

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC145512.9 - phase: 0
(918 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
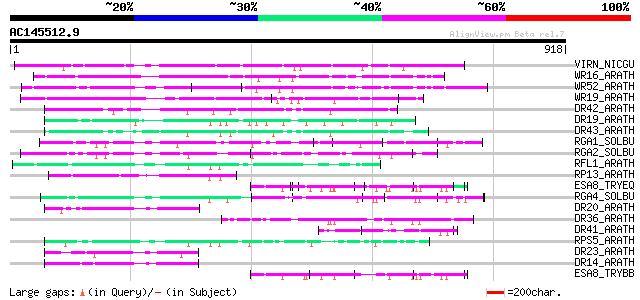
Score E
Sequences producing significant alignments: (bits) Value
VIRN_NICGU (Q40392) TMV resistance protein N 254 6e-67
WR16_ARATH (Q9FL92) Probable WRKY transcription factor 16 (WRKY ... 216 3e-55
WR52_ARATH (Q9FH83) Probable WRKY transcription factor 52 (WRKY ... 213 1e-54
WR19_ARATH (Q9SZ67) Probable WRKY transcription factor 19 (WRKY ... 197 8e-50
DR42_ARATH (Q9FKZ1) Probable disease resistance protein At5g66900 97 2e-19
DR19_ARATH (Q9C8T9) Putative disease resistance protein At1g63350 80 2e-14
DR43_ARATH (Q9FKZ0) Probable disease resistance protein At5g66910 80 2e-14
RGA1_SOLBU (Q7XA42) Putative disease resistance protein RGA1 (RG... 80 3e-14
RGA2_SOLBU (Q7XBQ9) Disease resistance protein RGA2 (RGA2-blb) (... 78 1e-13
RFL1_ARATH (Q8L3R3) Disease resistance protein RFL1 (RPS5-like p... 72 7e-12
RP13_ARATH (Q9M667) Disease resistance protein RPP13 (Resistance... 70 2e-11
ESA8_TRYEQ (P26337) Putative adenylate cyclase regulatory protein 70 3e-11
RGA4_SOLBU (Q7XA39) Putative disease resistance protein RGA4 (RG... 69 7e-11
DR20_ARATH (Q9SH22) Putative disease resistance protein At1g63360 68 1e-10
DR36_ARATH (Q8VZC7) Probable disease resistance protein At5g45510 66 5e-10
DR41_ARATH (Q9FKZ2) Probable disease resistance protein At5g66890 65 6e-10
RPS5_ARATH (O64973) Disease resistance protein RPS5 (Resistance ... 65 8e-10
DR23_ARATH (O82484) Putative disease resistance protein At4g10780 65 8e-10
DR14_ARATH (Q9SI85) Probable disease resistance protein At1g6263... 64 1e-09
ESA8_TRYBB (P23799) Putative adenylate cyclase regulatory protei... 63 3e-09
>VIRN_NICGU (Q40392) TMV resistance protein N
Length = 1144
Score = 254 bits (650), Expect = 6e-67
Identities = 230/803 (28%), Positives = 392/803 (48%), Gaps = 106/803 (13%)
Query: 8 DRDGYVHEFIEKIVEEVLRNIKPVALPAGDFLVGVEHQKQHVTLLLNVGSDDSIHKVGIL 67
+RD + I +IV+++ + ++L +VG++ + + LL +G + + +GI
Sbjct: 157 NRDKTDADCIRQIVDQISSKLCKISLSYLQNIVGIDTHLEKIESLLEIGIN-GVRIMGIW 215
Query: 68 GIGGIGKTTLALEVYNSIVS------QFECSCFLEKVRENSDKNGLIYLQKILLSHIFGE 121
G+GG+GKTT+A ++++++ QF+ +CFL+ ++EN K G+ LQ LLS + E
Sbjct: 216 GMGGVGKTTIARAIFDTLLGRMDSSYQFDGACFLKDIKEN--KRGMHSLQNALLSELLRE 273
Query: 122 KNTEITSVGEGISILKKRLPEKKVLLLLDDVDKKEQ-LKAIAGSSNWFRKGSRVIITTRY 180
K + +G + RL KKVL++LDD+D K+ L+ +AG +WF GSR+IITTR
Sbjct: 274 K-ANYNNEEDGKHQMASRLRSKKVLIVLDDIDNKDHYLEYLAGDLDWFGNGSRIIITTRD 332
Query: 181 KNLLISSGVERIYEVKGLNDEDAFDLVGWKTLKNDCSPRYKDVLLEQKYGRESDANKLRR 240
K+L+ + + IYEV L D ++ L + +G+E
Sbjct: 333 KHLIEKNDI--IYEVTALPDHESIQL-----------------FKQHAFGKEVP------ 367
Query: 241 LKDLKNDEGYANVLKRVVAYASGHPLALEVIGSHFSNKTIEQCNDALDRYKRVPHNMIQT 300
+E + + VV YA G PLAL+V GS N + + A++ K ++ I
Sbjct: 368 ------NENFEKLSLEVVNYAKGLPLALKVWGSLLHNLRLTEWKSAIEHMKNNSYSGIID 421
Query: 301 TLQLSYDSLQEEEKIVFLDIACCFKGWKLTMVEGILHAHHGHTMKDQINVLVEKYLIKIN 360
L++SYD L+ +++ +FLDIAC +G + + IL + H + + +L++K L+ I+
Sbjct: 422 KLKISYDGLEPKQQEMFLDIACFLRGEEKDYILQILESCH-IGAEYGLRILIDKSLVFIS 480
Query: 361 ESGNVTLHDLVEDMGKEIVRQEAPEDPGKRSRLWFSEDIIQVLEENTGTSKIEMIHFDGV 420
E V +HDL++DMGK IV + +DPG+RSRLW ++++ +V+ NTGT +E I
Sbjct: 481 EYNQVQMHDLIQDMGKYIVNFQ--KDPGERSRLWLAKEVEEVMSNNTGTMAMEAIWVSSY 538
Query: 421 IE-VQWDGEAFKKMENLKTL-IFSNDVFFSENPKHLPNSLRVLECRNHM*E-----LDLS 473
+++ +A K M+ L+ + + ++ + +LPN+LR C N+ E +L
Sbjct: 539 SSTLRFSNQAVKNMKRLRVFNMGRSSTHYAID--YLPNNLRCFVCTNYPWESFPSTFELK 596
Query: 474 YCINLE----SFSHV--VDGFGDKLRTMSVRGCFKLKSIPPLK-LDSLETMDLSCCFRLE 526
++L+ S H+ LR + + +L P + +LE ++L C LE
Sbjct: 597 MLVHLQLRHNSLRHLWTETKHLPSLRRIDLSWSKRLTRTPDFTGMPNLEYVNLYQCSNLE 656
Query: 527 SFPLVVDGILGKIKTLNVESCHNLRSIPPLKLDSLEKLDISYCGSLESFPQVEGRF---- 582
+ G K+ L + C +L+ P + ++SLE L + C SLE P++ GR
Sbjct: 657 EVHHSL-GCCSKVIGLYLNDCKSLKRFPCVNVESLEYLGLRSCDSLEKLPEIYGRMKPEI 715
Query: 583 ----------------------LGKLKTLNVKSCRIMISIPTLM--LSLLEELDLSYCLN 618
+ KL N+K+ ++++P+ + L L L +S C
Sbjct: 716 QIHMQGSGIRELPSSIFQYKTHVTKLLLWNMKN---LVALPSSICRLKSLVSLSVSGCSK 772
Query: 619 LENFPLVVDGFLGKLKTLSAKSCRNLRSIPSL----KLDLLETLDLSNCVSLESLPLVVD 674
LE+ P + G L L+ A LR S+ KL +L + V E P V +
Sbjct: 773 LESLPEEI-GDLDNLRVFDASDTLILRPPSSIIRLNKLIILMFRGFKDGVHFE-FPPVAE 830
Query: 675 GFLGKLKTLLVTNCHNLKSIPPLK---CDALETLDLS--CCYSLQSFSLVADRLWKLVLD 729
G L L+ L ++ C+ + P + +L+ LDLS L S L L L
Sbjct: 831 G-LHSLEYLNLSYCNLIDGGLPEEIGSLSSLKKLDLSRNNFEHLPSSIAQLGALQSLDLK 889
Query: 730 DCKELQEIKVIPPCLRMLSAVNC 752
DC+ L ++ +PP L L V+C
Sbjct: 890 DCQRLTQLPELPPELNELH-VDC 911
>WR16_ARATH (Q9FL92) Probable WRKY transcription factor 16 (WRKY
DNA-binding protein 16)
Length = 1372
Score = 216 bits (549), Expect = 3e-55
Identities = 204/722 (28%), Positives = 333/722 (45%), Gaps = 101/722 (13%)
Query: 40 VGVEHQKQHVTLLLNVGSDDSIHKVGILGIGGIGKTTLALEVYNSIVSQFECSCFLEKVR 99
+G+ + + ++N D I VGI G+ GIGKTTLA V++ + +F+ CF+E
Sbjct: 143 IGIYSKLLEIEKMINKQPLD-IRCVGIWGMPGIGKTTLAKAVFDQMSGEFDAHCFIEDYT 201
Query: 100 ENSDKNGLIYLQKILLSHIFGEKNTEITSVGEGISILKKRLPEKKVLLLLDDVDKKEQLK 159
+ + G+ LL F ++N + +S+L+ RL K+VL++LDDV ++
Sbjct: 202 KAIQEKGVY----CLLEEQFLKENAGASGTVTKLSLLRDRLNNKRVLVVLDDVRSPLVVE 257
Query: 160 AIAGSSNWFRKGSRVIITTRYKNLLISSGVERIYEVKGLNDEDAFDLVGWKTLKNDCSPR 219
+ G +WF S +IIT++ K++ V +IYEV+GLN+++A L +D +
Sbjct: 258 SFLGGFDWFGPKSLIIITSKDKSVFRLCRVNQIYEVQGLNEKEALQLFSLCASIDDMA-- 315
Query: 220 YKDVLLEQKYGRESDANKLRRLKDLKNDEGYANVLKRVVAYASGHPLALEVIGSHF-SNK 278
++ V +V+ YA+GHPLAL + G K
Sbjct: 316 ---------------------------EQNLHEVSMKVIKYANGHPLALNLYGRELMGKK 348
Query: 279 TIEQCNDALDRYKRVPHNMIQTTLQLSYDSLQEEEKIVFLDIACCFKGWKLTMVEGILHA 338
+ A + K P + ++ SYD+L + EK +FLDIAC F+G + V +L
Sbjct: 349 RPPEMEIAFLKLKECPPAIFVDAIKSSYDTLNDREKNIFLDIACFFQGENVDYVMQLLEG 408
Query: 339 HHGHTMKDQINVLVEKYLIKINESGNVTLHDLVEDMGKEIVRQEAPEDPGKRSRLWFSED 398
G I+VLVEK L+ I+E+ V +H+L++D+G++I+ +E + +RSRLW
Sbjct: 409 -CGFFPHVGIDVLVEKSLVTISEN-RVRMHNLIQDVGRQIINRETRQTK-RRSRLWEPCS 465
Query: 399 IIQVLEENTGT---------------SKIEMIHFD-GVIEVQWDGEAFKKMENLKTL-IF 441
I +LE+ +IE + D + AF M NL+ I+
Sbjct: 466 IKYLLEDKEQNENEEQKTTFERAQVPEEIEGMFLDTSNLSFDIKHVAFDNMLNLRLFKIY 525
Query: 442 SNDV-------FFSENPKHLPNSLRVLECRN-------------HM*ELDLSYCINLESF 481
S++ F + LPN LR+L N H+ E+++ Y
Sbjct: 526 SSNPEVHHVNNFLKGSLSSLPNVLRLLHWENYPLQFLPQNFDPIHLVEINMPY----SQL 581
Query: 482 SHVVDGFGDKLRTMSVRGCFKLKSI---PPLKLDSLETMDLSCCFRLESFPLVVDGILGK 538
+ G D ++R C + + LK +LE +DL C RL+SFP G L
Sbjct: 582 KKLWGGTKDLEMLKTIRLCHSQQLVDIDDLLKAQNLEVVDLQGCTRLQSFP--ATGQLLH 639
Query: 539 IKTLNVESCHNLRSIPPLKLDSLEKLDISYCGSLESFPQVEGRFLGKLKTLNVKSCRIMI 598
++ +N+ C ++S P + ++E L++ G +E P L +K + ++
Sbjct: 640 LRVVNLSGCTEIKSFPEIP-PNIETLNLQGTGIIE-LP------LSIVKPNYRELLNLLA 691
Query: 599 SIPTLM-LSLLEELDLSYCLNLENFPLVVDGFLGKLKTLSAKSCRNLRSIPSL-KLDLLE 656
IP L +S LE+ DL +L GKL L C LRS+P++ L+LL+
Sbjct: 692 EIPGLSGVSNLEQSDLKPLTSLMKISTSYQN-PGKLSCLELNDCSRLRSLPNMVNLELLK 750
Query: 657 TLDLSNCVSLESLPLVVDGFLGKLKTLLVTNCHNLKSIPPLKCDALETLDLSCCYSLQSF 716
LDLS C LE+ + GF LK L + ++ +P L +LE + C SL+S
Sbjct: 751 ALDLSGCSELET----IQGFPRNLKELYLVGT-AVRQVPQLP-QSLEFFNAHGCVSLKSI 804
Query: 717 SL 718
L
Sbjct: 805 RL 806
Score = 44.3 bits (103), Expect = 0.002
Identities = 30/113 (26%), Positives = 53/113 (46%), Gaps = 8/113 (7%)
Query: 279 TIEQCNDALDRYK--------RVPHNMIQTTLQLSYDSLQEEEKIVFLDIACCFKGWKLT 330
