

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC145331.13 + phase: 0 /partial
(365 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
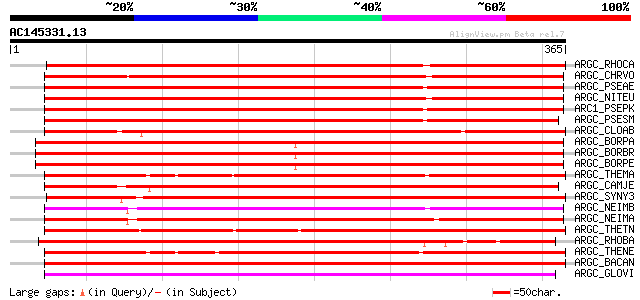
Score E
Sequences producing significant alignments: (bits) Value
ARGC_RHOCA (Q9LA02) N-acetyl-gamma-glutamyl-phosphate reductase ... 348 9e-96
ARGC_CHRVO (Q7NRT5) N-acetyl-gamma-glutamyl-phosphate reductase ... 311 2e-84
ARGC_PSEAE (Q9I5Q9) N-acetyl-gamma-glutamyl-phosphate reductase ... 303 3e-82
ARGC_NITEU (Q82UK2) N-acetyl-gamma-glutamyl-phosphate reductase ... 303 4e-82
ARC1_PSEPK (Q88QQ6) N-acetyl-gamma-glutamyl-phosphate reductase ... 301 2e-81
ARGC_PSESM (Q889Z3) N-acetyl-gamma-glutamyl-phosphate reductase ... 295 1e-79
ARGC_CLOAB (Q97GH7) N-acetyl-gamma-glutamyl-phosphate reductase ... 288 1e-77
ARGC_BORPA (Q7W3Z3) N-acetyl-gamma-glutamyl-phosphate reductase ... 287 3e-77
ARGC_BORBR (Q7WFC5) N-acetyl-gamma-glutamyl-phosphate reductase ... 287 3e-77
ARGC_BORPE (Q7VUW0) N-acetyl-gamma-glutamyl-phosphate reductase ... 286 4e-77
ARGC_THEMA (Q9X2A2) N-acetyl-gamma-glutamyl-phosphate reductase ... 283 5e-76
ARGC_CAMJE (Q9PIS0) N-acetyl-gamma-glutamyl-phosphate reductase ... 281 1e-75
ARGC_SYNY3 (P54899) N-acetyl-gamma-glutamyl-phosphate reductase ... 278 2e-74
ARGC_NEIMB (Q9JY18) N-acetyl-gamma-glutamyl-phosphate reductase ... 278 2e-74
ARGC_NEIMA (Q9JVU6) N-acetyl-gamma-glutamyl-phosphate reductase ... 278 2e-74
ARGC_THETN (Q8R7B8) N-acetyl-gamma-glutamyl-phosphate reductase ... 277 3e-74
ARGC_RHOBA (Q7UVL4) N-acetyl-gamma-glutamyl-phosphate reductase ... 273 4e-73
ARGC_THENE (Q9Z4S2) N-acetyl-gamma-glutamyl-phosphate reductase ... 272 8e-73
ARGC_BACAN (Q81M95) N-acetyl-gamma-glutamyl-phosphate reductase ... 268 2e-71
ARGC_GLOVI (Q7NE70) N-acetyl-gamma-glutamyl-phosphate reductase ... 267 3e-71
>ARGC_RHOCA (Q9LA02) N-acetyl-gamma-glutamyl-phosphate reductase (EC
1.2.1.38) (AGPR) (N-acetyl-glutamate semialdehyde
dehydrogenase) (NAGSA dehydrogenase)
Length = 342
Score = 348 bits (894), Expect = 9e-96
Identities = 170/342 (49%), Positives = 229/342 (66%), Gaps = 5/342 (1%)
Query: 25 RVGVLGASGYTGSEILRLLANHPQFGVALMTADRKAGQAISSVFPHLGTQDLPDLIAIKD 84
++ +LGASGYTG+E+ R++A HP+ + ++ DRKAG + VFPHL LPDL+ I++
Sbjct: 4 KIAILGASGYTGAELARIIATHPEMEITALSGDRKAGMRMGEVFPHLRHIGLPDLVKIEE 63
Query: 85 ANFSDVDAVFCCLPHGTTQEIIKGLPKHLKIVDLSADFRLRDVSEYEEWYGQPHRAPDLQ 144
+FS +D FC LPH T+Q +I LP+ LK+VDLSADFRLRD +EYE+WYG+PH A DLQ
Sbjct: 64 IDFSGIDLAFCALPHATSQAVIAELPRDLKVVDLSADFRLRDAAEYEKWYGKPHAAMDLQ 123
Query: 145 KEAIYGLTEVLREEIKNARLVANPGCYPTSIQLPLVPLIKANLIQTTNIIVDAKSGVSGA 204
EA+YGLTE REE+K ARL A GC + Q + PLI+A +I I++D K+GVSGA
Sbjct: 124 AEAVYGLTEFYREELKTARLCAGTGCNAAAGQYAIRPLIEAGVIDLDEIVIDLKAGVSGA 183
Query: 205 GRSAKENLLFTEVTEGMSSYGV-TRHRHAPEIEQGLADAAGSKVTISFTPHLIPMSRGMQ 263
GRS KENLL E+ G Y +HRH E +Q + AG V + FTPHL+P +RG+
Sbjct: 184 GRSLKENLLHAELAGGTMPYSAGGKHRHLGEFDQEFSKVAGRPVRVQFTPHLMPFNRGIL 243
Query: 264 STIYVEMAPGVRIDDLRHQLQLSYEDEEFVVLLENGVIPRTHSVKGSNYCLFNVFPDRIP 323
+T+YV P +D+ L YE+E F+ +L G + T SV GSN+ V DR+P
Sbjct: 244 ATVYVRGTP----EDIHAALASRYENEVFLEVLPFGQLASTRSVAGSNFVHLGVIGDRLP 299
Query: 324 GRAIIISVIDNLVKGASGQALQNLNLIMGFPENLGLQYLPLF 365
GRA++ +DNL KG+SGQA+QN NL++G PE LGL P+F
Sbjct: 300 GRAVVTVALDNLTKGSSGQAIQNANLMLGLPETLGLMLAPVF 341
>ARGC_CHRVO (Q7NRT5) N-acetyl-gamma-glutamyl-phosphate reductase (EC
1.2.1.38) (AGPR) (N-acetyl-glutamate semialdehyde
dehydrogenase) (NAGSA dehydrogenase)
Length = 342
Score = 311 bits (797), Expect = 2e-84
Identities = 163/343 (47%), Positives = 222/343 (64%), Gaps = 6/343 (1%)
Query: 24 VRVGVLGASGYTGSEILRLLANHPQFGVALMTADRKAGQAISSVFPHL-GTQDLPDLIAI 82
V+VG++G +GYTG E+LRLLA HPQ V +T+ + AG + +FP L G DL
Sbjct: 2 VKVGIVGGTGYTGVELLRLLAKHPQACVTAVTSRKDAGIRVDEMFPSLRGRIDLA-FTTP 60
Query: 83 KDANFSDVDAVFCCLPHGTT-QEIIKGLPKHLKIVDLSADFRLRDVSEYEEWYGQPHRAP 141
+A ++ D VF P+G Q+ L ++++DL+ADFR++DV E+E+WYG H +P
Sbjct: 61 DEARLAECDVVFFATPNGIAMQDAAALLDAGVRVIDLAADFRIKDVFEWEKWYGMQHASP 120
