

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC145218.2 + phase: 1 /pseudo/partial
(365 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
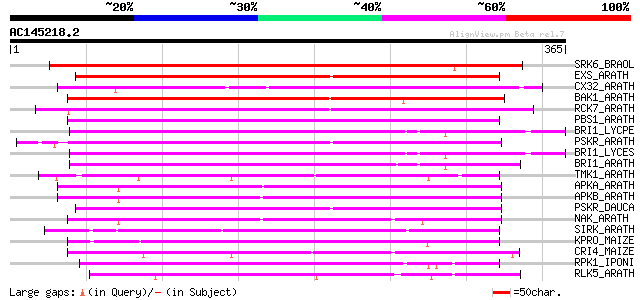
Score E
Sequences producing significant alignments: (bits) Value
SRK6_BRAOL (Q09092) Putative serine/threonine-protein kinase rec... 355 1e-97
EXS_ARATH (Q9LYN8) Leucine-rich repeat receptor protein kinase E... 226 5e-59
CX32_ARATH (P27450) Probable serine/threonine-protein kinase Cx3... 218 2e-56
BAK1_ARATH (Q94F62) BRASSINOSTEROID INSENSITIVE 1-associated rec... 215 2e-55
RCK7_ARATH (Q9LQQ8) Putative serine/threonine-protein kinase RLC... 210 4e-54
PBS1_ARATH (Q9FE20) Serine/threonine-protein kinase PBS1 (EC 2.7... 210 4e-54
BRI1_LYCPE (Q8L899) Systemin receptor SR160 precursor (EC 2.7.1.... 210 5e-54
PSKR_ARATH (Q9ZVR7) Putative phytosulfokine receptor precursor (... 209 9e-54
BRI1_LYCES (Q8GUQ5) Brassinosteroid LRR receptor kinase precurso... 207 3e-53
BRI1_ARATH (O22476) BRASSINOSTEROID INSENSITIVE 1 precursor (EC ... 203 6e-52
TMK1_ARATH (P43298) Putative receptor protein kinase TMK1 precur... 201 2e-51
APKA_ARATH (Q06548) Protein kinase APK1A, chloroplast precursor ... 199 7e-51
APKB_ARATH (P46573) Protein kinase APK1B, chloroplast precursor ... 197 3e-50
PSKR_DAUCA (Q8LPB4) Phytosulfokine receptor precursor (EC 2.7.1.... 195 1e-49
NAK_ARATH (P43293) Probable serine/threonine-protein kinase NAK ... 186 6e-47
SIRK_ARATH (O64483) Senescence-induced receptor-like serine/thre... 181 3e-45
KPRO_MAIZE (P17801) Putative receptor protein kinase ZmPK1 precu... 176 1e-43
CRI4_MAIZE (O24585) Putative receptor protein kinase CRINKLY4 pr... 173 7e-43
RPK1_IPONI (P93194) Receptor-like protein kinase precursor (EC 2... 169 8e-42
RLK5_ARATH (P47735) Receptor-like protein kinase 5 precursor (EC... 169 1e-41
>SRK6_BRAOL (Q09092) Putative serine/threonine-protein kinase
receptor precursor (EC 2.7.1.37) (S-receptor kinase)
(SRK)
Length = 849
