKFCVELTELRLAHNDIKSLPEELMHNSKLRNLDLGNN 208
+ LE + + C L L L N +K LP EL + + L LD+ N
Sbjct: 320 VDRNALEYLPLEIGQCANLGVLSLRDNKLKKLPPELGNCTVLHVLDVSGN 369
Score = 63.5 bits (153), Expect = 1e-09
Identities = 51/176 (28%), Positives = 93/176 (51%), Gaps = 5/176 (2%)
Query: 37 LASFNKLEKLDLKFNNLTSLEG-LRACVTLKWLSVVENKLESL-EGIQGLTKLTVLNAGK 94
L KL LD+ N L L + V+L L + +N LE+L +GI L++LT+L +
Sbjct: 217 LGLLTKLTYLDVSENRLEELPNEISGLVSLTDLDLAQNLLEALPDGIAKLSRLTILKLDQ 276
Query: 95 NKLKSM-DEIGSLSTIRALILNDNEIVSI-CNLDQMKELNTLVLSKNPIRKIGEALKKVK 152
N+L+ + D +G+ ++ LIL +N + + ++ QM +LN L + +N + + + +
Sbjct: 277 NRLQRLNDTLGNCENMQELILTENFLSELPASIGQMTKLNNLNVDRNALEYLPLEIGQCA 336
Query: 153 SITKLSLSHCQLEGIDTSLKFCVELTELRLAHNDIKSLPEELMHNSKLRNLDLGNN 208
++ LSL +L+ + L C L L ++ N + LP L+ N +L+ + L N
Sbjct: 337 NLGVLSLRDNKLKKLPPELGNCTVLHVLDVSGNQLLYLPYSLV-NLQLKAVWLSEN 391
Score = 58.5 bits (140), Expect = 3e-08
Identities = 62/243 (25%), Positives = 106/243 (43%), Gaps = 26/243 (10%)
Query: 17 NPSSISSLHLTHKALSDV-SCLASFNKLEKLDLKFNNLTSL-EGLRACVTLKWLSVVENK 74
N ++ L ++ + D+ + L+ D N + L G L L + +
Sbjct: 81 NFENLVELDVSRNDIPDIPDDIKHLQSLQVADFSSNPIPKLPSGFSQLKNLTVLGLNDMS 140
Query: 75 LESLEGIQG-LTKLTVLNAGKNKLKSMDE-IGSLSTIRALILNDNEIVSICN-LDQMKEL 131
L +L G LT+L L +N LK + E I L+ ++ L L DNEI + L + L
Sbjct: 141 LTTLPADFGSLTQLESLELRENLLKHLPETISQLTKLKRLDLGDNEIEDLPPYLGYLPGL 200
Query: 132 NTLVLSKNPIRKIGEALKKVKSITKLSLSHCQLEGIDTSLKFCVELTELRLAHNDIKSLP 191
+ L L N ++++ L + +T L +S +LE + + V LT+L LA N +++LP
Sbjct: 201 HELWLDHNQLQRLPPELGLLTKLTYLDVSENRLEELPNEISGLVSLTDLDLAQNLLEALP 260
Query: 192 EELMHNSKLRNLDLGNNVIAKWSDI---------------------KVLKSLTKLRNLNL 230
+ + S+L L L N + + +D + +TKL NLN+
Sbjct: 261 DGIAKLSRLTILKLDQNRLQRLNDTLGNCENMQELILTENFLSELPASIGQMTKLNNLNV 320
Query: 231 QGN 233
N
Sbjct: 321 DRN 323
Score = 54.7 bits (130), Expect = 5e-07
Identities = 44/157 (28%), Positives = 75/157 (47%), Gaps = 3/157 (1%)
Query: 57 EGLRACVTLKWLSVVENKLESL-EGIQGLTKLTVLNAGKNKLKSMD-EIGSLSTIRALIL 114
E LR TL+ L + N + L + L +L L N++ + +I + + L +
Sbjct: 31 EILRYSRTLEELFLDANHIRDLPKNFFRLHRLRKLGLSDNEIGRLPPDIQNFENLVELDV 90
Query: 115 NDNEIVSICN-LDQMKELNTLVLSKNPIRKIGEALKKVKSITKLSLSHCQLEGIDTSLKF 173
+ N+I I + + ++ L S NPI K+ ++K++T L L+ L +
Sbjct: 91 SRNDIPDIPDDIKHLQSLQVADFSSNPIPKLPSGFSQLKNLTVLGLNDMSLTTLPADFGS 150
Query: 174 CVELTELRLAHNDIKSLPEELMHNSKLRNLDLGNNVI 210
+L L L N +K LPE + +KL+ LDLG+N I
Sbjct: 151 LTQLESLELRENLLKHLPETISQLTKLKRLDLGDNEI 187