Query: 142 DLQKEAIYGLTEVLREEIKNARLVANPGCYPTSIQLPLVPLIKANLIQTTNIIVDAKSGV 201
L +A+YGL EV RE I++ARLVANPGCYPT++QL +PLI+A +I T ++I D KSGV
Sbjct: 121 HLVADAVYGLPEVNREAIRSARLVANPGCYPTAVQLGFLPLIEAGVIDTASLIADCKSGV 180
Query: 202 SGAGRSAKENLLFTEVTEGMSSYGVTRHRHAPEIEQGLADAAGSKVTISFTPHLIPMSRG 261
SGAGR + L E + +YGV HRH PEI QGLA A+G V ++F PHL PM RG
Sbjct: 181 SGAGRKGEIGALLAEAGDSFKAYGVAGHRHLPEIRQGLARASGKPVGLTFVPHLTPMIRG 240
Query: 262 MQSTIYVEMAPGVRIDDLRHQLQLSYEDEEFVVLLENGVIPRTHSVKGSNYCLFNVFPDR 321
+ +T+Y + V DL+ + Y DE FV +L G P T SV+G+N C V +
Sbjct: 241 IHATLYARLTKDV---DLQRLFESRYADESFVDVLPAGSHPETRSVRGANLCRIAVHRPQ 297
Query: 322 IPGRAIIISVIDNLVKGASGQALQNLNLIMGFPENLGLQYLPL 364
+++SVIDNLVKGA+GQA+QNLNL+ G PEN GL +PL
Sbjct: 298 GGDTVVVLSVIDNLVKGAAGQAVQNLNLMFGLPENEGLDVVPL 340
>ARGC_PSEAE (Q9I5Q9) N-acetyl-gamma-glutamyl-phosphate reductase (EC
1.2.1.38) (AGPR) (N-acetyl-glutamate semialdehyde
dehydrogenase) (NAGSA dehydrogenase)
Length = 344
Score = 303 bits (777), Expect = 3e-82
Identities = 161/343 (46%), Positives = 216/343 (62%), Gaps = 4/343 (1%)
Query: 24 VRVGVLGASGYTGSEILRLLANHPQFGVALMTADRKAGQAISSVFPHLGTQDLPDLIAIK 83
++VG++G +GYTG E+LRLLA HPQ V ++T+ +AG ++ ++P+L ++
Sbjct: 2 IKVGIVGGTGYTGVELLRLLAQHPQARVEVITSRSEAGVKVADMYPNLRGHYDDLQFSVP 61
Query: 84 DAN-FSDVDAVFCCLPHGTTQEII-KGLPKHLKIVDLSADFRLRDVSEYEEWYGQPHRAP 141
DA D VF PHG + + L +++DLSADFRL D E+ WYGQPH AP
Sbjct: 62 DAQRLGACDVVFFATPHGVAHALAGELLDAGTRVIDLSADFRLADAEEWARWYGQPHGAP 121
Query: 142 DLQKEAIYGLTEVLREEIKNARLVANPGCYPTSIQLPLVPLIKANLIQTTNIIVDAKSGV 201
L EA+YGL EV RE+I+ ARL+A PGCYPT+ QL L+PL++A L + +I D KSGV
Sbjct: 122 ALLDEAVYGLPEVNREKIRQARLIAVPGCYPTATQLGLIPLLEAGLADASRLIADCKSGV 181
Query: 202 SGAGRSAKENLLFTEVTEGMSSYGVTRHRHAPEIEQGLADAAGSKVTISFTPHLIPMSRG 261
SGAGR AK LF E E M +Y V HRH PEI QGL A+G V ++F PHL PM RG
Sbjct: 182 SGAGRGAKVGSLFCEAGESMMAYAVKGHRHLPEISQGLRRASGGDVGLTFVPHLTPMIRG 241
Query: 262 MQSTIYVEMAPGVRIDDLRHQLQLSYEDEEFVVLLENGVIPRTHSVKGSNYCLFNVFPDR 321
+ +T+Y +A R DL+ + Y DE FV ++ G P T SV+G+N C V +
Sbjct: 242 IHATLYAHVAD--RSVDLQALFEKRYADEPFVDVMPAGSHPETRSVRGANVCRIAVHRPQ 299
Query: 322 IPGRAIIISVIDNLVKGASGQALQNLNLIMGFPENLGLQYLPL 364
+++SVIDNLVKGASGQALQN+N++ G E LGL + L
Sbjct: 300 GGDLVVVLSVIDNLVKGASGQALQNMNILFGLDERLGLSHAAL 342
>ARGC_NITEU (Q82UK2) N-acetyl-gamma-glutamyl-phosphate reductase (EC
1.2.1.38) (AGPR) (N-acetyl-glutamate semialdehyde
dehydrogenase) (NAGSA dehydrogenase)
Length = 342
Score = 303 bits (776), Expect = 4e-82
Identities = 149/342 (43%), Positives = 221/342 (64%), Gaps = 4/342 (1%)
Query: 24 VRVGVLGASGYTGSEILRLLANHPQFGVALMTADRKAGQAISSVFPHLGTQDLPDLIAIK 83
+ G++G +GYTG E+LR+L HP+ + ++T+ ++AG + +F L Q
Sbjct: 2 INAGIVGGTGYTGVELLRILVQHPKVKLKVITSRQEAGTGVDELFSSLRGQIALKFSDPA 61
Query: 84 DANFSDVDAVFCCLPHGTTQEIIKGL-PKHLKIVDLSADFRLRDVSEYEEWYGQPHRAPD 142
+FS D VF P+G + K L +K++DL+ADFR++DV+E+E+WYG H AP+
Sbjct: 62 KVDFSKCDVVFFATPNGIAMQQAKALLDSGIKVIDLAADFRIKDVAEWEKWYGMTHAAPE 121
Query: 143 LQKEAIYGLTEVLREEIKNARLVANPGCYPTSIQLPLVPLIKANLIQTTNIIVDAKSGVS 202
L EA+YGL EV RE+I++ARL+ANPGCYPT++QL +PLI+A + ++I D KSGVS
Sbjct: 122 LVAEAVYGLPEVNREKIRDARLIANPGCYPTAVQLGFIPLIEAGAVDADHLIADTKSGVS 181
Query: 203 GAGRSAKENLLFTEVTEGMSSYGVTRHRHAPEIEQGLADAAGSKVTISFTPHLIPMSRGM 262
GAGR A+ + L+ E ++ SY V HRH PEI QGL++ + + ++F PHL PM RG+
Sbjct: 182 GAGRKAEIHTLYAEASDNFKSYAVPGHRHLPEIRQGLSERSNGPIGLTFVPHLTPMIRGI 241
Query: 263 QSTIYVEMAPGVRIDDLRHQLQLSYEDEEFVVLLENGVIPRTHSVKGSNYCLFNVFPDRI 322
+T+Y + V DL+ + Y +E FV +L G P T SV+GSN+C V
Sbjct: 242 HATLYARLTRDV---DLQTLYENRYANEPFVDVLPAGSHPETRSVRGSNFCRIAVHRPGN 298
Query: 323 PGRAIIISVIDNLVKGASGQALQNLNLIMGFPENLGLQYLPL 364
A+I+SV DNLVKGA+GQA+QN+N++ G PE G++++PL
Sbjct: 299 GDTAVILSVTDNLVKGAAGQAVQNMNIMYGLPETTGIRHVPL 340
>ARC1_PSEPK (Q88QQ6) N-acetyl-gamma-glutamyl-phosphate reductase 1
(EC 1.2.1.38) (AGPR 1) (N-acetyl-glutamate semialdehyde
dehydrogenase 1) (NAGSA dehydrogenase 1)
Length = 344
Score = 301 bits (771), Expect = 2e-81
Identities = 158/343 (46%), Positives = 219/343 (63%), Gaps = 4/343 (1%)
