

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC145330.4 + phase: 0
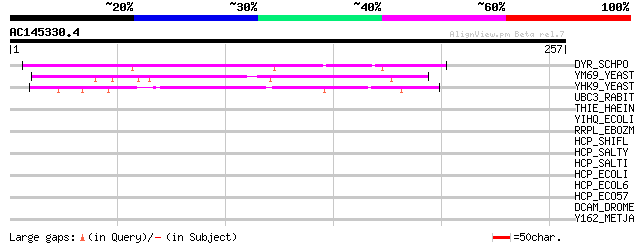
(257 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
DYR_SCHPO (P36591) Dihydrofolate reductase (EC 1.5.1.3) 94 3e-19
YM69_YEAST (Q05015) Hypothetical 24.5 kDa protein in ERG8-UBP8 i... 69 9e-12
YHK9_YEAST (P38777) Hypothetical 27.3 kDa protein in AAP1-SMF2 i... 66 7e-11
UBC3_RABIT (Q29503) Ubiquitin-conjugating enzyme E2-32 kDa compl... 32 1.2
THIE_HAEIN (P71350) Thiamine-phosphate pyrophosphorylase (EC 2.5... 32 1.5
YIHQ_ECOLI (P32138) Putative family 31 glucosidase yihQ 31 2.6
RRPL_EBOZM (Q05318) RNA-directed RNA polymerase (EC 2.7.7.48) (L... 30 4.4
HCP_SHIFL (Q83S05) Hydroxylamine reductase (EC 1.7.-.-) (Hybrid-... 30 4.4
HCP_SALTY (Q8ZQE8) Hydroxylamine reductase (EC 1.7.-.-) (Hybrid-... 30 4.4
HCP_SALTI (Q8Z829) Hydroxylamine reductase (EC 1.7.-.-) (Hybrid-... 30 4.4
HCP_ECOLI (P75825) Hydroxylamine reductase (EC 1.7.-.-) (Hybrid-... 30 4.4
HCP_ECOL6 (Q8FJE0) Hydroxylamine reductase (EC 1.7.-.-) (Hybrid-... 30 4.4
HCP_ECO57 (Q8X6L0) Hydroxylamine reductase (EC 1.7.-.-) (Hybrid-... 30 4.4
DCAM_DROME (P91931) S-adenosylmethionine decarboxylase proenzyme... 30 4.4
Y162_METJA (Q57626) Hypothetical protein MJ0162 30 5.7
>DYR_SCHPO (P36591) Dihydrofolate reductase (EC 1.5.1.3)
Length = 461
Score = 94.0 bits (232), Expect = 3e-19
Identities = 63/214 (29%), Positives = 96/214 (44%), Gaps = 20/214 (9%)
Query: 7 RKLKILCLHGFRTSGSFIKKQISKWDPSIFSQFHLEFPDGKFPAGGKSD----------- 55
+ LK+LCLHG+ SG K++ + L FP G A ++D
Sbjct: 3 KPLKVLCLHGWIQSGPVFSKKMGSVQKYLSKYAELHFPTGPVVADEEADPNDEEEKKRLA 62
Query: 56 -IEGIFPPPYFEWFQFDKDFTVYTNLDECISYLTEYIIANGPFDGFLGFSQGATLSALLI 114
+ G F WF+ + Y + DE + + +Y+ GPFDG +GFSQGA + A+L
Sbjct: 63 ALGGEQNGGKFGWFEVEDFKNTYGSWDESLECINQYMQEKGPFDGLIGFSQGAGIGAMLA 122
Query: 115 GYQAQGK---LLKEHPPIKFLVSISGSKFRDPSICDVAYKDPIKAKSVHFIGEKDWLKIP 171
G+ +HPP KF+V + G + P D Y + S+H G D L +P
Sbjct: 123 QMLQPGQPPNPYVQHPPFKFVVFVGGFRAEKPEF-DHFYNPKLTTPSLHIAGTSDTL-VP 180
Query: 172 ---SEELASAFDKPLIIRHPQGHTVPRLDEVSTG 202
S++L + ++ HP H VP+ TG
Sbjct: 181 LARSKQLVERCENAHVLLHPGQHIVPQQAVYKTG 214
>YM69_YEAST (Q05015) Hypothetical 24.5 kDa protein in ERG8-UBP8
intergenic region
Length = 223
Score = 69.3 bits (168), Expect = 9e-12
Identities = 57/206 (27%), Positives = 87/206 (41%), Gaps = 26/206 (12%)
Query: 11 ILCLHGFRTSGSFIKKQISKWDPSIFSQ-FHLEFPDG--KFPAGGKSDIEG--IFPPP-- 63
+L LHG SG + + + + + L +P +FP D G I P
Sbjct: 5 VLMLHGLAQSGDYFASKTKGFRAEMEKLGYKLYYPTAPNEFPPADVPDFLGEVIADAPGD 64