Score = 45.8 bits (107), Expect = 2e-04
Identities = 33/112 (29%), Positives = 54/112 (47%), Gaps = 9/112 (8%)
Query: 123 CNLDQMKE--------LNTLVLSKNPIRKIGEALKKVKSITKLSLSHCQLEGIDTSLKFC 174
C+L Q+ E L L L N IR + + ++ + KL LS ++ + ++
Sbjct: 23 CSLPQVPEEILRYSRTLEELFLDANHIRDLPKNFFRLHRLRKLGLSDNEIGRLPPDIQNF 82
Query: 175 VELTELRLAHNDIKSLPEELMHNSKLRNLDLGNNVIAKW-SDIKVLKSLTKL 225
L EL ++ NDI +P+++ H L+ D +N I K S LK+LT L
Sbjct: 83 ENLVELDVSRNDIPDIPDDIKHLQSLQVADFSSNPIPKLPSGFSQLKNLTVL 134
>CYAA_YEAST (P08678) Adenylate cyclase (EC 4.6.1.1) (ATP
pyrophosphate-lyase) (Adenylyl cyclase)
Length = 2026
Score = 66.2 bits (160), Expect = 2e-10
Identities = 60/212 (28%), Positives = 100/212 (46%), Gaps = 24/212 (11%)
Query: 43 LEKLDLKFNNLTSL-EGLRACVTLKWLSVVENKLESLEGIQGLTKLTVLNAGKNKLKSMD 101
L ++DL +N + SL + + V L +++ NKL + + +T L LN N++ S+
Sbjct: 934 LLQIDLSYNKIQSLPQSTKYLVKLAKMNLSHNKLNFIGDLSEMTDLRTLNLRYNRISSIK 993
Query: 102 EIGS------------------LSTIRALILNDNEIVSICNLD-QMKELNTLVLSKNPIR 142
S L +RAL + +N I SI D K + +L L+K +
Sbjct: 994 TNASNLQNLFLTDNRISNFEDTLPKLRALEIQENPITSISFKDFYPKNMTSLTLNKAQLS 1053
Query: 143 KI-GEALKKVKSITKLSLSHCQLEGIDTSLKFCVELTELRLAHNDIKSLPEELMHNSKLR 201
I GE L K+ + KL L+ L + + +L L +A N ++ +P EL LR
Sbjct: 1054 SIPGELLTKLSFLEKLELNQNNLTRLPQEISKLTKLVFLSVARNKLEYIPPELSQLKSLR 1113
Query: 202 NLDLGNNVIAKWSDIKVLKSLTKLRNLNLQGN 233
LDL +N I + D +++L +L +LN+ N
Sbjct: 1114 TLDLHSNNIRDFVD--GMENL-ELTSLNISSN 1142
Score = 58.9 bits (141), Expect = 3e-08
Identities = 56/223 (25%), Positives = 105/223 (46%), Gaps = 33/223 (14%)
Query: 41 NKLEKLDLKFNN-----LTSLEGLRACVTLKWLSVVENKLESLEGIQGLTKLTVLNAGKN 95
+++E LD+ N L +E ++L+ +++ +K S I KL L +N
Sbjct: 816 SEIESLDVSNNANIFLPLEFIESSIKLLSLRMVNIRASKFPS--NITKAYKLVSLELQRN 873
Query: 96 KLKSM-DEIGSLSTIRALILNDNEIVSI-CNLDQMKELNTLVLSKNPIRKIGEALKKVKS 153
++ + + I LS + L L NE+ S+ ++K L L LS N E + +
Sbjct: 874 FIRKVPNSIMKLSNLTILNLQCNELESLPAGFVELKNLQLLDLSSNKFMHYPEVINYCTN 933
Query: 154 ITKLSLSHCQLEGIDTSLKFCVELTELRLAHNDIKSLPE----------ELMHN------ 197
+ ++ LS+ +++ + S K+ V+L ++ L+HN + + + L +N
Sbjct: 934 LLQIDLSYNKIQSLPQSTKYLVKLAKMNLSHNKLNFIGDLSEMTDLRTLNLRYNRISSIK 993
Query: 198 ---SKLRNLDLGNNVIAKWSDIKVLKSLTKLRNLNLQGNPVAT 237
S L+NL L +N I+ + D +L KLR L +Q NP+ +
Sbjct: 994 TNASNLQNLFLTDNRISNFED-----TLPKLRALEIQENPITS 1031
Score = 53.5 bits (127), Expect = 1e-06
Identities = 45/174 (25%), Positives = 83/174 (46%), Gaps = 6/174 (3%)
