

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC145513.1 - phase: 0 /pseudo
(902 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
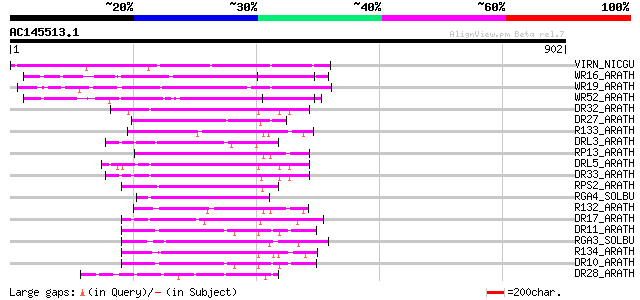
Score E
Sequences producing significant alignments: (bits) Value
VIRN_NICGU (Q40392) TMV resistance protein N 351 4e-96
WR16_ARATH (Q9FL92) Probable WRKY transcription factor 16 (WRKY ... 213 1e-54
WR19_ARATH (Q9SZ67) Probable WRKY transcription factor 19 (WRKY ... 191 7e-48
WR52_ARATH (Q9FH83) Probable WRKY transcription factor 52 (WRKY ... 189 4e-47
DR32_ARATH (Q9FG91) Probable disease resistance protein At5g43730 92 6e-18
DR27_ARATH (Q9T048) Putative disease resistance protein At4g27190 91 2e-17
R133_ARATH (Q9STE7) Putative disease resistance RPP13-like prote... 85 8e-16
DRL3_ARATH (Q9LMP6) Probable disease resistance protein At1g15890 85 1e-15
RP13_ARATH (Q9M667) Disease resistance protein RPP13 (Resistance... 84 2e-15
DRL5_ARATH (Q9C8K0) Probable disease resistance protein At1g51480 80 2e-14
DR33_ARATH (Q9FG90) Probable disease resistance protein At5g43740 80 2e-14
RPS2_ARATH (Q42484) Disease resistance protein RPS2 (Resistance ... 78 1e-13
RGA4_SOLBU (Q7XA39) Putative disease resistance protein RGA4 (RG... 78 1e-13
R132_ARATH (Q9STE5) Putative disease resistance RPP13-like prote... 77 3e-13
DR17_ARATH (O64790) Putative disease resistance protein At1g61300 77 3e-13
DR11_ARATH (Q94HW3) Probable disease resistance protein RDL6/RF9 77 3e-13
RGA3_SOLBU (Q7XA40) Putative disease resistance protein RGA3 (RG... 76 4e-13
R134_ARATH (Q38834) Putative disease resistance RPP13-like prote... 76 4e-13
DR10_ARATH (Q8W3J8) Probable disease resistance protein RDL5/RF45 75 6e-13
DR28_ARATH (O81825) Putative disease resistance protein At4g27220 75 8e-13
>VIRN_NICGU (Q40392) TMV resistance protein N
Length = 1144
Score = 351 bits (901), Expect = 4e-96
Identities = 212/538 (39%), Positives = 316/538 (58%), Gaps = 28/538 (5%)
Query: 1 MSSSSSRSRPQWIHDVFINFRGKDTRKTFVSHLYAALTDAGINTFLDDENLKKGEELGPE 60
M+SSSS SR W +DVF++FRG+DTRKTF SHLY L D GI TF DD+ L+ G + E
Sbjct: 1 MASSSSSSR--WSYDVFLSFRGEDTRKTFTSHLYEVLNDKGIKTFQDDKRLEYGATIPGE 58
Query: 61 LVRAIQGSQIAIVVFSKNYVNSSWCLNELEQIMKCKADNGQVVMPVFNGITPSNIRQHSP 120
L +AI+ SQ AIVVFS+NY S WCLNEL +IM+CK Q V+P+F + PS++R
Sbjct: 59 LCKAIEESQFAIVVFSENYATSRWCLNELVKIMECKTRFKQTVIPIFYDVDPSHVRNQKE 118
Query: 121 VIL------VDELDQIIFGKKR---ALRDVSYLTGWDMSNYSNQSKVVKEIVSQVLKNLD 171
+ + G +R AL + + L G + + +++IV Q+ L
Sbjct: 119 SFAKAFEEHETKYKDDVEGIQRWRIALNEAANLKGSCDNRDKTDADCIRQIVDQISSKLC 178
Query: 172 KKYLPLPNFQVGLKPRAEKPIRFLRQNTRKVCLVGIWGMGGIGKSTIAKVIYN------D 225
K L VG+ EK L V ++GIWGMGG+GK+TIA+ I++ D
Sbjct: 179 KISLSYLQNIVGIDTHLEKIESLLEIGINGVRIMGIWGMGGVGKTTIARAIFDTLLGRMD 238
Query: 226 LCYEFEDQSFVANIREVWEKDRGRIDLQEQLLSDILKTRKIKVLSVEQGKAMIKQRLRSK 285
Y+F+ F+ +I+ E RG LQ LLS++L+ K + E GK + RLRSK
Sbjct: 239 SSYQFDGACFLKDIK---ENKRGMHSLQNALLSELLR-EKANYNNEEDGKHQMASRLRSK 294
Query: 286 RILAVLDDVSELEQFNALCEGNS--VGPGSVIIITTRDLRVLNILEVDFIYEAEGLNASE 343
++L VLDD+ + + G+ G GS IIITTRD ++ + D IYE L E
Sbjct: 295 KVLIVLDDIDNKDHYLEYLAGDLDWFGNGSRIIITTRDKHLIE--KNDIIYEVTALPDHE 352
Query: 344 SLELFCGHAFRKVIPTEDFLILSRYVVAYCGGIPLALEVLGSYLLKRRKQEWQSVLSKLE 403
S++LF HAF K +P E+F LS VV Y G+PLAL+V GS L R EW+S + ++
Sbjct: 353 SIQLFKQHAFGKEVPNENFEKLSLEVVNYAKGLPLALKVWGSLLHNLRLTEWKSAIEHMK 412
Query: 404 KIPNDQIHEKLKISFNGLSDRMEKDIFLDVCCFFIGKDRAYVTKILNGCGLHADIGITVL 463
I +KLKIS++GL + ++++FLD+ CF G+++ Y+ +IL C + A+ G+ +L
Sbjct: 413 NNSYSGIIDKLKISYDGLEPK-QQEMFLDIACFLRGEEKDYILQILESCHIGAEYGLRIL 471
Query: 464 IERSLIKVEKNKKLGMHDLLRDMGREIVRESSPEEPEKRTRLWCHEDVVNVLEDHTTT 521
I++SL+ + + ++ MHDL++DMG+ IV + ++P +R+RLW ++V V+ ++T T
Sbjct: 472 IDKSLVFISEYNQVQMHDLIQDMGKYIV--NFQKDPGERSRLWLAKEVEEVMSNNTGT 527
>WR16_ARATH (Q9FL92) Probable WRKY transcription factor 16 (WRKY
DNA-binding protein 16)
Length = 1372
Score = 213 bits (543), Expect = 1e-54
Identities = 151/498 (30%), Positives = 265/498 (52%), Gaps = 42/498 (8%)
Query: 23 KDTRKTFVSHLYAALTDAGIN-TFLDDENLKKGEELGPELVRAIQGSQIAIVVFSKNYVN 81
