

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC145329.11 - phase: 0
(360 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
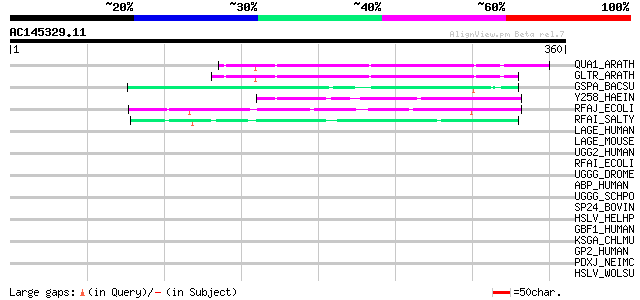
Score E
Sequences producing significant alignments: (bits) Value
QUA1_ARATH (Q9LSG3) Glycosyltransferase QUASIMODO1 (EC 2.4.1.-) 96 2e-19
GLTR_ARATH (Q9FWA4) Probable glycosyltransferase At3g02350 (EC 2... 93 1e-18
GSPA_BACSU (P25148) General stress protein A 57 7e-08
Y258_HAEIN (P43974) Putative glycosyl transferase HI0258 (EC 2.-... 51 4e-06
RFAJ_ECOLI (P27129) Lipopolysaccharide 1,2-glucosyltransferase (... 50 7e-06
RFAI_SALTY (P19816) Lipopolysaccharide 1,3-galactosyltransferase... 45 3e-04
LAGE_HUMAN (O95461) Glycosyltransferase-like protein LARGE (EC 2... 43 0.001
LAGE_MOUSE (Q9Z1M7) Glycosyltransferase-like protein LARGE (EC 2... 42 0.002
UGG2_HUMAN (Q9NYU1) UDP-glucose:glycoprotein glucosyltransferase... 38 0.045
RFAI_ECOLI (P27128) Lipopolysaccharide 1,3-galactosyltransferase... 36 0.13
UGGG_DROME (Q09332) UDP-glucose:glycoprotein glucosyltransferase... 36 0.17
ABP_HUMAN (P19801) Amiloride-sensitive amine oxidase [copper-con... 32 2.5
UGGG_SCHPO (Q09140) UDP-glucose:glycoprotein glucosyltransferase... 31 4.3
SP24_BOVIN (Q27967) Secreted phosphoprotein 24 precursor (Spp-24... 31 5.6
HSLV_HELHP (Q7VJD3) ATP-dependent protease hslV (EC 3.4.25.-) 31 5.6
GBF1_HUMAN (Q92538) Golgi-specific brefeldin A-resistance guanin... 31 5.6
KSGA_CHLMU (Q9PK40) Dimethyladenosine transferase (EC 2.1.1.-) (... 30 7.3
GP2_HUMAN (P55259) Pancreatic secretory granule membrane major g... 30 7.3
PDXJ_NEIMC (Q9RML6) Pyridoxal phosphate biosynthetic protein pdx... 30 9.5
HSLV_WOLSU (Q7M8Z6) ATP-dependent protease hslV (EC 3.4.25.-) 30 9.5
>QUA1_ARATH (Q9LSG3) Glycosyltransferase QUASIMODO1 (EC 2.4.1.-)
Length = 559
Score = 95.5 bits (236), Expect = 2e-19
Identities = 62/220 (28%), Positives = 107/220 (48%), Gaps = 11/220 (5%)
Query: 136 YYFDRNIVKNLISTSVRQALEQP-----LNYARNYLADLLESCVKRVIYLDSDLVLQDDI 190
+YF+ N ++N + P LN+ R YL ++ + R+++LD D+V+Q D+
Sbjct: 336 FYFE-NKLENATKDTTNMKFRNPKYLSILNHLRFYLPEMYPK-LHRILFLDDDVVVQKDL 393
Query: 191 AKLWNTDLGLNTIGAPQYCHANFTKYFTAAFWSDPVFSTTFEKRKACYFNTGVMVMDLVK 250
