

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
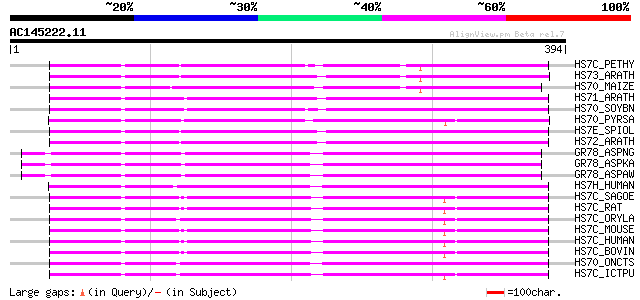
Query= AC145222.11 + phase: 0 /pseudo
(394 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
HS7C_PETHY (P09189) Heat shock cognate 70 kDa protein 186 7e-47
HS73_ARATH (O65719) Heat shock cognate 70 kDa protein 3 (Hsc70.3) 185 2e-46
HS70_MAIZE (P11143) Heat shock 70 kDa protein 184 3e-46
HS71_ARATH (P22953) Heat shock cognate 70 kDa protein 1 (Hsc70.1) 184 4e-46
HS70_SOYBN (P26413) Heat shock 70 kDa protein 183 6e-46
HS70_PYRSA (P37899) Heat shock 70 kDa protein 183 6e-46
HS7E_SPIOL (P29357) Chloroplast envelope membrane 70 kDa heat sh... 182 1e-45
HS72_ARATH (P22954) Heat shock cognate 70 kDa protein 2 (Hsc70.2) 182 2e-45
GR78_ASPNG (P83616) 78 kDa glucose-regulated protein homolog pre... 180 5e-45
GR78_ASPKA (P83617) 78 kDa glucose-regulated protein homolog pre... 180 5e-45
GR78_ASPAW (P59769) 78 kDa glucose-regulated protein homolog pre... 180 5e-45
HS7H_HUMAN (P34931) Heat shock 70 kDa protein 1-HOM (HSP70-HOM) 180 6e-45
HS7C_SAGOE (Q71U34) Heat shock cognate 71 kDa protein (Intracell... 179 8e-45
HS7C_RAT (P63018) Heat shock cognate 71 kDa protein 179 8e-45
HS7C_ORYLA (Q9W6Y1) Heat shock cognate 71 kDa protein (Hsc70.1) 179 8e-45
HS7C_MOUSE (P63017) Heat shock cognate 71 kDa protein 179 8e-45
HS7C_HUMAN (P11142) Heat shock cognate 71 kDa protein 179 8e-45
HS7C_BOVIN (P19120) Heat shock cognate 71 kDa protein 179 8e-45
HS70_ONCTS (Q91233) Heat shock 70 kDa protein (HSP70) 179 1e-44
HS7C_ICTPU (P47773) Heat shock cognate 71 kDa protein 179 1e-44
>HS7C_PETHY (P09189) Heat shock cognate 70 kDa protein
Length = 651
Score = 186 bits (473), Expect = 7e-47
Identities = 124/361 (34%), Positives = 198/361 (54%), Gaps = 20/361 (5%)
Query: 29 AIGIDIGTSQCSVAVWNGSEVELLKNKRNQKLMKSFVTFKDEAPSGGVTSQFSHEHEMLF 88
AIGID+GT+ V VW VE++ N + + S+V F D G ++ ++ M
Sbjct: 9 AIGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVGFTDTERLIGDAAK--NQVAMNP 66
Query: 89 GDTIFNMKRLIGRVDTDPVVHASKNL-PFLVQTLDIGVRPFIAALVNNVWRSTTPEEVLA 147
+T+F+ KRLIGR +DP V + L PF V G +P I + EE+ +
Sbjct: 67 INTVFDAKRLIGRRFSDPSVQSDIKLWPFKVIP-GPGDKPMIVVTYKGEEKQFAAEEISS 125
Query: 148 MFLVELRLMTETHLKRPIRNVVLTVPVSFSRFQLTRLERACAMAGLHVLRLMPEPTAVAL 207
M L +++ + E +L I+N V+TVP F+ Q + A +AGL+V+R++ EPTA A+
Sbjct: 126 MVLTKMKEIAEAYLGTTIKNAVVTVPAYFNDSQRQATKDAGVIAGLNVMRIINEPTAAAI 185
Query: 208 LYGQQQQKASQETMGSGSEKIALIFNMGAGYCDVAVTATAGGVSQIKALAGST-IGGEDL 266
YG +KAS S EK LIF++G G DV++ G+ ++KA AG T +GGED
Sbjct: 186 AYG-LDKKAS-----SAGEKNVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGEDF 239
Query: 267 LQNMMRHLLPDSENIFKRHGVEEI----KSMALLRVATQDAIHQLSTQSSVQVDVD-LGN 321
M+ H + + FKR ++I +++ LR A + A LS+ + +++D L
Sbjct: 240 DNRMVNHFVQE----FKRKNKKDISGNPRALRRLRTACERAKRTLSSTAQTTIEIDSLYE 295
Query: 322 SLKICKVVDRAEFEEVNKEVFEKCESLIIQCLHDAKVEVGDINDVILVGGCSYIPQGWRI 381
+ + RA FEE+N ++F KC + +CL DAK++ ++DV+LVGG + IP+ ++
Sbjct: 296 GIDFYSTITRARFEELNMDLFRKCMEPVEKCLRDAKMDKSSVHDVVLVGGSTRIPKVQQL 355
Query: 382 L 382
L
Sbjct: 356 L 356
>HS73_ARATH (O65719) Heat shock cognate 70 kDa protein 3 (Hsc70.3)
Length = 649
Score = 185 bits (470), Expect = 2e-46
Identities = 121/362 (33%), Positives = 200/362 (54%), Gaps = 20/362 (5%)
Query: 29 AIGIDIGTSQCSVAVWNGSEVELLKNKRNQKLMKSFVTFKDEAPSGGVTSQFSHEHEMLF 88
AIGID+GT+ V VW VE++ N + + S+V F D G ++ ++ M
Sbjct: 9 AIGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVAFTDSERLIGDAAK--NQVAMNP 66
Query: 89 GDTIFNMKRLIGRVDTDPVVHASKNL-PFLVQTLDIGVRPFIAALVNNVWRSTTPEEVLA 147
+T+F+ KRLIGR TD V + L PF +++ +P I + + EE+ +
Sbjct: 67 INTVFDAKRLIGRRFTDSSVQSDIKLWPFTLKS-GPAEKPMIVVNYKGEDKEFSAEEISS 125
Query: 148 MFLVELRLMTETHLKRPIRNVVLTVPVSFSRFQLTRLERACAMAGLHVLRLMPEPTAVAL 207
M L+++R + E +L I+N V+TVP F+ Q + A +AGL+V+R++ EPTA A+
Sbjct: 126 MILIKMREIAEAYLGTTIKNAVVTVPAYFNDSQRQATKDAGVIAGLNVMRIINEPTAAAI 185