Query: 24 VRVGVLGASGYTGSEILRLLANHPQFGVALMTADRKAGQAISSVFPHLGTQDLPDLIAIK 83
++VG++G +GYTG E+LRLLA HPQ VA++T+ +AG A++ ++P+L ++
Sbjct: 2 IKVGIVGGTGYTGVELLRLLAQHPQAEVAVITSRSEAGVAVADMYPNLRGHYDGLAFSVP 61
Query: 84 DAN-FSDVDAVFCCLPHGTTQEII-KGLPKHLKIVDLSADFRLRDVSEYEEWYGQPHRAP 141
D+ D VF PHG + + L K++DLSADFRL+D +E+ +WYGQPH AP
Sbjct: 62 DSKALGACDVVFFATPHGVAHALAGELLAAGTKVIDLSADFRLQDATEWGKWYGQPHGAP 121
Query: 142 DLQKEAIYGLTEVLREEIKNARLVANPGCYPTSIQLPLVPLIKANLIQTTNIIVDAKSGV 201
+L K+A+YGL EV RE+I+ ARL+A PGCYPT+ QL +PL++A L + +I D KSGV
Sbjct: 122 ELLKDAVYGLPEVNREKIRQARLIAVPGCYPTATQLGFLPLLEAGLADPSRLIADCKSGV 181
Query: 202 SGAGRSAKENLLFTEVTEGMSSYGVTRHRHAPEIEQGLADAAGSKVTISFTPHLIPMSRG 261
SGAGR A LF E E M +Y V HRH PEI QGL AAG + ++F PHL PM RG
Sbjct: 182 SGAGRGAAVGSLFCEAGESMKAYAVKGHRHLPEISQGLRLAAGKDIGLTFVPHLTPMIRG 241
Query: 262 MQSTIYVEMAPGVRIDDLRHQLQLSYEDEEFVVLLENGVIPRTHSVKGSNYCLFNVFPDR 321
+ +T+Y + DL+ + Y DE FV ++ G P T SV+G+N C V +
Sbjct: 242 IHATLYANVVD--TSVDLQALFEKRYADEPFVDVMPAGSHPETRSVRGANVCRIAVHRPQ 299
Query: 322 IPGRAIIISVIDNLVKGASGQALQNLNLIMGFPENLGLQYLPL 364
+++SVIDNLVKGASGQA+QNLN++ G E +GL + L
Sbjct: 300 GGDLVVVLSVIDNLVKGASGQAVQNLNILFGLDERMGLSHAGL 342
>ARGC_PSESM (Q889Z3) N-acetyl-gamma-glutamyl-phosphate reductase (EC
1.2.1.38) (AGPR) (N-acetyl-glutamate semialdehyde
dehydrogenase) (NAGSA dehydrogenase)
Length = 344
Score = 295 bits (755), Expect = 1e-79
Identities = 153/340 (45%), Positives = 214/340 (62%), Gaps = 4/340 (1%)
Query: 24 VRVGVLGASGYTGSEILRLLANHPQFGVALMTADRKAGQAISSVFPHLGTQDLPDLIAIK 83
V+VG++G +GYTG E+LRLLA HPQ V ++T+ +AG ++ ++P+L ++
Sbjct: 2 VKVGIVGGTGYTGVELLRLLAQHPQAEVVVITSRSEAGLPVADMYPNLRGHYDGLAFSVP 61
Query: 84 DAN-FSDVDAVFCCLPHGTTQEII-KGLPKHLKIVDLSADFRLRDVSEYEEWYGQPHRAP 141
D D VF PHG + + L K++DLSADFRL+D E+ +WYGQPH AP
Sbjct: 62 DVKTLGACDVVFFATPHGVAHALAGELLAAGTKVIDLSADFRLQDPVEWAKWYGQPHGAP 121
Query: 142 DLQKEAIYGLTEVLREEIKNARLVANPGCYPTSIQLPLVPLIKANLIQTTNIIVDAKSGV 201
+L ++A+YGL EV RE+I+NARL+A PGCYPT+ QL +PL++A + T +I D KSGV
Sbjct: 122 ELLQDAVYGLPEVNREQIRNARLIAVPGCYPTATQLGFLPLLEAGIADNTRLIADCKSGV 181
Query: 202 SGAGRSAKENLLFTEVTEGMSSYGVTRHRHAPEIEQGLADAAGSKVTISFTPHLIPMSRG 261
SGAGR L++E E +Y V HRH PEI QGL AA + ++F PHL+PM RG
Sbjct: 182 SGAGRGLNIGSLYSEANESFKAYAVKGHRHLPEITQGLRRAARGDIGLTFVPHLVPMIRG 241
Query: 262 MQSTIYVEMAPGVRIDDLRHQLQLSYEDEEFVVLLENGVIPRTHSVKGSNYCLFNVFPDR 321
+ ST+Y + R DL+ + Y DE FV ++ G P T SV+G+N C V +
Sbjct: 242 IHSTLYATVTD--RSVDLQALFEKRYADEPFVDVMPAGSHPETRSVRGANVCRIAVHRPQ 299
Query: 322 IPGRAIIISVIDNLVKGASGQALQNLNLIMGFPENLGLQY 361
+++SVIDNLVKGASGQA+QN+N++ G E GL +
Sbjct: 300 DGDLVVVLSVIDNLVKGASGQAVQNMNILFGLDERAGLSH 339
>ARGC_CLOAB (Q97GH7) N-acetyl-gamma-glutamyl-phosphate reductase (EC
1.2.1.38) (AGPR) (N-acetyl-glutamate semialdehyde
dehydrogenase) (NAGSA dehydrogenase)
Length = 345
Score = 288 bits (737), Expect = 1e-77
Identities = 140/347 (40%), Positives = 219/347 (62%), Gaps = 9/347 (2%)
Query: 24 VRVGVLGASGYTGSEILRLLANHPQFGVALMTADRKAGQAISSVFPHLGTQDLPDLIAIK 83
++ G++GA+GY G + LL NHP + + + + +S + + ++ + + I
Sbjct: 2 IQAGIIGATGYAGEVLTWLLNNHPNVNLVFLASHSHGNSSFNSTYGNF--TNVIENLCID 59
Query: 84 DA----NFSDVDAVFCCLPHGTTQEIIK-GLPKHLKIVDLSADFRLRDVSEYEEWYGQPH 138
D N ++D +F LP G T I+K + + +K++DL AD+RL+ Y+EWY H
Sbjct: 60 DNEVEDNLDNIDILFTALPSGKTFSIVKKAVEEKVKVIDLGADYRLKSAEVYKEWYNINH 119
Query: 139 RAPDLQKEAIYGLTEVLREEIKNARLVANPGCYPTSIQLPLVPLIKANLIQTTNIIVDAK 198
L K+A+YGL+E+ RE+IK + L+ANPGCYPT+ L L+PL+K NLI T++II+DAK
Sbjct: 120 ECKKLLKDAVYGLSEIYREKIKTSNLIANPGCYPTASSLALIPLLKNNLIDTSSIIIDAK 179
Query: 199 SGVSGAGRSAKENLLFTEVTEGMSSYGVTRHRHAPEIEQGLADAAGSKVTISFTPHLIPM 258
SGVSGAGR A N LF E + + +YGV HRH PEIEQ L + A K+T++FTPHL+PM
Sbjct: 180 SGVSGAGRKASTNNLFIECNDSIKAYGVASHRHTPEIEQTLTNEAKEKITLTFTPHLVPM 239
Query: 259 SRGMQSTIYVEMAPGVRIDDLRHQLQLSYEDEEFVVLLENGVIPRTHSVKGSNYCLFNVF 318
+RG+ S Y + + ++++ + Y++E F+ +++ +P T VKGSNYC +
Sbjct: 240 NRGILSVCYSNLKKEIELNEVYDIYKKFYKNEPFIRIID--TLPETRWVKGSNYCDIAIR 297
Query: 319 PDRIPGRAIIISVIDNLVKGASGQALQNLNLIMGFPENLGLQYLPLF 365