T CN +++ V N + L++ Y LQE K +FL IA F +
Sbjct: 1020 TAVNCNTSIENISPVLSLDPMEVSGNEDEEVLRVRYAGLQEIYKALFLYIAGLFNDEDVG 1079
Query: 331 MVEGILHAHHGHTMKDQINVLVEKYLIKINESGNVTLHDLVEDMGKEIVRQEA 383
+V ++ + + VL + LI+++ +G + +H L+ MGKEI+ E+
Sbjct: 1080 LVAPLIANIIDMDVSYGLKVLAYRSLIRVSSNGEIVMHYLLRQMGKEILHTES 1132
>WR52_ARATH (Q9FH83) Probable WRKY transcription factor 52 (WRKY
DNA-binding protein 52) (Disease resistance protein
RRS1) (Resistance to Ralstonia solanacearum 1 protein)
Length = 1378
Score = 213 bits (543), Expect = 1e-54
Identities = 242/815 (29%), Positives = 373/815 (45%), Gaps = 115/815 (14%)
Query: 20 IVEEVLRNIKPVALPAGDFLVGVEHQKQHVTLLLNVGSDDSIHKVGILGIGGIGKTTLAL 79
+VEE++R++ G +G+ + + ++N I VGI G+ GIGKTTLA
Sbjct: 134 LVEEIVRDVYETHFYVGR--IGIYSKLLEIENMVNK-QPIGIRCVGIWGMPGIGKTTLAK 190
Query: 80 EVYNSIVSQFECSCFLEKVRENSDKNGLIYLQKILLSHIFGEKNTEITSVGEGISILKKR 139
V++ + S F+ SCF+E ++ + GL L L + + I +S L+ R
Sbjct: 191 AVFDQMSSAFDASCFIEDYDKSIHEKGLYCL---LEEQLLPGNDATIMK----LSSLRDR 243
Query: 140 LPEKKVLLLLDDVDKKEQLKAIAGSSNWFRKGSRVIITTRYKNLLISSGVERIYEVKGLN 199
L K+VL++LDDV ++ +W GS +IIT+R K + G+ +IYEV+GLN
Sbjct: 244 LNSKRVLVVLDDVCNALVAESFLEGFDWLGPGSLIIITSRDKQVFRLCGINQIYEVQGLN 303
Query: 200 DEDAFDLVGWKTLKNDCSPRYKDVLLEQKYGRESDANKLRRLKDLKNDEGYANVLKRVVA 259
+++A L LL + L L RV++
Sbjct: 304 EKEARQLF----------------LLSASIMEDMGEQNLHELS------------VRVIS 335
Query: 260 YASGHPLALEVIGSHFSNKT-IEQCNDALDRYKRVPHNMIQTTLQLSYDSLQEEEKIVFL 318
YA+G+PLA+ V G K + + A + KR P I + SYD+L + EK +FL
Sbjct: 336 YANGNPLAISVYGRELKGKKKLSEMETAFLKLKRRPPFKIVDAFKSSYDTLSDNEKNIFL 395
Query: 319 DIACCFKGWKLTMVEGILHAHHGHTMKDQINVLVEKYLIKINESGNVTLHDLVEDMGKEI 378
DIAC F+G + V +L G +I+VLV+K L+ I+E+ V LH L +D+G+EI
Sbjct: 396 DIACFFQGENVNYVIQLLEG-CGFFPHVEIDVLVDKCLVTISEN-RVWLHKLTQDIGREI 453
Query: 379 VRQEAPEDPGKRSRLWFSEDIIQVLEEN---------------TGTSKIEMIHFD-GVIE 422
+ E + +R RLW I +LE N G+ +IE + D +
Sbjct: 454 INGETVQIE-RRRRLWEPWSIKYLLEYNEHKANGEPKTTFKRAQGSEEIEGLFLDTSNLR 512
Query: 423 VQWDGEAFKKMENLKTL-IFSND------VFFSENPKH-LPNSLRVLECRN--------- 465
AFK M NL+ L I+ ++ + F H LPN LR+L N
Sbjct: 513 FDLQPSAFKNMLNLRLLKIYCSNPEVHPVINFPTGSLHSLPNELRLLHWENYPLKSLPQN 572
Query: 466 ----HM*ELDLSYCINLESFSHVVDGFGDKLRTMSVRGCFKLKSIPP-LKLDSLETMDLS 520
H+ E+++ Y L+ + LRT+ + L I LK ++LE +DL
Sbjct: 573 FDPRHLVEINMPYS-QLQKLWGGTKNL-EMLRTIRLCHSQHLVDIDDLLKAENLEVIDLQ 630
Query: 521 CCFRLESFPLVVDGILGKIKTLNVESCHNLRS---IPPLKLDSLEKLDISYCGSLESFPQ 577
C RL++FP G L +++ +N+ C ++S IPP ++EKL + G L + P
Sbjct: 631 GCTRLQNFPAA--GRLLRLRVVNLSGCIKIKSVLEIPP----NIEKLHLQGTGIL-ALP- 682
Query: 578 VEGRFLGKLKTLNVKSCRIMISIPTLM-LSLLEELDLSYCLNLENFPLVVDGFLGKLKTL 636
+ +K + + + IP L S LE L LE+ D LGKL L
Sbjct: 683 -----VSTVKPNHRELVNFLTEIPGLSEASKLERLTSL----LESNSSCQD--LGKLICL 731
Query: 637 SAKSCRNLRSIPSLKLDLLETLDLSNCVSLESLPLVVDGFLGKLKTLLVTNCHNLKSIPP 696
K C L+S+P++ L LDLS C SL S + GF LK L + ++ +P
Sbjct: 732 ELKDCSCLQSLPNMANLDLNVLDLSGCSSLNS----IQGFPRFLKQLYLGGT-AIREVPQ 786
Query: 697 LKCDALETLDL--SCCYSLQSFSLVADRLWKLVLDDCKELQEIKVIPPCLRMLSAVNCTS 754
L +LE L+ SC SL + + + + L L L C EL+ I+ P L+ L T+
Sbjct: 787 LP-QSLEILNAHGSCLRSLPNMANL-EFLKVLDLSGCSELETIQGFPRNLKELYFAG-TT 843
Query: 755 LTSSCTSKLLNQELHKAGNTWFCLPRVPKIPEWFD 789
L L + L+ G+ LP K +FD
Sbjct: 844 LREVPQLPLSLEVLNAHGSDSEKLPMHYKFNNFFD 878
Score = 48.9 bits (115), Expect = 6e-05
Identities = 27/82 (32%), Positives = 46/82 (55%), Gaps = 1/82 (1%)
Query: 302 LQLSYDSLQEEEKIVFLDIACCFKGWKLTMVEGILHAHHGHTMKDQINVLVEKYLIKINE 361
L++SYD LQE +K++FL IA F + V ++ A + + VL + LI ++
Sbjct: 1089 LRVSYDDLQEMDKVLFLYIASLFNDEDVDFVAPLI-AGIDLDVSSGLKVLADVSLISVSS 1147
Query: 362 SGNVTLHDLVEDMGKEIVRQEA 383
+G + +H L MGKEI+ ++
Sbjct: 1148 NGEIVMHSLQRQMGKEILHGQS 1169
>WR19_ARATH (Q9SZ67) Probable WRKY transcription factor 19 (WRKY
DNA-binding protein 19)
Length = 1895
Score = 197 bits (502), Expect = 8e-50
Identities = 197/702 (28%), Positives = 330/702 (46%), Gaps = 83/702 (11%)
Query: 19 KIVEEVLRNIKPVALPAGDF-LVGVEHQKQHVTLLLNVGSDDSIHKVGILGIGGIGKTTL 77
++++E++R+ V A ++G++ Q + + LL + S D + +GI G GIGKTT+
Sbjct: 795 ELIDEIVRDALKVLCSADKVNMIGMDMQVEEILSLLCIESLD-VRSIGIWGTVGIGKTTI 853
Query: 78 ALEVYNSIVSQFECSCFLEKVRENSDKNGLIYLQKILLSHIFGEKNTEITSVGEGISILK 137
A E++ I Q+E L+ + + + G +++ LS + + I S L+
Sbjct: 854 AEEIFRKISVQYETCVVLKDLHKEVEVKGHDAVRENFLSEVLEVEPHVIRISDIKTSFLR 913
Query: 138 KRLPEKKVLLLLDDVDKKEQLKAIAGSSNWFRKGSRVIITTRYKNLLISSGVERIYEVKG 197
RL K++L++LDDV+ + G+ N+F GSR+I+T+R + + + ++ +YEVK
Sbjct: 914 SRLQRKRILVILDDVNDYRDVDTFLGTLNYFGPGSRIIMTSRNRRVFVLCKIDHVYEVKP 973
Query: 198 LNDEDAFDLVGWKTLKNDCSPR-YKDVLLEQKYGRESDANKLRRLKDLKNDEGYANVLKR 256
L+ + L+ T + SP YK + LE
Sbjct: 974 LDIPKSLLLLDRGTCQIVLSPEVYKTLSLE------------------------------ 1003
Query: 257 VVAYASGHPLALEVIGSHFSNKTIEQCNDALDRYKRVPHNMIQTTLQLSYDSLQEEEKIV 316
+V +++G+P L+ + S + N K I + S L + E+ +
Sbjct: 1004 LVKFSNGNPQVLQFLSS-----IDREWNKLSQEVKTTSPIYIPGIFEKSCCGLDDNERGI 1058
Query: 317 FLDIACCFKGWKLTMVEGILHAHHGHTMKDQINVLVEKYLIKINESGNVTLHDLVEDMGK 376
FLDIAC F V +L G + LV+K L+ I++ V + ++ G+
Sbjct: 1059 FLDIACFFNRIDKDNVAMLLDGC-GFSAHVGFRGLVDKSLLTISQHNLVDMLSFIQATGR 1117
Query: 377 EIVRQEAPEDPGKRSRLWFSEDIIQVLEENTGTSKIEMIHFDGVIEVQWDG--EAFKKME 434
EIVRQE+ + PG RSRLW ++ I V +TGTS IE I D ++ +++D F+KM
Sbjct: 1118 EIVRQESADRPGDRSRLWNADYIRHVFINDTGTSAIEGIFLD-MLNLKFDANPNVFEKMC 1176
Query: 435 NLKTLIF-------SNDVFFSENPKHLPNSLRVLECR-------------NHM*ELDL-S 473
NL+ L + V F + ++LP+ LR+L ++ EL+L S
Sbjct: 1177 NLRLLKLYCSKAEEKHGVSFPQGLEYLPSKLRLLHWEYYPLSSLPKSFNPENLVELNLPS 1236
Query: 474 YCI-----NLESFSHVVDGFGDKLRTMSVRGCFKLKSIPPL-KLDSLETMDLSCCFRLES 527
C ++ + +KL+ M + +L IP L +LE +DL C L S
Sbjct: 1237 SCAKKLWKGKKARFCTTNSSLEKLKKMRLSYSDQLTKIPRLSSATNLEHIDLEGCNSLLS 1296
Query: 528 FPLVVDGILGKIKTLNVESCHNLRSIPPL-KLDSLEKLDISYCGSLESFPQVEGRFLGKL 586
+ L K+ LN++ C L +IP + L+SLE L++S C L +FP++ +
Sbjct: 1297 LSQSI-SYLKKLVFLNLKGCSKLENIPSMVDLESLEVLNLSGCSKLGNFPEIS----PNV 1351
Query: 587 KTLNVKSCRIMISIPTLM--LSLLEELDLSYCLNLENFPLVVDGFLGKLKTLSAKSCRNL 644
K L + I IP+ + L LLE+LDL +L+N P + L L+TL+ C +L
Sbjct: 1352 KELYMGGTMIQ-EIPSSIKNLVLLEKLDLENSRHLKNLPTSIYK-LKHLETLNLSGCISL 1409
Query: 645 RSIP--SLKLDLLETLDLSNCVSLESLPLVVDGFLGKLKTLL 684
P S ++ L LDLS ++ LP + +L L LL
Sbjct: 1410 ERFPDSSRRMKCLRFLDLSR-TDIKELPSSI-SYLTALDELL 1449
Score = 58.2 bits (139), Expect = 1e-07
Identities = 59/232 (25%), Positives = 112/232 (47%), Gaps = 33/232 (14%)
Query: 433 MENLKTLIFSNDVFFSENPKHLPNSLRVLECRNHM*ELDLSYCINLESFSHVVDGFGDKL 492
+E LK + S +S+ +P L ++ +DL C +L S S + + KL
Sbjct: 1257 LEKLKKMRLS----YSDQLTKIPR----LSSATNLEHIDLEGCNSLLSLSQSIS-YLKKL 1307
Query: 493 RTMSVRGCFKLKSIPPL-KLDSLETMDLSCCFRLESFPLVVDGI---------------- 535
++++GC KL++IP + L+SLE ++LS C +L +FP + +
Sbjct: 1308 VFLNLKGCSKLENIPSMVDLESLEVLNLSGCSKLGNFPEISPNVKELYMGGTMIQEIPSS 1367
Query: 536 ---LGKIKTLNVESCHNLRSIPP--LKLDSLEKLDISYCGSLESFPQVEGRFLGKLKTLN 590
L ++ L++E+ +L+++P KL LE L++S C SLE FP R + L+ L+
Sbjct: 1368 IKNLVLLEKLDLENSRHLKNLPTSIYKLKHLETLNLSGCISLERFPD-SSRRMKCLRFLD 1426
Query: 591 VKSCRIMISIPTLMLSLLEELDLSYCLNLENFPLVVDGFLGKLKTLSAKSCR 642
+ I +P+ + L +L + + N P+V + + + ++S +
Sbjct: 1427 LSRTDIK-ELPSSISYLTALDELLFVDSRRNSPVVTNPNANSTELMPSESSK 1477
Score = 32.0 bits (71), Expect = 7.7
Identities = 28/85 (32%), Positives = 39/85 (44%), Gaps = 6/85 (7%)
Query: 677 LGKLKTLLVTNCHNLKSIPPLKCDA-LETLDLSCCYSLQSFSLVADRLWKLVLDDCKELQ 735
L KLK + ++ L IP L LE +DL C SL S S L KLV + K