Query: 208 LYGQQQQKASQETMGSGSEKIALIFNMGAGYCDVAVTATAGGVSQIKALAGST-IGGEDL 266
YG ++ S EK LIF++G G DV++ G+ ++KA AG T +GGED
Sbjct: 186 AYGLDKKATSV------GEKNVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGEDF 239
Query: 267 LQNMMRHLLPDSENIFKRHGVEEI----KSMALLRVATQDAIHQLSTQSSVQVDVD-LGN 321
M+ H + + FKR ++I +++ LR A + A LS+ + +++D L +
Sbjct: 240 DNRMVNHFVQE----FKRKNKKDISGNPRALRRLRTACERAKRTLSSTAQTTIEIDSLFD 295
Query: 322 SLKICKVVDRAEFEEVNKEVFEKCESLIIQCLHDAKVEVGDINDVILVGGCSYIPQGWRI 381
+ + RA FEE+N ++F KC + +CL DAK++ I+DV+LVGG + IP+ ++
Sbjct: 296 GIDFYAPITRARFEELNIDLFRKCMEPVEKCLRDAKMDKNSIDDVVLVGGSTRIPKVQQL 355
Query: 382 LL 383
L+
Sbjct: 356 LV 357
>HS70_MAIZE (P11143) Heat shock 70 kDa protein
Length = 645
Score = 184 bits (467), Expect = 3e-46
Identities = 117/355 (32%), Positives = 194/355 (53%), Gaps = 19/355 (5%)
Query: 29 AIGIDIGTSQCSVAVWNGSEVELLKNKRNQKLMKSFVTFKDEAPSGGVTSQFSHEHEMLF 88
AIGID+GT+ V +W VE++ N + + S+V F D G ++ ++ M
Sbjct: 8 AIGIDLGTTYSCVGLWQHDRVEIIANDQGNRTTPSYVGFTDTERLIGDAAK--NQVAMNP 65
Query: 89 GDTIFNMKRLIGRVDTDPVVHASKNLPFLVQTLDIGVRPFIAALVNNVWRSTTPEEVLAM 148
+T+F+ KRLIGR + P V +S L + + L +G +P I + EE+ +M
Sbjct: 66 TNTVFDAKRLIGRRFSSPAVQSSMKL-WPSRHLGLGDKPMIVFNYKGEEKQFAAEEISSM 124
Query: 149 FLVELRLMTETHLKRPIRNVVLTVPVSFSRFQLTRLERACAMAGLHVLRLMPEPTAVALL 208
L++++ + E +L I+N V+TVP F+ Q + A +AGL+V+R++ EPTA A+
Sbjct: 125 VLIKMKEIAEAYLGSTIKNAVVTVPAYFNDSQRQATKDAGVIAGLNVMRIINEPTAAAIA 184
Query: 209 YGQQQQKASQETMGSGSEKIALIFNMGAGYCDVAVTATAGGVSQIKALAGST-IGGEDLL 267
YG ++ S EK LIF++G G DV++ G+ ++KA AG T +GGED
Sbjct: 185 YGLDKKAT------SSGEKNVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGEDFD 238
Query: 268 QNMMRHLLPDSENIFKRHGVEEI----KSMALLRVATQDAIHQLSTQSSVQVDVD-LGNS 322
M+ H + + FKR ++I +++ LR A + A LS+ + +++D L
Sbjct: 239 NRMVNHFVQE----FKRKNKKDISGNPRALRRLRTACERAKRTLSSTAQTTIEIDSLFEG 294
Query: 323 LKICKVVDRAEFEEVNKEVFEKCESLIIQCLHDAKVEVGDINDVILVGGCSYIPQ 377
+ RA FEE+N ++F KC + +CL DAK++ ++DV+LVGG + IP+
Sbjct: 295 IDFTPRSSRARFEELNMDLFRKCMEPVEKCLRDAKMDKSSVHDVVLVGGSTRIPK 349
>HS71_ARATH (P22953) Heat shock cognate 70 kDa protein 1 (Hsc70.1)
Length = 651
Score = 184 bits (466), Expect = 4e-46
Identities = 116/357 (32%), Positives = 193/357 (53%), Gaps = 12/357 (3%)
Query: 29 AIGIDIGTSQCSVAVWNGSEVELLKNKRNQKLMKSFVTFKDEAPSGGVTSQFSHEHEMLF 88
AIGID+GT+ V VW VE++ N + + S+V F D G ++ ++ M
Sbjct: 9 AIGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVAFTDSERLIGDAAK--NQVAMNP 66
Query: 89 GDTIFNMKRLIGRVDTDPVVHASKNL-PFLVQTLDIGVRPFIAALVNNVWRSTTPEEVLA 147
+T+F+ KRLIGR +D V + L PF +Q +P I + EE+ +
Sbjct: 67 VNTVFDAKRLIGRRFSDSSVQSDMKLWPFKIQAGPAD-KPMIYVEYKGEEKEFAAEEISS 125
Query: 148 MFLVELRLMTETHLKRPIRNVVLTVPVSFSRFQLTRLERACAMAGLHVLRLMPEPTAVAL 207
M L+++R + E +L I+N V+TVP F+ Q + A +AGL+V+R++ EPTA A+
Sbjct: 126 MVLIKMREIAEAYLGVTIKNAVVTVPAYFNDSQRQATKDAGVIAGLNVMRIINEPTAAAI 185
Query: 208 LYGQQQQKASQETMGSGSEKIALIFNMGAGYCDVAVTATAGGVSQIKALAGST-IGGEDL 266
YG ++ S EK LIF++G G DV++ G+ ++KA AG T +GGED
Sbjct: 186 AYGLDKKATSV------GEKNVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGEDF 239
Query: 267 LQNMMRHLLPDSENIFKRHGVEEIKSMALLRVATQDAIHQLSTQSSVQVDVD-LGNSLKI 325
M+ H + + + K+ +++ LR + + A LS+ + +++D L +
Sbjct: 240 DNRMVNHFVQEFKRKSKKDITGNPRALRRLRTSCERAKRTLSSTAQTTIEIDSLYEGIDF 299
Query: 326 CKVVDRAEFEEVNKEVFEKCESLIIQCLHDAKVEVGDINDVILVGGCSYIPQGWRIL 382
+ RA FEE+N ++F KC + +CL DAK++ ++DV+LVGG + IP+ ++L
Sbjct: 300 YSTITRARFEELNMDLFRKCMEPVEKCLRDAKMDKSTVHDVVLVGGSTRIPKVQQLL 356
>HS70_SOYBN (P26413) Heat shock 70 kDa protein
Length = 645
Score = 183 bits (465), Expect = 6e-46
Identities = 118/357 (33%), Positives = 195/357 (54%), Gaps = 12/357 (3%)
Query: 29 AIGIDIGTSQCSVAVWNGSEVELLKNKRNQKLMKSFVTFKDEAPSGGVTSQFSHEHEMLF 88
AIGID+GT+ V VW VE++ N + + S+V F D G ++ ++ M
Sbjct: 8 AIGIDLGTTYSCVGVWQNDRVEIIPNDQGNRTTPSYVAFTDTERLIGDAAK--NQVAMNP 65