D+ + I++S IDNL+KGA+ QA+QN+N++ G E G+ P+F
Sbjct: 298 IDKRTNKIIVLSAIDNLMKGAASQAVQNMNIMYGLAEKTGIDLTPMF 344
>ARGC_BORPA (Q7W3Z3) N-acetyl-gamma-glutamyl-phosphate reductase (EC
1.2.1.38) (AGPR) (N-acetyl-glutamate semialdehyde
dehydrogenase) (NAGSA dehydrogenase)
Length = 354
Score = 287 bits (735), Expect = 3e-77
Identities = 145/350 (41%), Positives = 216/350 (61%), Gaps = 3/350 (0%)
Query: 18 KSSQNNVRVGVLGASGYTGSEILRLLANHPQFGVALMTADRKAGQAISSVFPHLGTQDLP 77
++S + ++VG++G +GYTG E+LRLL+ HP + +T+ ++ G ++ ++P+L +
Sbjct: 3 QASNSRIKVGIVGGTGYTGVELLRLLSQHPHVALTAITSRKEDGLPVADMYPNLRGRVKL 62
Query: 78 DLIAIKDANFSDVDAVFCCLPHGTTQ-EIIKGLPKHLKIVDLSADFRLRDVSEYEEWYGQ 136
A + A+ +D D VF PHG + + L +++DL+ADFRL+D + +E WY
Sbjct: 63 AFSAPEKASLTDCDVVFFATPHGVAMAQAAELLAAGTRVIDLAADFRLQDTAVFERWYKI 122
Query: 137 PHRAPDLQKEAIYGLTEVLREEIKNARLVANPGCYPTSIQLPLVPLIKAN--LIQTTNII 194
PH PD+ +++YGL E+ RE I AR++ NPGCYPT++ L L PLI+ L+ +I
Sbjct: 123 PHTCPDILADSVYGLVELNREAISKARVIGNPGCYPTTVLLGLAPLIEGGKQLVDVQTLI 182
Query: 195 VDAKSGVSGAGRSAKENLLFTEVTEGMSSYGVTRHRHAPEIEQGLADAAGSKVTISFTPH 254
D KSGVSGAGR A+ LF+E ++ +YGV HRH PEI L AG KV ++F PH
Sbjct: 183 ADCKSGVSGAGRKAEVGSLFSEASDNFKAYGVAGHRHQPEIVAQLEKLAGGKVGLTFVPH 242
Query: 255 LIPMSRGMQSTIYVEMAPGVRIDDLRHQLQLSYEDEEFVVLLENGVIPRTHSVKGSNYCL 314
L+PM RGM ST+Y + P R D + + Y DE FV ++ G +P T SV+ SN
Sbjct: 243 LVPMIRGMFSTLYARILPQARDTDFQALFEARYADEPFVDVMPAGSLPETRSVRASNNLR 302
Query: 315 FNVFPDRIPGRAIIISVIDNLVKGASGQALQNLNLIMGFPENLGLQYLPL 364
+V + +I+ V DNLVKGASGQA+QN+NL+ G PE+ GL + +
Sbjct: 303 ISVQRPGGGDQLVILVVQDNLVKGASGQAVQNMNLMFGLPESAGLDQVAI 352
>ARGC_BORBR (Q7WFC5) N-acetyl-gamma-glutamyl-phosphate reductase (EC
1.2.1.38) (AGPR) (N-acetyl-glutamate semialdehyde
dehydrogenase) (NAGSA dehydrogenase)
Length = 354
Score = 287 bits (735), Expect = 3e-77
Identities = 145/350 (41%), Positives = 216/350 (61%), Gaps = 3/350 (0%)
Query: 18 KSSQNNVRVGVLGASGYTGSEILRLLANHPQFGVALMTADRKAGQAISSVFPHLGTQDLP 77
++S + ++VG++G +GYTG E+LRLL+ HP + +T+ ++ G ++ ++P+L +
Sbjct: 3 QASNSRIKVGIVGGTGYTGVELLRLLSQHPHVALTAITSRKEDGLPVADMYPNLRGRVKL 62
Query: 78 DLIAIKDANFSDVDAVFCCLPHGTTQ-EIIKGLPKHLKIVDLSADFRLRDVSEYEEWYGQ 136
A + A+ +D D VF PHG + + L +++DL+ADFRL+D + +E WY
Sbjct: 63 AFSAPEKASLTDCDVVFFATPHGVAMAQAAELLAAGTRVIDLAADFRLQDTAVFERWYKI 122
Query: 137 PHRAPDLQKEAIYGLTEVLREEIKNARLVANPGCYPTSIQLPLVPLIKAN--LIQTTNII 194
PH PD+ +++YGL E+ RE I AR++ NPGCYPT++ L L PLI+ L+ +I
Sbjct: 123 PHTCPDILADSVYGLVELNREAISKARVIGNPGCYPTTVLLGLAPLIEGGKQLVDVQTLI 182
Query: 195 VDAKSGVSGAGRSAKENLLFTEVTEGMSSYGVTRHRHAPEIEQGLADAAGSKVTISFTPH 254
D KSGVSGAGR A+ LF+E ++ +YGV HRH PEI L AG KV ++F PH
Sbjct: 183 ADCKSGVSGAGRKAEVGSLFSEASDNFKAYGVAGHRHQPEIVAQLEKLAGGKVGLTFVPH 242
Query: 255 LIPMSRGMQSTIYVEMAPGVRIDDLRHQLQLSYEDEEFVVLLENGVIPRTHSVKGSNYCL 314
L+PM RGM ST+Y + P R D + + Y DE FV ++ G +P T SV+ SN
Sbjct: 243 LVPMIRGMFSTLYARILPQARDTDFQALFEARYADEPFVDVMPAGSLPETRSVRASNNLR 302
Query: 315 FNVFPDRIPGRAIIISVIDNLVKGASGQALQNLNLIMGFPENLGLQYLPL 364
+V + +I+ V DNLVKGASGQA+QN+NL+ G PE+ GL + +
Sbjct: 303 ISVQRPGGGDQLVILVVQDNLVKGASGQAVQNMNLMFGLPESAGLDQVAI 352
>ARGC_BORPE (Q7VUW0) N-acetyl-gamma-glutamyl-phosphate reductase (EC
1.2.1.38) (AGPR) (N-acetyl-glutamate semialdehyde
dehydrogenase) (NAGSA dehydrogenase)
Length = 354
Score = 286 bits (733), Expect = 4e-77
Identities = 145/350 (41%), Positives = 216/350 (61%), Gaps = 3/350 (0%)
Query: 18 KSSQNNVRVGVLGASGYTGSEILRLLANHPQFGVALMTADRKAGQAISSVFPHLGTQDLP 77
++S + ++VG++G +GYTG E+LRLL+ HP + +T+ ++ G ++ ++P+L +
Sbjct: 3 QASNSRIKVGIVGGTGYTGVELLRLLSQHPHVELTAITSRKEDGLPVADMYPNLRGRVKL 62
Query: 78 DLIAIKDANFSDVDAVFCCLPHGTTQ-EIIKGLPKHLKIVDLSADFRLRDVSEYEEWYGQ 136
A + A+ +D D VF PHG + + L +++DL+ADFRL+D + +E WY
Sbjct: 63 AFSAPEKASLTDCDVVFFATPHGVAMAQAAELLAAGTRVIDLAADFRLQDTAVFERWYKI 122
Query: 137 PHRAPDLQKEAIYGLTEVLREEIKNARLVANPGCYPTSIQLPLVPLIKAN--LIQTTNII 194
PH PD+ +++YGL E+ RE I AR++ NPGCYPT++ L L PLI+ L+ +I
Sbjct: 123 PHTCPDILADSVYGLVELNREAISKARVIGNPGCYPTTVLLGLAPLIEGGKQLVDVQTLI 182
Query: 195 VDAKSGVSGAGRSAKENLLFTEVTEGMSSYGVTRHRHAPEIEQGLADAAGSKVTISFTPH 254