LW D+ GA + C +F +Y +S P+ F KAC + G+ DL
Sbjct: 394 TGLWEIDMDGKVNGAVETCFGSFHRYAQYMNFSHPLIKEKFNP-KACAWAYGMNFFDLDA 452
Query: 251 WRKKGYTERIERWMEIQKVERIYELGSLPPFLLVFAGHVAAIEHRWNQHGLGGDNVKGSC 310
WR++ TE W + + +++LG+LPP L+ F ++ W+ GL G N S
Sbjct: 453 WRREKCTEEYHYWQNLNENRALWKLGTLPPGLITFYSTTKPLDKSWHVLGL-GYNPSISM 511
Query: 311 RDLHPGPVSLLHWSGSGKPWRRLDESKPCPLDALWEPFDL 350
++ V +H++G+ KPW + ++ PL +DL
Sbjct: 512 DEIRNAAV--VHFNGNMKPWLDIAMNQFRPLWTKHVDYDL 549
>GLTR_ARATH (Q9FWA4) Probable glycosyltransferase At3g02350 (EC
2.4.1.-)
Length = 561
Score = 92.8 bits (229), Expect = 1e-18
Identities = 56/204 (27%), Positives = 103/204 (50%), Gaps = 11/204 (5%)
Query: 132 RFKVYYFDRNIVKNLISTSVRQALEQP-----LNYARNYLADLLESCVKRVIYLDSDLVL 186
+ + +YF+ N +N S + P LN+ R YL ++ + ++++LD D+V+
Sbjct: 334 KLQKFYFE-NQAENATKDSHNLKFKNPKYLSMLNHLRFYLPEMYPK-LNKILFLDDDVVV 391
Query: 187 QDDIAKLWNTDLGLNTIGAPQYCHANFTKYFTAAFWSDPVFSTTFEKRKACYFNTGVMVM 246
Q D+ LW +L GA + C +F +Y +S P+ F AC + G+ +
Sbjct: 392 QKDVTGLWKINLDGKVNGAVETCFGSFHRYGQYLNFSHPLIKENFNP-SACAWAFGMNIF 450
Query: 247 DLVKWRKKGYTERIERWMEIQKVERIYELGSLPPFLLVFAGHVAAIEHRWNQHGLGGDNV 306
DL WR++ T++ W + + +++LG+LPP L+ F +++ W+ GL G N
Sbjct: 451 DLNAWRREKCTDQYHYWQNLNEDRTLWKLGTLPPGLITFYSKTKSLDKSWHVLGL-GYNP 509
Query: 307 KGSCRDLHPGPVSLLHWSGSGKPW 330
S ++ V +H++G+ KPW
Sbjct: 510 GVSMDEIRNAGV--IHYNGNMKPW 531
>GSPA_BACSU (P25148) General stress protein A
Length = 286
Score = 57.0 bits (136), Expect = 7e-08
Identities = 58/266 (21%), Positives = 104/266 (38%), Gaps = 28/266 (10%)
Query: 77 LVHVAITLDVEYLRGSIAAVHSILYHASCPENVFFHFLVTDTDLETLVRTTFPQLRFKVY 136
++H+ D Y R S+L + V + + + R L+F V
Sbjct: 6 IMHIVSCADDNYARHLGGMFVSLLTNMDQEREVKLYVIDGGIKPDNKKRLEETTLKFGVP 65
Query: 137 YFDRNIVKNLISTSVRQALEQPLNYARNYLADLL-ESCVKRVIYLDSDLVLQDDIAKLWN 195
+ N+ +V + Y R + DL+ + +KR+IY+D D ++ +DI+KLW+
Sbjct: 66 IEFLEVDTNMYEHAVESSHITKAAYYRISIPDLIKDESIKRMIYIDCDALVLEDISKLWD 125
Query: 196 TDLGLNTIGAPQYCHANFTKYFTAAFWSDPVFSTTFEKRKACYFNTGVMVMDLVKWRKKG 255
D+ T+ A + A + +D YFN+G+M++D WRK+
Sbjct: 126 LDIAPYTVAAVE--DAGQHERLKEMNVTD----------TGKYFNSGIMIIDFESWRKQN 173
Query: 256 YTERIERWMEIQKVERIYELGSLPPFLLVFAGHVAAIEHRWNQH-----------GLGGD 304
TE++ ++ E L + + RWN L G