Score = 355 bits (911), Expect = 1e-97
Identities = 175/318 (55%), Positives = 223/318 (70%), Gaps = 7/318 (2%)
Query: 27 GENDSESIEVPYYTFRSIQAATNNFSDSNKLGQGGYGPVYKGRFPGGQEIAIKRLSSVST 86
GE E +E+P ++ AT NFS NKLGQGG+G VYKGR G+EIA+KRLS S
Sbjct: 504 GEYKFEELELPLIEMETVVKATENFSSCNKLGQGGFGIVYKGRLLDGKEIAVKRLSKTSV 563
Query: 87 QGLQEFKNEIVLIAKLQHRNLVRLRGYCIKGDEKILLYEYMSNKSLDTFIFDRTRTVLLG 146
QG EF NE+ LIA+LQH NLV++ G CI+GDEK+L+YEY+ N SLD+++F +TR L
Sbjct: 564 QGTDEFMNEVTLIARLQHINLVQVLGCCIEGDEKMLIYEYLENLSLDSYLFGKTRRSKLN 623
Query: 147 WKLRFDIIVGIARGMLYLHQDSRLRVIHRDLKTSNILLDDEMIPKISDFGLAKIFGGKET 206
W RFDI G+ARG+LYLHQDSR R+IHRDLK SNILLD MIPKISDFG+A+IF ET
Sbjct: 624 WNERFDITNGVARGLLYLHQDSRFRIIHRDLKVSNILLDKNMIPKISDFGMARIFERDET 683
Query: 207 GASTQRVMGTYGYMSPEYALDGFFSIKSDVFSFGVVLLEILSGKKNTGFFRSQQISSLLG 266
A+T +V+GTYGYMSPEYA+ G FS KSDVFSFGV++LEI+SGKKN GF+ + LL
Sbjct: 684 EANTMKVVGTYGYMSPEYAMYGIFSEKSDVFSFGVIVLEIVSGKKNRGFYNLDYENDLLS 743
Query: 267 YAWRLWTENKLLDLMDSALSETCNE-------NEFVKCAQIGLLCVQDEPGNRPTMSNIL 319
Y W W E + L+++D + ++ + E +KC QIGLLCVQ+ +RP MS+++
Sbjct: 744 YVWSRWKEGRALEIVDPVIVDSLSSQPSIFQPQEVLKCIQIGLLCVQELAEHRPAMSSVV 803
Query: 320 TMLDGETATIPIPSQPTF 337
M E IP P P +
Sbjct: 804 WMFGSEATEIPQPKPPGY 821
>EXS_ARATH (Q9LYN8) Leucine-rich repeat receptor protein kinase EXS
precursor (EC 2.7.1.37) (Extra sporogenous cells protein)
(EXCESS MICROSPOROCYTES1 protein)
Length = 1192
Score = 226 bits (577), Expect = 5e-59
Identities = 118/281 (41%), Positives = 178/281 (62%), Gaps = 3/281 (1%)
Query: 44 IQAATNNFSDSNKLGQGGYGPVYKGRFPGGQEIAIKRLSSVSTQGLQEFKNEIVLIAKLQ 103
I AT++FS N +G GG+G VYK PG + +A+K+LS TQG +EF E+ + K++
Sbjct: 910 IVEATDHFSKKNIIGDGGFGTVYKACLPGEKTVAVKKLSEAKTQGNREFMAEMETLGKVK 969
Query: 104 HRNLVRLRGYCIKGDEKILLYEYMSNKSLDTFIFDRTRTV-LLGWKLRFDIIVGIARGML 162
H NLV L GYC +EK+L+YEYM N SLD ++ ++T + +L W R I VG ARG+
Sbjct: 970 HPNLVSLLGYCSFSEEKLLVYEYMVNGSLDHWLRNQTGMLEVLDWSKRLKIAVGAARGLA 1029
Query: 163 YLHQDSRLRVIHRDLKTSNILLDDEMIPKISDFGLAKIFGGKETGASTQRVMGTYGYMSP 222