Sbjct: 1257 LEKLKKMRLSYSDQLTKIPRLSSATNLEHIDLEGCNSLLSLSQSISYLKKLVFLNLKGCS 1316
Query: 736 EIKVIP-----PCLRMLSAVNCTSL 755
+++ IP L +L+ C+ L
Sbjct: 1317 KLENIPSMVDLESLEVLNLSGCSKL 1341
>DR42_ARATH (Q9FKZ1) Probable disease resistance protein At5g66900
Length = 809
Score = 97.4 bits (241), Expect = 2e-19
Identities = 144/625 (23%), Positives = 255/625 (40%), Gaps = 94/625 (15%)
Query: 58 DDSIHKVGILGIGGIGKTTLALEVYNSIVSQFECSCFLEKVRENSDKNGLIYLQKILLSH 117
DDS+ + + G GKTTL + + + + V N+ N + +Q +L +
Sbjct: 184 DDSVVTLVVSAPPGCGKTTLVSRLCDDPDIKGKFKHIFFNVVSNTP-NFRVIVQNLLQHN 242
Query: 118 IFGEKNTEITSVGE-GISILKKRLPEK-KVLLLLDDVDKKEQLKAIAGSSNWFRK----- 170
+ E S E G+ L + L E +LL+LDDV + G+ ++ +K
Sbjct: 243 GYNALTFENDSQAEVGLRKLLEELKENGPILLVLDDVWR--------GADSFLQKFQIKL 294
Query: 171 -GSRVIITTRYKNLLISSGVERIYEVKGLNDEDAFDL-VGWKTLKNDCSPRYKDVLLEQK 228
++++T+R+ S Y +K L D+DA L + W + + SP
Sbjct: 295 PNYKILVTSRFDFPSFDSN----YRLKPLEDDDARALLIHWASRPCNTSP---------- 340
Query: 229 YGRESDANKLRRLKDLKNDEGYANVLKRVVAYASGHPLALEVIGSHFSNKTIEQCNDALD 288
+ Y ++L++++ +G P+ +EV+G +++ ++
Sbjct: 341 -------------------DEYEDLLQKILKRCNGFPIVIEVVGVSLKGRSLNTWKGQVE 381
Query: 289 RYKR------VPHNMIQTTLQLSYDSLQEEEKIVFLDIACCFKGWKLTMVEGI-----LH 337
+ P+ + LQ S+D+L K FLD+ + K+ I L+
Sbjct: 382 SWSEGEKILGKPYPTVLECLQPSFDALDPNLKECFLDMGSFLEDQKIRASVIIDMWVELY 441
Query: 338 AHHGHTMKDQINVLVEKYLIKINESGN------------VTLHDLVEDMGKEIVRQEAPE 385
+ + L + L+K+ G VT HD++ ++ I + E E
Sbjct: 442 GKGSSILYMYLEDLASQNLLKLVPLGTNEHEDGFYNDFLVTQHDILRELA--ICQSEFKE 499
Query: 386 D-PGKRSRLWFSEDIIQVLEENTGTSKIEMIHFDGVIEVQWDGEAFKKMENLKTLIFSND 444
+ KR L E+ NT + + I D + +W +E L + S+D
Sbjct: 500 NLERKRLNLEILENTFPDWCLNTINASLLSISTDDLFSSKWLEMDCPNVEALVLNLSSSD 559
Query: 445 VFFSENPKHLPNSLRVLECRNHM*ELDLSYCINLESFSHVVDGFGDKLRTMSVRGCFKLK 504
+ L+VL NH Y L +FS + + R + L
Sbjct: 560 YALPSFISGM-KKLKVLTITNHG-----FYPARLSNFS-CLSSLPNLKRIRLEKVSITLL 612
Query: 505 SIPPLKLDSLETMDLSCCFRLESF----PLVVDGILGKIKTLNVESCHNLRSIP--PLKL 558
IP L+L SL+ + L C E F +VV L K++ ++++ C++L +P ++
Sbjct: 613 DIPQLQLSSLKKLSLVMCSFGEVFYDTEDIVVSNALSKLQEIDIDYCYDLDELPYWISEI 672
Query: 559 DSLEKLDISYCGSLESFPQVEGRFLGKLKTLNVKSCRIMISIP--TLMLSLLEELDLSYC 616
SL+ L I+ C L P+ G L +L+ L + S + +P T LS L LD+S+C
Sbjct: 673 VSLKTLSITNCNKLSQLPEAIGN-LSRLEVLRLCSSMNLSELPEATEGLSNLRFLDISHC 731
Query: 617 LNLENFPLVVDGFLGKLKTLSAKSC 641
L L P + G L LK +S + C
Sbjct: 732 LGLRKLPQEI-GKLQNLKKISMRKC 755
>DR19_ARATH (Q9C8T9) Putative disease resistance protein At1g63350
Length = 898
Score = 80.5 bits (197), Expect = 2e-14
Identities = 172/721 (23%), Positives = 285/721 (38%), Gaps = 162/721 (22%)
Query: 58 DDSIHKVGILGIGGIGKTTLALEVYNSI---VSQFECSCFLEKVRENSDKNGLIYL-QKI 113
+D + +G+ G+GG+GKTTL ++ N + F+ ++ +E + +N L + QK+
Sbjct: 169 EDGVGIMGLYGMGGVGKTTLLTQINNKFSKYMCGFDSVIWVVVSKEVNVENILDEIAQKV 228
Query: 114 LLSHIFGEKNTEITSVGEGISILKKRLPEKKVLLLLDDVDKKEQLKAIAGSSNWFRKGSR 173
HI GEK +G+ L L + + +L LDD+ +K L I + +
Sbjct: 229 ---HISGEKWDTKYKYQKGV-YLYNFLRKMRFVLFLDDIWEKVNLVEIGVPFPTIKNKCK 284
Query: 174 VIITTRYKNLLISSGVERIYEVKGLNDEDAFDL----VGWKTLKNDCSPRYKDVLLEQKY 229
V+ TTR ++ S GVE+ EV+ L D DA+DL VG TL +D P ++
Sbjct: 285 VVFTTRSLDVCTSMGVEKPMEVQCLADNDAYDLFQKKVGQITLGSD--PEIRE------- 335
Query: 230 GRESDANKLRRLKDLKNDEGYANVLKRVVA-YASGHPLALEVIGSHFS-NKTIEQCNDA- 286
L RVVA G PLAL V+ S +T+++ A
Sbjct: 336 ------------------------LSRVVAKKCCGLPLALNVVSETMSCKRTVQEWRHAI 371
Query: 287 --LDRYKRVPHNM---IQTTLQLSYDSLQ-EEEKIVFLDIACCFKGWKLT--------MV 332
L+ Y M I L+ SYDSL+ E+ K+ L A + K+ +
Sbjct: 372 YVLNSYAAKFSGMDDKILPLLKYSYDSLKGEDVKMCLLYCALFPEDAKIRKENLIEYWIC 431
Query: 333 EGILHAHHGHTMKDQ-----INVLVEKYL----IKINESGNVTLHDLVEDMGKEIVRQEA 383
E I+ G + I LV L ++++ + V LHD+V +M I
Sbjct: 432 EEIIDGSEGIDKAENQGYEIIGSLVRASLLMEEVELDGANIVCLHDVVREMALWIA---- 487
Query: 384 PEDPGKRSRLWF------SEDIIQVLEENT----GTSKIEMIHFDGVIEV---------- 423
D GK++ + +I++V N K + H DG ++
Sbjct: 488 -SDLGKQNEAFIVRASVGLREILKVENWNVVRRMSLMKNNIAHLDGRLDCMELTTLLLQS 546
Query: 424 ----QWDGEAFKKMENLKTLIFSNDVFFSENP-------------------KHLPNSLRV 460
+ E F M L L S + + SE P +HLP L+
Sbjct: 547 THLEKISSEFFNSMPKLAVLDLSGNYYLSELPNGISELVSLQYLNLSSTGIRHLPKGLQE 606
Query: 461 LECRNHM*ELDLSYCINLESFSHVVDGFG----DKLRTMSVRGCFKLKSIPPLK-LDSLE 515
L+ H + LE S + G L+ + + G + +K L++LE
Sbjct: 607 LKKLIH---------LYLERTSQLGSMVGISCLHNLKVLKLSGSSYAWDLDTVKELEALE 657
Query: 516 TMDL------SCCFRLESFPLVVDGILGKIKTLNVESCHN-----LRSIPPLKLDSLEKL 564
+++ C + F L ++ I+ L + + N R P+ +D L++
Sbjct: 658 HLEVLTTTIDDCTLGTDQF-LSSHRLMSCIRFLKISNNSNRNRNSSRISLPVTMDRLQEF 716
Query: 565 DISYCGSLESFPQVEGRF--LGKLKTLNVKSCRIMISIPTLM----LSLLEELDLSYCLN 618
I +C + E GR L +N+ +CR + + LM L L + + +
Sbjct: 717 TIEHCHTSEI---KMGRICSFSSLIEVNLSNCRRLRELTFLMFAPNLKRLHVVSSNQLED 773
Query: 619 LENFPLVVDGF------LGKLKTLSAKSCRNLRSI--PSLKLDLLETLDLSNCVSLESLP 670
+ N DG KL L + R L++I L LE +++ C +L+ LP
Sbjct: 774 IINKEKAHDGEKSGIVPFPKLNELHLYNLRELKNIYWSPLPFPCLEKINVMGCPNLKKLP 833
Query: 671 L 671
L
Sbjct: 834 L 834
>DR43_ARATH (Q9FKZ0) Probable disease resistance protein At5g66910
Length = 815
Score = 80.1 bits (196), Expect = 2e-14
Identities = 156/679 (22%), Positives = 273/679 (39%), Gaps = 111/679 (16%)
Query: 58 DDSIHKVGILGIGGIGKTTLALEVYNSIVSQFECSCFLEKVRENSDKNGLIYLQKILLSH 117
D+S+ V + G G GKTTL ++ + + E V N+ N +Q +L +
Sbjct: 188 DNSV--VVVSGPPGCGKTTLVTKLCDDPEIEGEFKKIFYSVVSNTP-NFRAIVQNLLQDN 244
Query: 118 IFGEKNTEITSVGE-GI-SILKKRLPEKKVLLLLDDVDKKEQLKAIAGSSNWFRKGSRVI 175
G + S E G+ +L++ + ++LL+LDDV + GS RK I
Sbjct: 245 GCGAITFDDDSQAETGLRDLLEELTKDGRILLVLDDVWQ--------GSEFLLRKFQ--I 294
Query: 176 ITTRYKNLLISSGVERIYEVKGLNDEDAFDLVG-WKTLKNDCSPRYKDVLLEQKYGRESD 234
YK L+ S FD W P Y V L+ +Y R
Sbjct: 295 DLPDYKILVTSQ----------------FDFTSLW--------PTYHLVPLKYEYARSLL 330
Query: 235 ANKLRRLKDLKNDEGYANVLKRVVAYASGHPLALEVIGSHFSNKTIEQCNDALDRYKR-- 292
DE Y ++L++++ +G PL +EV+G + + ++ +
Sbjct: 331 IQWASPPLHTSPDE-YEDLLQKILKRCNGFPLVIEVVGISLKGQALYLWKGQVESWSEGE 389
Query: 293 ----VPHNMIQTTLQLSYDSLQEEEKIVFLDIACCFKGWKL--TMVEGILHAHHGHTMKD 346
+ ++ LQ S++ L+ K F+D+ + K+ +++ I +G
Sbjct: 390 TILGNANPTVRQRLQPSFNVLKPHLKECFMDMGSFLQDQKIRASLIIDIWMELYGRGSSS 449
Query: 347 ------QINVLVEKYLIKINESGN------------VTLHDLVEDMGKEIVRQE-APEDP 387
+N L + L+K+ G VT H+++ ++ I + E P
Sbjct: 450 TNKFMLYLNELASQNLLKLVHLGTNKREDGFYNELLVTQHNILRELA--IFQSELEPIMQ 507
Query: 388 GKRSRLWFSEDII--QVLEENTGTSKIEMIHFDGVIEVQWDGEAFKKME----NLKTLIF 441
K+ L ED + L + +++ I+ D + +W +E N+ +L +
Sbjct: 508 RKKLNLEIREDNFPDECLNQPI-NARLLSIYTDDLFSSKWLEMDCPNVEALVLNISSLDY 566
Query: 442 SNDVFFSENPKHLPNSLRVLECRNHM*ELDLSYCINLESFSHVVDGFGDKLRTMSVRGCF 501
+ F +E K L+VL NH Y L +FS + + R +
Sbjct: 567 ALPSFIAEMKK-----LKVLTIANHG-----FYPARLSNFS-CLSSLPNLKRIRFEKVSV 615
Query: 502 KLKSIPPLKLDSLETMDLSCCFRLESF----PLVVDGILGKIKTLNVESCHNLRSIPPL- 556
L IP L+L SL+ + C E F + V L ++ ++++ C++L +P
Sbjct: 616 TLLDIPQLQLGSLKKLSFFMCSFGEVFYDTEDIDVSKALSNLQEIDIDYCYDLDELPYWI 675
Query: 557 -KLDSLEKLDISYCGSLESFPQVEGRFLGKLKTLNVKSCRIMISIP--TLMLSLLEELDL 613
++ SL+ L I+ C L P+ G L +L+ L + SC + +P T LS L LD+
Sbjct: 676 PEVVSLKTLSITNCNKLSQLPEAIGN-LSRLEVLRMCSCMNLSELPEATERLSNLRSLDI 734
Query: 614 SYCLNLENFPLVVDGFLGKLKTLSAKSCRNLRSIPSLKLDLLETLDLSNCVSLESLPLVV 673
S+CL L P + G L KL+ +S + C S++ LE+L +
Sbjct: 735 SHCLGLRKLPQEI-GKLQKLENISMRKCSGCELPDSVRY-------------LENLEVKC 780
Query: 674 DGFLGKLKTLLVTNCHNLK 692
D G L L+ NL+
Sbjct: 781 DEVTGLLWERLMPEMRNLR 799
>RGA1_SOLBU (Q7XA42) Putative disease resistance protein RGA1