D KSGVSGAGR A+ LF+E ++ +YGV HRH PEI L AG KV ++F PH
Sbjct: 183 ADCKSGVSGAGRKAEVGSLFSEASDNFKAYGVAGHRHQPEIVAQLEKLAGGKVGLTFVPH 242
Query: 255 LIPMSRGMQSTIYVEMAPGVRIDDLRHQLQLSYEDEEFVVLLENGVIPRTHSVKGSNYCL 314
L+PM RGM ST+Y + P R D + + Y DE FV ++ G +P T SV+ SN
Sbjct: 243 LVPMIRGMFSTLYARILPQARDTDFQALFEARYADEPFVDVMPAGSLPETRSVRASNNLR 302
Query: 315 FNVFPDRIPGRAIIISVIDNLVKGASGQALQNLNLIMGFPENLGLQYLPL 364
+V + +I+ V DNLVKGASGQA+QN+NL+ G PE+ GL + +
Sbjct: 303 ISVQRPGGGDQLVILVVQDNLVKGASGQAVQNMNLMFGLPESAGLDQVAI 352
>ARGC_THEMA (Q9X2A2) N-acetyl-gamma-glutamyl-phosphate reductase (EC
1.2.1.38) (AGPR) (N-acetyl-glutamate semialdehyde
dehydrogenase) (NAGSA dehydrogenase)
Length = 339
Score = 283 bits (724), Expect = 5e-76
Identities = 150/343 (43%), Positives = 216/343 (62%), Gaps = 7/343 (2%)
Query: 24 VRVGVLGASGYTGSEILRLLANHPQFGVALMTADRKAGQAISSVFPH-LGTQDLPDLIAI 82
+R G++GA+GYTG E++RLL NHP+ + +++ AG+ + +FP L L +
Sbjct: 2 IRAGIIGATGYTGLELVRLLKNHPEAKITYLSSRTYAGKKLEEIFPSTLENSILSEFDPE 61
Query: 83 KDANFSDVDAVFCCLPHGTTQEIIKGLPKHLKIVDLSADFRLRDVSEYEEWYGQPHRAPD 142
K + DV +F LP G + ++++ L K +KI+DL ADFR D Y EWYG+ +
Sbjct: 62 KVSKNCDV--LFTALPAGASYDLVREL-KGVKIIDLGADFRFDDPGVYREWYGKELSGYE 118
Query: 143 LQKEAIYGLTEVLREEIKNARLVANPGCYPTSIQLPLVPLIKANLIQTTNIIVDAKSGVS 202
K +YGL E+ REEIKNA++V NPGCYPTS+ L L P +K NL+ I+VDAKSGVS
Sbjct: 119 NIKR-VYGLPELHREEIKNAQVVGNPGCYPTSVILALAPALKHNLVDPETILVDAKSGVS 177
Query: 203 GAGRSAKENLLFTEVTEGMSSYGVTRHRHAPEIEQGLADAAGSKVTISFTPHLIPMSRGM 262
GAGR K + LF+EV E + Y V +HRH PE+EQ L +G KV + FTPHL+PM+RG+
Sbjct: 178 GAGRKEKVDYLFSEVNESLRPYNVAKHRHVPEMEQELGKISGKKVNVVFTPHLVPMTRGI 237
Query: 263 QSTIYVEMAPGVRIDDLRHQLQLSYEDEEFVVLLENGVIPRTHSVKGSNYCLFNVFPDRI 322
STIYV+ ++++ Y++E FV +L G+ P T GSN+ + +
Sbjct: 238 LSTIYVKTDKS--LEEIHEAYLEFYKNEPFVHVLPMGIYPSTKWCYGSNHVFIGMQMEER 295
Query: 323 PGRAIIISVIDNLVKGASGQALQNLNLIMGFPENLGLQYLPLF 365
I++S IDNLVKGASGQA+QN+N++ G E GL++ P++
Sbjct: 296 TNTLILMSAIDNLVKGASGQAVQNMNIMFGLDETKGLEFTPIY 338
>ARGC_CAMJE (Q9PIS0) N-acetyl-gamma-glutamyl-phosphate reductase (EC
1.2.1.38) (AGPR) (N-acetyl-glutamate semialdehyde
dehydrogenase) (NAGSA dehydrogenase)
Length = 342
Score = 281 bits (720), Expect = 1e-75
Identities = 144/341 (42%), Positives = 217/341 (63%), Gaps = 8/341 (2%)
Query: 24 VRVGVLGASGYTGSEILRLLANHPQFGVALMTADRKAGQAISSVFPHLGTQDLPDLIAIK 83
++VG+LGASGY G+E++R+L NHP+ ++ + + GQ ++P+ P + +
Sbjct: 3 IKVGILGASGYAGNELVRILLNHPKVEISYLGSSSSVGQNYQDLYPNT-----PLNLCFE 57
Query: 84 DANFSDV--DAVFCCLPHGTTQEII-KGLPKHLKIVDLSADFRLRDVSEYEEWYGQPHRA 140
+ N ++ D +F PH + +++ + L K +KI+DLSADFRL++ +YE WY H
Sbjct: 58 NKNLDELELDLLFLATPHKFSAKLLNENLLKKMKIIDLSADFRLKNPKDYELWYKFTHPN 117
Query: 141 PDLQKEAIYGLTEVLREEIKNARLVANPGCYPTSIQLPLVPLIKANLIQTTNIIVDAKSG 200
+L + A+YGL E+ +EEIK A LVANPGCY T L L PL K +I +++I+DAKSG
Sbjct: 118 QELLQNAVYGLCELYKEEIKKASLVANPGCYTTCSILSLYPLFKEKIIDFSSVIIDAKSG 177
Query: 201 VSGAGRSAKENLLFTEVTEGMSSYGVTRHRHAPEIEQGLADAAGSKVTISFTPHLIPMSR 260
VSGAGRSAK LF EV E + +Y + HRH PEIE+ L+ AA K+T+ FTPHL+PM R
Sbjct: 178 VSGAGRSAKVENLFCEVNENIKAYNLALHRHTPEIEEHLSYAAKEKITLQFTPHLVPMQR 237
Query: 261 GMQSTIYVEMAPGVRIDDLRHQLQLSYEDEEFVVLLENGVIPRTHSVKGSNYCLFNVFPD 320
G+ + Y + ++ D+R Y++ +F+ LL +P+T VK SN+ N D
Sbjct: 238 GILISAYANLKEDLQEQDIRDIYTKYYQNNKFIRLLPPQSLPQTRWVKSSNFADINFSVD 297
Query: 321 RIPGRAIIISVIDNLVKGASGQALQNLNLIMGFPENLGLQY 361
+ R I++ IDNL+KGA+GQA+QN+NL+ F E+ GL++
Sbjct: 298 QRTKRVIVLGAIDNLIKGAAGQAVQNMNLMFDFDEDEGLKF 338
>ARGC_SYNY3 (P54899) N-acetyl-gamma-glutamyl-phosphate reductase (EC
1.2.1.38) (AGPR) (N-acetyl-glutamate semialdehyde
dehydrogenase) (NAGSA dehydrogenase)
Length = 351
Score = 278 bits (711), Expect = 2e-74
Identities = 152/348 (43%), Positives = 212/348 (60%), Gaps = 11/348 (3%)
Query: 25 RVGVLGASGYTGSEILRLLANHPQFGVALMTADRKAGQAISSVFPHLG-----TQDLPDL 79
RVG++GASGY G +++RLL HPQ + + AG+ S ++PHL T + DL
Sbjct: 7 RVGIIGASGYGGIQLVRLLLEHPQVELTYLAGHSSAGKPYSDLYPHLTHRVNLTIEPIDL 66
Query: 80 IAIKDANFSDVDAVFCCLPHGTTQEIIKGL-PKHLKIVDLSADFRLRDVSEYEEWYGQPH 138