+LH +IHRD+K SNILLD + PK++DFGLA++ E+ ST + GT+GY+ P
Sbjct: 1030 FLHHGFIPHIIHRDIKASNILLDGDFEPKVADFGLARLISACESHVSTV-IAGTFGYIPP 1088
Query: 223 EYALDGFFSIKSDVFSFGVVLLEILSGKKNTG-FFRSQQISSLLGYAWRLWTENKLLDLM 281
EY + K DV+SFGV+LLE+++GK+ TG F+ + +L+G+A + + K +D++
Sbjct: 1089 EYGQSARATTKGDVYSFGVILLELVTGKEPTGPDFKESEGGNLVGWAIQKINQGKAVDVI 1148
Query: 282 DSALSETCNENEFVKCAQIGLLCVQDEPGNRPTMSNILTML 322
D L +N ++ QI +LC+ + P RP M ++L L
Sbjct: 1149 DPLLVSVALKNSQLRLLQIAMLCLAETPAKRPNMLDVLKAL 1189
>CX32_ARATH (P27450) Probable serine/threonine-protein kinase Cx32,
chloroplast precursor (EC 2.7.1.37)
Length = 419
Score = 218 bits (555), Expect = 2e-56
Identities = 128/330 (38%), Positives = 185/330 (55%), Gaps = 17/330 (5%)
Query: 32 ESIEVPYYTFRSIQAATNNFSDSNKLGQGGYGPVYKG----------RFPGGQEIAIKRL 81
ES + Y F ++ AT NF + LGQGG+G VY+G R G +AIKRL
Sbjct: 67 ESPNLKVYNFLDLKTATKNFKPDSMLGQGGFGKVYRGWVDATTLAPSRVGSGMIVAIKRL 126
Query: 82 SSVSTQGLQEFKNEIVLIAKLQHRNLVRLRGYCIKGDEKILLYEYMSNKSLDTFIFDRTR 141
+S S QG E+++E+ + L HRNLV+L GYC + E +L+YE+M SL++ +F R
Sbjct: 127 NSESVQGFAEWRSEVNFLGMLSHRNLVKLLGYCREDKELLLVYEFMPKGSLESHLFRRND 186
Query: 142 TVLLGWKLRFDIIVGIARGMLYLHQDSRLRVIHRDLKTSNILLDDEMIPKISDFGLAKIF 201
W LR I++G ARG+ +LH R VI+RD K SNILLD K+SDFGLAK+
Sbjct: 187 P--FPWDLRIKIVIGAARGLAFLHSLQR-EVIYRDFKASNILLDSNYDAKLSDFGLAKLG 243
Query: 202 GGKETGASTQRVMGTYGYMSPEYALDGFFSIKSDVFSFGVVLLEILSGKKNTGFFRSQQI 261
E T R+MGTYGY +PEY G +KSDVF+FGVVLLEI++G R +
Sbjct: 244 PADEKSHVTTRIMGTYGYAAPEYMATGHLYVKSDVFAFGVVLLEIMTGLTAHNTKRPRGQ 303
Query: 262 SSLLGYAW-RLWTENKLLDLMDSALSETCNENEFVKCAQIGLLCVQDEPGNRPTMSNILT 320
SL+ + L ++++ +MD + + A+I L C++ +P NRP M ++
Sbjct: 304 ESLVDWLRPELSNKHRVKQIMDKGIKGQYTTKVATEMARITLSCIEPDPKNRPHMKEVVE 363
Query: 321 MLDGETATIPIPSQPTFFTTKHQSCSSSSS 350
+L+ +P++ +TK +SS S
Sbjct: 364 VLEHIQGLNVVPNRS---STKQAVANSSRS 390
>BAK1_ARATH (Q94F62) BRASSINOSTEROID INSENSITIVE 1-associated
receptor kinase 1 precursor (EC 2.7.1.37)
(BRI1-associated receptor kinase 1) (Somatic