Query: 64 -----YFEWFQFDKDFTVYTNLDECISYLTEYIIANGPFDGFLGFSQGATLSALLIGYQA 118
W + D Y I YL Y++ NGPF G +GFSQGA ++ GY A
Sbjct: 65 GENTGVLAWLENDPSTGGYFIPQTTIDYLHNYVLENGPFAGIVGFSQGAGVA----GYLA 120
Query: 119 Q------GKLLKEHPPIKFLVSISGSKFRDPSICDVAYKDPIKAKSVHFIGEKDWLKIPS 172
G +E PP++F +++SG +F+ + PI S+H GE D + P+
Sbjct: 121 TDFNGLLGLTTEEQPPLEFFMAVSGFRFQPQQYQEQYDLHPISVPSLHVQGELDTITEPA 180
Query: 173 EEL----ASAFDKPLIIRHPQGHTVP 194
+ + D ++ H GH VP
Sbjct: 181 KVQGLYNSCTEDSRTLLMHSGGHFVP 206
>YHK9_YEAST (P38777) Hypothetical 27.3 kDa protein in AAP1-SMF2
intergenic region
Length = 243
Score = 66.2 bits (160), Expect = 7e-11
Identities = 63/231 (27%), Positives = 98/231 (42%), Gaps = 52/231 (22%)
Query: 10 KILCLHGFRTSG-------SFIKKQISKWD--------PSIFSQFHLEFP--DGKFPAGG 52
K+L LHGF +G S I+K + K + P + + L F D K+ A
Sbjct: 7 KLLFLHGFLQNGKVFSEKSSGIRKLLKKANVQCDYIDAPVLLEKKDLPFEMDDEKWQATL 66
Query: 53 KSDIEGIFPPPYFEWFQFDKDFTVYTNLDECISYLTEYIIANGPFDGFLGFSQGATLSAL 112
+D+ WF + + + ++ E + + ++I ANGP+DG +GFSQGA LS++
Sbjct: 67 DADVNRA-------WF-YHSEISHELDISEGLKSVVDHIKANGPYDGIVGFSQGAALSSI 118
Query: 113 LIGYQAQGKLLKEHPPIKFLVSISGSKFRDPS-------------ICDVAYKDPIKAKSV 159
+ ++ L+ +HP K V ISG F +P A K +K K +
Sbjct: 119 ITNKISE--LVPDHPQFKVSVVISGYSFTEPDPEHPGELRITEKFRDSFAVKPDMKTKMI 176
Query: 160 HFIGEKDWLKIPSEELASAFD-----------KPLIIRHPQGHTVPRLDEV 199
G D +PS +D K L HP GH VP ++
Sbjct: 177 FIYGASD-QAVPSVRSKYLYDIYLKAQNGNKEKVLAYEHPGGHMVPNKKDI 226
>UBC3_RABIT (Q29503) Ubiquitin-conjugating enzyme E2-32 kDa
complementing (EC 6.3.2.19) (Ubiquitin-protein ligase)
(Ubiquitin carrier protein) (E2-CDC34)
Length = 238
Score = 32.3 bits (72), Expect = 1.2
Identities = 35/153 (22%), Positives = 61/153 (38%), Gaps = 7/153 (4%)
Query: 25 KKQISKWDPSIFSQFHLEFPDGKFPAGGKSDIEGIFPPPYFEWFQFDKDFTVYTNLDECI 84
+ + W+ +IF + + G F A K I+ + PP F + +Y N D CI
Sbjct: 35 ESDLYNWEVAIFGPPNTLYEGGYFKAHIKFPIDYPYSPPTFRFLTKMWHPNIYENGDVCI 94
Query: 85 SYLTEYIIANGPFDGFL---GFSQGATLSALLIGYQAQGKLLKEHPPIKFLVSISGSKFR 141
S L + + P G L ++ + +L+ + P S+ K+R
Sbjct: 95 SILHPPV--DDPQSGELPSERWNPTQNVRTILLSVISLLNEPNTFSPANVDASVMFRKWR 152
Query: 142 DPSICDVAYKDPIK--AKSVHFIGEKDWLKIPS 172
D D Y + I+ + EKD +K+P+
Sbjct: 153 DSKGKDKEYAEIIRKQVSATKAEAEKDGVKVPT 185
>THIE_HAEIN (P71350) Thiamine-phosphate pyrophosphorylase (EC
2.5.1.3) (TMP pyrophosphorylase) (TMP-PPase)
(Thiamine-phosphate synthase)
Length = 226
Score = 32.0 bits (71), Expect = 1.5
Identities = 20/50 (40%), Positives = 30/50 (60%), Gaps = 8/50 (16%)
Query: 154 IKAKSVHFIGEKDWLKIPSEELASAFDKPLIIRHPQGHTVPRLDEVSTGQ 203
I+A +H +G+ D +P +E+ + DKPLII G +V RLDE G+