Sbjct: 174 ITEKVINFINEHPDEDFLVLHDQDALNAILYDQWYELHPRWNAQTYIMLKLKTPSTLLGR 233
Query: 305 NVKGSCRDLHPGPVSLLHWSGSGKPW 330
R+ +P +++H+ G KPW
Sbjct: 234 KQYNETRE-NP---AIVHFCGGEKPW 255
>Y258_HAEIN (P43974) Putative glycosyl transferase HI0258 (EC
2.-.-.-)
Length = 330
Score = 51.2 bits (121), Expect = 4e-06
Identities = 39/172 (22%), Positives = 72/172 (41%), Gaps = 12/172 (6%)
Query: 161 YARNYLADLLESCVKRVIYLDSDLVLQDDIAKLWNTDLGLNTIGAPQYCHANFTKYFTAA 220
YAR L +++ +++ IY+D D + + +LWN D+ + A C F A
Sbjct: 121 YARLNLTKYIKN-IEKAIYIDVDTLTNSSLQELWNIDITNYYLAA---CRDTFIDVKNEA 176
Query: 221 FWSDPVFSTTFEKRKACYFNTGVMVMDLVKWRKKGYTERIERWMEIQKVERIYELGSLPP 280
+ T YFN G+++++L KW+++ ++ WM K + +
Sbjct: 177 Y------KKTIGLEGYSYFNAGILLINLNKWKEENIFQKSINWM--NKYNNVMKYQDQDI 228
Query: 281 FLLVFAGHVAAIEHRWNQHGLGGDNVKGSCRDLHPGPVSLLHWSGSGKPWRR 332
+ G V I +R+N D +K P+ + H+ G K W +
Sbjct: 229 LNGICKGKVKFINNRFNFTPTDRDLIKKKNLLCVKMPIVISHYCGPNKFWHK 280
>RFAJ_ECOLI (P27129) Lipopolysaccharide 1,2-glucosyltransferase (EC
2.4.1.58)
Length = 338
Score = 50.4 bits (119), Expect = 7e-06
Identities = 61/263 (23%), Positives = 112/263 (42%), Gaps = 24/263 (9%)
Query: 78 VHVAITLDVEYLRGSIAAVHSILYHASCPENVFFHFLV---TDTDLETLVRTTFP-QLRF 133
++VA +D YL G ++ SI+ + + N+ F+ + D + + + QLR
Sbjct: 28 LNVAYGVDANYLDGVGVSITSIVLN-NRHINLDFYIIADVYNDGFFQKIAKLAEQNQLRI 86
Query: 134 KVYYFDRNIVKNLISTSVRQALEQPLNYARNYLADLLESCVKRVIYLDSDLVLQDDIAKL 193
+Y + + ++ L T V Y R + LL + R++YLD+D+V + DI++L
Sbjct: 87 TLYRINTDKLQCLPCTQVWSRAM----YFRLFAFQLLGLTLDRLLYLDADVVCKGDISQL 142
Query: 194 WNTDLGLNTIGAPQYCHANFTKYFTAAFWSDPVFSTTFEKRKACYFNTGVMVMDLVKWRK 253
+ LGLN A + + SDP + YFN+GV+ +DL KW
Sbjct: 143 LH--LGLNGAVAAVVKDVEPMQEKAVSRLSDP-------ELLGQYFNSGVVYLDLKKWAD 193
Query: 254 KGYTERIERWMEIQKVERIYELGSLPPFLLVFAGHVAAIEHRWNQ----HGLGGDNVKGS 309
TE+ + + +Y+ ++ G + +N D +
Sbjct: 194 AKLTEK--ALSILMSKDNVYKYPDQDVMNVLLKGMTLFLPREYNTIYTIKSELKDKTHQN 251
Query: 310 CRDLHPGPVSLLHWSGSGKPWRR 332
+ L L+H++G+ KPW +
Sbjct: 252 YKKLITESTLLIHYTGATKPWHK 274
>RFAI_SALTY (P19816) Lipopolysaccharide 1,3-galactosyltransferase
(EC 2.4.1.44) (Lipopolysaccharide
3-alpha-galactosyltransferase)
Length = 337
Score = 45.1 bits (105), Expect = 3e-04