++ R +FVSHL AL G+N F+D + + L E ++ +++++++ N
Sbjct: 14 EEVRYSFVSHLSKALQRKGVNDVFIDSD-----DSLSNESQSMVERARVSVMILPGNRTV 68
Query: 82 SSWCLNELEQIMKCKADNGQVVMPVFNGITPSNIRQHSPVILVDELDQIIFGKKRALRDV 141
S L++L +++ C+ + QVV+PV G+ S S AL
Sbjct: 69 S---LDKLVKVLDCQKNKDQVVVPVLYGVRSSETEWLS-----------------ALDSK 108
Query: 142 SYLTGWDMSNYSNQSKVVKEIVSQVLKNLDKKYLPLPNFQVGLKPRAEKPIRFLRQNTRK 201
+ + + S++VKE V V + L Y+ ++G+ + + + + +
Sbjct: 109 GFSSVHHSRKECSDSQLVKETVRDVYEKLF--YME----RIGIYSKLLEIEKMINKQPLD 162
Query: 202 VCLVGIWGMGGIGKSTIAKVIYNDLCYEFEDQSFVANIREVWEKDRGRIDLQEQLLSDIL 261
+ VGIWGM GIGK+T+AK +++ + EF+ F+ + + ++ L+EQ L +
Sbjct: 163 IRCVGIWGMPGIGKTTLAKAVFDQMSGEFDAHCFIEDYTKAIQEKGVYCLLEEQFLKE-- 220
Query: 262 KTRKIKVLSVEQGKAMIKQRLRSKRILAVLDDV-SELEQFNALCEGNSVGPGSVIIITTR 320
++++ RL +KR+L VLDDV S L + L + GP S+IIIT++
Sbjct: 221 ---NAGASGTVTKLSLLRDRLNNKRVLVVLDDVRSPLVVESFLGGFDWFGPKSLIIITSK 277
Query: 321 DLRVLNILEVDFIYEAEGLNASESLELFCGHAFRKVIPTEDFLILSRYVVAYCGGIPLAL 380
D V + V+ IYE +GLN E+L+LF A + ++ +S V+ Y G PLAL
Sbjct: 278 DKSVFRLCRVNQIYEVQGLNEKEALQLFSLCASIDDMAEQNLHEVSMKVIKYANGHPLAL 337
Query: 381 EVLGSYLL-KRRKQEWQSVLSKLEKIPNDQIHEKLKISFNGLSDRMEKDIFLDVCCFFIG 439
+ G L+ K+R E + KL++ P + +K S++ L+DR EK+IFLD+ CFF G
Sbjct: 338 NLYGRELMGKKRPPEMEIAFLKLKECPPAIFVDAIKSSYDTLNDR-EKNIFLDIACFFQG 396
Query: 440 KDRAYVTKILNGCGLHADIGITVLIERSLIKVEKNKKLGMHDLLRDMGREIVRESSPEEP 499
++ YV ++L GCG +GI VL+E+SL+ + +N ++ MH+L++D+GR+I+ + +
Sbjct: 397 ENVDYVMQLLEGCGFFPHVGIDVLVEKSLVTISEN-RVRMHNLIQDVGRQIINRET-RQT 454
Query: 500 EKRTRLWCHEDVVNVLED 517
++R+RLW + +LED
Sbjct: 455 KRRSRLWEPCSIKYLLED 472
Score = 48.5 bits (114), Expect = 8e-05
Identities = 30/93 (32%), Positives = 51/93 (54%), Gaps = 2/93 (2%)
Query: 404 KIPNDQIHEKLKISFNGLSDRMEKDIFLDVCCFFIGKDRAYVTKIL-NGCGLHADIGITV 462
++ ++ E L++ + GL + + K +FL + F +D V ++ N + G+ V
Sbjct: 1041 EVSGNEDEEVLRVRYAGLQE-IYKALFLYIAGLFNDEDVGLVAPLIANIIDMDVSYGLKV 1099
Query: 463 LIERSLIKVEKNKKLGMHDLLRDMGREIVRESS 495
L RSLI+V N ++ MH LLR MG+EI+ S
Sbjct: 1100 LAYRSLIRVSSNGEIVMHYLLRQMGKEILHTES 1132
>WR19_ARATH (Q9SZ67) Probable WRKY transcription factor 19 (WRKY
DNA-binding protein 19)
Length = 1895
Score = 191 bits (485), Expect = 7e-48
Identities = 149/518 (28%), Positives = 268/518 (50%), Gaps = 43/518 (8%)
Query: 14 HDVFINFRGKD-TRKTFVSHLYAALTDAGINTFLDDENLKKGEELGPELVRAIQGSQIAI 72
+DV I + D + + F+SHL A+L GI+ + +K E V A+ ++ I
Sbjct: 668 YDVVIRYGRADISNEDFISHLRASLCRRGISVY------EKFNE-----VDALPKCRVLI 716
Query: 73 VVFSKNYVNSSWCLNELEQIMKCKADNGQVVMPVFNGITP----SNIRQHSPVILVDELD 128
+V + YV S+ L I++ + +VV P+F ++P N + + L DE
Sbjct: 717 IVLTSTYVPSN-----LLNILEHQHTEDRVVYPIFYRLSPYDFVCNSKNYERFYLQDEPK 771
Query: 129 QIIFGKKRALRDVSYLTGWDMSNYSNQSKVVKEIVSQVLKNL---DKKYLPLPNFQVGLK 185
+ + AL++++ + G+ +++ S +S+++ EIV LK L DK + +G+
Sbjct: 772 KW----QAALKEITQMPGYTLTDKS-ESELIDEIVRDALKVLCSADKVNM------IGMD 820
Query: 186 PRAEKPIRFLRQNTRKVCLVGIWGMGGIGKSTIAKVIYNDLCYEFEDQSFVANIREVWEK 245
+ E+ + L + V +GIWG GIGK+TIA+ I+ + ++E + ++ + E
Sbjct: 821 MQVEEILSLLCIESLDVRSIGIWGTVGIGKTTIAEEIFRKISVQYETCVVLKDLHKEVEV 880
Query: 246 DRGRIDLQEQLLSDILKTRKIKVLSVEQGKAMIKQRLRSKRILAVLDDVSELEQFNA-LC 304
+G ++E LS++L+ + + + ++ RL+ KRIL +LDDV++ + L
Sbjct: 881 -KGHDAVRENFLSEVLEVEPHVIRISDIKTSFLRSRLQRKRILVILDDVNDYRDVDTFLG 939
Query: 305 EGNSVGPGSVIIITTRDLRVLNILEVDFIYEAEGLNASESLELFCGHAFRKVIPTEDFLI 364
N GPGS II+T+R+ RV + ++D +YE + L+ +SL L + V+ E +
Sbjct: 940 TLNYFGPGSRIIMTSRNRRVFVLCKIDHVYEVKPLDIPKSLLLLDRGTCQIVLSPEVYKT 999
Query: 365 LSRYVVAYCGGIPLALEVLGSYLLKRRKQEWQSVLSKLEKIPNDQIHEKLKISFNGLSDR 424
LS +V + G P L+ L S +EW + +++ I + S GL D
Sbjct: 1000 LSLELVKFSNGNPQVLQFLSSI-----DREWNKLSQEVKTTSPIYIPGIFEKSCCGLDDN 1054
Query: 425 MEKDIFLDVCCFFIGKDRAYVTKILNGCGLHADIGITVLIERSLIKVEKNKKLGMHDLLR 484
E+ IFLD+ CFF D+ V +L+GCG A +G L+++SL+ + ++ + M ++
Sbjct: 1055 -ERGIFLDIACFFNRIDKDNVAMLLDGCGFSAHVGFRGLVDKSLLTISQHNLVDMLSFIQ 1113
Query: 485 DMGREIVRESSPEEPEKRTRLWCHEDVVNVLEDHTTTN 522
GREIVR+ S + P R+RLW + + +V + T T+
Sbjct: 1114 ATGREIVRQESADRPGDRSRLWNADYIRHVFINDTGTS 1151