AI S DAVF LP+G ++ L K K++DLSAD+R R+++ Y EWY +
Sbjct: 67 EAIA----SRCDAVFLGLPNGLACDMAPALLAKGCKVLDLSADYRFRNLNTYTEWYKKDR 122
Query: 139 RAPDLQKEAIYGLTEVLREEIKNARLVANPGCYPTSIQLPLVPLIKANLIQTTNIIVDAK 198
+ +A+YGL E+ R+ I++A+L+ PGCYPT+ L L PL+K I I+DAK
Sbjct: 123 QDQATNNQAVYGLPELYRDAIRDAQLIGCPGCYPTASLLALSPLLKQGFIVPETAIIDAK 182
Query: 199 SGVSGAGRSAKENLLFTEVTEGMSSYGVTRHRHAPEIEQGLADAAGSKVTISFTPHLIPM 258
SG SG GR AK N+L E + +YGV +HRH PEIEQ +D AG+++ + FTPHLIPM
Sbjct: 183 SGTSGGGREAKVNMLLAEAEGSLGAYGVAKHRHTPEIEQIASDLAGTELRVQFTPHLIPM 242
Query: 259 SRGMQSTIYVEMA-PGVRIDDLRHQLQLSYEDEEFVVLLENGVIPRTHSVKGSNYCLFNV 317
RG+ +T+Y + PG+ DDL Y FV +L +GV P+T G+N C +
Sbjct: 243 VRGILATVYATLRDPGLVRDDLITIYSAFYRASPFVKILPHGVYPQTKWAWGTNLCYIGI 302
Query: 318 FPDRIPGRAIIISVIDNLVKGASGQALQNLNLIMGFPENLGLQYLPLF 365
D GR I++S IDNLVKG +GQA+Q LNL+MG+ E+LGL L +
Sbjct: 303 EVDPRTGRVIVLSAIDNLVKGQAGQAVQCLNLMMGWEESLGLPQLAFY 350
>ARGC_NEIMB (Q9JY18) N-acetyl-gamma-glutamyl-phosphate reductase (EC
1.2.1.38) (AGPR) (N-acetyl-glutamate semialdehyde
dehydrogenase) (NAGSA dehydrogenase)
Length = 347
Score = 278 bits (711), Expect = 2e-74
Identities = 153/349 (43%), Positives = 211/349 (59%), Gaps = 16/349 (4%)
Query: 24 VRVGVLGASGYTGSEILRLLANHPQFGVALMTADRKAGQAISSVFPHL-GTQDL----PD 78
++VG++GA+GYTG E+LRLLA HP VA +T+ +AG A++ FP L G L PD
Sbjct: 5 IKVGIVGATGYTGVELLRLLAAHPDVEVAAVTSRSEAGTAVADYFPSLRGVYGLAFQTPD 64
Query: 79 LIAIKDANFSDVDAVFCCLPHGTT-QEIIKGLPKHLKIVDLSADFRLRDVSEYEEWYGQP 137
+A D VF P+G ++ + + + ++++DLSADFR+RD+ +E WYG
Sbjct: 65 -----EAGLEQCDIVFFATPNGIAMKDAPRLIEQGVRVIDLSADFRIRDIPTWEHWYGMT 119
Query: 138 HRAPDLQKEAIYGLTEVLREEIKNARLVANPGCYPTSIQLPLVPLIK-ANLIQTTNIIVD 196
H APDL +A+YGL+E+ RE + ARLVANPGCYPT + LPLVPL++ L +I D
Sbjct: 120 HAAPDLVSQAVYGLSELNREAVAQARLVANPGCYPTCVSLPLVPLLRQCRLKPGMPLIAD 179
Query: 197 AKSGVSGAGRSAKENLLFTEVTEGMSSYGVTRHRHAPEIEQGLADAA-GSKVTISFTPHL 255
KSGVSGAGR L E + +YG+ HRH PEI Q +A G FTPHL
Sbjct: 180 CKSGVSGAGRKGNVGSLLCEAGDNFKAYGIAGHRHLPEIRQTIAGLQDGIAEGFVFTPHL 239
Query: 256 IPMSRGMQSTIYVEMAPGVRIDDLRHQLQLSYEDEEFVVLLENGVIPRTHSVKGSNYCLF 315
PM RGM +T+Y+ ++ G D L+ Y D FV +L G P T SV+G+N C
Sbjct: 240 APMIRGMHATVYLHLSDG---SDPETVLRDYYRDSPFVDILPTGSAPETRSVRGANLCRI 296
Query: 316 NVFPDRIPGRAIIISVIDNLVKGASGQALQNLNLIMGFPENLGLQYLPL 364
++ +++SVIDNLVKGA+GQA+QN+N++ G E GL +PL
Sbjct: 297 SIQQAAQSDVWVVLSVIDNLVKGAAGQAVQNMNIMFGLEETHGLDAIPL 345
>ARGC_NEIMA (Q9JVU6) N-acetyl-gamma-glutamyl-phosphate reductase (EC
1.2.1.38) (AGPR) (N-acetyl-glutamate semialdehyde
dehydrogenase) (NAGSA dehydrogenase)
Length = 347
Score = 278 bits (710), Expect = 2e-74
Identities = 153/349 (43%), Positives = 214/349 (60%), Gaps = 16/349 (4%)
Query: 24 VRVGVLGASGYTGSEILRLLANHPQFGVALMTADRKAGQAISSVFPHL-GTQDL----PD 78
++VG++GA+GYTG E+LRLLA HP VA +T+ +AG A++ FP L G L PD
Sbjct: 5 IKVGIVGATGYTGVELLRLLAAHPDVEVAAVTSRSEAGTAVADYFPSLRGVYGLAFQTPD 64
Query: 79 LIAIKDANFSDVDAVFCCLPHGTT-QEIIKGLPKHLKIVDLSADFRLRDVSEYEEWYGQP 137
+A D VF P+G ++ + + + ++++DLSADFR+RD+ +E WYG
Sbjct: 65 -----EAGLEQCDIVFFATPNGIAMKDAPRLIEQGVRVIDLSADFRIRDIPTWEHWYGMT 119
Query: 138 HRAPDLQKEAIYGLTEVLREEIKNARLVANPGCYPTSIQLPLVPLIK-ANLIQTTNIIVD 196
H APDL +A+YGL+E+ RE + ARLVANPGCYPT + LPLVPL++ L +I D
Sbjct: 120 HAAPDLVSQAVYGLSELNREAVAQARLVANPGCYPTCVSLPLVPLLRQCRLKPGMPLIAD 179
Query: 197 AKSGVSGAGRSAKENLLFTEVTEGMSSYGVTRHRHAPEIEQGLADAA-GSKVTISFTPHL 255
KSGVSGAGR L EV + +YG+ HRH PEI Q +A G FTPHL
Sbjct: 180 CKSGVSGAGRKGNVGSLLCEVGDNFKAYGIAGHRHLPEIRQTIAGLQDGIAEGFVFTPHL 239
Query: 256 IPMSRGMQSTIYVEMAPGVRIDDLRHQLQLSYEDEEFVVLLENGVIPRTHSVKGSNYCLF 315
PM RGM +T+Y+ ++ G+ + + L+ Y D FV +L G P T SV+G+N C
Sbjct: 240 APMIRGMHATVYLHLSDGICPETI---LRDYYRDSLFVDILPAGSTPETRSVRGANLCRI 296
Query: 316 NVFPDRIPGRAIIISVIDNLVKGASGQALQNLNLIMGFPENLGLQYLPL 364
++ +++SVIDNLVKGA+GQA+QN+N++ G E GL +PL
Sbjct: 297 SIQQAAQSDVWVVLSVIDNLVKGAAGQAVQNMNIMFGLKETHGLGAIPL 345
>ARGC_THETN (Q8R7B8) N-acetyl-gamma-glutamyl-phosphate reductase (EC
1.2.1.38) (AGPR) (N-acetyl-glutamate semialdehyde
dehydrogenase) (NAGSA dehydrogenase)