Identities = 59/261 (22%), Positives = 105/261 (39%), Gaps = 26/261 (9%)
Query: 79 HVAITLDVEYLRGSIAAVHSILYHASCPENVFFHFLVTD-------TDLETLVRTTFPQL 131
++A +D +L G ++ S+L + PE + TD E L + Q+
Sbjct: 29 NIAYGIDKNFLFGCGVSIASVLL--ANPEKALAFHVFTDFFDSEDQQRFEALAKQYATQI 86
Query: 132 RFKVYYFDRNIVKNLISTSVRQALEQPLNYARNYLADLLESCVKRVIYLDSDLVLQDDIA 191
VY D +K+L ST Y R +AD RV+YLD+D+ + I
Sbjct: 87 --VVYLIDCERLKSLPSTKNWTYA----TYFRFIIADYFSDKTDRVLYLDADIACKGSIQ 140
Query: 192 KLWNTDLGLNTIGAPQYCHANFTKYFTAAFWSDPVFSTTFEKRKACYFNTGVMVMDLVKW 251
+L + + N I A +W+ S + YFN G +++++ W
Sbjct: 141 ELIDLNFAENEIAA-------VVAEGELEWWTKRSVSLATPGLVSGYFNAGFILINIPLW 193
Query: 252 RKKGYTER-IERWMEIQKVERIYELGSLPPFLLVFAGHVAA-IEHRWNQHGLGGDNVKGS 309
+ +++ IE + + V+RI L L +F + A ++ ++N +K S
Sbjct: 194 TAENISKKAIEMLKDPEVVQRITHLDQ--DVLNIFLVNKARFVDKKFNTQFSLNYELKDS 251
Query: 310 CRDLHPGPVSLLHWSGSGKPW 330
+ +H+ G KPW
Sbjct: 252 VINPVDAETVFVHYIGPTKPW 272
>LAGE_HUMAN (O95461) Glycosyltransferase-like protein LARGE (EC
2.4.-.-) (Acetylglucosaminyltransferase-like protein)
Length = 756
Score = 42.7 bits (99), Expect = 0.001
Identities = 50/209 (23%), Positives = 88/209 (41%), Gaps = 35/209 (16%)
Query: 59 NADQC--EPISREIGACHPSLVHVAITLDVEYLRGSIAAVHSILYHASCPENVFFHFLVT 116
N+ +C +P+ + H ++V R + V S+L+H P + FH L+
Sbjct: 122 NSSECGQQPVVEKCETIHVAIVCAGYNAS----RDVVTLVKSVLFHRRNP--LHFH-LIA 174
Query: 117 DTDLETLVRTTF-----PQLRFKVYYFDRNIVKNLISTSVRQALEQPLNYARNYLADLLE 171
D+ E ++ T F P +R Y D +K+ +S + + L L
Sbjct: 175 DSIAEQILATLFQTWMVPAVRVDFYNADE--LKSEVSWIPNKHYSGIYGLMKLVLTKTLP 232
Query: 172 SCVKRVIYLDSDLVLQDDIAKLW---NTDLGLNTIGAPQYCHANFTKYFTAAFWSD---- 224
+ ++RVI LD+D+ DIA+LW + G +G + N + ++ W +
Sbjct: 233 ANLERVIVLDTDITFATDIAELWAVFHKFKGQQVLGLVE----NQSDWYLGNLWKNHRPW 288
Query: 225 PVFSTTFEKRKACYFNTGVMVMDLVKWRK 253
P +NTGV+++ L K RK
Sbjct: 289 PALGRG--------YNTGVILLLLDKLRK 309
>LAGE_MOUSE (Q9Z1M7) Glycosyltransferase-like protein LARGE (EC
2.4.-.-) (Acetylglucosaminyltransferase-like protein)
Length = 756
Score = 42.4 bits (98), Expect = 0.002
Identities = 49/189 (25%), Positives = 81/189 (41%), Gaps = 30/189 (15%)
Query: 78 VHVAITL-DVEYLRGSIAAVHSILYHASCPENVFFHFLVTDTDLETLVRTTF-----PQL 131
+HVAI R + V S+L+H P + FH L+ D+ E ++ T F P +