Query: 75 LESLEGIQGLTKLTVLNAGKNKLKSMDEIGSLSTIRALILNDNEIVSICN-LDQMKELNT 133
LE +E L L ++N +K S I + +L L N I + N + ++ L
Sbjct: 833 LEFIESSIKLLSLRMVNIRASKFPS--NITKAYKLVSLELQRNFIRKVPNSIMKLSNLTI 890
Query: 134 LVLSKNPIRKIGEALKKVKSITKLSLSHCQLEGIDTSLKFCVELTELRLAHNDIKSLPEE 193
L L N + + ++K++ L LS + + +C L ++ L++N I+SLP+
Sbjct: 891 LNLQCNELESLPAGFVELKNLQLLDLSSNKFMHYPEVINYCTNLLQIDLSYNKIQSLPQS 950
Query: 194 LMHNSKLRNLDLGNNVIAKWSDIKVLKSLTKLRNLNLQGNPVATNEKVIRKIKN 247
+ KL ++L +N K + I L +T LR LNL+ N +++ + ++N
Sbjct: 951 TKYLVKLAKMNLSHN---KLNFIGDLSEMTDLRTLNLRYNRISSIKTNASNLQN 1001
Score = 48.9 bits (115), Expect = 3e-05
Identities = 73/262 (27%), Positives = 109/262 (40%), Gaps = 50/262 (19%)
Query: 18 PSSISSLHLTHKALSDV--SCLASFNKLEKLDLKFNNLTSL-EGLRACVTLKWLSVVENK 74
P +++SL L LS + L + LEKL+L NNLT L + + L +LSV NK
Sbjct: 1039 PKNMTSLTLNKAQLSSIPGELLTKLSFLEKLELNQNNLTRLPQEISKLTKLVFLSVARNK 1098
Query: 75 LESL------------------------EGIQGLTKLTVLNAGKNKLKSMDEIGSL---- 106
LE + +G++ L +LT LN N + S
Sbjct: 1099 LEYIPPELSQLKSLRTLDLHSNNIRDFVDGMENL-ELTSLNISSNAFGNSSLENSFYHNM 1157
Query: 107 -------STIRALILNDNEIVSIC--NLDQMKELNTLVLSKNPIRKIGEALKKVKSITKL 157
++ I DN+ + L L LS N + K++SIT+L
Sbjct: 1158 SYGSKLSKSLMFFIAADNQFDDAMWPLFNCFVNLKVLNLSYNNFSDVSHM--KLESITEL 1215
Query: 158 SLSHCQLEGI--DTSLKFCVELTELRLAHNDIKSLPEELMHNSKLRNLDLGNNV----IA 211
LS +L + DT LK+ L L L N + SLP EL + S+L D+G N I+
Sbjct: 1216 YLSGNKLTTLSGDTVLKWS-SLKTLMLNSNQMLSLPAELSNLSQLSVFDVGANQLKYNIS 1274
Query: 212 KWSDIKVLKSLTKLRNLNLQGN 233
+ ++ +L+ LN GN
Sbjct: 1275 NYHYDWNWRNNKELKYLNFSGN 1296
>FLIH_CAEEL (P34268) Flightless-I protein homolog
Length = 1257
Score = 65.9 bits (159), Expect = 2e-10
Identities = 58/209 (27%), Positives = 107/209 (50%), Gaps = 16/209 (7%)
Query: 65 LKWLSVVENKLESL-EGIQGLTKLTVLNAGKNKLKSMD-EIGSLSTIRALILNDNEIVSI 122
+ WL + ++KLE + + + L L N+L S+ E+ L +R++I+ DN + +
Sbjct: 34 MTWLKLNDSKLEQVPDELSRCANLEHLQMAHNQLISVHGELSDLPRLRSVIVRDNNLKTA 93
Query: 123 ---CNLDQMKELNTLVLSKNPIRKIGEALKKVKSITKLSLSHCQLEGIDTSL-KFCVELT 178
++ +MK+L + LS+N +R++ L+ K L+LS+ +E I S+ ++L
Sbjct: 94 GIPTDIFRMKDLTIIDLSRNQLREVPTNLEYAKGSIVLNLSYNNIETIPNSVCANLIDLL 153
Query: 179 ELRLAHNDIKSLPEELMHNSKLRNLDLGNNVIAKWSDIKVLKSLTKLRNLNLQGNPVATN 238
L L++N + LP ++ S L++L L NN + + +K L S+T L L++ TN
Sbjct: 154 FLDLSNNKLDMLPPQIRRLSMLQSLKLSNNPLNHF-QLKQLPSMTSLSVLHMSN----TN 208
Query: 239 EKVIRKIKNALPKL-QVFNAKPIDKDTKN 266