Query: 89 GDTIFNMKRLIGRVDTDPVVHASKNL-PFLVQTLDIGVRPFIAALVNNVWRSTTPEEVLA 147
+T+F+ KRLIGR +D V L PF V +P I + + EE+ +
Sbjct: 66 QNTVFDAKRLIGRRFSDSSVQNDMKLWPFKVGGSPCD-KPMIVVNYKGEEKKFSAEEISS 124
Query: 148 MFLVELRLMTETHLKRPIRNVVLTVPVSFSRFQLTRLERACAMAGLHVLRLMPEPTAVAL 207
M LV++R + E L ++N V+TVP F+ Q + A A++GL+VLR++ EPTA A+
Sbjct: 125 MVLVKMREVAEAFLGHAVKNAVVTVPAYFNDSQRQATKDAGAISGLNVLRIINEPTAAAI 184
Query: 208 LYGQQQQKASQETMGSGSEKIALIFNMGAGYCDVAVTATAGGVSQIKALAGST-IGGEDL 266
YG +KAS++ E+ LIF++G G DV++ G+ ++KA AG T +GGED
Sbjct: 185 AYG-LDKKASRK-----GEQNVLIFDLGGGTFDVSILTIEEGIFEVKATAGDTHLGGEDF 238
Query: 267 LQNMMRHLLPDSENIFKRHGVEEIKSMALLRVATQDAIHQLSTQSSVQVDVD-LGNSLKI 325
M+ H + + + K+ +++ LR A + A LS+ + +++D L +
Sbjct: 239 DNRMVNHFVSEFKRKNKKDISGNARALRRLRTACERAKRTLSSTAQTTIEIDSLYEGIDF 298
Query: 326 CKVVDRAEFEEVNKEVFEKCESLIIQCLHDAKVEVGDINDVILVGGCSYIPQGWRIL 382
+ RA FEE+N ++F KC + +CL DAK++ +++V+LVGG + IP+ ++L
Sbjct: 299 YATITRARFEEMNMDLFRKCMEPVEKCLRDAKIDKSQVHEVVLVGGSTRIPKVHQLL 355
>HS70_PYRSA (P37899) Heat shock 70 kDa protein
Length = 649
Score = 183 bits (465), Expect = 6e-46
Identities = 118/360 (32%), Positives = 201/360 (55%), Gaps = 13/360 (3%)
Query: 28 IAIGIDIGTSQCSVAVWNGSEVELLKNKRNQKLMKSFVTFKDEAPSGGVTSQFSHEHEML 87
+AIGID+GT+ V VW VE++ N + + S+V F + G +++ ++ M
Sbjct: 8 VAIGIDLGTTYSCVGVWLHDRVEVIANDQGNRTTPSYVAFTETERLIGDSAK--NQVAMN 65
Query: 88 FGDTIFNMKRLIGRVDTDPVVHAS-KNLPFLVQTLDIGVRPFIAALVNNVWRSTTPEEVL 146
+T+F+ KRLIGR DP V K PF V D G +P + + PEE+
Sbjct: 66 PDNTVFDAKRLIGRRFQDPAVQEDVKPFPFKVICKD-GDKPAVEVKYKGETKIFAPEEIS 124
Query: 147 AMFLVELRLMTETHLKRPIRNVVLTVPVSFSRFQLTRLERACAMAGLHVLRLMPEPTAVA 206
+M L++++ + E+ L + ++N V+TVP F+ Q + A A+ GL+VLR++ EPTA A
Sbjct: 125 SMVLLKMKEIAESFLGKEVKNAVITVPAYFNDSQRQATKDAGAITGLNVLRIINEPTAAA 184
Query: 207 LLYGQQQQKASQETMGSGSEKIALIFNMGAGYCDVAVTATAGGVSQIKALAGST-IGGED 265
+ YG ++T GS SE+ LIF++G G DV++ G+ ++KA AG T +GGED
Sbjct: 185 IAYG-----LDKKTSGSKSERNVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGED 239
Query: 266 LLQNMMRHLLPDSENIFKRHGVEEIKSMALLRVATQDAIHQLS--TQSSVQVDVDLGNSL 323
++ + + + +K+ +S+ LR A + A LS TQ++V++D L + +
Sbjct: 240 FDSRLVNFFVSEFKRKYKKDVTSNARSLRRLRTACERAKRTLSSGTQTTVEID-SLIDGI 298
Query: 324 KICKVVDRAEFEEVNKEVFEKCESLIIQCLHDAKVEVGDINDVILVGGCSYIPQGWRILL 383
+ RA+FEE+ ++F + + L D+K+ +I+DV+LVGG + IP+ ++L+
Sbjct: 299 DYYASITRAKFEELCMDLFRGTSEPVEKVLRDSKISKSEIHDVVLVGGSTRIPKVQQLLI 358
>HS7E_SPIOL (P29357) Chloroplast envelope membrane 70 kDa heat
shock-related protein
Length = 652
Score = 182 bits (463), Expect = 1e-45
Identities = 118/357 (33%), Positives = 194/357 (54%), Gaps = 12/357 (3%)
Query: 29 AIGIDIGTSQCSVAVWNGSEVELLKNKRNQKLMKSFVTFKDEAPSGGVTSQFSHEHEMLF 88
AIGID+GT+ V VW VE++ N + + S+V F D G ++ ++ M
Sbjct: 8 AIGIDLGTTYSRVGVWQHDRVEIIANDQGNRTTPSYVAFTDSERLIGDAAK--NQVAMNP 65
Query: 89 GDTIFNMKRLIGRVDTDPVVHAS-KNLPFLVQTLDIGVRPFIAALVNNVWRSTTPEEVLA 147
+T+F+ KRLIGR +D V A K+ PF V + G +P I + EE+ +
Sbjct: 66 INTVFDAKRLIGRRFSDASVQADMKHRPFKVVS-GPGEKPMIGVNYKGEEKQFAAEEISS 124
Query: 148 MFLVELRLMTETHLKRPIRNVVLTVPVSFSRFQLTRLERACAMAGLHVLRLMPEPTAVAL 207
M L +++ + E +L ++N V+TVP F+ Q + A ++GL+V+R++ EPTA A+
Sbjct: 125 MVLTKMKEIAEAYLGSTVKNAVVTVPAYFNDSQRQATKDAGVISGLNVMRIINEPTAAAI 184
Query: 208 LYGQQQQKASQETMGSGSEKIALIFNMGAGYCDVAVTATAGGVSQIKALAGST-IGGEDL 266
YG ++ S EK LIF++G G DV++ G+ ++KA AG T +GGED
Sbjct: 185 AYGLDKKATSV------GEKNVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGEDF 238
Query: 267 LQNMMRHLLPDSENIFKRHGVEEIKSMALLRVATQDAIHQLSTQSSVQVDVD-LGNSLKI 325
M+ H L + + K+ +E + LR A + A LS+ + +++D L +
Sbjct: 239 DNRMVNHSLQEFKRKNKKDIMETPGHIRRLRTACERAKRTLSSTAQTTIEIDSLYEGVDF 298
Query: 326 CKVVDRAEFEEVNKEVFEKCESLIIQCLHDAKVEVGDINDVILVGGCSYIPQGWRIL 382
+ RA FEE+N ++F KC + +CL DAK++ ++DV+LVGG + IP+ ++L
Sbjct: 299 YSPITRARFEELNIDLFRKCMEPVEKCLRDAKMDKSTVHDVVLVGGSTRIPKVQQLL 355
>HS72_ARATH (P22954) Heat shock cognate 70 kDa protein 2 (Hsc70.2)