>WR52_ARATH (Q9FH83) Probable WRKY transcription factor 52 (WRKY
DNA-binding protein 52) (Disease resistance protein
RRS1) (Resistance to Ralstonia solanacearum 1 protein)
Length = 1378
Score = 189 bits (479), Expect = 4e-47
Identities = 152/494 (30%), Positives = 261/494 (52%), Gaps = 52/494 (10%)
Query: 23 KDTRKTFVSHLYAALTDAGINTFLDDENLKKGEELGPELVRAIQGSQIAIVVFSKNYVNS 82
++ R +FVSHL AL GIN + D ++ + L E I+ + ++++V N S
Sbjct: 17 EEVRYSFVSHLSEALRRKGINNVVVDVDID--DLLFKESQAKIEKAGVSVMVLPGNCDPS 74
Query: 83 SWCLNELEQIMKCKADN-GQVVMPVFNGITPSNIRQHSPVILVDELDQIIFGKKRALRDV 141
L++ ++++C+ +N Q V+ V G D L LRD
Sbjct: 75 EVWLDKFAKVLECQRNNKDQAVVSVLYG---------------DSL----------LRD- 108
Query: 142 SYLTGWDMSNYSNQSKVVKE-----IVSQVLKNLDKKYLPLPNFQVGLKPRAEKPIRFLR 196
+L+ D S + KE +V ++++++ + + + ++G+ + + +
Sbjct: 109 QWLSELDFRGLSRIHQSRKECSDSILVEEIVRDVYETHFYVG--RIGIYSKLLEIENMVN 166
Query: 197 QNTRKVCLVGIWGMGGIGKSTIAKVIYNDLCYEFEDQSFVANI-REVWEKDRGRIDLQEQ 255
+ + VGIWGM GIGK+T+AK +++ + F+ F+ + + + EK + L+EQ
Sbjct: 167 KQPIGIRCVGIWGMPGIGKTTLAKAVFDQMSSAFDASCFIEDYDKSIHEKGLYCL-LEEQ 225
Query: 256 LLSDILKTRKIKVLSVEQGKAMIKQRLRSKRILAVLDDVSELEQFNALCEG-NSVGPGSV 314
LL T I LS ++ RL SKR+L VLDDV + EG + +GPGS+
Sbjct: 226 LLPGNDAT--IMKLS------SLRDRLNSKRVLVVLDDVCNALVAESFLEGFDWLGPGSL 277
Query: 315 IIITTRDLRVLNILEVDFIYEAEGLNASESLELFCGHA-FRKVIPTEDFLILSRYVVAYC 373
IIIT+RD +V + ++ IYE +GLN E+ +LF A + + ++ LS V++Y
Sbjct: 278 IIITSRDKQVFRLCGINQIYEVQGLNEKEARQLFLLSASIMEDMGEQNLHELSVRVISYA 337
Query: 374 GGIPLALEVLGSYLLKRRK-QEWQSVLSKLEKIPNDQIHEKLKISFNGLSDRMEKDIFLD 432
G PLA+ V G L ++K E ++ KL++ P +I + K S++ LSD EK+IFLD
Sbjct: 338 NGNPLAISVYGRELKGKKKLSEMETAFLKLKRRPPFKIVDAFKSSYDTLSDN-EKNIFLD 396
Query: 433 VCCFFIGKDRAYVTKILNGCGLHADIGITVLIERSLIKVEKNKKLGMHDLLRDMGREIVR 492
+ CFF G++ YV ++L GCG + I VL+++ L+ + +N ++ +H L +D+GREI+
Sbjct: 397 IACFFQGENVNYVIQLLEGCGFFPHVEIDVLVDKCLVTISEN-RVWLHKLTQDIGREII- 454
Query: 493 ESSPEEPEKRTRLW 506
+ E+R RLW
Sbjct: 455 NGETVQIERRRRLW 468
Score = 54.7 bits (130), Expect = 1e-06
Identities = 30/85 (35%), Positives = 49/85 (57%), Gaps = 1/85 (1%)
Query: 411 HEKLKISFNGLSDRMEKDIFLDVCCFFIGKDRAYVTKILNGCGLHADIGITVLIERSLIK 470
+E L++S++ L + M+K +FL + F +D +V ++ G L G+ VL + SLI
Sbjct: 1086 YEVLRVSYDDLQE-MDKVLFLYIASLFNDEDVDFVAPLIAGIDLDVSSGLKVLADVSLIS 1144
Query: 471 VEKNKKLGMHDLLRDMGREIVRESS 495
V N ++ MH L R MG+EI+ S
Sbjct: 1145 VSSNGEIVMHSLQRQMGKEILHGQS 1169
>DR32_ARATH (Q9FG91) Probable disease resistance protein At5g43730
Length = 848
Score = 92.0 bits (227), Expect = 6e-18
Identities = 104/365 (28%), Positives = 173/365 (46%), Gaps = 46/365 (12%)
Query: 165 QVLKNLD--KKYLPLPNFQVGLK---PRAEKP-------------IRFLRQNTRKVCLVG 206
+V+KNL+ K+ L NF+V + P+AEK I + ++ +G
Sbjct: 117 KVMKNLEEVKELLSKKNFEVVAQKIIPKAEKKHIQTTVGLDTMVGIAWESLIDDEIRTLG 176
Query: 207 IWGMGGIGKSTIAKVIYNDLCYEFEDQSFVANIREVWEKDRGRIDLQEQLLSDILKTRKI 266
++GMGGIGK+T+ + + N E E + F I V KD +Q+Q+L + ++
Sbjct: 177 LYGMGGIGKTTLLESLNNKFV-ELESE-FDVVIWVVVSKDFQLEGIQDQILGRLRPDKEW 234
Query: 267 KVLSVEQGKAMIKQRLRSKRILAVLDDV-SELEQFNALCEGNSVGPGSVIIITTRDLRVL 325
+ + + ++I L+ K+ + +LDD+ SE++ S GS I+ TTR V
Sbjct: 235 ERETESKKASLINNNLKRKKFVLLLDDLWSEVDLIKIGVPPPSRENGSKIVFTTRSKEVC 294
Query: 326 NILEVDFIYEAEGLNASESLELFCGHAFRKVIPT-EDFLILSRYVVAYCGGIPLALEVLG 384
++ D + + L+ E+ ELF ++ + +D L+R V A C G+PLAL V+G
Sbjct: 295 KHMKADKQIKVDCLSPDEAWELFRLTVGDIILRSHQDIPALARIVAAKCHGLPLALNVIG 354
Query: 385 SYLL-KRRKQEWQSVLSKL----EKIP--NDQIHEKLKISFNGLSDRMEKDIFLDVCCFF 437
++ K QEW+ ++ L K P ++I LK S++ L + K FL C F
Sbjct: 355 KAMVCKETVQEWRHAINVLNSPGHKFPGMEERILPILKFSYDSLKNGEIKLCFL-YCSLF 413
Query: 438 -----IGKDRAYVTKILNGC---------GLHADIGITVLIERS--LIKVEKNKKLGMHD 481
I KD+ I G G + I L+ R+ LI+ E K+ MHD
Sbjct: 414 PEDFEIEKDKLIEYWICEGYINPNRYEDGGTNQGYDIIGLLVRAHLLIECELTDKVKMHD 473
Query: 482 LLRDM 486
++R+M
Sbjct: 474 VIREM 478
>DR27_ARATH (Q9T048) Putative disease resistance protein At4g27190
Length = 985
Score = 90.5 bits (223), Expect = 2e-17
Identities = 70/259 (27%), Positives = 132/259 (50%), Gaps = 11/259 (4%)