R + N P L + N + +D N
Sbjct: 209 ----RTLDNIPPTLDDMHNLRDVDFSENN 233
Score = 56.6 bits (135), Expect = 1e-07
Identities = 92/400 (23%), Positives = 156/400 (39%), Gaps = 79/400 (19%)
Query: 46 LDLKFNNLTSLEGLRAC--VTLKWLSVVENKLESLEG-IQGLTKLTVLNAGKN-----KL 97
L+L +NN+ ++ + L +L + NKL+ L I+ L+ L L N +L
Sbjct: 131 LNLSYNNIETIPNSVCANLIDLLFLDLSNNKLDMLPPQIRRLSMLQSLKLSNNPLNHFQL 190
Query: 98 KSMDEIGSLSTIRALILNDNEIVSICNLDQMKELNTLVLSKNPIRKIGEALKKVKSITKL 157
K + + SLS + N LD M L + S+N + + EAL K++++ KL
Sbjct: 191 KQLPSMTSLSVLHMSNTNRTLDNIPPTLDDMHNLRDVDFSENNLPIVPEALFKLRNLRKL 250
Query: 158 SLSHCQLEGIDTSLKFCVELTELRLAHNDIKSLPEELMHNSKLRNLDLGNNV-------- 209
+LS ++E ++ + L L ++HN + LP+ ++ ++L L NN
Sbjct: 251 NLSGNKIEKLNMTEGEWENLETLNMSHNQLTVLPDCVVKLTRLTKLYAANNQLTFEGIPS 310
Query: 210 -IAKWSDIKVL--------------KSLTKLRNLNLQGNPVAT----------------- 237
I K + VL KL+ L L N + T
Sbjct: 311 GIGKLIQLTVLHLSYNKLELVPEGISRCVKLQKLKLDHNRLITLPEGIHLLPDLKVLDLH 370
Query: 238 -NEKVIR--KIKNALPKLQVFNAKPIDKDTKNEKGHMTDDAHDFSFDHVDQNEDDHLEAA 294
NE ++ K +A KL +N + + G MT + S H D L A
Sbjct: 371 ENENLVMPPKPNDARKKLAFYNIDFSLEHQRKIAGQMTSPSSSISSVHSGGARQDAL--A 428
Query: 295 DKRKSNKKRKETADASEKEAGVYDKENTGHNKDNGNKKKDKLTGTVDPDTKNKSTKKKLK 354
+++ ++RK+ AD DK G +K G L
Sbjct: 429 RRKEFIRRRKQQADQQSA-----DKVIQGMSKIAG-------------------VGAALT 464
Query: 355 KDDNKPSEKALALEENVNRTEKKKKNRKNKEQSEFDIIDD 394
+ + E+ L + VN + +KNR++ + S DI D+
Sbjct: 465 EKQQEEEERQLEAKSAVNWKKNIEKNRRHIDYS--DIFDE 502
Score = 36.6 bits (83), Expect = 0.14
Identities = 27/94 (28%), Positives = 48/94 (50%), Gaps = 3/94 (3%)
Query: 148 LKKVKSITKLSLSHCQLEGIDTSLKFCVELTELRLAHNDIKSLPEELMHNSKLRNLDLGN 207
++++ +T L L+ +LE + L C L L++AHN + S+ EL +LR++ + +
Sbjct: 28 VEQMTQMTWLKLNDSKLEQVPDELSRCANLEHLQMAHNQLISVHGELSDLPRLRSVIVRD 87
Query: 208 NVIAKW---SDIKVLKSLTKLRNLNLQGNPVATN 238
N + +DI +K LT + Q V TN
Sbjct: 88 NNLKTAGIPTDIFRMKDLTIIDLSRNQLREVPTN 121
Score = 33.9 bits (76), Expect = 0.92
Identities = 18/87 (20%), Positives = 44/87 (49%), Gaps = 2/87 (2%)
Query: 124 NLDQMKELNTLVLSKNPIRKIGEALKKVKSITKLSLSHCQLEGIDTSLKFCVELTELRLA 183
+++QM ++ L L+ + + ++ + L + ++ L ++H QL + L L + +
Sbjct: 27 DVEQMTQMTWLKLNDSKLEQVPDELSRCANLEHLQMAHNQLISVHGELSDLPRLRSVIVR 86
Query: 184 HNDIKS--LPEELMHNSKLRNLDLGNN 208
N++K+ +P ++ L +DL N
Sbjct: 87 DNNLKTAGIPTDIFRMKDLTIIDLSRN 113
>LUM_MOUSE (P51885) Lumican precursor (Keratan sulfate proteoglycan
lumican) (KSPG lumican)
Length = 338