Sbjct: 138 IHVAIVCAGYNASRDVVTLVKSVLFHRRNP--LHFH-LIADSIAEQILATLFQTWMVPAV 194
Query: 132 RFKVYYFDRNIVKNLISTSVRQALEQPLNYARNYLADLLESCVKRVIYLDSDLVLQDDIA 191
R Y D +K+ +S + + L L + ++RVI LD+D+ DIA
Sbjct: 195 RVDFYNADE--LKSEVSWIPNKHYSGIYGLMKLVLTKTLPANLERVIVLDTDITFATDIA 252
Query: 192 KLW---NTDLGLNTIGAPQYCHANFTKYFTAAFWSD----PVFSTTFEKRKACYFNTGVM 244
+LW + G +G + N + ++ W + P +NTGV+
Sbjct: 253 ELWAVFHKFKGQQVLGLVE----NQSDWYLGNLWKNHRPWPALGRG--------YNTGVI 300
Query: 245 VMDLVKWRK 253
++ L K RK
Sbjct: 301 LLLLDKLRK 309
>UGG2_HUMAN (Q9NYU1) UDP-glucose:glycoprotein glucosyltransferase 2
precursor (EC 2.4.1.-) (UDP--Glc:glycoprotein
glucosyltransferase 2) (UGT 2) (HUGT2)
Length = 1516
Score = 37.7 bits (86), Expect = 0.045
Identities = 32/130 (24%), Positives = 62/130 (47%), Gaps = 6/130 (4%)
Query: 134 KVYYFDRNIVKNLISTSVRQALEQPL---NYARNYLADLLESCVKRVIYLDSDLVLQDDI 190
K Y F +V+ +RQ E+ Y +L L V ++I++D+D +++ D+
Sbjct: 1283 KEYGFRYELVQYRWPRWLRQQTERQRIIWGYKILFLDVLFPLAVDKIIFVDADQIVRHDL 1342
Query: 191 AKLWNTDLGLNTIGAPQYCHANFTKYFTAAFWSDPVFSTTFEKRKACYFNTGVMVMDLVK 250
+L + DL G +C + + FW +++ +RK Y + + V+DL K
Sbjct: 1343 KELRDFDLDGAPYGYTPFCDSR-REMDGYRFWKTGYWASHLLRRK--YHISALYVVDLKK 1399
Query: 251 WRKKGYTERI 260
+R+ G +R+
Sbjct: 1400 FRRIGAGDRL 1409
>RFAI_ECOLI (P27128) Lipopolysaccharide 1,3-galactosyltransferase
(EC 2.4.1.44) (Lipopolysaccharide
3-alpha-galactosyltransferase)
Length = 339
Score = 36.2 bits (82), Expect = 0.13
Identities = 40/200 (20%), Positives = 78/200 (39%), Gaps = 13/200 (6%)
Query: 132 RFKVYYFDRNIVKNLIST-SVRQALEQPLNYARNYLADLLESCVKRVIYLDSDLVLQDDI 190
R K+Y + + +++L ST + A+ Y R +AD + +V+YLD+D++ Q I
Sbjct: 86 RIKIYLINGDRLRSLPSTKNWTHAI-----YFRFVIADYFINKAPKVLYLDADIICQGTI 140
Query: 191 AKLWNTDLGLNTIGAPQYCHANFTKYFTAAFWSDPVFSTTFEKRKACYFNTGVMVMDLVK 250
L N P A A +W S YFN+G ++++ +
Sbjct: 141 EPLIN-------FSFPDDKVAMVVTEGQADWWEKRAHSLGVAGIAKGYFNSGFLLINTAQ 193
Query: 251 WRKKGYTERIERWMEIQKVERIYELGSLPPFLLVFAGHVAAIEHRWNQHGLGGDNVKGSC 310
W + + R + ++ + ++ A + + ++N +K S
Sbjct: 194 WAAQQVSARAIAMLNEPEIIKKITHPDQDVLNMLLADKLIFADIKYNTQFSLNYQLKESF 253
Query: 311 RDLHPGPVSLLHWSGSGKPW 330
+ +H+ G KPW
Sbjct: 254 INPVTNDTIFIHYIGPTKPW 273