Length = 344
Score = 277 bits (708), Expect = 3e-74
Identities = 145/345 (42%), Positives = 219/345 (63%), Gaps = 6/345 (1%)
Query: 24 VRVGVLGASGYTGSEILRLLANHPQFGVALMTADRKAGQAISSVFPHL-GTQDLP-DLIA 81
++VG+ GA+GYTG E++RLL+ H + + +++ AG++I+ V+P L G D+ D +
Sbjct: 2 IKVGIFGATGYTGVELMRLLSKHEKVKIEYISSQSYAGKSIADVYPSLTGFYDISLDDVE 61
Query: 82 IKDANFSDVDAVFCCLPHGTTQEIIK-GLPKHLKIVDLSADFRLRDVSEYEEWYGQPHRA 140
++ A S D VF LP G +I + + K +K++DL ADFR D S Y+EWYG +
Sbjct: 62 VEKA-VSKCDVVFTALPAGYASKIAREAVKKGVKVIDLGADFRFDDYSTYKEWYGGEYDG 120
Query: 141 PDLQKEAIYGLTEVLREEIKNARLVANPGCYPTSIQLPLVPLIKANLIQTTNIIVDAKSG 200
D K A YGL E+ R+ IK A +V NPGCYPTS+ L LVPL+K +I NII+D+KSG
Sbjct: 121 YDGIKRA-YGLPEIYRDSIKEADVVGNPGCYPTSVILGLVPLLKEKVIDG-NIIIDSKSG 178
Query: 201 VSGAGRSAKENLLFTEVTEGMSSYGVTRHRHAPEIEQGLADAAGSKVTISFTPHLIPMSR 260
+SGAG S + ++ E E + +Y V HRH PE+ Q L+ A+G KV++ F PHL PM+R
Sbjct: 179 ISGAGHSPSQKNIYAECNESIRAYNVANHRHVPEMVQELSKASGEKVSVVFAPHLTPMTR 238
Query: 261 GMQSTIYVEMAPGVRIDDLRHQLQLSYEDEEFVVLLENGVIPRTHSVKGSNYCLFNVFPD 320
G+ ST+Y ++ + +++ Y++E FV +L+ G P T +V GSN+C D
Sbjct: 239 GILSTMYCKLRENLNQEEIYEVFAEFYKNEYFVKVLKPGAYPSTKNVYGSNFCHIGFEVD 298
Query: 321 RIPGRAIIISVIDNLVKGASGQALQNLNLIMGFPENLGLQYLPLF 365
+ G ++++ IDNLVKGASGQA+QN+N++ E GL +P++
Sbjct: 299 KATGTLVVMTAIDNLVKGASGQAIQNMNIMFEIEEKAGLDIVPVY 343
>ARGC_RHOBA (Q7UVL4) N-acetyl-gamma-glutamyl-phosphate reductase (EC
1.2.1.38) (AGPR) (N-acetyl-glutamate semialdehyde
dehydrogenase) (NAGSA dehydrogenase)
Length = 346
Score = 273 bits (699), Expect = 4e-73
Identities = 149/347 (42%), Positives = 213/347 (60%), Gaps = 11/347 (3%)
Query: 20 SQNNVRVGVLGASGYTGSEILRLLANHPQFGVALMTADRKAGQAISSVFPHL-GTQDLPD 78
S +N+RV ++G++GYT E+ RLL HP + + T+ + G+ +S + P L G D+
Sbjct: 2 SSSNLRVALVGSTGYTALEVARLLLTHPGADLVVATSRQDEGKPLSEIHPMLAGRCDVTL 61
Query: 79 LIAIKDANFSDVDAVFCCLPHGTTQEIIKGLPKH-LKIVDLSADFRLRDVSEYEEWYGQP 137
D D CCLPHG + E +K L + ++++D SADFRL + Y+ WYG
Sbjct: 62 QPLDADVIAKSADVAMCCLPHGASAESVKQLAEAGMRVIDFSADFRLSSLETYQHWYGVK 121
Query: 138 HRAPDLQKEAIYGLTEVLREEIKNARLVANPGCYPTSIQLPLVPLIKANLIQTTNIIVDA 197
H P+ + +YG+ E +EI++A +VANPGCYPTS +PL PL+KA LI+T +IIVD+
Sbjct: 122 HPWPERIGDVVYGMPEFFADEIRSADIVANPGCYPTSAIMPLAPLVKAGLIETDDIIVDS 181
Query: 198 KSGVSGAGRSAKENLLFTEVTEGMSSYGVTRHRHAPEIEQGLADAAGSKVTISFTPHLIP 257
KSGVSGAGRS K L+ E E +S+Y V HRHAPEI + AG+ + + FTPHL P
Sbjct: 182 KSGVSGAGRSPKLGTLYCETNESISAYAVGTHRHAPEIADLVERIAGAPIEVMFTPHLTP 241
Query: 258 MSRGMQSTIYVEMA--PGVRIDDLRHQLQL---SYEDEEFVVLLENGVIPRTHSVKGSNY 312
M RG+ STIYV+ G D +R + L +Y D+ V ++++ +P T V G+N+
Sbjct: 242 MDRGILSTIYVKPVGKAGSVEDAVRAMMSLLRDTYSDQPCVHVVDH--LPATKYVAGTNH 299
Query: 313 CLFNVFPDRIPGRAIIISVIDNLVKGASGQALQNLNLIMGFPENLGL 359
+V P RA+I+ IDNL KGASG A+QN+N++ G PE GL
Sbjct: 300 VQISVRPS--GKRAVIVCAIDNLTKGASGAAVQNMNVMFGLPETAGL 344
>ARGC_THENE (Q9Z4S2) N-acetyl-gamma-glutamyl-phosphate reductase (EC
1.2.1.38) (AGPR) (N-acetyl-glutamate semialdehyde
dehydrogenase) (NAGSA dehydrogenase)
Length = 339
Score = 272 bits (696), Expect = 8e-73
Identities = 150/344 (43%), Positives = 210/344 (60%), Gaps = 9/344 (2%)
Query: 24 VRVGVLGASGYTGSEILRLLANHPQFGVALMTADRKAGQAISSVFPH-LGTQDLPDLIAI 82
+RVG++GA+GYTG E+ RLL HP + M++ AG+ + V+P L L D +
Sbjct: 2 IRVGIVGATGYTGIELFRLLKKHPSAKIVYMSSRTYAGKKMMEVYPSTLEEAVLEDFDSR 61
Query: 83 KDANFSDVDAVFCCLPHGTTQEIIKGLPKHLKIVDLSADFRLRDVSEYEEWYGQPHRAPD 142
K + DV VF LP G + + L +++KI+DL ADFR D + Y EWYG ++ D
Sbjct: 62 KISESCDV--VFTALPAGVSYRLAMEL-QNVKIIDLGADFRFDDPNTYAEWYG--NKLED 116
Query: 143 LQK-EAIYGLTEVLREEIKNARLVANPGCYPTSIQLPLVPLIKANLIQTTNIIVDAKSGV 201
E +YGL E+ R+ IKNAR+V NPGCYPTS+ L L P +K+ +++ I+VD+KSGV
Sbjct: 117 YNSTERVYGLPELYRDSIKNARIVGNPGCYPTSVILALTPALKSGIVEVDTIVVDSKSGV 176
Query: 202 SGAGRSAKENLLFTEVTEGMSSYGVTRHRHAPEIEQGLADAAGSKVTISFTPHLIPMSRG 261
SGAGR + F+EV E + Y V +HRH PE+EQ L AG V + FTPHL+PM+RG
Sbjct: 177 SGAGRKETLDYTFSEVNENLRPYNVAKHRHVPEMEQELKKIAGKDVKVIFTPHLVPMTRG 236
Query: 262 MQSTIYVEMAPGVRIDDLRHQLQLSYEDEEFVVLLENGVIPRTHSVKGSNYCLFNVFPDR 321