embryogenesis receptor-like kinase 3)
Length = 615
Score = 215 bits (547), Expect = 2e-55
Identities = 119/291 (40%), Positives = 179/291 (60%), Gaps = 5/291 (1%)
Query: 39 YTFRSIQAATNNFSDSNKLGQGGYGPVYKGRFPGGQEIAIKRLSSVSTQGLQ-EFKNEIV 97
++ R +Q A++NFS+ N LG+GG+G VYKGR G +A+KRL TQG + +F+ E+
Sbjct: 277 FSLRELQVASDNFSNKNILGRGGFGKVYKGRLADGTLVAVKRLKEERTQGGELQFQTEVE 336
Query: 98 LIAKLQHRNLVRLRGYCIKGDEKILLYEYMSNKSLDTFIFDRTRTVL-LGWKLRFDIIVG 156
+I+ HRNL+RLRG+C+ E++L+Y YM+N S+ + + +R + L W R I +G
Sbjct: 337 MISMAVHRNLLRLRGFCMTPTERLLVYPYMANGSVASCLRERPESQPPLDWPKRQRIALG 396
Query: 157 IARGMLYLHQDSRLRVIHRDLKTSNILLDDEMIPKISDFGLAKIFGGKETGASTQRVMGT 216
ARG+ YLH ++IHRD+K +NILLD+E + DFGLAK+ K+T +T V GT
Sbjct: 397 SARGLAYLHDHCDPKIIHRDVKAANILLDEEFEAVVGDFGLAKLMDYKDTHVTT-AVRGT 455
Query: 217 YGYMSPEYALDGFFSIKSDVFSFGVVLLEILSGKKNTGFFR--SQQISSLLGYAWRLWTE 274
G+++PEY G S K+DVF +GV+LLE+++G++ R + LL + L E
Sbjct: 456 IGHIAPEYLSTGKSSEKTDVFGYGVMLLELITGQRAFDLARLANDDDVMLLDWVKGLLKE 515
Query: 275 NKLLDLMDSALSETCNENEFVKCAQIGLLCVQDEPGNRPTMSNILTMLDGE 325
KL L+D L + E + Q+ LLC Q P RP MS ++ ML+G+
Sbjct: 516 KKLEALVDVDLQGNYKDEEVEQLIQVALLCTQSSPMERPKMSEVVRMLEGD 566
>RCK7_ARATH (Q9LQQ8) Putative serine/threonine-protein kinase
RLCKVII (EC 2.7.1.37)
Length = 423
Score = 210 bits (535), Expect = 4e-54
Identities = 126/345 (36%), Positives = 199/345 (57%), Gaps = 19/345 (5%)
Query: 18 RDLIGLGNIGENDSESIEVP--------------YYTFRSIQAATNNFSDSNKLGQGGYG 63
RD+ G +G+ D S++V +TF+ + AT NF LG+GG+G
Sbjct: 56 RDVNNEGGVGKEDQLSLDVKGLNLNDQVTGKKAQTFTFQELAEATGNFRSDCFLGEGGFG 115
Query: 64 PVYKGRFPG-GQEIAIKRLSSVSTQGLQEFKNEIVLIAKLQHRNLVRLRGYCIKGDEKIL 122
V+KG Q +AIK+L QG++EF E++ ++ H NLV+L G+C +GD+++L
Sbjct: 116 KVFKGTIEKLDQVVAIKQLDRNGVQGIREFVVEVLTLSLADHPNLVKLIGFCAEGDQRLL 175
Query: 123 LYEYMSNKSLDTFIFDR-TRTVLLGWKLRFDIIVGIARGMLYLHQDSRLRVIHRDLKTSN 181
+YEYM SL+ + + L W R I G ARG+ YLH VI+RDLK SN
Sbjct: 176 VYEYMPQGSLEDHLHVLPSGKKPLDWNTRMKIAAGAARGLEYLHDRMTPPVIYRDLKCSN 235