(RGA3-blb)
Length = 1025
Score = 79.7 bits (195), Expect = 3e-14
Identities = 161/695 (23%), Positives = 284/695 (40%), Gaps = 136/695 (19%)
Query: 49 VTLLLNVGSD-DSIHKVGILGIGGIGKTTLALEVYNS--IVSQFECSCFLEKVRENSDKN 105
V +L+N SD + + ILG+GG+GKTTL+ V+N + +F ++ + ++K
Sbjct: 208 VKILINTASDAQKLSVLPILGMGGLGKTTLSQMVFNDQRVTERFYPKIWICISDDFNEKR 267
Query: 106 GLIYLQKILLSHIFGEKNTEITSVGEGISILKKRLPE----KKVLLLLDDVDKKEQ---- 157
L K ++ I G+ +++ ++ L+K+L E K+ L+LDDV ++Q
Sbjct: 268 ----LIKAIVESIEGKSLSDM-----DLAPLQKKLQELLNGKRYFLVLDDVWNEDQHKWA 318
Query: 158 -LKAI--AGSSNWFRKGSRVIITTRYKNLLISSGVERIYEVKGLNDEDAFDLVGWKTLKN 214
L+A+ G+S G+ V+ TTR + + G + YE+ L+ ED + L
Sbjct: 319 NLRAVLKVGAS-----GAFVLTTTRLEKVGSIMGTLQPYELSNLSPEDCWFL-------- 365
Query: 215 DCSPRYKDVLLEQKYGRESDANKLRRLKDLKNDEGYANVLKRVVAYASGHPLALEVIGSH 274
+++ +G + + N + K +V G PLA + +G
Sbjct: 366 ---------FMQRAFGHQEEIN-----------PNLMAIGKEIVKKCGGVPLAAKTLGGI 405
Query: 275 F----SNKTIEQCNDALDRYKRVPHNMIQTTLQLSYDSLQEEEKIVFLDIACCFKGWKLT 330
+ E D+ + I L+LSY L + + F+ A K K+
Sbjct: 406 LRFKREEREWEHVRDSPIWNLPQDESSILPALRLSYHHLPLDLRQCFVYCAVFPKDTKMA 465
Query: 331 MVEGILHAHHGHTMKDQINVLVEKYLIKINESGNVTLHDLVEDMGKEI-VRQEAPEDPGK 389
E ++ H L+ K GN+ L D+ ++ E+ +R E +
Sbjct: 466 K-ENLIAFWMAH------GFLLSK--------GNLELEDVGNEVWNELYLRSFFQEIEVE 510
Query: 390 RSRLWFS-EDII-----QVLEENTGTSKIEMIHFDGVIEVQWDGEAFKKMENLKTLIFSN 443
+ +F D+I + NT +S I I+ +DG I
Sbjct: 511 SGKTYFKMHDLIHDLATSLFSANTSSSNIREIN------ANYDGYMMS--------IGFA 556
Query: 444 DVFFSENPKHLPN--SLRVLECRNHM*ELDLSYCINLESFSHVVDGFGD--KLRTMSVRG 499
+V S +P L SLRVL RN + + + GD LR + + G
Sbjct: 557 EVVSSYSPSLLQKFVSLRVLNLRN-------------SNLNQLPSSIGDLVHLRYLDLSG 603
Query: 500 CFKLKSIPP--LKLDSLETMDLSCCFRLESFPLVVDGILGKIKTLNVESCHNLRSIPPL- 556
F+++++P KL +L+T+DL C L P LG ++ L ++ C +L S PP
Sbjct: 604 NFRIRNLPKRLCKLQNLQTLDLHYCDSLSCLPKQTSK-LGSLRNLLLDGC-SLTSTPPRI 661
Query: 557 -KLDSLEKLDISYCGSLESFPQVEGRFLGKLKTLNVKSCRIMISIPTLMLSL-LEELDLS 614
L L+ L G + +G LG+LK LN+ + + + +E +LS
Sbjct: 662 GLLTCLKSLSCFVIG------KRKGHQLGELKNLNLYGSISITKLDRVKKDTDAKEANLS 715
Query: 615 YCLNLENFPLVVDGFLGKLKTLSAKSCRNLRSIPSLK-LDLLETLDLSNCVSLESLPLVV 673
NL + L D L K + + +LK L+ L+++ + +
Sbjct: 716 AKANLHSLCLSWD--------LDGKHRYDSEVLEALKPHSNLKYLEINGFGGIRLPDWMN 767
Query: 674 DGFLGKLKTLLVTNCHNLKSIPPL-KCDALETLDL 707
L + ++ + C N +PP + LE+L+L
Sbjct: 768 QSVLKNVVSIRIRGCENCSCLPPFGELPCLESLEL 802
Score = 54.3 bits (129), Expect = 1e-06
Identities = 71/265 (26%), Positives = 121/265 (44%), Gaps = 32/265 (12%)
Query: 535 ILGKIKTLNVESCHNLRSIPPL-KLDSLEKLDISYCGSLESFPQVE-----GRF--LGKL 586
+L + ++ + C N +PP +L LE L++ + GS + VE GRF L KL
Sbjct: 770 VLKNVVSIRIRGCENCSCLPPFGELPCLESLEL-HTGSAD-VEYVEDNVHPGRFPSLRKL 827
Query: 587 KTLNVKSCRIMISIP-TLMLSLLEELDLSYCLNLENFPLVVDGFLGKLKTLSA--KSCRN 643
+ + + ++ + +LEE+ +C P+ V L +KTL
Sbjct: 828 VIWDFSNLKGLLKMEGEKQFPVLEEMTFYWC------PMFVIPTLSSVKTLKVIVTDATV 881
Query: 644 LRSIPSLKLDLLETLDLSNCVSLESLPLVVDGFLGKLKTLLVTNCHNLKSIPP--LKCDA 701
LRSI +L+ L +LD+S+ V SLP + L LK L ++ NLK +P +A
Sbjct: 882 LRSISNLRA--LTSLDISDNVEATSLPEEMFKSLANLKYLKISFFRNLKELPTSLASLNA 939
Query: 702 LETLDLSCCYSLQSFSLVADR----LWKLVLDDCKELQEIKVIPPCLRMLSAVNCTSLTS 757
L++L C +L+S + L +L + +C L K +P L+ L+A+ T+LT
Sbjct: 940 LKSLKFEFCDALESLPEEGVKGLTSLTELSVSNCMML---KCLPEGLQHLTAL--TTLTI 994
Query: 758 SCTSKLLNQELHKAGNTWFCLPRVP 782
+ + + G W + +P
Sbjct: 995 TQCPIVFKRCERGIGEDWHKIAHIP 1019
Score = 45.4 bits (106), Expect = 7e-04
Identities = 33/118 (27%), Positives = 60/118 (49%), Gaps = 6/118 (5%)
Query: 503 LKSIPPLKLDSLETMDLSCCFRLESFPLVVDGILGKIKTLNVESCHNLRSIPP--LKLDS 560
L+SI L+ +L ++D+S S P + L +K L + NL+ +P L++
Sbjct: 882 LRSISNLR--ALTSLDISDNVEATSLPEEMFKSLANLKYLKISFFRNLKELPTSLASLNA 939
Query: 561 LEKLDISYCGSLESFPQVEGRFLGKLKTLNVKSCRIMISIPTLM--LSLLEELDLSYC 616
L+ L +C +LES P+ + L L L+V +C ++ +P + L+ L L ++ C
Sbjct: 940 LKSLKFEFCDALESLPEEGVKGLTSLTELSVSNCMMLKCLPEGLQHLTALTTLTITQC 997
>RGA2_SOLBU (Q7XBQ9) Disease resistance protein RGA2 (RGA2-blb)
(Blight resistance protein RPI)
Length = 970
Score = 77.8 bits (190), Expect = 1e-13
Identities = 178/743 (23%), Positives = 307/743 (40%), Gaps = 165/743 (22%)
Query: 18 EKIVEEVLRNIKPVALPAGDFLVGVEHQKQHVTLLL--NVGSDDSIHKVGILGIGGIGKT 75
EKIVE + ++ + G + +K + +L NV + + ILG+GG+GKT
Sbjct: 130 EKIVERQAVRRETGSVLTEPQVYGRDKEKDEIVKILINNVSDAQHLSVLPILGMGGLGKT 189
Query: 76 TLALEVYNS--IVSQFECSCFLEKVRENSDKNGLIYLQKILLSHIFGEKNTEITSVGE-G 132
TLA V+N + F ++ V E+ D+ LI K ++ I G +GE
Sbjct: 190 TLAQMVFNDQRVTEHFHSKIWI-CVSEDFDEKRLI---KAIVESIEGR-----PLLGEMD 240
Query: 133 ISILKKRLPE----KKVLLLLDDVDKKEQ-----LKAI--AGSSNWFRKGSRVIITTRYK 181
++ L+K+L E K+ LL+LDDV ++Q L+A+ G+S G+ V+ TTR +
Sbjct: 241 LAPLQKKLQELLNGKRYLLVLDDVWNEDQQKWANLRAVLKVGAS-----GASVLTTTRLE 295
Query: 182 NLLISSGVERIYEVKGLNDEDAFDLVGWKTLKNDCSPRYKDVLLEQKYGRESDANKLRRL 241
+ G + YE+ L+ ED + L +++ +G + + N
Sbjct: 296 KVGSIMGTLQPYELSNLSQEDCWLL-----------------FMQRAFGHQEEIN----- 333
Query: 242 KDLKNDEGYANVLKRVVAYASGHPLALEVIGSHFSNKTIEQCNDALDRYKRVP------- 294
+ K +V + G PLA + +G K E+ A + + P
Sbjct: 334 ------PNLVAIGKEIVKKSGGVPLAAKTLGGILCFKREER---AWEHVRDSPIWNLPQD 384
Query: 295 HNMIQTTLQLSYDSLQEEEKIVFLDIACCFKGWKLTMVEGI-LHAHHGHTMKDQINVLVE 353
+ I L+LSY L + K F A K K+ + I L HG +
Sbjct: 385 ESSILPALRLSYHQLPLDLKQCFAYCAVFPKDAKMEKEKLISLWMAHGFLL--------- 435
Query: 354 KYLIKINESGNVTLHDLVEDMGKEI-VRQEAPEDPGKRSRLWFS-EDII-----QVLEEN 406
GN+ L D+ +++ KE+ +R E K + +F D+I + N
Sbjct: 436 -------SKGNMELEDVGDEVWKELYLRSFFQEIEVKDGKTYFKMHDLIHDLATSLFSAN 488
Query: 407 TGTSKIEMIHFDGVIEVQWDGEAFKKMENLKTLIFSNDVFFSENPKHLPNSLRVLECRNH 466
T +S I I+ + G A ++F F++ P SLRVL
Sbjct: 489 TSSSNIREINKHSYTHMMSIGFA--------EVVF----FYTLPPLEKFISLRVLN---- 532
Query: 467 M*ELDLSYCINLESFSHVVDGFGD--KLRTMSVRGCFKLKSIPP--LKLDSLETMDLSCC 522
+ +F+ + GD LR +++ G ++S+P KL +L+T+DL C
Sbjct: 533 ---------LGDSTFNKLPSSIGDLVHLRYLNLYGS-GMRSLPKQLCKLQNLQTLDLQYC 582
Query: 523 FRLESFPLVVDGILGKIKTLNVESCHNLRSIPPLKLDSLEKLDISYCGSLESF--PQVEG 580
+L P LG ++ L ++ +L +PP ++ SL L +L F + +G
Sbjct: 583 TKLCCLPKETSK-LGSLRNLLLDGSQSLTCMPP-RIGSLTCLK-----TLGQFVVGRKKG 635
Query: 581 RFLGKLKTLNVKSCRIMISIPTLMLSLLE---------ELDLSYCLNLENFPLVVDGF-- 629
LG+L LN+ ++ +S LE E +LS NL + + + F
Sbjct: 636 YQLGELGNLNLYG--------SIKISHLERVKNDKDAKEANLSAKGNLHSLSMSWNNFGP 687
Query: 630 ----LGKLKTLSAKSCRNLRSIPSLKLDLLETLDLSNCVSLESLPLVVDGFLGKLKTLLV 685
++K L A + ++ SLK+ + L ++ L +V ++L+
Sbjct: 688 HIYESEEVKVLEALKPHS--NLTSLKIYGFRGIHLPEWMNHSVLKNIV--------SILI 737
Query: 686 TNCHNLKSIPPL-KCDALETLDL 707
+N N +PP LE+L+L
Sbjct: 738 SNFRNCSCLPPFGDLPCLESLEL 760
Score = 52.0 bits (123), Expect = 7e-06
Identities = 68/278 (24%), Positives = 116/278 (41%), Gaps = 39/278 (14%)
Query: 399 IIQVLEENTGTSKIEMIHFDGVIEVQWDGEAFKKMENLKTLIFSNDVFFSENPKHLP--N 456
+++ L+ ++ + +++ F G+ +W + K N+ +++ SN N LP
Sbjct: 697 VLEALKPHSNLTSLKIYGFRGIHLPEWMNHSVLK--NIVSILISN----FRNCSCLPPFG 750
Query: 457 SLRVLECRN-HM*ELDLSYCINLESFSHVVDGFGDKLRTMSVRGCFKLKSIPPLKLDSLE 515
L LE H D+ Y ++ H GF ++R S+R KLD +
Sbjct: 751 DLPCLESLELHWGSADVEYVEEVDIDVH--SGFPTRIRFPSLR-----------KLDIWD 797
Query: 516 TMDLSCCFRLES---FPLVVDGILGKIKTLNVESCHNLRSIPPLKLDSLEKLDISYCGSL 572
L + E FP++ + I+ + L + S NLR++ L+ I Y
Sbjct: 798 FGSLKGLLKKEGEEQFPVLEEMIIHECPFLTLSS--NLRALTSLR--------ICYNKVA 847
Query: 573 ESFPQVEGRFLGKLKTLNVKSCRIMISIPTLMLSL--LEELDLSYCLNLENFPLVVDGFL 630
SFP+ + L LK L + C + +PT + SL L+ L + C LE+ P L