Score = 65.5 bits (158), Expect = 3e-10
Identities = 69/233 (29%), Positives = 104/233 (44%), Gaps = 18/233 (7%)
Query: 17 NPSSISSLHLTHKALSDVSC----LASFNKLEKLDLKFNNLTSLEGLRACVTLKWLSVVE 72
N + + L L H L + + +L+KL + +NNLT G +L+ L +
Sbjct: 88 NVTDLQWLILDHNLLENSKIKGKVFSKLKQLKKLHINYNNLTESVGPLP-KSLQDLQLTN 146
Query: 73 NKLESLEGIQGLTKLTVLNAGKNKLKSMDEIGSLSTIRALILNDNEIVSICNLDQ--MKE 130
NK+ L GL LT + N+LK SL +++L D + L
Sbjct: 147 NKISKLGSFDGLVNLTFIYLQHNQLKEDAVSASLKGLKSLEYLDLSFNQMSKLPAGLPTS 206
Query: 131 LNTLVLSKNPIRKI-GEALKKVKSITKLSLSHCQL--EGIDTSLKFCVELTELRLAHNDI 187
L TL L N I I E K+ + L LSH +L G+ + L EL L++N +
Sbjct: 207 LLTLYLDNNKISNIPDEYFKRFTGLQYLRLSHNELADSGVPGNSFNISSLLELDLSYNKL 266
Query: 188 KSLPEELMHNSKLRNLDLGNNVIAKW---SDIKVL--KSLTKLRNLNLQGNPV 235
KS+P N L N L N + K+ S K+L S +K+++L L GNP+
Sbjct: 267 KSIPTV---NENLENYYLEVNELEKFDVKSFCKILGPLSYSKIKHLRLDGNPL 316
Score = 34.3 bits (77), Expect = 0.71
Identities = 41/175 (23%), Positives = 81/175 (45%), Gaps = 15/175 (8%)
Query: 3 RLSSEQVLKDNNAVNPSSISSLHLTHKALSDVSCLASFN---KLEKLDLKFNNLTSL-EG 58
+L++ ++ K + +++ ++L H L + + AS LE LDL FN ++ L G
Sbjct: 143 QLTNNKISKLGSFDGLVNLTFIYLQHNQLKEDAVSASLKGLKSLEYLDLSFNQMSKLPAG 202
Query: 59 LRACVTLKWLSVVENKLESL--EGIQGLTKLTVLNAGKNKLKSMDEIG---SLSTIRALI 113
L + +L NK+ ++ E + T L L N+L G ++S++ L
Sbjct: 203 LPTSLLTLYLD--NNKISNIPDEYFKRFTGLQYLRLSHNELADSGVPGNSFNISSLLELD 260
Query: 114 LNDNEIVSICNLDQMKELNTLVLSKNPIRK--IGEALKKVKSITKLSLSHCQLEG 166
L+ N++ SI +++ L L N + K + K + ++ + H +L+G
Sbjct: 261 LSYNKLKSIPTVNE--NLENYYLEVNELEKFDVKSFCKILGPLSYSKIKHLRLDG 313
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.308 0.128 0.345
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 53,179,347
Number of Sequences: 164201
Number of extensions: 2277046
Number of successful extensions: 13725
Number of sequences better than 10.0: 920
Number of HSP's better than 10.0 without gapping: 176
Number of HSP's successfully gapped in prelim test: 764
Number of HSP's that attempted gapping in prelim test: 9869
Number of HSP's gapped (non-prelim): 2620
length of query: 468
length of database: 59,974,054
effective HSP length: 114
effective length of query: 354
effective length of database: 41,255,140
effective search space: 14604319560
effective search space used: 14604319560
T: 11
A: 40
X1: 16 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.7 bits)
S2: 68 (30.8 bits)
Medicago: description of AC146332.10