Query: 182 ILLDDEMIPKISDFGLAKIF-GGKETGASTQRVMGTYGYMSPEYALDGFFSIKSDVFSFG 240
ILL ++ PK+SDFGLAK+ G +T ST RVMGTYGY +P+YA+ G + KSD++SFG
Sbjct: 236 ILLGEDYQPKLSDFGLAKVGPSGDKTHVST-RVMGTYGYCAPDYAMTGQLTFKSDIYSFG 294
Query: 241 VVLLEILSGKKNTGFFRSQQISSLLGYAWRLWTENK-LLDLMDSALSETCNENEFVKCAQ 299
VVLLE+++G+K ++++ +L+G+A L+ + + ++D L +
Sbjct: 295 VVLLELITGRKAIDNTKTRKDQNLVGWARPLFKDRRNFPKMVDPLLQGQYPVRGLYQALA 354
Query: 300 IGLLCVQDEPGNRPTMSNILTMLDGETATIPIPSQPTFFTTKHQS 344
I +CVQ++P RP +S+++ L+ ++ P+ P+ + K+ S
Sbjct: 355 ISAMCVQEQPTMRPVVSDVVLALNFLASSKYDPNSPSSSSGKNPS 399
>PBS1_ARATH (Q9FE20) Serine/threonine-protein kinase PBS1 (EC
2.7.1.37) (AvrPphB susceptible protein 1)
Length = 456
Score = 210 bits (535), Expect = 4e-54
Identities = 113/287 (39%), Positives = 171/287 (59%), Gaps = 3/287 (1%)
Query: 39 YTFRSIQAATNNFSDSNKLGQGGYGPVYKGRFPG-GQEIAIKRLSSVSTQGLQEFKNEIV 97
+ FR + AAT NF LG+GG+G VYKGR GQ +A+K+L QG +EF E++
Sbjct: 74 FAFRELAAATMNFHPDTFLGEGGFGRVYKGRLDSTGQVVAVKQLDRNGLQGNREFLVEVL 133
Query: 98 LIAKLQHRNLVRLRGYCIKGDEKILLYEYMSNKSLDTFIFDRTRTV-LLGWKLRFDIIVG 156
+++ L H NLV L GYC GD+++L+YE+M SL+ + D L W +R I G
Sbjct: 134 MLSLLHHPNLVNLIGYCADGDQRLLVYEFMPLGSLEDHLHDLPPDKEALDWNMRMKIAAG 193
Query: 157 IARGMLYLHQDSRLRVIHRDLKTSNILLDDEMIPKISDFGLAKIFGGKETGASTQRVMGT 216
A+G+ +LH + VI+RD K+SNILLD+ PK+SDFGLAK+ + + RVMGT
Sbjct: 194 AAKGLEFLHDKANPPVIYRDFKSSNILLDEGFHPKLSDFGLAKLGPTGDKSHVSTRVMGT 253
Query: 217 YGYMSPEYALDGFFSIKSDVFSFGVVLLEILSGKKNTGFFRSQQISSLLGYAWRLWTE-N 275
YGY +PEYA+ G ++KSDV+SFGVV LE+++G+K +L+ +A L+ +
Sbjct: 254 YGYCAPEYAMTGQLTVKSDVYSFGVVFLELITGRKAIDSEMPHGEQNLVAWARPLFNDRR 313
Query: 276 KLLDLMDSALSETCNENEFVKCAQIGLLCVQDEPGNRPTMSNILTML 322
K + L D L + + +C+Q++ RP +++++T L
Sbjct: 314 KFIKLADPRLKGRFPTRALYQALAVASMCIQEQAATRPLIADVVTAL 360
>BRI1_LYCPE (Q8L899) Systemin receptor SR160 precursor (EC 2.7.1.37)
(Brassinosteroid LRR receptor kinase)
Length = 1207
Score = 210 bits (534), Expect = 5e-54
Identities = 123/329 (37%), Positives = 187/329 (56%), Gaps = 8/329 (2%)