Sbjct: 848 TSFPEEMFKNLANLKYLTISRCNNLKELPTSLASLNALKSLKIQLCCALESLPEEGLEGL 907
Query: 631 GKLKTLSAKSCRNLRSIPS--LKLDLLETLDLSNCVSL 666
L L + C L+ +P L L +L + C L
Sbjct: 908 SSLTELFVEHCNMLKCLPEGLQHLTTLTSLKIRGCPQL 945
>RFL1_ARATH (Q8L3R3) Disease resistance protein RFL1 (RPS5-like
protein 1) (pNd13/pNd14)
Length = 885
Score = 72.0 bits (175), Expect = 7e-12
Identities = 142/638 (22%), Positives = 240/638 (37%), Gaps = 126/638 (19%)
Query: 5 LLLDRDGYVHEFIEKIVEEVLRNIKPVALPAGDFLVGVEHQKQHVTLLLNVGSDDSIHKV 64
LL + +G + + IV E + LP +VG Q + + N +D + V
Sbjct: 123 LLREVEGLSSQGVFDIVTEAAPIAEVEELPIQSTIVG---QDSMLDKVWNCLMEDKVWIV 179
Query: 65 GILGIGGIGKTTLALEVYNS---IVSQFECSCFLEKVRENSDKNGLIYLQKILLSHIFG- 120
G+ G+GG+GKTTL ++ N + F+ ++ KN ++ + + G
Sbjct: 180 GLYGMGGVGKTTLLTQINNKFSKLGGGFDVVIWVVV-----SKNATVHKIQKSIGEKLGL 234
Query: 121 -EKNTEITSVGEGISILKKRLPEKKVLLLLDDVDKKEQLKAIAGSSNWFRKGSRVIITTR 179
KN + + + + L KK +LLLDD+ +K +LK I G +V TT
Sbjct: 235 VGKNWDEKNKNQRALDIHNVLRRKKFVLLLDDIWEKVELKVIGVPYPSGENGCKVAFTTH 294
Query: 180 YKNLLISSGVERIYEVKGLNDEDAFDLVGWKTLKNDCSPRYKDVLLEQKYGRESDANKLR 239
K + GV+ E+ L+ +A+DL+ K + E G D +L
Sbjct: 295 SKEVCGRMGVDNPMEISCLDTGNAWDLL-------------KKKVGENTLGSHPDIPQLA 341
Query: 240 RLKDLKNDEGYANVLKRVVAYASGHPLALEVIGSHFS-NKTIEQCNDALDRYKRVP---- 294
R +V G PLAL VIG S +TI++ A +
Sbjct: 342 R---------------KVSEKCCGLPLALNVIGETMSFKRTIQEWRHATEVLTSATDFSG 386
Query: 295 -HNMIQTTLQLSYDSLQ-EEEKIVFLDIACCFKGWKLT--------MVEGILHAHHGHTM 344
+ I L+ SYDSL E+ K FL + + +++ + EG + G
Sbjct: 387 MEDEILPILKYSYDSLNGEDAKSCFLYCSLFPEDFEIRKEMLIEYWICEGFIKEKQGREK 446
Query: 345 K-----DQINVLVEKYLI--KINESGNVTLHDLVEDMGKEIVRQEAPEDPGKRSRLWFSE 397
D + LV L+ + V++HD+V +M I D GK
Sbjct: 447 AFNQGYDILGTLVRSSLLLEGAKDKDVVSMHDMVREMALWIF-----SDLGKHKERC--- 498
Query: 398 DIIQVLEENTGTSKI-EMIHFDGVIEVQWDGEAFKKMENLKTLIFSNDVFFSENPKHLPN 456
+++ G ++ E+ ++ V + F+K+ + +F N K +
Sbjct: 499 ----IVQAGIGLDELPEVENWRAVKRMSLMNNNFEKILGSPECVELITLFLQNNYKLVDI 554
Query: 457 SLRVLECRNHM*ELDLSYCINLESFSHVVDGFGDKLRTMSVRGCFKLKSIPPLKLDSLET 516
S+ C + LDLS +L + +L SL+
Sbjct: 555 SMEFFRCMPSLAVLDLSENHSLSELPEEIS-----------------------ELVSLQY 591
Query: 517 MDLSCCFRLESFPLVVDGILGKIKTLNVESCHNLRSIPPLKLDSLEKLDISYCGSLESFP 576
+DLS + +E P H LR + LKL+ + LES
Sbjct: 592 LDLSGTY-IERLP---------------HGLHELRKLVHLKLERTRR--------LESIS 627
Query: 577 QVEGRFLGKLKTLNVKSCRIMISIPTLM-LSLLEELDL 613
+ +L L+TL ++ + + + L LLE L+L
Sbjct: 628 GIS--YLSSLRTLRLRDSKTTLDTGLMKELQLLEHLEL 663
>RP13_ARATH (Q9M667) Disease resistance protein RPP13 (Resistance to
Peronospora parasitica protein 13)
Length = 835
Score = 70.5 bits (171), Expect = 2e-11
Identities = 88/340 (25%), Positives = 154/340 (44%), Gaps = 61/340 (17%)
Query: 64 VGILGIGGIGKTTLALEVYNS--IVSQFECSCFLEKVRENSDKNGLIYLQKIL-LSHIFG 120
+ I G+GG+GKT LA ++YNS + +FE + +E K G I ++ I L G
Sbjct: 188 ISIFGMGGLGKTALARKLYNSRDVKERFEYRAWTYVSQEY--KTGDILMRIIRSLGMTSG 245
Query: 121 EKNTEITSVGEGIS--ILKKRLPEKKVLLLLDDVDKKEQLKAIAGSSNWFRKGSRVIITT 178
E+ +I E L L KK L+++DD+ ++E ++ + +GSRVIITT
Sbjct: 246 EELEKIRKFAEEELEVYLYGLLEGKKYLVVVDDIWEREAWDSLKRALPCNHEGSRVIITT 305
Query: 179 RYKNLLISSGVE-RIY--EVKGLNDEDAFDLVGWKTLKNDCSPRYKDVLLEQKYGRESDA 235
R K ++ GV+ R Y +++ L E++++L + +N +++K
Sbjct: 306 RIK--AVAEGVDGRFYAHKLRFLTFEESWELFEQRAFRN----------IQRK------- 346
Query: 236 NKLRRLKDLKNDEGYANVLKRVVAYASGHPLALEVIGSHFSNKTIEQCNDALDR-YKRVP 294
DE K +V G PL + V+ S KT + ND + ++R+
Sbjct: 347 -----------DEDLLKTGKEMVQKCRGLPLCIVVLAGLLSRKTPSEWNDVCNSLWRRLK 395
Query: 295 HNMIQT---TLQLSYDSLQEEEKIVFLDIACCFKGWK--------LTMVEGILHAHHGHT 343
+ I LS+ L+ E K+ FL ++ + ++ L + EG +
Sbjct: 396 DDSIHVAPIVFDLSFKELRHESKLCFLYLSIFPEDYEIDLEKLIHLLVAEGFIQGDEEMM 455
Query: 344 MKD----QINVLVEKYLIKI--NESGNV---TLHDLVEDM 374
M+D I L+++ L++ E G V +HDL+ D+
Sbjct: 456 MEDVARYYIEELIDRSLLEAVRRERGKVMSCRIHDLLRDV 495
>ESA8_TRYEQ (P26337) Putative adenylate cyclase regulatory protein
Length = 630
Score = 69.7 bits (169), Expect = 3e-11
Identities = 88/315 (27%), Positives = 135/315 (41%), Gaps = 55/315 (17%)
Query: 398 DIIQVLEENTGTSKIEMIHFDGVIEVQWDGEAFKKMENLKTLIFSNDVFFSE-------- 449
++ + LEE S + + G + V K + NLK L SN F +
Sbjct: 289 NVTKGLEELCKFSNLRELDISGCL-VLGSAVVLKNLINLKVLSVSNCKNFKDLNGLERLV 347
Query: 450 -------NPKHLPNSLRVLECRNHM*ELDLSYCINLESFSHVVDGFGD--KLRTMSVRGC 500
+ H +SL + +++ ELD+S C +L F DG D L + +R
Sbjct: 348 NLDKLNLSGCHGVSSLGFVANLSNLKELDISGCESLVCF----DGLQDLNNLEVLYLRDV 403
Query: 501 FKLKSIPPLK-LDSLETMDLSCCFRLESFPLVVDGI--LGKIKTLNVESCHNLRSIPPL- 556
++ +K L + +DLS C R+ S + G+ L ++ L++E C + S P+
Sbjct: 404 KSFTNVGAIKNLSKMRELDLSGCERITS----LSGLETLKGLEELSLEGCGEIMSFDPIW 459
Query: 557 KLDSLEKLDISYCGSLESFPQVEGRFLGKLKTLNVKSCRIMISI-PTLMLSLLEELDLSY 615
L L L +S CG+LE +EG + L+ L + CR + P L + ++LS
Sbjct: 460 SLHHLRVLYVSECGNLEDLSGLEG--ITGLEELYLHGCRKCTNFGPIWNLRNVCVVELSC 517
Query: 616 CLNLENF---------------------PLVVDGFLGKLKTLSAKSCRNLRSIPSL-KLD 653
C NLE+ P+ V G L LK LS C NL+ + L +L
Sbjct: 518 CENLEDLSGLQCLTGLEELYLIGCEEITPIGVVGNLRNLKCLSTCWCANLKELGGLDRLV 577
Query: 654 LLETLDLSNCVSLES 668
LE LDLS C L S
Sbjct: 578 NLEKLDLSGCCGLSS 592
Score = 54.7 bits (130), Expect = 1e-06
Identities = 86/346 (24%), Positives = 134/346 (37%), Gaps = 86/346 (24%)
Query: 469 ELDLSYCINLESFSHVVDGFGDKLRTM--------------------------------- 495
+LDLS C NLE +V LR +
Sbjct: 138 DLDLSECANLELRELMVVLTLRNLRKLRMKRTMVNDMWCSSIGLLKFLVHLEVDGSRGVT 197
Query: 496 SVRGCFKLKSIPPLKLDS----LETMDLSCCF-RLESFPLVVDGIL----------GKIK 540
+ G +LK++ L LDS + D C +L S L + GK+K
Sbjct: 198 DITGLCRLKTLEALSLDSCINITKGFDKICALPQLTSLSLCQTNVTDKDLRCIHPDGKLK 257
Query: 541 TLNVESCHNLRSIPPL-KLDSLEKLDISYCGSLESFPQVEGRFLGKLKTLNVKSCRIMIS 599
L SCH + + + + SLEKL +S C ++ + +F
Sbjct: 258 VLRYSSCHEITDLTAIGGMRSLEKLSLSGCWNVTKGLEELCKF----------------- 300
Query: 600 IPTLMLSLLEELDLSYCLNLENFPLVVDGFLGKLKTLSAKSCRNLRSIPSL-KLDLLETL 658
S L ELD+S CL L + VV L LK LS +C+N + + L +L L+ L
Sbjct: 301 ------SNLRELDISGCLVLGS--AVVLKNLINLKVLSVSNCKNFKDLNGLERLVNLDKL 352
Query: 659 DLSNCVSLESLPLVVDGFLGKLKTLLVTNCHNLKSIPPLKCDALETLDLSCCYSLQSFSL 718
+LS C + SL V + L LK L ++ C +L L+ L L++ ++SF+
Sbjct: 353 NLSGCHGVSSLGFVAN--LSNLKELDISGCESLVCFDGLQ--DLNNLEVLYLRDVKSFTN 408
Query: 719 VA-----DRLWKLVLDDCKELQEIKVIPPC--LRMLSAVNCTSLTS 757
V ++ +L L C+ + + + L LS C + S
Sbjct: 409 VGAIKNLSKMRELDLSGCERITSLSGLETLKGLEELSLEGCGEIMS 454
Score = 52.4 bits (124), Expect = 6e-06
Identities = 75/293 (25%), Positives = 133/293 (44%), Gaps = 45/293 (15%)
Query: 479 ESFSHVVDGFGDKLRTMSVRGC-FKLKSIPPLK-LDSLETMDLSCCFRLESFPLVVD--- 533
E F + + + +++ GC +L+ + L+ L++LE +DLS C LE L+V
Sbjct: 99 EIFRRLEGSKNGRWKILNLSGCGSELQDLTALRDLEALEDLDLSECANLELRELMVVLTL 158
Query: 534 ---------------------GILGKIKTLNVESCHNLRSIPPL-KLDSLEKLDISYCGS 571
G+L + L V+ + I L +L +LE L + C +
Sbjct: 159 RNLRKLRMKRTMVNDMWCSSIGLLKFLVHLEVDGSRGVTDITGLCRLKTLEALSLDSCIN 218
Query: 572 L-ESFPQVEGRFLGKLKTLNVKSCRIMISIPTLML----SLLEELDLSYCLNLENFPLVV 626
+ + F ++ L +L +L++ C+ ++ L L+ L S C + + +
Sbjct: 219 ITKGFDKICA--LPQLTSLSL--CQTNVTDKDLRCIHPDGKLKVLRYSSCHEITDLTAI- 273
Query: 627 DGFLGKLKTLSAKSCRNL-RSIPSL-KLDLLETLDLSNCVSLESLPLVVDGFLGKLKTLL 684
G + L+ LS C N+ + + L K L LD+S C+ L S ++ + L LK L
Sbjct: 274 -GGMRSLEKLSLSGCWNVTKGLEELCKFSNLRELDISGCLVLGSAVVLKN--LINLKVLS 330
Query: 685 VTNCHNLKSIPPL-KCDALETLDLSCCYSLQSFSLVAD--RLWKLVLDDCKEL 734
V+NC N K + L + L+ L+LS C+ + S VA+ L +L + C+ L
Sbjct: 331 VSNCKNFKDLNGLERLVNLDKLNLSGCHGVSSLGFVANLSNLKELDISGCESL 383
Score = 48.1 bits (113), Expect = 1e-04