Query: 199 TRKVCLVGIWGMGGIGKSTIAKVIYNDLCYEFEDQSFVANIREVWEKDRGRIDLQEQLLS 258
+ K +G+WGMGG+GK+T+ + + N L E Q F I + K+ ++Q+Q+
Sbjct: 161 SEKAQKIGVWGMGGVGKTTLVRTLNNKLREEGATQPFGLVIFVIVSKEFDPREVQKQIAE 220
Query: 259 DILKTRKIKVLSVEQGKAMIKQRLRSKRILAVLDDVSELEQFNAL-CEGNSVGPGSVIII 317
+ +++ + + + ++ ++ L +LDDV + + L GS +I+
Sbjct: 221 RLDIDTQMEESEEKLARRIYVGLMKERKFLLILDDVWKPIDLDLLGIPRTEENKGSKVIL 280
Query: 318 TTRDLRVLNILEVDFIYEAEGLNASESLELFCGHAFRKVIPTEDFLILSRYVVAYCGGIP 377
T+R L V ++ D + L ++ ELFC +A V+ ++ +++ V CGG+P
Sbjct: 281 TSRFLEVCRSMKTDLDVRVDCLLEEDAWELFCKNA-GDVVRSDHVRKIAKAVSQECGGLP 339
Query: 378 LALEVLGSYLL-KRRKQEWQSVLSKLEK-IP-----NDQIHEKLKISFNGLSDRMEKDIF 430
LA+ +G+ + K+ + W VLSKL K +P ++I + LK+S++ L D+ K F
Sbjct: 340 LAIITVGTAMRGKKNVKLWNHVLSKLSKSVPWIKSIEEKIFQPLKLSYDFLEDK-AKFCF 398
Query: 431 LDVCCFFIGKDRAYVTKIL 449
L +C F VT+++
Sbjct: 399 L-LCALFPEDYSIEVTEVV 416
>R133_ARATH (Q9STE7) Putative disease resistance RPP13-like protein
3
Length = 847
Score = 85.1 bits (209), Expect = 8e-16
Identities = 89/339 (26%), Positives = 158/339 (46%), Gaps = 47/339 (13%)
Query: 192 IRFLRQNTR-KVCLVGIWGMGGIGKSTIAKVIYN--DLCYEFEDQSFVANIREVWEKDRG 248
++ L N + K ++ I+GMGG+GK+ +A+ +YN D+ F+ +++ +E +D
Sbjct: 174 VKLLSDNEKDKSYIISIFGMGGLGKTALARKLYNSGDVKRRFDCRAWTYVSQEYKTRDIL 233
Query: 249 RIDLQEQLLSDILKTRKIKVLSV-EQGKAMIKQRLRSKRILAVLDDVSELEQFNAL---- 303
++ + + KIK+ E+ + + L K + V+DDV + + + +L
Sbjct: 234 IRIIRSLGIVSAEEMEKIKMFEEDEELEVYLYGLLEGKNYMVVVDDVWDPDAWESLKRAL 293
Query: 304 -CEGNSVGPGSVIIITTRDLRVLNILE-VDFIYEAEGLNASESLELFCGHAFRKVIPT-E 360
C+ GS +IITTR + +E + ++ L ES LF AF + E
Sbjct: 294 PCDHR----GSKVIITTRIRAIAEGVEGTVYAHKLRFLTFEESWTLFERKAFSNIEKVDE 349
Query: 361 DFLILSRYVVAYCGGIPLALEVLGSYLLKRRKQEWQSVLSKL-EKIPNDQIH-------- 411
D + +V CGG+PLA+ VL L ++R EW V + L ++ ++ IH
Sbjct: 350 DLQRTGKEMVKKCGGLPLAIVVLSGLLSRKRTNEWHEVCASLWRRLKDNSIHISTVFDLS 409
Query: 412 -----EKLKISF-------NGLSDRMEKDIFLDVCCFFIGKDRAYVTKILNGCGLHADIG 459
+LK+ F ++EK I L V FI +D + + + C
Sbjct: 410 FKEMRHELKLCFLYFSVFPEDYEIKVEKLIHLLVAEGFIQEDEEMMMEDVARC------Y 463
Query: 460 ITVLIERSLIKVEKNKK-----LGMHDLLRDMGREIVRE 493
I L++RSL+K E+ ++ +HDLLRD+ + +E
Sbjct: 464 IDELVDRSLVKAERIERGKVMSCRIHDLLRDLAIKKAKE 502
>DRL3_ARATH (Q9LMP6) Probable disease resistance protein At1g15890
Length = 921
Score = 84.7 bits (208), Expect = 1e-15
Identities = 90/297 (30%), Positives = 141/297 (47%), Gaps = 27/297 (9%)
Query: 156 SKVVKEIVSQVLK--NLDKKYLPLPNFQVGLKPRAEKPIRFLRQNTRKVCLVGIWGMGGI 213
+K V E+V++ + ++KK++ VGL + L ++ R+ +G++GMGG+
Sbjct: 201 AKGVFEVVAEKIPAPKVEKKHIQTT---VGLDAMVGRAWNSLMKDERRT--LGLYGMGGV 255
Query: 214 GKSTIAKVIYNDLCYEFEDQSFVANIREVWEKDRGRIDLQEQLLSDILKTRKIKVLSVEQ 273
GK+T+ I N V I V KD +QEQ+L + R K ++ ++
Sbjct: 256 GKTTLLASINNKFLEGMNGFDLV--IWVVVSKDLQNEGIQEQILGRLGLHRGWKQVTEKE 313
Query: 274 GKAMIKQRLRSKRILAVLDDV-SELEQFNALCEGNSVGPGSVIIITTRDLRVLNILEVDF 332
+ I L K+ + +LDD+ SE++ + GS I+ TTR V +EVD
Sbjct: 314 KASYICNILNVKKFVLLLDDLWSEVDLEKIGVPPLTRENGSKIVFTTRSKDVCRDMEVDG 373
Query: 333 IYEAEGLNASESLELFCGHAFRKVIPT-----EDFLILSRYVVAYCGGIPLALEVLGSYL 387
+ + L E+ ELF +KV P ED L+R V C G+PLAL V+G +
Sbjct: 374 EMKVDCLPPDEAWELFQ----KKVGPIPLQSHEDIPTLARKVAEKCCGLPLALSVIGKAM 429
Query: 388 LKRRK-QEWQSVL----SKLEKIPN--DQIHEKLKISFNGLSDRMEKDIFLDVCCFF 437
R QEWQ V+ S + P+ ++I LK S++ L D K FL C F
Sbjct: 430 ASRETVQEWQHVIHVLNSSSHEFPSMEEKILPVLKFSYDDLKDEKVKLCFL-YCSLF 485
>RP13_ARATH (Q9M667) Disease resistance protein RPP13 (Resistance to
Peronospora parasitica protein 13)
Length = 835
Score = 83.6 bits (205), Expect = 2e-15
Identities = 90/315 (28%), Positives = 148/315 (46%), Gaps = 38/315 (12%)
Query: 204 LVGIWGMGGIGKSTIAKVIYN--DLCYEFEDQSFVANIREVWEKDRGRIDLQEQLLSDIL 261
++ I+GMGG+GK+ +A+ +YN D+ FE +++ +E D ++ ++
Sbjct: 187 IISIFGMGGLGKTALARKLYNSRDVKERFEYRAWTYVSQEYKTGDILMRIIRSLGMTSGE 246
Query: 262 KTRKIKVLSVEQGKAMIKQRLRSKRILAVLDDVSELEQFNALCEGNSVG-PGSVIIITTR 320
+ KI+ + E+ + + L K+ L V+DD+ E E +++L GS +IITTR
Sbjct: 247 ELEKIRKFAEEELEVYLYGLLEGKKYLVVVDDIWEREAWDSLKRALPCNHEGSRVIITTR 306