Query: 40 TFRSIQAATNNFSDSNKLGQGGYGPVYKGRFPGGQEIAIKRLSSVSTQGLQEFKNEIVLI 99
TF + ATN F + + +G GG+G VYK + G +AIK+L VS QG +EF E+ I
Sbjct: 877 TFADLLEATNGFHNDSLVGSGGFGDVYKAQLKDGSVVAIKKLIHVSGQGDREFTAEMETI 936
Query: 100 AKLQHRNLVRLRGYCIKGDEKILLYEYMSNKSLDTFIFDRTRT-VLLGWKLRFDIIVGIA 158
K++HRNLV L GYC G+E++L+YEYM SL+ + DR +T + L W R I +G A
Sbjct: 937 GKIKHRNLVPLLGYCKVGEERLLVYEYMKYGSLEDVLHDRKKTGIKLNWPARRKIAIGAA 996
Query: 159 RGMLYLHQDSRLRVIHRDLKTSNILLDDEMIPKISDFGLAKIFGGKETGASTQRVMGTYG 218
RG+ +LH + +IHRD+K+SN+LLD+ + ++SDFG+A++ +T S + GT G
Sbjct: 997 RGLAFLHHNCIPHIIHRDMKSSNVLLDENLEARVSDFGMARLMSAMDTHLSVSTLAGTPG 1056
Query: 219 YMSPEYALDGFFSIKSDVFSFGVVLLEILSGKKNTGFFRSQQISSLLGYAWRLWTENKLL 278
Y+ PEY S K DV+S+GVVLLE+L+GK+ T ++L+G+ +L + K+
Sbjct: 1057 YVPPEYYQSFRCSTKGDVYSYGVVLLELLTGKQPTDSADFGD-NNLVGWV-KLHAKGKIT 1114
Query: 279 DLMDSAL--SETCNENEFVKCAQIGLLCVQDEPGNRPTMSNILTMLDGETATIPIPSQPT 336
D+ D L + E E ++ ++ C+ D RPTM ++ M A + S T
Sbjct: 1115 DVFDRELLKEDASIEIELLQHLKVACACLDDRHWKRPTMIQVMAMFKEIQAGSGMDSTST 1174
Query: 337 FFTTKHQSCSSSSSKLEISMQIDSSYQEG 365
+ S + I M I+ S +EG
Sbjct: 1175 IGA---DDVNFSGVEGGIEMGINGSIKEG 1200
>PSKR_ARATH (Q9ZVR7) Putative phytosulfokine receptor precursor (EC
2.7.1.37) (Phytosulfokine LRR receptor kinase)
Length = 1008
Score = 209 bits (532), Expect = 9e-54
Identities = 116/328 (35%), Positives = 186/328 (56%), Gaps = 18/328 (5%)
Query: 5 QIQESLHDSEKHVRDLIGLGNIGE--------NDSESIEVPYYTFRSIQAATNNFSDSNK 56
++ + +SE R LG IG ND E ++ + +TN+F +N
Sbjct: 688 EVDPEIEESESMNRK--ELGEIGSKLVVLFQSNDKE------LSYDDLLDSTNSFDQANI 739
Query: 57 LGQGGYGPVYKGRFPGGQEIAIKRLSSVSTQGLQEFKNEIVLIAKLQHRNLVRLRGYCIK 116
+G GG+G VYK P G+++AIK+LS Q +EF+ E+ +++ QH NLV LRG+C
Sbjct: 740 IGCGGFGMVYKATLPDGKKVAIKKLSGDCGQIEREFEAEVETLSRAQHPNLVLLRGFCFY 799
Query: 117 GDEKILLYEYMSNKSLDTFIFDRT-RTVLLGWKLRFDIIVGIARGMLYLHQDSRLRVIHR 175
++++L+Y YM N SLD ++ +R LL WK R I G A+G+LYLH+ ++HR
Sbjct: 800 KNDRLLIYSYMENGSLDYWLHERNDGPALLKWKTRLRIAQGAAKGLLYLHEGCDPHILHR 859