Identities = 70/242 (28%), Positives = 96/242 (38%), Gaps = 64/242 (26%)
Query: 571 SLESFPQVEGRFLGKLKTLNVKSCRIMISIPTLM--LSLLEELDLSYCLNLENFPLVV-- 626
S E F ++EG G+ K LN+ C + T + L LE+LDLS C NLE L+V
Sbjct: 97 SSEIFRRLEGSKNGRWKILNLSGCGSELQDLTALRDLEALEDLDLSECANLELRELMVVL 156
Query: 627 ----------------------DGFLGKLKTLSAKSCRNLRSIPSL-KLDLLETLDLSNC 663
G L L L R + I L +L LE L L +C
Sbjct: 157 TLRNLRKLRMKRTMVNDMWCSSIGLLKFLVHLEVDGSRGVTDITGLCRLKTLEALSLDSC 216
Query: 664 VS-------------LESLPL----VVDGFL------GKLKTLLVTNCHNLKSIPPL-KC 699
++ L SL L V D L GKLK L ++CH + + +
Sbjct: 217 INITKGFDKICALPQLTSLSLCQTNVTDKDLRCIHPDGKLKVLRYSSCHEITDLTAIGGM 276
Query: 700 DALETLDLSCCYS-------LQSFSLVADRLWKLVLDDCKELQEIKVIPPC--LRMLSAV 750
+LE L LS C++ L FS L +L + C L V+ L++LS
Sbjct: 277 RSLEKLSLSGCWNVTKGLEELCKFS----NLRELDISGCLVLGSAVVLKNLINLKVLSVS 332
Query: 751 NC 752
NC
Sbjct: 333 NC 334
Score = 46.2 bits (108), Expect = 4e-04
Identities = 45/146 (30%), Positives = 67/146 (45%), Gaps = 25/146 (17%)
Query: 465 NHM*ELDLSYCINLESFSHVVDGFGDKLRTMSVRGCFKLKSIPPL-KLDSLETMDLSCCF 523
+H+ L +S C NLE S + G L + + GC K + P+ L ++ ++LSCC
Sbjct: 462 HHLRVLYVSECGNLEDLSGLEGITG--LEELYLHGCRKCTNFGPIWNLRNVCVVELSCCE 519
Query: 524 RLESF---------------------PLVVDGILGKIKTLNVESCHNLRSIPPL-KLDSL 561
LE P+ V G L +K L+ C NL+ + L +L +L
Sbjct: 520 NLEDLSGLQCLTGLEELYLIGCEEITPIGVVGNLRNLKCLSTCWCANLKELGGLDRLVNL 579
Query: 562 EKLDISYCGSLESFPQVEGRFLGKLK 587
EKLD+S C L S +E L KL+
Sbjct: 580 EKLDLSGCCGLSSSVFMELMSLPKLQ 605
>RGA4_SOLBU (Q7XA39) Putative disease resistance protein RGA4
(RGA4-blb)
Length = 988
Score = 68.6 bits (166), Expect = 7e-11
Identities = 156/686 (22%), Positives = 267/686 (38%), Gaps = 122/686 (17%)
Query: 51 LLLNVGSDDSIHKVGILGIGGIGKTTLALEVYNS--IVSQFECSCFLEKVRENSDKNGLI 108
L+ NV + + I+G+GG+GKTTLA ++N + F ++ V ++ D+ LI
Sbjct: 167 LINNVNVAEELPVFPIIGMGGLGKTTLAQMIFNDERVTKHFNPKIWV-CVSDDFDEKRLI 225
Query: 109 YLQKILLSHIFGEKNTEITSVGEGISILKKRLPEKKVLLLLDDV--DKKEQLKAIAGSSN 166
K ++ +I + + + L++ L K+ LL+LDDV D E+ +
Sbjct: 226 ---KTIIGNI-ERSSPHVEDLASFQKKLQELLNGKRYLLVLDDVWNDDLEKWAKLRAVLT 281
Query: 167 WFRKGSRVIITTRYKNLLISSGVERIYEVKGLNDEDAFDLVGWKTLKNDCSPRYKDVLLE 226
+G+ ++ TTR + + G + Y + L+ D+ L ++
Sbjct: 282 VGARGASILATTRLEKVGSIMGTLQPYHLSNLSPHDSLLL-----------------FMQ 324
Query: 227 QKYGRESDANKLRRLKDLKNDEGYANVLKRVVAYASGHPLALEVIGS--HFSNKTIEQCN 284
+ +G++ +AN + K +V G PLA + +G F + E +
Sbjct: 325 RAFGQQKEAN-----------PNLVAIGKEIVKKCGGVPLAAKTLGGLLRFKREESEWEH 373
Query: 285 DALDRYKRVP--HNMIQTTLQLSYDSLQEEEKIVFLDIACCFKGWK--------LTMVEG 334
+ +P + I L+LSY L + + F A K K L M G
Sbjct: 374 VRDNEIWSLPQDESSILPALRLSYHHLPLDLRQCFAYCAVFPKDTKMIKENLITLWMAHG 433
Query: 335 ILHAHHGHTMKDQIN-VLVEKYLIKI-----NESGNV--TLHDLVEDMGKEIVRQEAPED 386
L + ++D N V E YL +SGN +HDL+ D+ + A
Sbjct: 434 FLLSKGNLELEDVGNEVWNELYLRSFFQEIEAKSGNTYFKIHDLIHDLATSLFSASA--- 490
Query: 387 PGKRSRLWFSEDIIQVLEENTGTSKIEM-IHFDGVIEVQWDGEAFKKMENLKTLIFSNDV 445
+ E N K + I F V+ + KK +L+ L S
Sbjct: 491 -----------SCGNIREINVKDYKHTVSIGFAAVVS-SYSPSLLKKFVSLRVLNLSYSK 538
Query: 446 FFSENPKHLPNSLRVLECRNHM*ELDLSYCINLESFSHVVDGFGDKLRTMSVRGCFKLKS 505
+ LP+S+ L H+ LDLS C N R++ R C
Sbjct: 539 L-----EQLPSSIGDL---LHLRYLDLS-CNN--------------FRSLPERLC----- 570
Query: 506 IPPLKLDSLETMDLSCCFRLESFPLVVDGILGKIKTLNVESCHNLRSIPPL--KLDSLEK 563
KL +L+T+D+ C+ L P L ++ L V+ C L S PP L L+
Sbjct: 571 ----KLQNLQTLDVHNCYSLNCLPKQTSK-LSSLRHLVVDGC-PLTSTPPRIGLLTCLKT 624
Query: 564 LDISYCGSLESFPQVEGRFLGKLKTLNVKSCRIMISIPTLMLSLLEELDLSYCLNLENFP 623
L GS + + LG+LK LN+ + + + E +LS NL++
Sbjct: 625 LGFFIVGSKKGYQ------LGELKNLNLCGSISITHLERVKNDTDAEANLSAKANLQSLS 678
Query: 624 LVVDG-FLGKLKTLSAKSCRNLRSIPSLKLDLLETLDLSNCVSLESLPLVVDGFLGKLKT 682
+ D + ++ K L+ P+LK L++ + L K+ +
Sbjct: 679 MSWDNDGPNRYESKEVKVLEALKPHPNLKY-----LEIIAFGGFRFPSWINHSVLEKVIS 733
Query: 683 LLVTNCHNLKSIPPL-KCDALETLDL 707
+ + +C N +PP + LE L+L
Sbjct: 734 VRIKSCKNCLCLPPFGELPCLENLEL 759
Score = 63.5 bits (153), Expect = 2e-09
Identities = 94/338 (27%), Positives = 147/338 (42%), Gaps = 39/338 (11%)
Query: 469 ELDLSYCINLESFSHVVDGFG-DKLRTMSVRGCFKLKSIPPLKLDSLETMDLSCCFRLES 527
E +LS NL+S S D G ++ + V+ LK P LK LE + FR S
Sbjct: 665 EANLSAKANLQSLSMSWDNDGPNRYESKEVKVLEALKPHPNLKY--LEIIAFGG-FRFPS 721
Query: 528 FPLVVDGILGKIKTLNVESCHNLRSIPPL-KLDSLEKLDISYCGSLE----SFPQVEGRF 582
+ + +L K+ ++ ++SC N +PP +L LE L++ GS E V RF
Sbjct: 722 W--INHSVLEKVISVRIKSCKNCLCLPPFGELPCLENLELQN-GSAEVEYVEEDDVHSRF 778
Query: 583 LGKLKTLNVKSCRIMI--SIPTLM-------LSLLEELDLSYCLNLENFPLVVDGFLGKL 633
+ ++K RI S+ LM +LEE+ + YC PL V L +
Sbjct: 779 STRRSFPSLKKLRIWFFRSLKGLMKEEGEEKFPMLEEMAILYC------PLFVFPTLSSV 832
Query: 634 KTLSAKSCRNLRSIPSL-KLDLLETLDLSNCVSLESLPLVVDGFLGKLKTLLVTNCHNLK 692
K L N R + S+ L L +L + SLP + L L+ L + NLK
Sbjct: 833 KKLEVHGNTNTRGLSSISNLSTLTSLRIGANYRATSLPEEMFTSLTNLEFLSFFDFKNLK 892
Query: 693 SIPP--LKCDALETLDLSCCYSLQSFSLVA----DRLWKLVLDDCKELQEIKVIPPCLRM 746
+P +AL+ L + C SL+SF L +L + CK L K +P L+
Sbjct: 893 DLPTSLTSLNALKRLQIESCDSLESFPEQGLEGLTSLTQLFVKYCKML---KCLPEGLQH 949
Query: 747 LSAVNCTSLTSSCTSKLLNQELHKAGNTWFCLPRVPKI 784
L+A+ T+L S ++ + + G W + +P +
Sbjct: 950 LTAL--TNLGVSGCPEVEKRCDKEIGEDWHKIAHIPNL 985
Score = 47.0 bits (110), Expect = 2e-04
Identities = 54/228 (23%), Positives = 102/228 (44%), Gaps = 23/228 (10%)
Query: 400 IQVLEENTGTSKIEMIHFDGVIEVQWDGEAFKKMENLKTLIFSN--DVFFSENPKHLPNS 457
++ LE G++++E + D V +F ++ L+ F + + E + P
Sbjct: 754 LENLELQNGSAEVEYVEEDDVHSRFSTRRSFPSLKKLRIWFFRSLKGLMKEEGEEKFP-- 811
Query: 458 LRVLECRNHM*ELDLSYCINLESFSHVVDGFGDKLRTMSVRGCFKLKSIPPLK-LDSLET 516
+LE E+ + YC L F + ++ + V G + + + L +L +
Sbjct: 812 --MLE------EMAILYC-PLFVFPTL-----SSVKKLEVHGNTNTRGLSSISNLSTLTS 857
Query: 517 MDLSCCFRLESFPLVVDGILGKIKTLNVESCHNLRSIPP--LKLDSLEKLDISYCGSLES 574
+ + +R S P + L ++ L+ NL+ +P L++L++L I C SLES
Sbjct: 858 LRIGANYRATSLPEEMFTSLTNLEFLSFFDFKNLKDLPTSLTSLNALKRLQIESCDSLES 917
Query: 575 FPQVEGRFLGKLKTLNVKSCRIMISIPTLM--LSLLEELDLSYCLNLE 620
FP+ L L L VK C+++ +P + L+ L L +S C +E
Sbjct: 918 FPEQGLEGLTSLTQLFVKYCKMLKCLPEGLQHLTALTNLGVSGCPEVE 965
Score = 45.8 bits (107), Expect = 5e-04
Identities = 71/268 (26%), Positives = 107/268 (39%), Gaps = 74/268 (27%)
Query: 584 GKLKTLNVKSCRIMISI----------PTLMLSL--LEELDLSYCLNLENFPLVVDGFLG 631
G ++ +NVK + +SI P+L+ L L+LSY LE P + G L
Sbjct: 493 GNIREINVKDYKHTVSIGFAAVVSSYSPSLLKKFVSLRVLNLSYS-KLEQLPSSI-GDLL 550
Query: 632 KLKTLSAKSCRNLRSIPS--LKLDLLETLDLSNCVSLESLP-----------LVVDGF-- 676
L+ L SC N RS+P KL L+TLD+ NC SL LP LVVDG
Sbjct: 551 HLRYLDL-SCNNFRSLPERLCKLQNLQTLDVHNCYSLNCLPKQTSKLSSLRHLVVDGCPL 609
Query: 677 ----------------------------LGKLKTLLVTNCHNLKSIPPLKCDALETLDLS 708
LG+LK L + ++ + +K D +LS
Sbjct: 610 TSTPPRIGLLTCLKTLGFFIVGSKKGYQLGELKNLNLCGSISITHLERVKNDTDAEANLS 669
Query: 709 CCYSLQSFSLVADRLWKLVLDDCKELQEIKVIP-----PCLRMLSAVNC------TSLTS 757
+LQS S+ W + E +E+KV+ P L+ L + + +
Sbjct: 670 AKANLQSLSM----SWDNDGPNRYESKEVKVLEALKPHPNLKYLEIIAFGGFRFPSWINH 725
Query: 758 SCTSKLLNQELHKAGNTWFCLPRVPKIP 785
S K+++ + N CLP ++P
Sbjct: 726 SVLEKVISVRIKSCKNC-LCLPPFGELP 752
>DR20_ARATH (Q9SH22) Putative disease resistance protein At1g63360
Length = 884
Score = 68.2 bits (165), Expect = 1e-10
Identities = 77/277 (27%), Positives = 121/277 (42%), Gaps = 63/277 (22%)
Query: 58 DDSIHKVGILGIGGIGKTTLALEVYNS--------------IVSQFECSCFLEKVRENSD 103
+D + +G+ G+GG+GKTTL ++YN +VSQ +EKV++
Sbjct: 169 EDGVGIMGMYGMGGVGKTTLLTQLYNMFNKDKCGFDIGIWVVVSQ---EFHVEKVQDE-- 223