Length = 653
Score = 182 bits (461), Expect = 2e-45
Identities = 114/357 (31%), Positives = 193/357 (53%), Gaps = 12/357 (3%)
Query: 29 AIGIDIGTSQCSVAVWNGSEVELLKNKRNQKLMKSFVTFKDEAPSGGVTSQFSHEHEMLF 88
AIGID+GT+ V VW VE++ N + + S+V F D G ++ ++ M
Sbjct: 9 AIGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVAFTDSERLIGDAAK--NQVAMNP 66
Query: 89 GDTIFNMKRLIGRVDTDPVVHASKNL-PFLVQTLDIGVRPFIAALVNNVWRSTTPEEVLA 147
+T+F+ KRLIGR +D V + + L PF + + +P I + EE+ +
Sbjct: 67 VNTVFDAKRLIGRRFSDASVQSDRQLWPFTIIS-GTAEKPMIVVEYKGEEKQFAAEEISS 125
Query: 148 MFLVELRLMTETHLKRPIRNVVLTVPVSFSRFQLTRLERACAMAGLHVLRLMPEPTAVAL 207
M L+++R + E L ++N V+TVP F+ Q + A +AGL+VLR++ EPTA A+
Sbjct: 126 MVLIKMREIAEAFLGTTVKNAVVTVPAYFNDSQRQATKDAGVIAGLNVLRIINEPTAAAI 185
Query: 208 LYGQQQQKASQETMGSGSEKIALIFNMGAGYCDVAVTATAGGVSQIKALAGST-IGGEDL 266
YG ++ S EK LIF++G G DV++ G+ ++KA AG T +GGED
Sbjct: 186 AYGLDKKATSV------GEKNVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGEDF 239
Query: 267 LQNMMRHLLPDSENIFKRHGVEEIKSMALLRVATQDAIHQLSTQSSVQVDVD-LGNSLKI 325
M+ H + + + K+ + +++ LR A + A LS+ + +++D L
Sbjct: 240 DNRMVNHFVQEFKRKNKQDITGQPRALRRLRTACERAKRTLSSTAQTTIEIDSLYGGADF 299
Query: 326 CKVVDRAEFEEVNKEVFEKCESLIIQCLHDAKVEVGDINDVILVGGCSYIPQGWRIL 382
+ RA FEE+N ++F KC + +CL DAK++ +++++LVGG + IP+ ++L
Sbjct: 300 YSPITRARFEEMNMDLFRKCMEPVEKCLRDAKMDKSTVHEIVLVGGSTRIPKVQQLL 356
>GR78_ASPNG (P83616) 78 kDa glucose-regulated protein homolog
precursor (GRP 78) (Immunoglobulin heavy chain binding
protein homolog) (BiP)
Length = 672
Score = 180 bits (457), Expect = 5e-45
Identities = 119/372 (31%), Positives = 205/372 (54%), Gaps = 20/372 (5%)
Query: 9 TVASDSETTGEEKSSPLPEIAIGIDIGTSQCSVAVWNGSEVELLKNKRNQKLMKSFVTFK 68
T + ET+ +E + IGID+GT+ V V +VE+L N + ++ S+V F
Sbjct: 35 TAHAQDETSPQESYGTV----IGIDLGTTYSCVGVMQNGKVEILVNDQGNRITPSYVAFT 90
Query: 69 DEAPSGGVTSQFSHEHEMLFGDTIFNMKRLIGR-VDTDPVVHASKNLPFLVQTLDIGVRP 127
DE G ++ +++ TIF++KRLIGR D V +K+ P+ V D +P
Sbjct: 91 DEERLVGDAAK--NQYAANPRRTIFDIKRLIGRKFDDKDVQKDAKHFPYKVVNKD--GKP 146
Query: 128 FIAALVNNVWRSTTPEEVLAMFLVELRLMTETHLKRPIRNVVLTVPVSFSRFQLTRLERA 187
+ VN ++ TPEEV AM L +++ + E +L + + + V+TVP F+ Q + A
Sbjct: 147 HVKVDVNQTPKTLTPEEVSAMVLGKMKEIAEGYLGKKVTHAVVTVPAYFNDAQRQATKDA 206
Query: 188 CAMAGLHVLRLMPEPTAVALLYGQQQQKASQETMGSGSEKIALIFNMGAGYCDVAVTATA 247
+AGL+VLR++ EPTA A+ YG + +G E+ +++++G G DV++ +
Sbjct: 207 GTIAGLNVLRVVNEPTAAAIAYGLDK---------TGDERQVIVYDLGGGTFDVSLLSID 257
Query: 248 GGVSQIKALAGST-IGGEDLLQNMMRHLLPDSENIFKRHGVEEIKSMALLRVATQDAIHQ 306
GV ++ A AG T +GGED Q +M H + +++K+M L+ + A
Sbjct: 258 NGVFEVLATAGDTHLGGEDFDQRVMDHFVKLYNKKNNVDVTKDLKAMGKLKREVEKAKRT 317
Query: 307 LSTQSSVQVDVD-LGNSLKICKVVDRAEFEEVNKEVFEKCESLIIQCLHDAKVEVGDIND 365
LS+Q S +++++ N + + RA+FEE+N ++F+K + Q L DAKV+ +++D
Sbjct: 318 LSSQMSTRIEIEAFHNGEDFSETLTRAKFEELNMDLFKKTLKPVEQVLKDAKVKKSEVDD 377
Query: 366 VILVGGCSYIPQ 377
++LVGG + IP+
Sbjct: 378 IVLVGGSTRIPK 389
>GR78_ASPKA (P83617) 78 kDa glucose-regulated protein homolog
precursor (GRP 78) (Immunoglobulin heavy chain binding
protein homolog) (BiP)
Length = 672
Score = 180 bits (457), Expect = 5e-45
Identities = 119/372 (31%), Positives = 205/372 (54%), Gaps = 20/372 (5%)
Query: 9 TVASDSETTGEEKSSPLPEIAIGIDIGTSQCSVAVWNGSEVELLKNKRNQKLMKSFVTFK 68
T + ET+ +E + IGID+GT+ V V +VE+L N + ++ S+V F
Sbjct: 35 TAHAQDETSPQESYGTV----IGIDLGTTYSCVGVMQNGKVEILVNDQGNRITPSYVAFT 90
Query: 69 DEAPSGGVTSQFSHEHEMLFGDTIFNMKRLIGR-VDTDPVVHASKNLPFLVQTLDIGVRP 127
DE G ++ +++ TIF++KRLIGR D V +K+ P+ V D +P
Sbjct: 91 DEERLVGDAAK--NQYAANPRRTIFDIKRLIGRKFDDKDVQKDAKHFPYKVVNKD--GKP 146
Query: 128 FIAALVNNVWRSTTPEEVLAMFLVELRLMTETHLKRPIRNVVLTVPVSFSRFQLTRLERA 187
+ VN ++ TPEEV AM L +++ + E +L + + + V+TVP F+ Q + A
Sbjct: 147 HVKVDVNQTPKTLTPEEVSAMVLGKMKEIAEGYLGKKVTHAVVTVPAYFNDAQRQATKDA 206
Query: 188 CAMAGLHVLRLMPEPTAVALLYGQQQQKASQETMGSGSEKIALIFNMGAGYCDVAVTATA 247