+ STIYV+ ++ + Q Y E FV +L GV P T GSN+ + +
Sbjct: 237 ILSTIYVK--TNASLEKIHRIYQEFYAGEPFVHVLPLGVFPSTKWCYGSNHVFIGMQLEE 294
Query: 322 IPGRAIIISVIDNLVKGASGQALQNLNLIMGFPENLGLQYLPLF 365
G I++S IDNLVKGASGQA+QN+N++ E GL+ LP++
Sbjct: 295 RTGTPILMSAIDNLVKGASGQAIQNMNIMFDLKETEGLESLPIY 338
>ARGC_BACAN (Q81M95) N-acetyl-gamma-glutamyl-phosphate reductase (EC
1.2.1.38) (AGPR) (N-acetyl-glutamate semialdehyde
dehydrogenase) (NAGSA dehydrogenase)
Length = 345
Score = 268 bits (684), Expect = 2e-71
Identities = 136/344 (39%), Positives = 212/344 (61%), Gaps = 2/344 (0%)
Query: 24 VRVGVLGASGYTGSEILRLLANHPQFGVALMTADRKAGQAISSVFPHLGTQDLPDLIAIK 83
++V ++GA+GY G E++RLL HP F +A + + + G+ I++V+PH + L I
Sbjct: 1 MKVAIIGATGYGGIELIRLLEQHPYFSIASLHSFSQVGECITNVYPHFQNVLVHTLQEID 60
Query: 84 DANFS-DVDAVFCCLPHGTTQEII-KGLPKHLKIVDLSADFRLRDVSEYEEWYGQPHRAP 141
+ + VF P G + E+ K L LK++DLS DFR++D YE+WY +
Sbjct: 61 VEEIEKEAEIVFLATPAGVSAELTPKLLAVGLKVIDLSGDFRMKDPFIYEQWYKRAAAKE 120
Query: 142 DLQKEAIYGLTEVLREEIKNARLVANPGCYPTSIQLPLVPLIKANLIQTTNIIVDAKSGV 201
+ +EA+YGL+E R EI+ A L+ANPGC+ T+ L ++PL+++ +I+ +II+DAKSGV
Sbjct: 121 GVLREAVYGLSEWKRSEIQKANLIANPGCFATAALLAILPLVRSGIIEEDSIIIDAKSGV 180
Query: 202 SGAGRSAKENLLFTEVTEGMSSYGVTRHRHAPEIEQGLADAAGSKVTISFTPHLIPMSRG 261
SGAG++ F E+ + + Y V H+H PEIEQ LA+ I+F+ HLIP+SRG
Sbjct: 181 SGAGKTPTTMTHFPELYDNLRIYKVNEHQHIPEIEQMLAEWNRETKPITFSTHLIPISRG 240
Query: 262 MQSTIYVEMAPGVRIDDLRHQLQLSYEDEEFVVLLENGVIPRTHSVKGSNYCLFNVFPDR 321
+ T+Y ++ + I+ L+ + +YE F+ + G P V+GSNYC + D
Sbjct: 241 IMVTLYAKVKREMEIEQLQQLYEEAYEQSAFIRIRMQGEFPSPKEVRGSNYCDMGIAYDE 300
Query: 322 IPGRAIIISVIDNLVKGASGQALQNLNLIMGFPENLGLQYLPLF 365
GR I+SVIDN++KGA+GQA+QN N++ G E GLQ++PL+
Sbjct: 301 RTGRVTIVSVIDNMMKGAAGQAIQNANIVAGLEETTGLQHMPLY 344
>ARGC_GLOVI (Q7NE70) N-acetyl-gamma-glutamyl-phosphate reductase (EC
1.2.1.38) (AGPR) (N-acetyl-glutamate semialdehyde
dehydrogenase) (NAGSA dehydrogenase)
Length = 350
Score = 267 bits (683), Expect = 3e-71
Identities = 141/339 (41%), Positives = 205/339 (59%), Gaps = 3/339 (0%)
Query: 24 VRVGVLGASGYTGSEILRLLANHPQFGVALMTADRKAGQAISSVFPHLGTQDLPDLIAIK 83
+RVG++GASGY G +++RLL +HP+ +A + A++ AG ++P L + A++
Sbjct: 5 LRVGIVGASGYGGVQLVRLLLDHPRVEIAYLGANQNAGTPFGELYPQLAHRIDRVCEAVE 64
Query: 84 -DANFSDVDAVFCCLPHGTTQEIIKGL-PKHLKIVDLSADFRLRDVSEYEEWYGQPHRAP 141
D VF P+G + GL L++ DLSAD+R ++ Y+ WYG
Sbjct: 65 LDRIVEACSVVFLATPNGIAHTLAPGLLAGGLRVFDLSADYRFVNLETYQSWYGGDRHDA 124
Query: 142 DLQKEAIYGLTEVLREEIKNARLVANPGCYPTSIQLPLVPLIKANLIQTTNIIVDAKSGV 201
+ +EA+YGL E+ RE I+ ARLV PGCYPT+ L PL+K LI ++I+DAKSGV
Sbjct: 125 AVAREAVYGLPELYRERIRTARLVGCPGCYPTASLLAAAPLLKQGLIDPRSLIIDAKSGV 184
Query: 202 SGAGRSAKENLLFTEVTEGMSSYGVTRHRHAPEIEQGLADAAGSKVTISFTPHLIPMSRG 261
SGAGR+ K LF E +++Y V RHRH PEIEQ +D AG +V + FTPHLIPM+RG
Sbjct: 185 SGAGRALKTGSLFAEADSSVAAYSVARHRHIPEIEQVCSDLAGMRVQVQFTPHLIPMARG 244
Query: 262 MQSTIYVEMA-PGVRIDDLRHQLQLSYEDEEFVVLLENGVIPRTHSVKGSNYCLFNVFPD 320
M T+Y ++ PG+ +D+ + Y V +L +G+ P+T G+N C + D
Sbjct: 245 MLVTLYAQLRDPGLVSEDMLTIYEAFYRQAPAVRVLGSGIYPQTKWASGTNTCFIGLEVD 304
Query: 321 RIPGRAIIISVIDNLVKGASGQALQNLNLIMGFPENLGL 359
+ R +++S +DNLVKG SGQA+Q +NL G+ E LGL
Sbjct: 305 QRTERVVVLSALDNLVKGQSGQAIQAMNLTQGWEEMLGL 343
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.319 0.138 0.398
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 42,349,223
Number of Sequences: 164201
Number of extensions: 1791547
Number of successful extensions: 4806
Number of sequences better than 10.0: 155
Number of HSP's better than 10.0 without gapping: 122
Number of HSP's successfully gapped in prelim test: 33
Number of HSP's that attempted gapping in prelim test: 4292
Number of HSP's gapped (non-prelim): 159
length of query: 365
length of database: 59,974,054
effective HSP length: 112
effective length of query: 253
effective length of database: 41,583,542
effective search space: 10520636126
effective search space used: 10520636126
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 66 (30.0 bits)
Medicago: description of AC145331.13