Query: 104 KNGLIYLQKILLSHIFGEKNTEITSVGEGISILKKRLPEKKVLLLLDDVDKKEQLKAIAG 163
QK+ L G++ T+ +GI L L EK +L LDD+ +K L I
Sbjct: 224 -----IAQKLGLG---GDEWTQKDKSQKGIC-LYNILREKSFVLFLDDIWEKVDLAEIGV 274
Query: 164 SSNWFRKGSRVIITTRYKNLLISSGVERIYEVKGLNDEDAFDLVGWKTLKNDCSPRYKDV 223
+KG ++ TTR + + GVE EV+ L + AFDL
Sbjct: 275 PDPRTKKGRKLAFTTRSQEVCARMGVEHPMEVQCLEENVAFDL----------------- 317
Query: 224 LLEQKYGRESDANKLRRLKDLKNDEGYANVLKRVVAYASGHPLALEVIGSHFS-NKTIEQ 282
++K G+ + L +D G + + V G PLAL VIG S +TI++
Sbjct: 318 -FQKKVGQTT----------LGSDPGIPQLARIVAKKCCGLPLALNVIGETMSCKRTIQE 366
Query: 283 CNDA---LDRYKRV---PHNMIQTTLQLSYDSLQEEE 313
A L+ Y + + L+ SYD+L+ E+
Sbjct: 367 WRHAIHVLNSYAAEFIGMEDKVLPLLKYSYDNLKGEQ 403
>DR36_ARATH (Q8VZC7) Probable disease resistance protein At5g45510
Length = 1202
Score = 65.9 bits (159), Expect = 5e-10
Identities = 121/475 (25%), Positives = 195/475 (40%), Gaps = 96/475 (20%)
Query: 351 LVEKYLIKINESGNVTLHDLVEDMGKEIVRQEAPEDPGKRSRLWFSEDIIQVLEENTGTS 410
L+++ ++KI E+ NV + ++ + ++ GK SRL FS + + G
Sbjct: 514 LIDRGMLKIQEN-NVVVPEMAM---RNVIDPRRGGHLGK-SRLGFSR--VYGGNKRKGIG 566
Query: 411 KIEMIHFDGVIEVQWDGEAFKKMENLKTLIFSNDVFFSENPKHLPNSLRVLECRNHM*EL 470
KI + D + VQ KK + + T++ S D PK +L+ LE
Sbjct: 567 KITQLD-DMIKTVQ-----AKKGDKITTILVSGDRLRRVTPKKFFKNLKELEVLG----- 615
Query: 471 DLSYCINLESFSHVVDGFGDKL---RTMSVRGCFKLKSIPPLK-LDSLETMDLSCCFRL- 525
+ + V F D+L R + +R C LKSI LK L L T+++S L
Sbjct: 616 -----LFEPTVKPFVPSFSDQLKLLRVLIIRDCDLLKSIEELKALTKLNTLEVSGASSLS 670
Query: 526 -------ESFP----LVVDGI-----------LGKIKTLNVESCHNLRSIPPLK-LDSLE 562
ESFP L + G+ L ++ L ++ C L+ +P ++ L +LE
Sbjct: 671 KISEKFFESFPELRSLHLSGLKIESSPPSISGLKELHCLIIKDCPLLQDLPNIQELVNLE 730
Query: 563 KLDISYCGSLES-FPQVEGRFLGKLKTLNVKSCRIMISIPTLMLSLLEELDLSYCLNLEN 621
+D+S L + F +G K K N +L+ L+ LD S +E
Sbjct: 731 VVDVSGASGLRTCFDNADGAKKNKSKNKNF-----------YLLTKLQHLDFSGS-QIER 778
Query: 622 FPLVVDGF----LGKLKTLSAKSCRNLRSIPSLK-LDLLETLDLSNCVS----------- 665
P+ D L L L ++C LR +PSLK L L+ LDLS S
Sbjct: 779 LPIFQDSAVAAKLHSLTRLLLRNCSKLRRLPSLKPLSGLQILDLSGTTSLVEMLEVCFED 838
Query: 666 -LESLPLVVDGF-----------LGKLKTLLVTNCHNLKSIPPL-KCDALETLDLSCCYS 712
LE L + G L L LL+ +C NL +IP + K + LE +D+S
Sbjct: 839 KLELKTLNLSGTNLSELATTIEDLSSLNELLLRDCINLDAIPNIEKLENLEVIDVSGSAK 898
Query: 713 LQSFSLVADRLWKLVLDDCKELQEIKVIPPCLRMLSAVNCTSLTSSCTSKLLNQE 767
L ++++ L + D L +V P L + ++C + K ++
Sbjct: 899 LAKIEGSFEKMFYLRVVD---LSGTQVETPELPADTKIHCLKRFTRADGKCFERD 950
Score = 35.0 bits (79), Expect = 0.92
Identities = 22/63 (34%), Positives = 35/63 (54%), Gaps = 2/63 (3%)
Query: 652 LDLLETLDLSNCVSLESLPLVVDGFLGKLKTLLVTNCHNLKSIPPLKCDALETLDLSCCY 711
L LETL ++N +SLE++ + L LK L + C +K+I P +L L+L C
Sbjct: 1101 LKSLETLSITNLLSLETISFIAK--LENLKNLSLDCCPKIKTIFPEMPASLPVLNLKHCE 1158
Query: 712 SLQ 714
+L+
Sbjct: 1159 NLE 1161
Score = 32.0 bits (71), Expect = 7.7
Identities = 16/63 (25%), Positives = 35/63 (55%), Gaps = 4/63 (6%)
Query: 492 LRTMSVRGCFKLKSIPPL-KLDSLETMDLSCCFRLESFPLVVDGILGKIKTLNVESCHNL 550
L T+S+ L++I + KL++L+ + L CC ++++ + + + LN++ C NL
Sbjct: 1104 LETLSITNLLSLETISFIAKLENLKNLSLDCCPKIKT---IFPEMPASLPVLNLKHCENL 1160
Query: 551 RSI 553
+
Sbjct: 1161 EKV 1163
>DR41_ARATH (Q9FKZ2) Probable disease resistance protein At5g66890
Length = 415
Score = 65.5 bits (158), Expect = 6e-10
Identities = 68/242 (28%), Positives = 106/242 (43%), Gaps = 51/242 (21%)
Query: 511 LDSLETMDLSCCFRLESFPLVVDGILGKIKTLNVESCH-NLRSIPPLKLDSLEKLDISYC 569
L+ + +LSC L S P +K + E +L IP L L SLEKL + +C
Sbjct: 190 LEPAKLTNLSC---LSSLP--------NLKRIRFEKVSISLLDIPKLGLKSLEKLSLWFC 238
Query: 570 GSLESFPQVEGRFLGKLKTLNVKSCRIMISIPTLMLSLLEELDLSYCLNLENFPLVVDGF 629
+++ ++E + L L+E+++ YC NL+ P +
Sbjct: 239 HVVDALNELED--------------------VSETLQSLQEIEIDYCYNLDELPYWISQV 278
Query: 630 LGKLKTLSAKSCRNLRSIPSLKLDL--LETLDLSNCVSLESLPLVVDGFLGKLKTLLVTN 687
+ LK LS +C L + DL LETL LS+C SL LP +D L L+ L V+
Sbjct: 279 V-SLKKLSVTNCNKLCRVIEAIGDLRDLETLRLSSCASLLELPETIDR-LDNLRFLDVSG 336
Query: 688 CHNLKSIPPL--KCDALETLDLSCCY---------SLQSFSLVADR----LWKLVLDDCK 732
LK++P K LE + + CY +L++ + D LWK++ + K
Sbjct: 337 GFQLKNLPLEIGKLKKLEKISMKDCYRCELPDSVKNLENLEVKCDEDTAFLWKILKPEMK 396
Query: 733 EL 734
L
Sbjct: 397 NL 398
Score = 45.8 bits (107), Expect = 5e-04
Identities = 50/162 (30%), Positives = 72/162 (43%), Gaps = 24/162 (14%)
Query: 583 LGKLKTLNVKSCRI-MISIPTLMLSLLEELDLSYCLNLENFPLVVDGFLGKLKTLSAKSC 641
L LK + + I ++ IP L L LE+L L +C VVD L +L+ +S
Sbjct: 204 LPNLKRIRFEKVSISLLDIPKLGLKSLEKLSLWFCH-------VVDA-LNELEDVSET-- 253
Query: 642 RNLRSIPSLKLDLLETLDLSNCVSLESLPLVVDGFLGKLKTLLVTNCHNLKSIPPLKCDA 701
L L+ +++ C +L+ LP + + LK L VTNC+ L + D
Sbjct: 254 ----------LQSLQEIEIDYCYNLDELPYWISQVVS-LKKLSVTNCNKLCRVIEAIGDL 302
Query: 702 --LETLDLSCCYSLQSFSLVADRLWKLVLDDCKELQEIKVIP 741
LETL LS C SL DRL L D ++K +P
Sbjct: 303 RDLETLRLSSCASLLELPETIDRLDNLRFLDVSGGFQLKNLP 344
Score = 42.4 bits (98), Expect = 0.006
Identities = 33/132 (25%), Positives = 63/132 (47%), Gaps = 28/132 (21%)
Query: 460 VLECRNHM*ELDLSYCINLESFSH---------------------VVDGFGD--KLRTMS 496
V E + E+++ YC NL+ + V++ GD L T+
Sbjct: 250 VSETLQSLQEIEIDYCYNLDELPYWISQVVSLKKLSVTNCNKLCRVIEAIGDLRDLETLR 309
Query: 497 VRGCFKLKSIPPL--KLDSLETMDLSCCFRLESFPLVVDGILGKIKTLNVESCHNLRSIP 554
+ C L +P +LD+L +D+S F+L++ PL + G L K++ ++++ C+ R
Sbjct: 310 LSSCASLLELPETIDRLDNLRFLDVSGGFQLKNLPLEI-GKLKKLEKISMKDCY--RCEL 366
Query: 555 PLKLDSLEKLDI 566
P + +LE L++
Sbjct: 367 PDSVKNLENLEV 378
>RPS5_ARATH (O64973) Disease resistance protein RPS5 (Resistance to
Pseudomonas syringae protein 5) (pNd3/pNd10)
Length = 889
Score = 65.1 bits (157), Expect = 8e-10
Identities = 159/691 (23%), Positives = 267/691 (38%), Gaps = 156/691 (22%)
Query: 58 DDSIHKVGILGIGGIGKTTLALEVYNS---IVSQFEC--------SCFLEKV-RENSDKN 105
+D +G+ G+GG+GKTTL ++ N I +F+ S + K+ R+ ++K
Sbjct: 173 EDGSGILGLYGMGGVGKTTLLTKINNKFSKIDDRFDVVIWVVVSRSSTVRKIQRDIAEKV 232
Query: 106 GLIYLQKILLSHIFGEKNTEITSVGEGISILKKRLPEKKVLLLLDDVDKKEQLKAIAGSS 165
GL ++ + EKN +V + L +K +LLLDD+ +K LKA+
Sbjct: 233 GLGGME-------WSEKNDNQIAVD-----IHNVLRRRKFVLLLDDIWEKVNLKAVGVPY 280
Query: 166 NWFRKGSRVIITTRYKNLLISSGVERIYEVKGLNDEDAFDLVGWKTLKNDCSPRYKDVLL 225
G +V TTR +++ GV+ EV L E+++DL K KN
Sbjct: 281 PSKDNGCKVAFTTRSRDVCGRMGVDDPMEVSCLQPEESWDLFQMKVGKNTL--------- 331
Query: 226 EQKYGRESDANKLRRLKDLKNDEGYANVLKRVVAYASGHPLALEVIGSHFS-NKTIEQCN 284
G D L R +V G PLAL VIG + +T+ +
Sbjct: 332 ----GSHPDIPGLAR---------------KVARKCRGLPLALNVIGEAMACKRTVHEWC 372
Query: 285 DALDRYKRVP------HNMIQTTLQLSYDSLQEE-EKIVFLDIACCFKGWKLT------- 330
A+D + I L+ SYD+L E K FL + + + +
Sbjct: 373 HAIDVLTSSAIDFSGMEDEILHVLKYSYDNLNGELMKSCFLYCSLFPEDYLIDKEGLVDY 432
Query: 331 -MVEGILHAHHGHTMK-----DQINVLVEKYLIKINE--SGNVTLHDLVEDMGKEIVRQE 382
+ EG ++ G + I LV L+ E NV +HD+V +M I
Sbjct: 433 WISEGFINEKEGRERNINQGYEIIGTLVRACLLLEEERNKSNVKMHDVVREMALWI---- 488
Query: 383 APEDPGKRSRLWF------SEDIIQVLEENTGTSKIEMIHFDGVIEVQWDGEAFKKMENL 436
D GK+ ++ +V + NT KI +++ + IE +D + L
Sbjct: 489 -SSDLGKQKEKCIVRAGVGLREVPKVKDWNT-VRKISLMNNE--IEEIFDSHECAALTTL 544
Query: 437 KTLIFSNDVFFSENPKHLPNSLRVLECRNHM*ELDLSYCINLESFSHVVDGFGDKLRTMS 496
+ NDV + S C H+ LDLS +L + LR +
Sbjct: 545 --FLQKNDV--------VKISAEFFRCMPHLVVLDLSENQSLNELPEEISELA-SLRYFN 593
Query: 497 VR---------GCFKLKSIPPLKLDSLETMDLSCCFRLESFPLVVDGILGKIKTLNVESC 547
+ G + LK + L L+ + + LG I L + +
Sbjct: 594 LSYTCIHQLPVGLWTLKKLIHLNLEHMSS-------------------LGSI--LGISNL 632