+AGL+VLR++ EPTA A+ YG + +G E+ +++++G G DV++ +
Sbjct: 207 GTIAGLNVLRVVNEPTAAAIAYGLDK---------TGDERQVIVYDLGGGTFDVSLLSID 257
Query: 248 GGVSQIKALAGST-IGGEDLLQNMMRHLLPDSENIFKRHGVEEIKSMALLRVATQDAIHQ 306
GV ++ A AG T +GGED Q +M H + +++K+M L+ + A
Sbjct: 258 NGVFEVLATAGDTHLGGEDFDQRVMDHFVKLYNKKNNVDVTKDLKAMGKLKREVEKAKRT 317
Query: 307 LSTQSSVQVDVD-LGNSLKICKVVDRAEFEEVNKEVFEKCESLIIQCLHDAKVEVGDIND 365
LS+Q S +++++ N + + RA+FEE+N ++F+K + Q L DAKV+ +++D
Sbjct: 318 LSSQMSTRIEIEAFHNGEDFSETLTRAKFEELNMDLFKKTLKPVEQVLKDAKVKKSEVDD 377
Query: 366 VILVGGCSYIPQ 377
++LVGG + IP+
Sbjct: 378 IVLVGGSTRIPK 389
>GR78_ASPAW (P59769) 78 kDa glucose-regulated protein homolog
precursor (GRP 78) (Immunoglobulin heavy chain binding
protein homolog) (BiP)
Length = 672
Score = 180 bits (457), Expect = 5e-45
Identities = 119/372 (31%), Positives = 205/372 (54%), Gaps = 20/372 (5%)
Query: 9 TVASDSETTGEEKSSPLPEIAIGIDIGTSQCSVAVWNGSEVELLKNKRNQKLMKSFVTFK 68
T + ET+ +E + IGID+GT+ V V +VE+L N + ++ S+V F
Sbjct: 35 TAHAQDETSPQESYGTV----IGIDLGTTYSCVGVMQNGKVEILVNDQGNRITPSYVAFT 90
Query: 69 DEAPSGGVTSQFSHEHEMLFGDTIFNMKRLIGR-VDTDPVVHASKNLPFLVQTLDIGVRP 127
DE G ++ +++ TIF++KRLIGR D V +K+ P+ V D +P
Sbjct: 91 DEERLVGDAAK--NQYAANPRRTIFDIKRLIGRKFDDKDVQKDAKHFPYKVVNKD--GKP 146
Query: 128 FIAALVNNVWRSTTPEEVLAMFLVELRLMTETHLKRPIRNVVLTVPVSFSRFQLTRLERA 187
+ VN ++ TPEEV AM L +++ + E +L + + + V+TVP F+ Q + A
Sbjct: 147 HVKVDVNQTPKTLTPEEVSAMVLGKMKEIAEGYLGKKVTHAVVTVPAYFNDAQRQATKDA 206
Query: 188 CAMAGLHVLRLMPEPTAVALLYGQQQQKASQETMGSGSEKIALIFNMGAGYCDVAVTATA 247
+AGL+VLR++ EPTA A+ YG + +G E+ +++++G G DV++ +
Sbjct: 207 GTIAGLNVLRVVNEPTAAAIAYGLDK---------TGDERQVIVYDLGGGTFDVSLLSID 257
Query: 248 GGVSQIKALAGST-IGGEDLLQNMMRHLLPDSENIFKRHGVEEIKSMALLRVATQDAIHQ 306
GV ++ A AG T +GGED Q +M H + +++K+M L+ + A
Sbjct: 258 NGVFEVLATAGDTHLGGEDFDQRVMDHFVKLYNKKNNVDVTKDLKAMGKLKREVEKAKRT 317
Query: 307 LSTQSSVQVDVD-LGNSLKICKVVDRAEFEEVNKEVFEKCESLIIQCLHDAKVEVGDIND 365
LS+Q S +++++ N + + RA+FEE+N ++F+K + Q L DAKV+ +++D
Sbjct: 318 LSSQMSTRIEIEAFHNGEDFSETLTRAKFEELNMDLFKKTLKPVEQVLKDAKVKKSEVDD 377
Query: 366 VILVGGCSYIPQ 377
++LVGG + IP+
Sbjct: 378 IVLVGGSTRIPK 389
>HS7H_HUMAN (P34931) Heat shock 70 kDa protein 1-HOM (HSP70-HOM)
Length = 641
Score = 180 bits (456), Expect = 6e-45
Identities = 117/358 (32%), Positives = 193/358 (53%), Gaps = 15/358 (4%)
Query: 28 IAIGIDIGTSQCSVAVWNGSEVELLKNKRNQKLMKSFVTFKDEAPSGGVTSQFSHEHEML 87
IAIGID+GT+ V V+ +VE++ N + + S+V F D G ++ ++ M
Sbjct: 7 IAIGIDLGTTYSCVGVFQHGKVEIIANDQGNRTTPSYVAFTDTERLIGDAAK--NQVAMN 64
Query: 88 FGDTIFNMKRLIGRVDTDPVVHASKNL-PFLVQTLDIGVRPFIAALVNNVWRSTTPEEVL 146
+T+F+ KRLIGR DPVV A L PF Q ++ G +P + ++ PEE+
Sbjct: 65 PQNTVFDAKRLIGRKFNDPVVQADMKLWPF--QVINEGGKPKVLVSYKGENKAFYPEEIS 122
Query: 147 AMFLVELRLMTETHLKRPIRNVVLTVPVSFSRFQLTRLERACAMAGLHVLRLMPEPTAVA 206
+M L +L+ E L P+ N V+TVP F+ Q + A +AGL+VLR++ EPTA A
Sbjct: 123 SMVLTKLKETAEAFLGHPVTNAVITVPAYFNDSQRQATKDAGVIAGLNVLRIINEPTAAA 182
Query: 207 LLYGQQQQKASQETMGSGSEKIALIFNMGAGYCDVAVTATAGGVSQIKALAGST-IGGED 265
+ YG + G E+ LIF++G G DV++ G+ ++KA AG T +GGED
Sbjct: 183 IAYGLDK--------GGQGERHVLIFDLGGGTFDVSILTIDDGIFEVKATAGDTHLGGED 234
Query: 266 LLQNMMRHLLPDSENIFKRHGVEEIKSMALLRVATQDAIHQLSTQSSVQVDVD-LGNSLK 324
++ H + + + K+ + +++ LR A + A LS+ + +++D L +
Sbjct: 235 FDNRLVSHFVEEFKRKHKKDISQNKRAVRRLRTACERAKRTLSSSTQANLEIDSLYEGID 294
Query: 325 ICKVVDRAEFEEVNKEVFEKCESLIIQCLHDAKVEVGDINDVILVGGCSYIPQGWRIL 382
+ RA FEE+ ++F + + L DAK++ I+D++LVGG + IP+ R+L
Sbjct: 295 FYTSITRARFEELCADLFRGTLEPVEKALRDAKMDKAKIHDIVLVGGSTRIPKVQRLL 352
>HS7C_SAGOE (Q71U34) Heat shock cognate 71 kDa protein
(Intracellular vitamin D binding protein 1)
Length = 646
Score = 179 bits (455), Expect = 8e-45
Identities = 119/358 (33%), Positives = 199/358 (55%), Gaps = 17/358 (4%)
Query: 29 AIGIDIGTSQCSVAVWNGSEVELLKNKRNQKLMKSFVTFKDEAPSGGVTSQFSHEHEMLF 88
A+GID+GT+ V V+ +VE++ N + + S+V F D G ++ ++ M