Query: 321 DLRVLNILEVDFI-YEAEGLNASESLELFCGHAFRKVI-PTEDFLILSRYVVAYCGGIPL 378
V ++ F ++ L ES ELF AFR + ED L + +V C G+PL
Sbjct: 307 IKAVAEGVDGRFYAHKLRFLTFEESWELFEQRAFRNIQRKDEDLLKTGKEMVQKCRGLPL 366
Query: 379 ALEVLGSYLLKRRKQEWQSVLSKL-EKIPNDQIH--------------EKLKISFNGLS- 422
+ VL L ++ EW V + L ++ +D IH + K+ F LS
Sbjct: 367 CIVVLAGLLSRKTPSEWNDVCNSLWRRLKDDSIHVAPIVFDLSFKELRHESKLCFLYLSI 426
Query: 423 ------DRMEKDIFLDVCCFFIGKDRAYVTKILNGCGLHADIGITVLIERSLIKV---EK 473
+EK I L V FI D + + + A I LI+RSL++ E+
Sbjct: 427 FPEDYEIDLEKLIHLLVAEGFIQGDEEMMMEDV------ARYYIEELIDRSLLEAVRRER 480
Query: 474 NKKLG--MHDLLRDM 486
K + +HDLLRD+
Sbjct: 481 GKVMSCRIHDLLRDV 495
>DRL5_ARATH (Q9C8K0) Probable disease resistance protein At1g51480
Length = 854
Score = 80.1 bits (196), Expect = 2e-14
Identities = 104/379 (27%), Positives = 172/379 (44%), Gaps = 48/379 (12%)
Query: 149 MSNYSNQSKVVKEIVSQVLKNLDKKY-------LPLPNFQ-------VGLKPRAEKPIRF 194
+S+Y+ KV+K + +V + L KK+ +P+P + VGL E +
Sbjct: 109 ISSYNYGEKVMKNL-EEVKELLSKKHFEVVAHKIPVPKVEEKNIHTTVGLYAMVEMAWKS 167
Query: 195 LRQNT-RKVCLVGIWGMGGIGKSTIAKVIYNDLCYEFEDQSFVANIREVWEKDRGRIDLQ 253
L + R +CL GMGG+GK+T+ I N E E + F I V KD +Q
Sbjct: 168 LMNDEIRTLCL---HGMGGVGKTTLLACINNKFV-ELESE-FDVVIWVVVSKDFQLEGIQ 222
Query: 254 EQLLSDILKTRKIKVLSVEQGKAMIKQRLRSKRILAVLDDV-SELEQFNALCEGNSVGPG 312
+Q+L + ++ + + + ++I L+ K+ + +LDD+ SE++ + G
Sbjct: 223 DQILGRLRLDKEWERETENKKASLINNNLKRKKFVLLLDDLWSEVDLNKIGVPPPTRENG 282
Query: 313 SVIIITTRDLRVLNILEVDFIYEAEGLNASESLELFCGHAFRKVIPT-EDFLILSRYVVA 371
+ I+ T R V ++ D + L+ E+ ELF ++ + ED L+R V A
Sbjct: 283 AKIVFTKRSKEVSKYMKADMQIKVSCLSPDEAWELFRITVDDVILSSHEDIPALARIVAA 342
Query: 372 YCGGIPLALEVLGSYL-LKRRKQEWQSVLSKL-----EKIP--NDQIHEKLKISFNGLSD 423
C G+PLAL V+G + K QEW ++ L K P ++I LK S++ L +
Sbjct: 343 KCHGLPLALIVIGEAMACKETIQEWHHAINVLNSPAGHKFPGMEERILLVLKFSYDSLKN 402
Query: 424 RMEKDIFLDVCCFF-----IGKDRAYVTKILNGC---------GLHADIGITVLIERS-- 467
K FL C F I K++ I G G + I L+ R+
Sbjct: 403 GEIKLCFL-YCSLFPEDFEIEKEKLIEYWICEGYINPNRYEDGGTNQGYDIIGLLVRAHL 461
Query: 468 LIKVEKNKKLGMHDLLRDM 486
LI+ E K+ MH ++R+M
Sbjct: 462 LIECELTTKVKMHYVIREM 480
>DR33_ARATH (Q9FG90) Probable disease resistance protein At5g43740
Length = 862
Score = 80.1 bits (196), Expect = 2e-14
Identities = 92/355 (25%), Positives = 162/355 (44%), Gaps = 33/355 (9%)
Query: 157 KVVKEIVSQVLKNLDKKYLPLPNFQVGLKPRAEKPIRFLRQNTRKVCLVGIWGMGGIGKS 216
K + + +++ ++KK + VGL E L + ++ +G++GMGG+GK+
Sbjct: 131 KDFRMVAQEIIHKVEKKLIQTT---VGLDKLVEMAWSSLMND--EIGTLGLYGMGGVGKT 185
Query: 217 TIAKVIYNDLCYEFEDQSFVANIREVWEKDRGRIDLQEQLLSDILKTRKIKVLSVEQGKA 276
T+ + + N E E + F I V KD +Q+Q+L + ++ + + + +
Sbjct: 186 TLLESLNNKFV-ELESE-FDVVIWVVVSKDFQFEGIQDQILGRLRSDKEWERETESKKAS 243
Query: 277 MIKQRLRSKRILAVLDDV-SELEQFNALCEGNSVGPGSVIIITTRDLRVLNILEVDFIYE 335
+I L K+ + +LDD+ SE++ + GS I+ TTR V ++ D +
Sbjct: 244 LIYNNLERKKFVLLLDDLWSEVDMTKIGVPPPTRENGSKIVFTTRSTEVCKHMKADKQIK 303
Query: 336 AEGLNASESLELFCGHAFRKVIPT-EDFLILSRYVVAYCGGIPLALEVLGSYL-LKRRKQ 393
L+ E+ ELF ++ + +D L+R V A C G+PLAL V+G + K Q
Sbjct: 304 VACLSPDEAWELFRLTVGDIILRSHQDIPALARIVAAKCHGLPLALNVIGKAMSCKETIQ 363
Query: 394 EWQSVLSKLEKIPN------DQIHEKLKISFNGLSDRMEKDIFLDVCCFF-----IGKDR 442
EW ++ L + ++I LK S++ L + K FL C F I K++
Sbjct: 364 EWSHAINVLNSAGHEFPGMEERILPILKFSYDSLKNGEIKLCFL-YCSLFPEDSEIPKEK 422
Query: 443 AYVTKILNGC---------GLHADIGITVLIERS--LIKVEKNKKLGMHDLLRDM 486
I G G + I L+ R+ LI+ E + MHD++R+M
Sbjct: 423 WIEYWICEGFINPNRYEDGGTNHGYDIIGLLVRAHLLIECELTDNVKMHDVIREM 477
>RPS2_ARATH (Q42484) Disease resistance protein RPS2 (Resistance to
Pseudomonas syringae protein 2)
Length = 909
Score = 77.8 bits (190), Expect = 1e-13
Identities = 66/264 (25%), Positives = 121/264 (45%), Gaps = 13/264 (4%)
Query: 182 VGLKPRAEKPIRFLRQNTRKVCLVGIWGMGGIGKSTIAKVIYNDLCYEFEDQSFVANIRE 241
VG E+ + FL + + ++G++G GG+GK+T+ + I N+L + + ++
Sbjct: 156 VGNTTMMEQVLEFLSEEEERG-IIGVYGPGGVGKTTLMQSINNELITKGHQYDVLIWVQ- 213
Query: 242 VWEKDRGRIDLQEQLLSDILKTRKIKVLSVEQGKAMIKQRLRSKRILAVLDDV-SELEQF 300
++ G +Q+ + + + + K E I + LR KR L +LDDV E++