>UGGG_DROME (Q09332) UDP-glucose:glycoprotein glucosyltransferase
precursor (EC 2.4.1.-) (UDP--Glc:glycoprotein
glucosyltransferase) (dUGT)
Length = 1548
Score = 35.8 bits (81), Expect = 0.17
Identities = 21/87 (24%), Positives = 46/87 (52%), Gaps = 3/87 (3%)
Query: 174 VKRVIYLDSDLVLQDDIAKLWNTDLGLNTIGAPQYCHANFTKYFTAAFWSDPVFSTTFEK 233
V+++I++D+D +++ DI +L++ DLG +C + + FW + +
Sbjct: 1335 VRKIIFVDADAIVRTDIKELYDMDLGGAPYAYTPFCDSR-KEMEGFRFWKQGYWRSHLMG 1393
Query: 234 RKACYFNTGVMVMDLVKWRKKGYTERI 260
R+ Y + + V+DL ++RK +R+
Sbjct: 1394 RR--YHISALYVVDLKRFRKIAAGDRL 1418
>ABP_HUMAN (P19801) Amiloride-sensitive amine oxidase
[copper-containing] precursor (EC 1.4.3.6) (Diamine
oxidase) (DAO) (Amiloride-binding protein) (ABP)
(Histaminase) (Kidney amine oxidase) (KAO)
Length = 751
Score = 32.0 bits (71), Expect = 2.5
Identities = 48/192 (25%), Positives = 75/192 (39%), Gaps = 29/192 (15%)
Query: 70 IGACHPSLVHVAITLDVEYLRGSIAAVHSILYHASCPENVFFHFLVTDTDLETLVRTTFP 129
IG H LVH + LDV + S + L + + P + H +V TL +T +
Sbjct: 506 IGNIHTHLVHYRVDLDVAGTKNSFQTLQMKLENITNPWSP-RHRVVQ----PTLEQTQYS 560
Query: 130 QLRFKVYYFDRNIVKNLISTSVRQALEQPLNYARNYLADLLESCVKRVIYLDSDLVL--- 186
R + F R + K L+ TS + E P + R+Y + I+ +D VL
Sbjct: 561 WERQAAFRFKRKLPKYLLFTSPQ---ENPWGHKRSY---------RLQIHSMADQVLPPG 608
Query: 187 -QDDIAKLW-NTDLGLNTIGAPQYCHANFTKYFTAAFWSDPVFSTTFEKRKACYFNTGVM 244
Q++ A W L + + C ++ Y W PV F + N +
Sbjct: 609 WQEEQAITWARYPLAVTKYRESELCSSSI--YHQNDPWHPPVVFEQF-----LHNNENIE 661
Query: 245 VMDLVKWRKKGY 256
DLV W G+
Sbjct: 662 NEDLVAWVTVGF 673
>UGGG_SCHPO (Q09140) UDP-glucose:glycoprotein glucosyltransferase
precursor (EC 2.4.1.-) (UDP--Glc:glycoprotein
glucosyltransferase) (UGT)
Length = 1448
Score = 31.2 bits (69), Expect = 4.3
Identities = 22/91 (24%), Positives = 43/91 (47%), Gaps = 3/91 (3%)
Query: 176 RVIYLDSDLVLQDDIAKLWNTDLGLNTIGAPQYCHANFTKYFTAAFWSDPVFSTTFEKRK 235
+VIY+D+D +++ D+ +L + DL G C + + FW + K
Sbjct: 1254 KVIYVDADQIVRADLQELMDMDLHGAPYGYTPMCDSR-EEMEGFRFWKKGYWKKFLRGLK 1312
Query: 236 ACYFNTGVMVMDLVKWRKKGYTERIERWMEI 266
Y + + V+DL ++RK G + + R ++
Sbjct: 1313 --YHISALYVVDLDRFRKMGAGDLLRRQYQL 1341
>SP24_BOVIN (Q27967) Secreted phosphoprotein 24 precursor (Spp-24)
(Secreted phosphoprotein 2)
Length = 200