Sbjct: 6 AVGIDLGTTYSCVGVFQHGKVEIIANDQGNRTTPSYVAFTDTERLIGDAAK--NQVAMNP 63
Query: 89 GDTIFNMKRLIGRVDTDPVVHAS-KNLPFLVQTLDIGVRPFIAALVNNVWRSTTPEEVLA 147
+T+F+ KRLIGR D VV + K+ PF+V D G RP + +S PEEV +
Sbjct: 64 TNTVFDAKRLIGRRFDDAVVQSDMKHWPFMVVN-DAG-RPKVQVEYKGETKSFYPEEVSS 121
Query: 148 MFLVELRLMTETHLKRPIRNVVLTVPVSFSRFQLTRLERACAMAGLHVLRLMPEPTAVAL 207
M L +++ + E +L + + N V+TVP F+ Q + A +AGL+VLR++ EPTA A+
Sbjct: 122 MVLTKMKEIAEAYLGKTVTNAVVTVPAYFNDSQRQATKDAGTIAGLNVLRIINEPTAAAI 181
Query: 208 LYGQQQQKASQETMGSGSEKIALIFNMGAGYCDVAVTATAGGVSQIKALAGST-IGGEDL 266
YG ++ G+E+ LIF++G G DV++ G+ ++K+ AG T +GGED
Sbjct: 182 AYGLDKK--------VGAERNVLIFDLGGGTFDVSILTIEDGIFEVKSTAGDTHLGGEDF 233
Query: 267 LQNMMRHLLPDSENIFKRHGVEEIKSMALLRVATQDAIHQL--STQSSVQVDVDLGNSLK 324
M+ H + + + K+ E +++ LR A + A L STQ+S+++D L +
Sbjct: 234 DNRMVNHFIAEFKRKHKKDISENKRAVRRLRTACERAKRTLSSSTQASIEID-SLYEGID 292
Query: 325 ICKVVDRAEFEEVNKEVFEKCESLIIQCLHDAKVEVGDINDVILVGGCSYIPQGWRIL 382
+ RA FEE+N ++F + + L DAK++ I+D++LVGG + IP+ ++L
Sbjct: 293 FYTSITRARFEELNADLFRGTLDPVEKALRDAKLDKSQIHDIVLVGGSTRIPKIQKLL 350
>HS7C_RAT (P63018) Heat shock cognate 71 kDa protein
Length = 646
Score = 179 bits (455), Expect = 8e-45
Identities = 119/358 (33%), Positives = 199/358 (55%), Gaps = 17/358 (4%)
Query: 29 AIGIDIGTSQCSVAVWNGSEVELLKNKRNQKLMKSFVTFKDEAPSGGVTSQFSHEHEMLF 88
A+GID+GT+ V V+ +VE++ N + + S+V F D G ++ ++ M
Sbjct: 6 AVGIDLGTTYSCVGVFQHGKVEIIANDQGNRTTPSYVAFTDTERLIGDAAK--NQVAMNP 63
Query: 89 GDTIFNMKRLIGRVDTDPVVHAS-KNLPFLVQTLDIGVRPFIAALVNNVWRSTTPEEVLA 147
+T+F+ KRLIGR D VV + K+ PF+V D G RP + +S PEEV +
Sbjct: 64 TNTVFDAKRLIGRRFDDAVVQSDMKHWPFMVVN-DAG-RPKVQVEYKGETKSFYPEEVSS 121
Query: 148 MFLVELRLMTETHLKRPIRNVVLTVPVSFSRFQLTRLERACAMAGLHVLRLMPEPTAVAL 207
M L +++ + E +L + + N V+TVP F+ Q + A +AGL+VLR++ EPTA A+
Sbjct: 122 MVLTKMKEIAEAYLGKTVTNAVVTVPAYFNDSQRQATKDAGTIAGLNVLRIINEPTAAAI 181
Query: 208 LYGQQQQKASQETMGSGSEKIALIFNMGAGYCDVAVTATAGGVSQIKALAGST-IGGEDL 266
YG ++ G+E+ LIF++G G DV++ G+ ++K+ AG T +GGED
Sbjct: 182 AYGLDKK--------VGAERNVLIFDLGGGTFDVSILTIEDGIFEVKSTAGDTHLGGEDF 233
Query: 267 LQNMMRHLLPDSENIFKRHGVEEIKSMALLRVATQDAIHQL--STQSSVQVDVDLGNSLK 324
M+ H + + + K+ E +++ LR A + A L STQ+S+++D L +
Sbjct: 234 DNRMVNHFIAEFKRKHKKDISENKRAVRRLRTACERAKRTLSSSTQASIEID-SLYEGID 292
Query: 325 ICKVVDRAEFEEVNKEVFEKCESLIIQCLHDAKVEVGDINDVILVGGCSYIPQGWRIL 382
+ RA FEE+N ++F + + L DAK++ I+D++LVGG + IP+ ++L
Sbjct: 293 FYTSITRARFEELNADLFRGTLDPVEKALRDAKLDKSQIHDIVLVGGSTRIPKIQKLL 350
>HS7C_ORYLA (Q9W6Y1) Heat shock cognate 71 kDa protein (Hsc70.1)
Length = 686
Score = 179 bits (455), Expect = 8e-45
Identities = 117/358 (32%), Positives = 199/358 (54%), Gaps = 17/358 (4%)
Query: 29 AIGIDIGTSQCSVAVWNGSEVELLKNKRNQKLMKSFVTFKDEAPSGGVTSQFSHEHEMLF 88
A+GID+GT+ V V+ +VE++ N + + S+V F D G ++ ++ M
Sbjct: 6 AVGIDLGTTYSCVGVFQHGKVEIIANDQGNRTTPSYVAFTDTERLIGDAAK--NQVAMNP 63
Query: 89 GDTIFNMKRLIGRVDTDPVVHASKN-LPFLVQTLDIGVRPFIAALVNNVWRSTTPEEVLA 147
+T+F+ KRLIGR D VV + N PF V ++ RP + +S PEEV +
Sbjct: 64 TNTVFDAKRLIGRRFDDHVVQSDMNDWPFNV--INDNTRPKVQVEYKGETKSFYPEEVSS 121
Query: 148 MFLVELRLMTETHLKRPIRNVVLTVPVSFSRFQLTRLERACAMAGLHVLRLMPEPTAVAL 207
M L +++ + E +L + + N V+TVP F+ Q + A ++GL+VLR++ EPTA A+
Sbjct: 122 MVLTKMKEIAEAYLGKTVNNAVITVPAYFNDSQRQATKDAGTISGLNVLRIINEPTAAAI 181
Query: 208 LYGQQQQKASQETMGSGSEKIALIFNMGAGYCDVAVTATAGGVSQIKALAGST-IGGEDL 266
YG ++ GSE+ LIF++G G DV++ G+ ++K+ AG T +GGED
Sbjct: 182 AYGLDKK--------VGSERNVLIFDLGGGTFDVSILTIEDGIFEVKSTAGDTHLGGEDF 233
Query: 267 LQNMMRHLLPDSENIFKRHGVEEIKSMALLRVATQDAIHQL--STQSSVQVDVDLGNSLK 324
M+ H + + + +K+ + +++ LR A + A L STQ+S+++D L +
Sbjct: 234 DNRMVNHFIAEFKRKYKKDISDNKRAVRRLRSACERAKRTLSSSTQASIEID-SLYEGVD 292
Query: 325 ICKVVDRAEFEEVNKEVFEKCESLIIQCLHDAKVEVGDINDVILVGGCSYIPQGWRIL 382
+ RA FEE+N ++F + + L DAK++ G I+D++LVGG + IP+ ++L
Sbjct: 293 FYTSITRARFEELNADLFRGTLDPVEKSLRDAKMDKGQIHDIVLVGGSTRIPKIQKLL 350