Sbjct: 214 -MSREFGECTIQQAVGARLGLSWDEKETG-ENRALKIYRALRQKRFLLLLDDVWEEIDLE 271
Query: 301 NALCEGNSVGPGSVIIITTRDLRVLNILEVDFIYEAEGLNASESLELFCGHAFRK-VIPT 359
++ TTR + + N + ++ E L + ELFC +RK ++ +
Sbjct: 272 KTGVPRPDRENKCKVMFTTRSIALCNNMGAEYKLRVEFLEKKHAWELFCSKVWRKDLLES 331
Query: 360 EDFLILSRYVVAYCGGIPLALEVLGSYLLKRR-KQEWQSVLSKLEKIPNDQ-----IHEK 413
L+ +V+ CGG+PLAL LG + R ++EW L + P + +
Sbjct: 332 SSIRRLAEIIVSKCGGLPLALITLGGAMAHRETEEEWIHASEVLTRFPAEMKGMNYVFAL 391
Query: 414 LKISFNGLSDRMEKDIFLDVCCFF 437
LK S++ L + + FL C F
Sbjct: 392 LKFSYDNLESDLLRSCFL-YCALF 414
>RGA4_SOLBU (Q7XA39) Putative disease resistance protein RGA4
(RGA4-blb)
Length = 988
Score = 77.8 bits (190), Expect = 1e-13
Identities = 69/223 (30%), Positives = 118/223 (51%), Gaps = 12/223 (5%)
Query: 207 IWGMGGIGKSTIAKVIYNDLCYEFEDQSFVANIREVWEKDRGRIDLQEQLLSDILKTRKI 266
I GMGG+GK+T+A++I+ND E + F I D L + ++ +I ++
Sbjct: 182 IIGMGGLGKTTLAQMIFND---ERVTKHFNPKIWVCVSDDFDEKRLIKTIIGNIERSSP- 237
Query: 267 KVLSVEQGKAMIKQRLRSKRILAVLDDV--SELEQFNALCEGNSVGP-GSVIIITTRDLR 323
V + + +++ L KR L VLDDV +LE++ L +VG G+ I+ TTR +
Sbjct: 238 HVEDLASFQKKLQELLNGKRYLLVLDDVWNDDLEKWAKLRAVLTVGARGASILATTRLEK 297
Query: 324 VLNILEVDFIYEAEGLNASESLELFCGHAF-RKVIPTEDFLILSRYVVAYCGGIPLALEV 382
V +I+ Y L+ +SL LF AF ++ + + + + +V CGG+PLA +
Sbjct: 298 VGSIMGTLQPYHLSNLSPHDSLLLFMQRAFGQQKEANPNLVAIGKEIVKKCGGVPLAAKT 357
Query: 383 LGSYL-LKRRKQEWQSVL-SKLEKIPNDQ--IHEKLKISFNGL 421
LG L KR + EW+ V +++ +P D+ I L++S++ L
Sbjct: 358 LGGLLRFKREESEWEHVRDNEIWSLPQDESSILPALRLSYHHL 400
>R132_ARATH (Q9STE5) Putative disease resistance RPP13-like protein
2
Length = 847
Score = 76.6 bits (187), Expect = 3e-13
Identities = 88/327 (26%), Positives = 149/327 (44%), Gaps = 64/327 (19%)
Query: 201 KVCLVGIWGMGGIGKSTIAKVIYN--DLCYEFEDQSFVANIREVWEKDRGRI---DLQEQ 255
K+ ++ I+GM G+GK+++A+ ++N D+ FE + VW G D+ +
Sbjct: 183 KIYMISIFGMEGLGKTSLARKLFNSSDVKESFEYR--------VWTNVSGECNTRDILMR 234
Query: 256 LLSDILKTRK--IKVLSVEQGKAMIKQRLRSKRILAVLDDVSELEQFNALCEGNSVG-PG 312
++S + +T + ++ ++ ++ + + L+ KR L V+DD+ E E +L G
Sbjct: 235 IISSLEETSEGELEKMAQQELEVYLHDILQEKRYLVVVDDIWESEALESLKRALPCSYQG 294
Query: 313 SVIIITT--------RDLRVLNILEVDFIYEAEGLNASESLELFCGHAFRKVIPTEDFLI 364
S +IITT RD RV + + L ES LF AFR ++ + L
Sbjct: 295 SRVIITTSIRVVAEGRDKRV-------YTHNIRFLTFKESWNLFEKKAFRYILKVDQELQ 347
Query: 365 -LSRYVVAYCGGIPLALEVLGSYLLKRRKQEWQSVLSKLEKIPNDQIH------------ 411
+ + +V CGG+P VL + +++ EW V S L ++ +D IH
Sbjct: 348 KIGKEMVQKCGGLPRTTVVLAGLMSRKKPNEWNDVWSSL-RVKDDNIHVSSLFDLSFKDM 406
Query: 412 -EKLKISFNGLS-------DRMEKDIFLDVCCFFIGKDRAYVTKILNGCGLHADIGITVL 463
+LK+ F LS +EK I L V FI +D + + A I L
Sbjct: 407 GHELKLCFLYLSVFPEDYEVDVEKLIQLLVAEGFIQEDEEMTMEDV------ARYYIEDL 460
Query: 464 IERSLIKVEKNKK-----LGMHDLLRD 485
+ SL++V K KK +HDL+R+
Sbjct: 461 VYISLVEVVKRKKGKLMSFRIHDLVRE 487
>DR17_ARATH (O64790) Putative disease resistance protein At1g61300
Length = 766
Score = 76.6 bits (187), Expect = 3e-13
Identities = 91/358 (25%), Positives = 152/358 (42%), Gaps = 38/358 (10%)
Query: 182 VGLKPRAEKPIRFLRQNTRKVCLVGIWGMGGIGKSTIAKVIYNDLCYEFEDQSFVANIRE 241
+G + EK L ++ +V ++G+ GMGG+GK+T+ K I+N + F I
Sbjct: 43 IGQEEMLEKAWNRLMED--RVGIMGLHGMGGVGKTTLFKKIHNK--FAKMSSRFDIVIWI 98
Query: 242 VWEKDRGRIDLQEQLLSDILKTRKIKVLSVEQGKAM-IKQRLRSKRILAVLDDVSELEQF 300
V K LQE + + + E KA I + L+ KR + +LDD+ E
Sbjct: 99 VVSKGAKLSKLQEDIAEKLHLCDDLWKNKNESDKATDIHRVLKGKRFVLMLDDIWEKVDL 158
Query: 301 NALCEGNSVGPGSV----IIITTRDLRVLNILEVDFIYEAEGLNASESLELFCGHAFRKV 356
A+ P V + TTRD +V + + + L ++ ELF
Sbjct: 159 EAI---GVPYPSEVNKCKVAFTTRDQKVCGEMGDHKPMQVKCLEPEDAWELFKNKVGDNT 215
Query: 357 IPTEDFLI-LSRYVVAYCGGIPLALEVLGSYLL-KRRKQEWQSVLSKLEKIP------ND 408
+ ++ ++ L+R V C G+PLAL V+G + K QEW+ + L + +
Sbjct: 216 LRSDPVIVELAREVAQKCRGLPLALSVIGETMASKTMVQEWEHAIDVLTRSAAEFSNMGN 275
Query: 409 QIHEKLKISFNGLSDRMEKDIFLDVCCFFIGKDRAYVTKILNGCGLHADIGITVLIER-- 466
+I LK S++ L D K FL C F D Y K+++ IG +I+R
Sbjct: 276 KILPILKYSYDSLGDEHIKSCFL-YCALFPEDDEIYNEKLIDYWICEGFIGEDQVIKRAR 334
Query: 467 -------------SLIKVEKNKKLGMHDLLRDMGREIVRESSPEEPE--KRTRLWCHE 509