Score = 30.8 bits (68), Expect = 5.6
Identities = 24/77 (31%), Positives = 33/77 (42%), Gaps = 3/77 (3%)
Query: 133 FKVYYFDRNIVKNLISTSVRQALEQPLNYARNYLADLLESCVKRVIYLDSDLVLQDDIAK 192
F VY +D +K +S SV + Q L+ YL S VKRV LD D + D +
Sbjct: 21 FPVYDYDPASLKEALSASVAKVNSQSLS---PYLFRAFRSSVKRVNALDEDSLTMDLEFR 77
Query: 193 LWNTDLGLNTIGAPQYC 209
+ T + P C
Sbjct: 78 IQETTCRRESEADPATC 94
>HSLV_HELHP (Q7VJD3) ATP-dependent protease hslV (EC 3.4.25.-)
Length = 180
Score = 30.8 bits (68), Expect = 5.6
Identities = 20/65 (30%), Positives = 28/65 (42%), Gaps = 7/65 (10%)
Query: 250 KWRKKGYTERIERWMEIQKVERIYELGSLPPFLLVFAGHVAAIEHRWNQHGLGGDNVKGS 309
+WRK Y ++E M + E IY L + G +AAI G GG+ +
Sbjct: 89 EWRKDKYLRKLEAMMIVLNTEHIYILSGTGDVVEPEDGTIAAI-------GSGGNYALSA 141
Query: 310 CRDLH 314
R LH
Sbjct: 142 ARALH 146
>GBF1_HUMAN (Q92538) Golgi-specific brefeldin A-resistance guanine
nucleotide exchange factor 1 (BFA-resistant GEF 1)
Length = 1859
Score = 30.8 bits (68), Expect = 5.6
Identities = 15/76 (19%), Positives = 35/76 (45%)
Query: 99 ILYHASCPENVFFHFLVTDTDLETLVRTTFPQLRFKVYYFDRNIVKNLISTSVRQALEQP 158
+L+ + PE +F DL+ T P + Y FD+++ + +I ++ +
Sbjct: 903 LLHRGATPEGIFLRVPTASYDLDLFTMTWGPTIAALSYVFDKSLEETIIQKAISGFRKCA 962
Query: 159 LNYARNYLADLLESCV 174
+ A L+D+ ++ +
Sbjct: 963 MISAHYGLSDVFDNLI 978
>KSGA_CHLMU (Q9PK40) Dimethyladenosine transferase (EC 2.1.1.-)
(S-adenosylmethionine-6-N', N'-adenosyl(rRNA)
dimethyltransferase) (16S rRNA dimethylase) (High level
kasugamycin resistance protein ksgA) (Kasugamycin
dimethyltransferase)
Length = 277
Score = 30.4 bits (67), Expect = 7.3
Identities = 15/46 (32%), Positives = 25/46 (53%), Gaps = 3/46 (6%)
Query: 2 SSIIKFSRFFSAVMTVIIISPSLHSSHYPALAIRSSIIHHIPHHDY 47
SS+ F RFF+ V +SP+ YP ++ S+++H H D+
Sbjct: 164 SSLTVFLRFFADVQYAFKVSPNCF---YPKPSVSSAVVHMRVHEDF 206
>GP2_HUMAN (P55259) Pancreatic secretory granule membrane major
glycoprotein GP2 precursor (Pancreatic zymogen granule
membrane protein GP-2) (ZAP75)
Length = 527
Score = 30.4 bits (67), Expect = 7.3
Identities = 22/67 (32%), Positives = 31/67 (45%), Gaps = 3/67 (4%)
Query: 151 VRQALEQPLNYARNYLADLLESCVKRVIYLDSDLVLQDDIAKLWNTDLGLNTIGAPQYCH 210
VR AL Q NY Y D +E V+ V+Y+ + +L+ +N L N P