Query: 176 DLKTSNILLDDEMIPKISDFGLAKIFGGKETGASTQRVMGTYGYMSPEYALDGFFSIKSD 235
D+K+SNILLD+ ++DFGLA++ ET ST ++GT GY+ PEY + K D
Sbjct: 860 DIKSSNILLDENFNSHLADFGLARLMSPYETHVSTD-LVGTLGYIPPEYGQASVATYKGD 918
Query: 236 VFSFGVVLLEILSGKKNTGFFRSQQISSLLGYAWRLWTENKLLDLMDSALSETCNENEFV 295
V+SFGVVLLE+L+ K+ + + L+ + ++ E++ ++ D + N+ E
Sbjct: 919 VYSFGVVLLELLTDKRPVDMCKPKGCRDLISWVVKMKHESRASEVFDPLIYSKENDKEMF 978
Query: 296 KCAQIGLLCVQDEPGNRPTMSNILTMLD 323
+ +I LC+ + P RPT +++ LD
Sbjct: 979 RVLEIACLCLSENPKQRPTTQQLVSWLD 1006
>BRI1_LYCES (Q8GUQ5) Brassinosteroid LRR receptor kinase precursor (EC
2.7.1.37) (tBRI1) (Altered brassinolide sensitivity 1)
(Systemin receptor SR160)
Length = 1207
Score = 207 bits (528), Expect = 3e-53
Identities = 122/329 (37%), Positives = 186/329 (56%), Gaps = 8/329 (2%)
Query: 40 TFRSIQAATNNFSDSNKLGQGGYGPVYKGRFPGGQEIAIKRLSSVSTQGLQEFKNEIVLI 99
TF + ATN F + + +G GG+G VYK + G +AIK+L VS QG +EF E+ I
Sbjct: 877 TFADLLEATNGFHNDSLVGSGGFGDVYKAQLKDGSVVAIKKLIHVSGQGDREFTAEMETI 936
Query: 100 AKLQHRNLVRLRGYCIKGDEKILLYEYMSNKSLDTFIFDRTRT-VLLGWKLRFDIIVGIA 158
K++HRNLV L GYC G+E++L+YEYM SL+ + DR + + L W R I +G A
Sbjct: 937 GKIKHRNLVPLLGYCKVGEERLLVYEYMKYGSLEDVLHDRKKIGIKLNWPARRKIAIGAA 996
Query: 159 RGMLYLHQDSRLRVIHRDLKTSNILLDDEMIPKISDFGLAKIFGGKETGASTQRVMGTYG 218
RG+ +LH + +IHRD+K+SN+LLD+ + ++SDFG+A++ +T S + GT G
Sbjct: 997 RGLAFLHHNCIPHIIHRDMKSSNVLLDENLEARVSDFGMARLMSAMDTHLSVSTLAGTPG 1056
Query: 219 YMSPEYALDGFFSIKSDVFSFGVVLLEILSGKKNTGFFRSQQISSLLGYAWRLWTENKLL 278
Y+ PEY S K DV+S+GVVLLE+L+GK+ T ++L+G+ +L + K+
Sbjct: 1057 YVPPEYYQSFRCSTKGDVYSYGVVLLELLTGKQPTDSADFGD-NNLVGWV-KLHAKGKIT 1114
Query: 279 DLMDSAL--SETCNENEFVKCAQIGLLCVQDEPGNRPTMSNILTMLDGETATIPIPSQPT 336
D+ D L + E E ++ ++ C+ D RPTM ++ M A + S T
Sbjct: 1115 DVFDRELLKEDASIEIELLQHLKVACACLDDRHWKRPTMIQVMAMFKEIQAGSGMDSTST 1174
Query: 337 FFTTKHQSCSSSSSKLEISMQIDSSYQEG 365
+ S + I M I+ S +EG
Sbjct: 1175 IGA---DDVNFSGVEGGIEMGINGSIKEG 1200
>BRI1_ARATH (O22476) BRASSINOSTEROID INSENSITIVE 1 precursor (EC