+L+ + + MHD++R+M I + ++ R R+ HE
Sbjct: 335 NKGYEMLGTLTLANLLTKVGTEHVVMHDVVREMALWIASDFGKQKENFVVRARVGLHE 392
>DR11_ARATH (Q94HW3) Probable disease resistance protein RDL6/RF9
Length = 1049
Score = 76.6 bits (187), Expect = 3e-13
Identities = 91/364 (25%), Positives = 166/364 (45%), Gaps = 61/364 (16%)
Query: 182 VGLKPRAEKPIRFLRQNTRKVCLVGIWGMGGIGKSTIAKVIYN--DLCYEFEDQSFVANI 239
VGL+ +K + +L V +V I GMGG+GK+T+AK ++N D+ ++F+ S+V
Sbjct: 164 VGLEANVKKLVGYLVDEAN-VQVVSITGMGGLGKTTLAKQVFNHEDVKHQFDGLSWVC-- 220
Query: 240 REVWEKDRGRIDLQEQLLSDIL---KTRKIKVLSVEQGKAMIKQRLRSKRILAVLDDVSE 296
+D R+++ +++L D+ + +KI ++ + + + + L + + L VLDD+ E
Sbjct: 221 ---VSQDFTRMNVWQKILRDLKPKEEEKKIMEMTQDTLQGELIRLLETSKSLIVLDDIWE 277
Query: 297 LEQFNALCEGNSVGPGSVIIITTRDLRVLNILEVDFI-YEAEGLNASESLELFCGHAFRK 355
E + + G +++T+R+ V +I ++ E L +S LF R
Sbjct: 278 KEDWELIKPIFPPTKGWKVLLTSRNESVAMRRNTSYINFKPECLTTEDSWTLF----QRI 333
Query: 356 VIPTED---FLI------LSRYVVAYCGGIPLALEVLGSYLL-KRRKQEWQSVLSKL--- 402
+P +D F I L + ++ +CGG+PLA+ VLG L K +W+ + +
Sbjct: 334 ALPMKDAAEFKIDEEKEELGKLMIKHCGGLPLAIRVLGGMLAEKYTSHDWRRLSENIGSH 393
Query: 403 --------EKIPNDQIHEKLKISFNGLSDRMEKDIFLDVCCF------------------ 436
N+ + L +SF L + K FL + F
Sbjct: 394 LVGGRTNFNDDNNNTCNYVLSLSFEELPSYL-KHCFLYLAHFPDDYEINVKNLSYYWAAE 452
Query: 437 FIGKDRAYVTKILNGCGLHADIGITVLIERSLIKVEKNKKLGMHDL--LRDMGREIVRES 494
I + R Y +I+ G D+ I L+ R+++ E++ K + L DM RE+
Sbjct: 453 GIFQPRHYDGEIIRDVG---DVYIEELVRRNMVISERDVKTSRFETCHLHDMMREVCLLK 509
Query: 495 SPEE 498
+ EE
Sbjct: 510 AKEE 513
>RGA3_SOLBU (Q7XA40) Putative disease resistance protein RGA3
(RGA1-blb) (Blight resistance protein B149)
Length = 947
Score = 76.3 bits (186), Expect = 4e-13
Identities = 88/374 (23%), Positives = 176/374 (46%), Gaps = 53/374 (14%)
Query: 183 GLKPRAEKPIRFLRQN---TRKVCLVGIWGMGGIGKSTIAKVIYNDLCYEFEDQSFVANI 239
G + ++ ++ L N + +V ++ I GMGG+GK+T+A++++N DQ +
Sbjct: 153 GREKEEDEIVKILINNVSYSEEVPVLPILGMGGLGKTTLAQMVFN-------DQRITEHF 205
Query: 240 R-EVWEKDRGRIDLQEQLLSDILKT---RKIKVLSVEQGKAMIKQRLRSKRILAVLDDV- 294
++W D +++L+ I+++ + + + + + +++ L KR VLDDV
Sbjct: 206 NLKIWVCVSDDFD-EKRLIKAIVESIEGKSLGDMDLAPLQKKLQELLNGKRYFLVLDDVW 264
Query: 295 -SELEQFNALCEGNSVG-PGSVIIITTRDLRVLNILEVDFIYEAEGLNASESLELFCGHA 352
+ E+++ L +G G+ I+ITTR ++ +I+ +Y+ L+ + LF A
Sbjct: 265 NEDQEKWDNLRAVLKIGASGASILITTRLEKIGSIMGTLQLYQLSNLSQEDCWLLFKQRA 324
Query: 353 FRKVIPTEDFLI-LSRYVVAYCGGIPLALEVLGSYL-LKRRKQEWQSVL-SKLEKIPNDQ 409
F T L+ + + +V CGG+PLA + LG L KR + EW+ V S++ +P D+
Sbjct: 325 FCHQTETSPKLMEIGKEIVKKCGGVPLAAKTLGGLLRFKREESEWEHVRDSEIWNLPQDE 384
Query: 410 --IHEKLKISFNGL----------------SDRMEKDIFLDVCCFFIGKDRAYVTKILNG 451
+ L++S++ L ++EK+ + I A+ + G
Sbjct: 385 NSVLPALRLSYHHLPLDLRQCFAYCAVFPKDTKIEKE-------YLIALWMAHSFLLSKG 437
Query: 452 CGLHADIGITVLIERSL------IKVEKNKK-LGMHDLLRDMGREIVRESSPEEPEKRTR 504
D+G V E L I+V+ K MHDL+ D+ + S+ ++
Sbjct: 438 NMELEDVGNEVWNELYLRSFFQEIEVKSGKTYFKMHDLIHDLATSMFSASASSRSIRQIN 497
Query: 505 LWCHEDVVNVLEDH 518
+ ED++ ++ ++
Sbjct: 498 VKDDEDMMFIVTNY 511
>R134_ARATH (Q38834) Putative disease resistance RPP13-like protein
4
Length = 852
Score = 76.3 bits (186), Expect = 4e-13
Identities = 87/364 (23%), Positives = 156/364 (41%), Gaps = 61/364 (16%)
Query: 182 VGLKPRAEKPIRFL-RQNTRKVCLVGIWGMGGIGKSTIAKVIYND--LCYEFEDQSFVAN 238
VGL+ K +L R N ++ ++ GMGG+GK+TIA+ ++ND + + FE
Sbjct: 161 VGLEGDKRKIKEWLFRSNDSQLLIMAFVGMGGLGKTTIAQEVFNDKEIEHRFE------- 213
Query: 239 IREVWEKDRGRIDLQEQLLSDILKTRKIKVLSVEQGKAM--IKQRLRSKRILAVLDDV-- 294
R +W + +EQ++ IL+ + + G + I+Q L KR L V+DDV
Sbjct: 214 -RRIW-VSVSQTFTEEQIMRSILRNLGDASVGDDIGTLLRKIQQYLLGKRYLIVMDDVWD 271
Query: 295 SELEQFNALCEGNSVGPGSVIIITTRDLRVLNILEV--DFIYEAEGLNASESLELFCGHA 352
L ++ + +G G G +I+TTR V ++ D + E L+ S LFC A
Sbjct: 272 KNLSWWDKIYQGLPRGQGGSVIVTTRSESVAKRVQARDDKTHRPELLSPDNSWLLFCNVA 331
Query: 353 FRK---VIPTEDFLILSRYVVAYCGGIPLALEVLGSYLLKRRK--QEWQSVLSKLE---- 403
F + + + +V C G+PL ++ +G LL + EW+ + +
Sbjct: 332 FAANDGTCERPELEDVGKEIVTKCKGLPLTIKAVGGLLLCKDHVYHEWRRIAEHFQDELR 391