Sbjct: 343 VRMALFQDQNYTNPYEGDAVELSVESVLYVGA--ILEQGDTSRFNLVL-RNCYATPTEDK 399
Query: 211 ANFTKYF 217
A+ KYF
Sbjct: 400 ADLVKYF 406
>PDXJ_NEIMC (Q9RML6) Pyridoxal phosphate biosynthetic protein pdxJ
(PNP synthase)
Length = 242
Score = 30.0 bits (66), Expect = 9.5
Identities = 37/160 (23%), Positives = 71/160 (44%), Gaps = 13/160 (8%)
Query: 139 DRNIVKNLISTSVRQALEQPLNYARNYLADLLESCVKRVIYLDSDLVLQDDIAKLWNTDL 198
DR +K+ +V+ A+ LN ++LE+ +K + D+ + + + T+
Sbjct: 46 DRRHIKDADVFAVKNAIRTRLNLEMALTEEMLENALK---VMPEDVCIVPEKRQEITTEG 102
Query: 199 GLNTIGAPQYCHANFTKYFTAAFWSDPVFSTTFEKRKACYFNTGVMVMDLVKWRKKGYTE 258
GL+ + A Q A FTK T A +F +++ ++ G V++L Y +
Sbjct: 103 GLDVL-AQQEKIAGFTKILTDAGIRVSLFIDADDRQIQAAYDVGAPVVEL---HTGAYAD 158
Query: 259 RIERWMEIQKVERI----YELGSLPPFLLVFAGHVAAIEH 294
++++ ER+ + G L L+V AGH I +
Sbjct: 159 ARSHAEQLKQFERLQNGAHFAGDLG--LVVNAGHGLTIHN 196
>HSLV_WOLSU (Q7M8Z6) ATP-dependent protease hslV (EC 3.4.25.-)
Length = 176
Score = 30.0 bits (66), Expect = 9.5
Identities = 28/95 (29%), Positives = 39/95 (40%), Gaps = 11/95 (11%)
Query: 220 AFWSDPVFSTTFEKRKACYFNTGVMVMDLVK-WRKKGYTERIERWMEIQKVERIYELGSL 278
AF +F E RK + V+D K WRK Y R+E M + ER++ L
Sbjct: 57 AFTLFEMFERILENRKGDLVKS---VIDFSKEWRKDKYLRRLEAMMIVMDRERLFILSGT 113
Query: 279 PPFLLVFAGHVAAIEHRWNQHGLGGDNVKGSCRDL 313
+ G +AAI G GG+ + R L
Sbjct: 114 GDVVEPEDGKIAAI-------GSGGNYALSAARAL 141
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.326 0.140 0.449
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 44,981,132
Number of Sequences: 164201
Number of extensions: 1980960
Number of successful extensions: 5077
Number of sequences better than 10.0: 22
Number of HSP's better than 10.0 without gapping: 7
Number of HSP's successfully gapped in prelim test: 15
Number of HSP's that attempted gapping in prelim test: 5059
Number of HSP's gapped (non-prelim): 23
length of query: 360
length of database: 59,974,054
effective HSP length: 111
effective length of query: 249
effective length of database: 41,747,743
effective search space: 10395188007
effective search space used: 10395188007
T: 11
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 66 (30.0 bits)
Medicago: description of AC145329.11