Query: 548 HNLRSIPPLKLDSLEKLDISYCGSLESFPQVEGRFLGKLKTLNVKSCRIMISIPTL---- 603
NLR++ DS LD+S L+ +E + TL++ S +++ P L
Sbjct: 633 WNLRTLG--LRDSRLLLDMSLVKELQLLEHLE------VITLDISSS--LVAEPLLCSQR 682
Query: 604 MLSLLEELDLSYCLNLENFPLVVDGFLGKLKTLSAKSCRNLRSIPSLKLDLLETLDLSNC 663
++ ++E+D Y L E+ ++ +G L+ L K C + +K++ T S+
Sbjct: 683 LVECIKEVDFKY-LKEESVRVLTLPTMGNLRKLGIKRC----GMREIKIE--RTTSSSSR 735
Query: 664 VSLESLPLVVDGFLGKLKTLLVTNCHNLKSI 694
+ P L + + CH LK +
Sbjct: 736 NKSPTTPC-----FSNLSRVFIAKCHGLKDL 761
>DR23_ARATH (O82484) Putative disease resistance protein At4g10780
Length = 892
Score = 65.1 bits (157), Expect = 8e-10
Identities = 72/279 (25%), Positives = 114/279 (40%), Gaps = 69/279 (24%)
Query: 58 DDSIHKVGILGIGGIGKTTLALEVYNSIVSQFECSCFLEKVRENSDKNGL-IYLQKILLS 116
DD + +G+ G+GG+GKTTL +++N++ + KNG+ I + ++ S
Sbjct: 170 DDGVGTMGLYGMGGVGKTTLLTQIHNTL---------------HDTKNGVDIVIWVVVSS 214
Query: 117 HIFGEKNTEITSVGEGISILKKR----------------LPEKKVLLLLDDVDKKEQLKA 160
+ K E +GE + + K L +K+ +LLLDD+ KK L
Sbjct: 215 DLQIHKIQE--DIGEKLGFIGKEWNKKQESQKAVDILNCLSKKRFVLLLDDIWKKVDLTK 272
Query: 161 IAGSSNWFRKGSRVIITTRYKNLLISSGVERIYEVKGLNDEDAFDLVGWKTLKNDCSPRY 220
I S +V+ TTR ++ GV EV+ L+ DA++
Sbjct: 273 IGIPSQTRENKCKVVFTTRSLDVCARMGVHDPMEVQCLSTNDAWE--------------- 317
Query: 221 KDVLLEQKYGRESDANKLRRLKDLKNDEGYANVLKRVVAYASGHPLALEVIGSHFSNKTI 280
L ++K G+ S L + + K+V G PLAL VIG + K
Sbjct: 318 ---LFQEKVGQIS----------LGSHPDILELAKKVAGKCRGLPLALNVIGETMAGKRA 364
Query: 281 EQ----CNDALDRYKRVPHNM---IQTTLQLSYDSLQEE 312
Q D L Y M I L+ SYD+L ++
Sbjct: 365 VQEWHHAVDVLTSYAAEFSGMDDHILLILKYSYDNLNDK 403
>DR14_ARATH (Q9SI85) Probable disease resistance protein At1g62630
(pNd4)
Length = 893
Score = 64.3 bits (155), Expect = 1e-09
Identities = 74/266 (27%), Positives = 118/266 (43%), Gaps = 43/266 (16%)
Query: 58 DDSIHKVGILGIGGIGKTTLALEVYNSIVSQ---FECSCFLEKVRE-NSDKNGLIYLQKI 113
+D +G+ G+GG+GKTTL +++N F+ ++ +E N +K QK+
Sbjct: 169 EDGTGIMGMYGMGGVGKTTLLTQLFNMFNKDKCGFDIGIWVVVSQEVNVEKIQDEIAQKL 228
Query: 114 LLSHIFGEKNTEITSVGEGISILKKRLPEKKVLLLLDDVDKKEQLKAIAGSSNWFRKGSR 173
L G + T+ +G+ + L KK +L LDD+ K +L I +KG +
Sbjct: 229 GLG---GHEWTQRDISQKGVHLFNF-LKNKKFVLFLDDLWDKVELANIGVPDPRTQKGCK 284
Query: 174 VIITTRYKNLLISSGVERIYEVKGLNDEDAFDLVGWKTLKNDCSPRYKDVLLEQKYGRES 233
+ T+R N+ S G E EV+ L + AFDL ++K G+
Sbjct: 285 LAFTSRSLNVCTSMGDEEPMEVQCLEENVAFDL------------------FQKKVGQ-- 324
Query: 234 DANKLRRLKDLKNDEGYANVLKRVVAYASGHPLALEVIGSHFS-NKTIEQCNDA---LDR 289
K L +D G + + V G PLAL VIG S +TI++ +A L+
Sbjct: 325 --------KTLGSDPGIPQLARIVAKKCCGLPLALNVIGETMSCKRTIQEWRNAIHVLNS 376
Query: 290 YKRV---PHNMIQTTLQLSYDSLQEE 312
Y + I L+ SYD+L+ E
Sbjct: 377 YAAEFIGMEDKILPLLKYSYDNLKGE 402
>ESA8_TRYBB (P23799) Putative adenylate cyclase regulatory protein
(Leucine repeat protein) (VSG expression site-associated
protein F14.9)
Length = 630
Score = 63.2 bits (152), Expect = 3e-09
Identities = 76/285 (26%), Positives = 123/285 (42%), Gaps = 53/285 (18%)
Query: 497 VRGCFKLKSIPPLKLDSL----ETMDLSCCF-RLESFPLVVDGIL----------GKIKT 541
+ G F+LK++ L LD+ + D C +L S L + GK+K
Sbjct: 199 ITGLFRLKTLEALSLDNCINITKGFDKICALPQLTSLSLCQTNVTDKDLRCIHPDGKLKM 258
Query: 542 LNVESCHNLRSIPPL-KLDSLEKLDISYCGSLESFPQVEGRFLGKLKTLNVKSCRIMISI 600
L++ SCH + + + + SLEKL +S C ++ + +F
Sbjct: 259 LDISSCHEITDLTAIGGVRSLEKLSLSGCWNVTKGLEELCKF------------------ 300
Query: 601 PTLMLSLLEELDLSYCLNLENFPLVVDGFLGKLKTLSAKSCRNLRSIPSL-KLDLLETLD 659
S L ELD+S CL L + VV L LK LS +C+N + + L +L LE L+
Sbjct: 301 -----SNLRELDISGCLVLGS--AVVLKNLINLKVLSVSNCKNFKDLNGLERLVNLEKLN 353
Query: 660 LSNCVSLESLPLVVDGFLGKLKTLLVTNCHNLKSIPPLKCDALETLDLSCCYSLQSFSLV 719
LS C + SL V + L LK L ++ C +L L+ L L++ ++SF+ V
Sbjct: 354 LSGCHGVSSLGFVAN--LSNLKELDISGCESLVCFDGLQ--DLNNLEVLYLRDVKSFTNV 409
Query: 720 A-----DRLWKLVLDDCKELQEIKVIPPC--LRMLSAVNCTSLTS 757
++ +L L C+ + + + L LS C + S
Sbjct: 410 GAIKNLSKMRELDLSGCERITSLSGLETLKGLEELSLEGCGEIMS 454
Score = 61.6 bits (148), Expect = 9e-09
Identities = 83/311 (26%), Positives = 129/311 (40%), Gaps = 47/311 (15%)
Query: 398 DIIQVLEENTGTSKIEMIHFDGVIEVQWDGEAFKKMENLKTLIFSNDVFFSE-------- 449
++ + LEE S + + G + V K + NLK L SN F +
Sbjct: 289 NVTKGLEELCKFSNLRELDISGCL-VLGSAVVLKNLINLKVLSVSNCKNFKDLNGLERLV 347
Query: 450 -------NPKHLPNSLRVLECRNHM*ELDLSYCINLESFSHVVD---------------- 486
+ H +SL + +++ ELD+S C +L F + D
Sbjct: 348 NLEKLNLSGCHGVSSLGFVANLSNLKELDISGCESLVCFDGLQDLNNLEVLYLRDVKSFT 407
Query: 487 GFG-----DKLRTMSVRGCFKLKSIPPLK-LDSLETMDLSCCFRLESFPLVVDGILGKIK 540
G K+R + + GC ++ S+ L+ L LE + L C + SF + L ++
Sbjct: 408 NVGAIKNLSKMRELDLSGCERITSLSGLETLKGLEELSLEGCGEIMSFDPIWS--LYHLR 465
Query: 541 TLNVESCHNLRSIPPLK-LDSLEKLDISYCGSLESFPQVEGRFLGKLKTLNVKSCRIMIS 599
L V C NL + L+ L LE++ + C +F + L + L + C +
Sbjct: 466 VLYVSECGNLEDLSGLQCLTGLEEMYLHGCRKCTNFGPIWN--LRNVCVLELSCCENLDD 523
Query: 600 IPTLM-LSLLEELDLSYCLNLENFPLVVDGFLGKLKTLSAKSCRNLRSIPSL-KLDLLET 657
+ L L+ LEEL L C + +V G L LK LS C NL+ + L +L LE
Sbjct: 524 LSGLQCLTGLEELYLIGCEEITTIGVV--GNLRNLKCLSTCWCANLKELGGLERLVNLEK 581
Query: 658 LDLSNCVSLES 668
LDLS C L S
Sbjct: 582 LDLSGCCGLSS 592
Score = 50.4 bits (119), Expect = 2e-05
Identities = 71/242 (29%), Positives = 97/242 (39%), Gaps = 64/242 (26%)
Query: 571 SLESFPQVEGRFLGKLKTLNVKSCRIMISIPTLM--LSLLEELDLSYCLNLENFPLVV-- 626
S E F ++EG G+ K LN+ C + T + L LE+LDLS C NLE L+V
Sbjct: 97 SSEIFRRLEGSKNGRWKILNLSGCGSELQDLTALRDLEALEDLDLSECANLELRELMVVL 156
Query: 627 ----------------------DGFLGKLKTLSAKSCRNLRSIPSL-KLDLLETLDLSNC 663
G L L L R + I L +L LE L L NC
Sbjct: 157 TLRNLRKLRMKRTMVNDMWCSSIGLLKFLVHLEVDGSRGVTDITGLFRLKTLEALSLDNC 216
Query: 664 VS-------------LESLPL----VVDGFL------GKLKTLLVTNCHNLKSIPPL-KC 699
++ L SL L V D L GKLK L +++CH + + +
Sbjct: 217 INITKGFDKICALPQLTSLSLCQTNVTDKDLRCIHPDGKLKMLDISSCHEITDLTAIGGV 276
Query: 700 DALETLDLSCCYS-------LQSFSLVADRLWKLVLDDCKELQEIKVIPPC--LRMLSAV 750
+LE L LS C++ L FS L +L + C L V+ L++LS
Sbjct: 277 RSLEKLSLSGCWNVTKGLEELCKFS----NLRELDISGCLVLGSAVVLKNLINLKVLSVS 332
Query: 751 NC 752
NC
Sbjct: 333 NC 334
Score = 43.5 bits (101), Expect = 0.003
Identities = 43/145 (29%), Positives = 66/145 (44%), Gaps = 25/145 (17%)
Query: 466 HM*ELDLSYCINLESFSHVVDGFGDKLRTMSVRGCFKLKSIPPL-KLDSLETMDLSCCFR 524
H+ L +S C NLE S + G L M + GC K + P+ L ++ ++LSCC
Sbjct: 463 HLRVLYVSECGNLEDLSGLQCLTG--LEEMYLHGCRKCTNFGPIWNLRNVCVLELSCCEN 520
Query: 525 LESFP------------------LVVDGILGKIKTLNVES---CHNLRSIPPL-KLDSLE 562
L+ + G++G ++ L S C NL+ + L +L +LE
Sbjct: 521 LDDLSGLQCLTGLEELYLIGCEEITTIGVVGNLRNLKCLSTCWCANLKELGGLERLVNLE 580
Query: 563 KLDISYCGSLESFPQVEGRFLGKLK 587
KLD+S C L S +E L KL+
Sbjct: 581 KLDLSGCCGLSSSVFMELMSLPKLQ 605
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.322 0.140 0.416
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 108,036,856
Number of Sequences: 164201
Number of extensions: 4724691
Number of successful extensions: 13389
Number of sequences better than 10.0: 127
Number of HSP's better than 10.0 without gapping: 39
Number of HSP's successfully gapped in prelim test: 90
Number of HSP's that attempted gapping in prelim test: 12942
Number of HSP's gapped (non-prelim): 301
length of query: 918
length of database: 59,974,054
effective HSP length: 119
effective length of query: 799
effective length of database: 40,434,135
effective search space: 32306873865
effective search space used: 32306873865
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 71 (32.0 bits)
Medicago: description of AC145512.9