Sbjct: 92 IEADGIH-VGQSD---MPVQEIRAKTDKPLII----GWSVNRLDEAKIGE 133
>YIHQ_ECOLI (P32138) Putative family 31 glucosidase yihQ
Length = 678
Score = 31.2 bits (69), Expect = 2.6
Identities = 21/57 (36%), Positives = 33/57 (57%), Gaps = 2/57 (3%)
Query: 70 FDKDFTVYTNLDECISYLTEYIIANGPFDGFLGFSQGATLSALL-IGYQAQGKLLKE 125
F +F++ L E I+ LT+ I++ P + FS+G+ +SA L I QG+LL E
Sbjct: 51 FRGNFSIKDKLQEKIA-LTDAIVSQSPDGWLIHFSRGSDISATLNISADDQGRLLLE 106
>RRPL_EBOZM (Q05318) RNA-directed RNA polymerase (EC 2.7.7.48) (Large
structural protein) (L protein)
Length = 2212
Score = 30.4 bits (67), Expect = 4.4
Identities = 20/78 (25%), Positives = 41/78 (51%), Gaps = 9/78 (11%)
Query: 146 CDVAYKDPI-KAKSVHFIGEKDWLKIP---SEELASAFDKPLIIRHPQGHTVPRLDEVST 201
C++++ +P + K ++ I E D +++P ELA + +I T D +S+
Sbjct: 1392 CNISFDNPFFQGKRLNII-EDDLIRLPHLSGWELAKTIMQSIISDSNNSST----DPISS 1446
Query: 202 GQLQNFVAEILSQPKVGV 219
G+ ++F L+ PK+G+
Sbjct: 1447 GETRSFTTHFLTYPKIGL 1464
>HCP_SHIFL (Q83S05) Hydroxylamine reductase (EC 1.7.-.-)
(Hybrid-cluster protein) (HCP)
Length = 552
Score = 30.4 bits (67), Expect = 4.4
Identities = 12/40 (30%), Positives = 27/40 (67%), Gaps = 2/40 (5%)
Query: 3 GENGRKLKILCLHGFRTSGSFIKKQ--ISKWDPSIFSQFH 40
GEN L++LCL+G + + ++++ + ++D I++Q+H
Sbjct: 147 GENILGLRLLCLYGLKGAAAYMEHAHVLGQYDNDIYAQYH 186
>HCP_SALTY (Q8ZQE8) Hydroxylamine reductase (EC 1.7.-.-)
(Hybrid-cluster protein) (HCP)
Length = 550
Score = 30.4 bits (67), Expect = 4.4
Identities = 12/40 (30%), Positives = 27/40 (67%), Gaps = 2/40 (5%)
Query: 3 GENGRKLKILCLHGFRTSGSFIKKQ--ISKWDPSIFSQFH 40
GEN L++LCL+G + + ++++ + ++D I++Q+H
Sbjct: 145 GENILGLRLLCLYGLKGAAAYMEHAHVLGQYDNDIYAQYH 184
>HCP_SALTI (Q8Z829) Hydroxylamine reductase (EC 1.7.-.-)
(Hybrid-cluster protein) (HCP)
Length = 550
Score = 30.4 bits (67), Expect = 4.4
Identities = 12/40 (30%), Positives = 27/40 (67%), Gaps = 2/40 (5%)
Query: 3 GENGRKLKILCLHGFRTSGSFIKKQ--ISKWDPSIFSQFH 40
GEN L++LCL+G + + ++++ + ++D I++Q+H
Sbjct: 145 GENILGLRLLCLYGLKGAAAYMEHAHVLGQYDNDIYAQYH 184
>HCP_ECOLI (P75825) Hydroxylamine reductase (EC 1.7.-.-)
(Hybrid-cluster protein) (HCP)
Length = 550
Score = 30.4 bits (67), Expect = 4.4
Identities = 12/40 (30%), Positives = 27/40 (67%), Gaps = 2/40 (5%)
Query: 3 GENGRKLKILCLHGFRTSGSFIKKQ--ISKWDPSIFSQFH 40
GEN L++LCL+G + + ++++ + ++D I++Q+H
Sbjct: 145 GENILGLRLLCLYGLKGAAAYMEHAHVLGQYDNDIYAQYH 184
>HCP_ECOL6 (Q8FJE0) Hydroxylamine reductase (EC 1.7.-.-)
(Hybrid-cluster protein) (HCP)
Length = 550
Score = 30.4 bits (67), Expect = 4.4
Identities = 12/40 (30%), Positives = 27/40 (67%), Gaps = 2/40 (5%)