>HS7C_MOUSE (P63017) Heat shock cognate 71 kDa protein
Length = 646
Score = 179 bits (455), Expect = 8e-45
Identities = 119/358 (33%), Positives = 199/358 (55%), Gaps = 17/358 (4%)
Query: 29 AIGIDIGTSQCSVAVWNGSEVELLKNKRNQKLMKSFVTFKDEAPSGGVTSQFSHEHEMLF 88
A+GID+GT+ V V+ +VE++ N + + S+V F D G ++ ++ M
Sbjct: 6 AVGIDLGTTYSCVGVFQHGKVEIIANDQGNRTTPSYVAFTDTERLIGDAAK--NQVAMNP 63
Query: 89 GDTIFNMKRLIGRVDTDPVVHAS-KNLPFLVQTLDIGVRPFIAALVNNVWRSTTPEEVLA 147
+T+F+ KRLIGR D VV + K+ PF+V D G RP + +S PEEV +
Sbjct: 64 TNTVFDAKRLIGRRFDDAVVQSDMKHWPFMVVN-DAG-RPKVQVEYKGETKSFYPEEVSS 121
Query: 148 MFLVELRLMTETHLKRPIRNVVLTVPVSFSRFQLTRLERACAMAGLHVLRLMPEPTAVAL 207
M L +++ + E +L + + N V+TVP F+ Q + A +AGL+VLR++ EPTA A+
Sbjct: 122 MVLTKMKEIAEAYLGKTVTNAVVTVPAYFNDSQRQATKDAGTIAGLNVLRIINEPTAAAI 181
Query: 208 LYGQQQQKASQETMGSGSEKIALIFNMGAGYCDVAVTATAGGVSQIKALAGST-IGGEDL 266
YG ++ G+E+ LIF++G G DV++ G+ ++K+ AG T +GGED
Sbjct: 182 AYGLDKK--------VGAERNVLIFDLGGGTFDVSILTIEDGIFEVKSTAGDTHLGGEDF 233
Query: 267 LQNMMRHLLPDSENIFKRHGVEEIKSMALLRVATQDAIHQL--STQSSVQVDVDLGNSLK 324
M+ H + + + K+ E +++ LR A + A L STQ+S+++D L +
Sbjct: 234 DNRMVNHFIAEFKRKHKKDISENKRAVRRLRTACERAKRTLSSSTQASIEID-SLYEGID 292
Query: 325 ICKVVDRAEFEEVNKEVFEKCESLIIQCLHDAKVEVGDINDVILVGGCSYIPQGWRIL 382
+ RA FEE+N ++F + + L DAK++ I+D++LVGG + IP+ ++L
Sbjct: 293 FYTSITRARFEELNADLFRGTLDPVEKALRDAKLDKSQIHDIVLVGGSTRIPKIQKLL 350
>HS7C_HUMAN (P11142) Heat shock cognate 71 kDa protein
Length = 646
Score = 179 bits (455), Expect = 8e-45
Identities = 119/358 (33%), Positives = 199/358 (55%), Gaps = 17/358 (4%)
Query: 29 AIGIDIGTSQCSVAVWNGSEVELLKNKRNQKLMKSFVTFKDEAPSGGVTSQFSHEHEMLF 88
A+GID+GT+ V V+ +VE++ N + + S+V F D G ++ ++ M
Sbjct: 6 AVGIDLGTTYSCVGVFQHGKVEIIANDQGNRTTPSYVAFTDTERLIGDAAK--NQVAMNP 63
Query: 89 GDTIFNMKRLIGRVDTDPVVHAS-KNLPFLVQTLDIGVRPFIAALVNNVWRSTTPEEVLA 147
+T+F+ KRLIGR D VV + K+ PF+V D G RP + +S PEEV +
Sbjct: 64 TNTVFDAKRLIGRRFDDAVVQSDMKHWPFMVVN-DAG-RPKVQVEYKGETKSFYPEEVSS 121
Query: 148 MFLVELRLMTETHLKRPIRNVVLTVPVSFSRFQLTRLERACAMAGLHVLRLMPEPTAVAL 207
M L +++ + E +L + + N V+TVP F+ Q + A +AGL+VLR++ EPTA A+
Sbjct: 122 MVLTKMKEIAEAYLGKTVTNAVVTVPAYFNDSQRQATKDAGTIAGLNVLRIINEPTAAAI 181
Query: 208 LYGQQQQKASQETMGSGSEKIALIFNMGAGYCDVAVTATAGGVSQIKALAGST-IGGEDL 266
YG ++ G+E+ LIF++G G DV++ G+ ++K+ AG T +GGED
Sbjct: 182 AYGLDKK--------VGAERNVLIFDLGGGTFDVSILTIEDGIFEVKSTAGDTHLGGEDF 233
Query: 267 LQNMMRHLLPDSENIFKRHGVEEIKSMALLRVATQDAIHQL--STQSSVQVDVDLGNSLK 324
M+ H + + + K+ E +++ LR A + A L STQ+S+++D L +
Sbjct: 234 DNRMVNHFIAEFKRKHKKDISENKRAVRRLRTACERAKRTLSSSTQASIEID-SLYEGID 292
Query: 325 ICKVVDRAEFEEVNKEVFEKCESLIIQCLHDAKVEVGDINDVILVGGCSYIPQGWRIL 382
+ RA FEE+N ++F + + L DAK++ I+D++LVGG + IP+ ++L
Sbjct: 293 FYTSITRARFEELNADLFRGTLDPVEKALRDAKLDKSQIHDIVLVGGSTRIPKIQKLL 350
>HS7C_BOVIN (P19120) Heat shock cognate 71 kDa protein
Length = 650
Score = 179 bits (455), Expect = 8e-45
Identities = 119/358 (33%), Positives = 199/358 (55%), Gaps = 17/358 (4%)
Query: 29 AIGIDIGTSQCSVAVWNGSEVELLKNKRNQKLMKSFVTFKDEAPSGGVTSQFSHEHEMLF 88
A+GID+GT+ V V+ +VE++ N + + S+V F D G ++ ++ M
Sbjct: 6 AVGIDLGTTYSCVGVFQHGKVEIIANDQGNRTTPSYVAFTDTERLIGDAAK--NQVAMNP 63
Query: 89 GDTIFNMKRLIGRVDTDPVVHAS-KNLPFLVQTLDIGVRPFIAALVNNVWRSTTPEEVLA 147
+T+F+ KRLIGR D VV + K+ PF+V D G RP + +S PEEV +
Sbjct: 64 TNTVFDAKRLIGRRFDDAVVQSDMKHWPFMVVN-DAG-RPKVQVEYKGETKSFYPEEVSS 121
Query: 148 MFLVELRLMTETHLKRPIRNVVLTVPVSFSRFQLTRLERACAMAGLHVLRLMPEPTAVAL 207
M L +++ + E +L + + N V+TVP F+ Q + A +AGL+VLR++ EPTA A+
Sbjct: 122 MVLTKMKEIAEAYLGKTVTNAVVTVPAYFNDSQRQATKDAGTIAGLNVLRIINEPTAAAI 181
Query: 208 LYGQQQQKASQETMGSGSEKIALIFNMGAGYCDVAVTATAGGVSQIKALAGST-IGGEDL 266
YG ++ G+E+ LIF++G G DV++ G+ ++K+ AG T +GGED
Sbjct: 182 AYGLDKK--------VGAERNVLIFDLGGGTFDVSILTIEDGIFEVKSTAGDTHLGGEDF 233
Query: 267 LQNMMRHLLPDSENIFKRHGVEEIKSMALLRVATQDAIHQL--STQSSVQVDVDLGNSLK 324
M+ H + + + K+ E +++ LR A + A L STQ+S+++D L +