Query: 404 --KIPNDQIHEKLKISFNGLSDRMEK-----DIFLDVCC--------------FFIGKDR 442
D + L++S++ L ++ ++ + C F + ++
Sbjct: 392 GNTSETDNVMSSLQLSYDELPSHLKSCILTLSLYPEDCVIPKQQLVHGWIGEGFVMWRNG 451
Query: 443 AYVTKILNGCGLHADIGITVLIERSLIKVEKNKKLG------MHDLLRDMGREIVRESSP 496
T+ C + L R LI+V G +HD++RD+ +I ++ S
Sbjct: 452 RSATESGEDC-------FSGLTNRCLIEVVDKTYSGTIITCKIHDMVRDLVIDIAKKDSF 504
Query: 497 EEPE 500
PE
Sbjct: 505 SNPE 508
>DR10_ARATH (Q8W3J8) Probable disease resistance protein RDL5/RF45
Length = 855
Score = 75.5 bits (184), Expect = 6e-13
Identities = 90/364 (24%), Positives = 165/364 (44%), Gaps = 61/364 (16%)
Query: 182 VGLKPRAEKPIRFLRQNTRKVCLVGIWGMGGIGKSTIAKVIYN--DLCYEFEDQSFVANI 239
VGL+ +K + +L V +V I GMGG+GK+T+AK ++N D+ ++F+ S+V
Sbjct: 164 VGLEANVKKLVGYLVDEAN-VQVVSITGMGGLGKTTLAKQVFNHEDVKHQFDGLSWVC-- 220
Query: 240 REVWEKDRGRIDLQEQLLSDIL---KTRKIKVLSVEQGKAMIKQRLRSKRILAVLDDVSE 296
+D R+++ +++L D+ + +KI ++ + + + + L + + L VLDD+ E
Sbjct: 221 ---VSQDFTRMNVWQKILRDLKPKEEEKKIMEMTQDTLQGELIRLLETSKSLIVLDDIWE 277
Query: 297 LEQFNALCEGNSVGPGSVIIITTRDLRVLNILEVDFI-YEAEGLNASESLELFCGHAFRK 355
E + + G +++T+R+ V +I ++ E L +S LF R
Sbjct: 278 KEDWELIKPIFPPTKGWKVLLTSRNESVAMRRNTSYINFKPECLTTEDSWTLF----QRI 333
Query: 356 VIPTED---FLI------LSRYVVAYCGGIPLALEVLGSYLL-KRRKQEWQSVLSKL--- 402
+P +D F I L + ++ +CGG+PLA+ VLG L K +W+ + +
Sbjct: 334 ALPMKDAAEFKIDEEKEELGKLMIKHCGGLPLAIRVLGGMLAEKYTSHDWRRLSENIGSH 393
Query: 403 --------EKIPNDQIHEKLKISFNGLSDRMEKDIFLDVCCF------------------ 436
N+ + L +SF L + K FL + F
Sbjct: 394 LVGGRTNFNDDNNNTCNNVLSLSFEELPSYL-KHCFLYLAHFPEDYEIKVENLSYYWAAE 452
Query: 437 FIGKDRAYVTKILNGCGLHADIGITVLIERSLIKVEKNKKLGMHDL--LRDMGREIVRES 494
I + R Y + + G D+ I L+ R+++ E++ K + L DM RE+
Sbjct: 453 GIFQPRHYDGETIRDVG---DVYIEELVRRNMVISERDVKTSRFETCHLHDMMREVCLLK 509
Query: 495 SPEE 498
+ EE
Sbjct: 510 AKEE 513
>DR28_ARATH (O81825) Putative disease resistance protein At4g27220
Length = 919
Score = 75.1 bits (183), Expect = 8e-13
Identities = 93/338 (27%), Positives = 158/338 (46%), Gaps = 41/338 (12%)
Query: 116 RQHSPVILVDELDQIIFGKKRALRDVSYLTGWDMSNYSNQSKVVKEIVSQVLKNLDKKYL 175
++ S I + + D I K + L + G D+ + +K +EIV +VL
Sbjct: 61 KRSSCAIWLSDKDVEILEKVKRLEE----QGQDLIKKISVNKSSREIVERVLG------- 109
Query: 176 PLPNF---QVGLKPRAEKPIRFLRQNTRKVCLVGIWGMGGIGKSTIAKVIYNDLCYEFED 232
P+F + L+ + ++N +K+ G+WGMGG+GK+T+ + + NDL
Sbjct: 110 --PSFHPQKTALEMLDKLKDCLKKKNVQKI---GVWGMGGVGKTTLVRTLNNDLLKYAAT 164
Query: 233 QSFVANIREVWEKDRGRIDLQEQLLSDILKTRKIKVLSVEQ----GKAMIKQRLRSKRIL 288
Q F I KD DL +++ DI K R K + EQ G + ++ + K L
Sbjct: 165 QQFALVIWVTVSKD---FDL-KRVQMDIAK-RLGKRFTREQMNQLGLTICERLIDLKNFL 219
Query: 289 AVLDDV---SELEQFNALCEGNSVGPGSVIIITTRDLRVLNILEVDFIYEAEGLNASESL 345
+LDDV +L+Q + S +++T+R L V + + + L E+
Sbjct: 220 LILDDVWHPIDLDQL-GIPLALERSKDSKVVLTSRRLEVCQQMMTNENIKVACLQEKEAW 278
Query: 346 ELFCGHAFRKVIPTEDFLILSRYVVAYCGGIPLALEVLGSYLL-KRRKQEWQSVLSKLEK 404
ELFC H +V +++ +++ V C G+PLA+ +G L K + + W+ L+ L++
Sbjct: 279 ELFC-HNVGEVANSDNVKPIAKDVSHECCGLPLAIITIGRTLRGKPQVEVWKHTLNLLKR 337
Query: 405 -IPNDQIHEK----LKISFNGLSDRMEKDIFLDVCCFF 437
P+ EK LK+S++ L D M K FL C F
Sbjct: 338 SAPSIDTEEKIFGTLKLSYDFLQDNM-KSCFL-FCALF 373
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.345 0.153 0.504
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 91,998,799
Number of Sequences: 164201
Number of extensions: 3641835
Number of successful extensions: 20058
Number of sequences better than 10.0: 116
Number of HSP's better than 10.0 without gapping: 42
Number of HSP's successfully gapped in prelim test: 74
Number of HSP's that attempted gapping in prelim test: 19852
Number of HSP's gapped (non-prelim): 151
length of query: 902
length of database: 59,974,054
effective HSP length: 119
effective length of query: 783
effective length of database: 40,434,135
effective search space: 31659927705
effective search space used: 31659927705
T: 11
A: 40
X1: 15 ( 7.5 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 38 (21.6 bits)
S2: 70 (31.6 bits)
Medicago: description of AC145513.1