Query: 3 GENGRKLKILCLHGFRTSGSFIKKQ--ISKWDPSIFSQFH 40
GEN L++LCL+G + + ++++ + ++D I++Q+H
Sbjct: 145 GENILGLRLLCLYGLKGAAAYMEHAHVLGQYDNDIYAQYH 184
>HCP_ECO57 (Q8X6L0) Hydroxylamine reductase (EC 1.7.-.-)
(Hybrid-cluster protein) (HCP)
Length = 550
Score = 30.4 bits (67), Expect = 4.4
Identities = 12/40 (30%), Positives = 27/40 (67%), Gaps = 2/40 (5%)
Query: 3 GENGRKLKILCLHGFRTSGSFIKKQ--ISKWDPSIFSQFH 40
GEN L++LCL+G + + ++++ + ++D I++Q+H
Sbjct: 145 GENILGLRLLCLYGLKGAAAYMEHAHVLGQYDNDIYAQYH 184
>DCAM_DROME (P91931) S-adenosylmethionine decarboxylase proenzyme
(EC 4.1.1.50) (AdoMetDC) (SamDC) [Contains:
S-adenosylmethionine decarboxylase alpha chain;
S-adenosylmethionine decarboxylase beta chain]
Length = 347
Score = 30.4 bits (67), Expect = 4.4
Identities = 32/136 (23%), Positives = 55/136 (39%), Gaps = 11/136 (8%)
Query: 32 DPSIFSQFHLEFPDGKFPAGGKSDIEGIFPPPYFEWFQFDKDFTVYTNLDECISYLTEYI 91
DP S F+ + A KS I+ I P + + F FD +++ Y+T +I
Sbjct: 193 DPETMSIFYKNKFNDANGATVKSGIDTILPTMHIDDFLFDPCGYSMNGINDKGEYMTIHI 252
Query: 92 IANGPFDGFLGFSQGATLSALLIGYQAQGKLLKEHPPIKFLVSISGSKFRDPSICDVAYK 151
F ++ F LS + +++ P KF+V+I +K C +AY
Sbjct: 253 TPENQF-SYVSFETNVALSNY---RKLINQVINTFKPGKFIVTIFANK------CSLAY- 301
Query: 152 DPIKAKSVHFIGEKDW 167
+ +K V + W
Sbjct: 302 ETMKELEVEYSQGSHW 317
>Y162_METJA (Q57626) Hypothetical protein MJ0162
Length = 421
Score = 30.0 bits (66), Expect = 5.7
Identities = 21/72 (29%), Positives = 31/72 (42%), Gaps = 7/72 (9%)
Query: 73 DFTVYTNLDECISYLTEYIIANGPFDGFLGFSQGATLSALLIGYQAQGKL-------LKE 125
D ++ N + CI T ++ GP +L + +L GYQA+G L KE
Sbjct: 285 DESLVFNKEPCIIVSTSGMVQGGPVLKYLKLLKDPKNKLILTGYQAEGTLGRELEEGAKE 344
Query: 126 HPPIKFLVSISG 137
P K + I G
Sbjct: 345 IQPFKNKIPIRG 356
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.318 0.140 0.422
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 34,384,698
Number of Sequences: 164201
Number of extensions: 1537206
Number of successful extensions: 3166
Number of sequences better than 10.0: 15
Number of HSP's better than 10.0 without gapping: 5
Number of HSP's successfully gapped in prelim test: 10
Number of HSP's that attempted gapping in prelim test: 3155
Number of HSP's gapped (non-prelim): 15
length of query: 257
length of database: 59,974,054
effective HSP length: 108
effective length of query: 149
effective length of database: 42,240,346
effective search space: 6293811554
effective search space used: 6293811554
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 64 (29.3 bits)
Medicago: description of AC145330.4