Sbjct: 234 DNRMVNHFIAEFKRKHKKDISENKRAVRRLRTACERAKRTLSSSTQASIEID-SLYEGID 292
Query: 325 ICKVVDRAEFEEVNKEVFEKCESLIIQCLHDAKVEVGDINDVILVGGCSYIPQGWRIL 382
+ RA FEE+N ++F + + L DAK++ I+D++LVGG + IP+ ++L
Sbjct: 293 FYTSITRARFEELNADLFRGTLDPVEKALRDAKLDKSQIHDIVLVGGSTRIPKIQKLL 350
>HS70_ONCTS (Q91233) Heat shock 70 kDa protein (HSP70)
Length = 644
Score = 179 bits (454), Expect = 1e-44
Identities = 116/357 (32%), Positives = 194/357 (53%), Gaps = 15/357 (4%)
Query: 29 AIGIDIGTSQCSVAVWNGSEVELLKNKRNQKLMKSFVTFKDEAPSGGVTSQFSHEHEMLF 88
+IGID+GT+ V V+ +VE++ N + + S+V F D G ++ ++ M
Sbjct: 8 SIGIDLGTTYSCVGVFQHGKVEIIANDQGNRTTPSYVAFTDTERLIGDAAK--NQVAMNP 65
Query: 89 GDTIFNMKRLIGRVDTDPVVHAS-KNLPFLVQTLDIGVRPFIAALVNNVWRSTTPEEVLA 147
+T+F+ KRLIGR D VV A K+ PF V + G +P + +S PEE+ +
Sbjct: 66 NNTVFDAKRLIGRKFNDQVVQADMKHWPFKV--VSDGGKPKVQVDYKGENKSFNPEEISS 123
Query: 148 MFLVELRLMTETHLKRPIRNVVLTVPVSFSRFQLTRLERACAMAGLHVLRLMPEPTAVAL 207
M LV++R + E +L + + N V+TVP F+ Q + A +AGL+VLR++ EPTA A+
Sbjct: 124 MVLVKMREIAEAYLGQKVSNAVITVPAYFNDSQRQATKDAGVIAGLNVLRIINEPTAAAI 183
Query: 208 LYGQQQQKASQETMGSGSEKIALIFNMGAGYCDVAVTATAGGVSQIKALAGST-IGGEDL 266
YG + G E+ LIF++G G DV++ G+ ++KA AG T +GGED
Sbjct: 184 AYGMDK--------GMSRERNVLIFDLGGGTFDVSILTIEDGIFEVKATAGDTHLGGEDF 235
Query: 267 LQNMMRHLLPDSENIFKRHGVEEIKSMALLRVATQDAIHQLSTQSSVQVDVD-LGNSLKI 325
++ H + + + K+ + +++ LR A + A LS+ S +++D L +
Sbjct: 236 DNRLVSHFVEEFKRKHKKDISQNKRALRRLRTACERAKRTLSSSSQASIEIDSLFEGIDF 295
Query: 326 CKVVDRAEFEEVNKEVFEKCESLIIQCLHDAKVEVGDINDVILVGGCSYIPQGWRIL 382
+ RA FEE+ ++F + + L DAK++ I+DV+LVGG + IP+ ++L
Sbjct: 296 YTSITRARFEEMCSDLFRGTLEPVEKALRDAKMDKAQIHDVVLVGGSTRIPKVQKLL 352
>HS7C_ICTPU (P47773) Heat shock cognate 71 kDa protein
Length = 649
Score = 179 bits (453), Expect = 1e-44
Identities = 118/358 (32%), Positives = 198/358 (54%), Gaps = 17/358 (4%)
Query: 29 AIGIDIGTSQCSVAVWNGSEVELLKNKRNQKLMKSFVTFKDEAPSGGVTSQFSHEHEMLF 88
A+GID+GT+ V V+ +VE++ N + + S+V F D G ++ ++ M
Sbjct: 6 AVGIDLGTTYSCVGVFQHGKVEIIANDQGNRTTPSYVAFTDSERLIGDAAK--NQVAMNP 63
Query: 89 GDTIFNMKRLIGRVDTDPVVHAS-KNLPFLVQTLDIGVRPFIAALVNNVWRSTTPEEVLA 147
+TIF+ KRLIGR D VV A K+ PF V + G RP + ++ PEE+ +
Sbjct: 64 TNTIFDAKRLIGRRFEDSVVQADMKHWPFKV--ISDGGRPRLEVEYKGEAKNFYPEEISS 121
Query: 148 MFLVELRLMTETHLKRPIRNVVLTVPVSFSRFQLTRLERACAMAGLHVLRLMPEPTAVAL 207
M LV+++ + E +L + I N V+TVP F+ Q R + A ++GL+VLR++ EPTA A+
Sbjct: 122 MVLVKMKEIAEAYLGKSINNAVITVPAYFNDSQRQRTKDAGTISGLNVLRIINEPTAAAI 181
Query: 208 LYGQQQQKASQETMGSGSEKIALIFNMGAGYCDVAVTATAGGVSQIKALAGST-IGGEDL 266
YG ++ GSE+ LIF++G G DV++ G+ +K+ AG T +GGED
Sbjct: 182 AYGLDKK--------VGSERNVLIFDLGGGTFDVSILTIEDGIFDLKSTAGDTHLGGEDF 233
Query: 267 LQNMMRHLLPDSENIFKRHGVEEIKSMALLRVATQDAIHQL--STQSSVQVDVDLGNSLK 324
M+ H + + + K+ + +++ L A + A L STQ+S+++D L +
Sbjct: 234 DNRMVNHFIAEFKRKHKKDISDNKRAVRRLATACERAKRTLSSSTQASIEID-SLYEGVD 292
Query: 325 ICKVVDRAEFEEVNKEVFEKCESLIIQCLHDAKVEVGDINDVILVGGCSYIPQGWRIL 382
+ RA FEE+N ++F + + L DAK++ ++D++LVGG + IP+ ++L
Sbjct: 293 FYTSITRARFEELNADLFRGTLDPVEKALRDAKMDKAQVHDIVLVGGSTRIPKMEKLL 350
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.319 0.134 0.381
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 43,690,167
Number of Sequences: 164201
Number of extensions: 1772803
Number of successful extensions: 6298
Number of sequences better than 10.0: 415
Number of HSP's better than 10.0 without gapping: 401
Number of HSP's successfully gapped in prelim test: 14
Number of HSP's that attempted gapping in prelim test: 4445
Number of HSP's gapped (non-prelim): 498
length of query: 394
length of database: 59,974,054
effective HSP length: 112
effective length of query: 282
effective length of database: 41,583,542
effective search space: 11726558844
effective search space used: 11726558844
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 67 (30.4 bits)
Medicago: description of AC145222.11