2.7.1.37) (AtBRI1) (Brassinosteroid LRR receptor kinase)
Length = 1196
Score = 203 bits (516), Expect = 6e-52
Identities = 115/300 (38%), Positives = 174/300 (57%), Gaps = 5/300 (1%)
Query: 40 TFRSIQAATNNFSDSNKLGQGGYGPVYKGRFPGGQEIAIKRLSSVSTQGLQEFKNEIVLI 99
TF + ATN F + + +G GG+G VYK G +AIK+L VS QG +EF E+ I
Sbjct: 872 TFADLLQATNGFHNDSLIGSGGFGDVYKAILKDGSAVAIKKLIHVSGQGDREFMAEMETI 931
Query: 100 AKLQHRNLVRLRGYCIKGDEKILLYEYMSNKSLDTFIFDRTRT-VLLGWKLRFDIIVGIA 158
K++HRNLV L GYC GDE++L+YE+M SL+ + D + V L W R I +G A
Sbjct: 932 GKIKHRNLVPLLGYCKVGDERLLVYEFMKYGSLEDVLHDPKKAGVKLNWSTRRKIAIGSA 991
Query: 159 RGMLYLHQDSRLRVIHRDLKTSNILLDDEMIPKISDFGLAKIFGGKETGASTQRVMGTYG 218
RG+ +LH + +IHRD+K+SN+LLD+ + ++SDFG+A++ +T S + GT G
Sbjct: 992 RGLAFLHHNCSPHIIHRDMKSSNVLLDENLEARVSDFGMARLMSAMDTHLSVSTLAGTPG 1051
Query: 219 YMSPEYALDGFFSIKSDVFSFGVVLLEILSGKKNTGFFRSQQISSLLGYAWRLWTENKLL 278
Y+ PEY S K DV+S+GVVLLE+L+GK+ T ++L+G+ + + ++
Sbjct: 1052 YVPPEYYQSFRCSTKGDVYSYGVVLLELLTGKRPTD-SPDFGDNNLVGWV-KQHAKLRIS 1109
Query: 279 DLMDSAL--SETCNENEFVKCAQIGLLCVQDEPGNRPTMSNILTMLDGETATIPIPSQPT 336
D+ D L + E E ++ ++ + C+ D RPTM ++ M A I SQ T
Sbjct: 1110 DVFDPELMKEDPALEIELLQHLKVAVACLDDRAWRRPTMVQVMAMFKEIQAGSGIDSQST 1169
>TMK1_ARATH (P43298) Putative receptor protein kinase TMK1 precursor
(EC 2.7.1.-)
Length = 942
Score = 201 bits (511), Expect = 2e-51
Identities = 117/316 (37%), Positives = 186/316 (58%), Gaps = 19/316 (6%)
Query: 20 LIGLGNIGEN----DSESIEVPYYTFRSIQAATNNFSDSNKLGQGGYGPVYKGRFPGGQE 75
L G +G+N ++ ++ + RS+ TNNFS N LG GG+G VYKG G +
Sbjct: 556 LPGTSEVGDNIQMVEAGNMLISIQVLRSV---TNNFSSDNILGSGGFGVVYKGELHDGTK 612
Query: 76 IAIKRLSS--VSTQGLQEFKNEIVLIAKLQHRNLVRLRGYCIKGDEKILLYEYMSNKSLD 133
IA+KR+ + ++ +G EFK+EI ++ K++HR+LV L GYC+ G+EK+L+YEYM +L
Sbjct: 613 IAVKRMENGVIAGKGFAEFKSEIAVLTKVRHRHLVTLLGYCLDGNEKLLVYEYMPQGTLS 672
Query: 134 TFIFDRTRTVL--LGWKLRFDIIVGIARGMLYLHQDSRLRVIHRDLKTSNILLDDEMIPK 191
+F+ + L L WK R + + +ARG+ YLH + IHRDLK SNILL D+M K
Sbjct: 673 RHLFEWSEEGLKPLLWKQRLTLALDVARGVEYLHGLAHQSFIHRDLKPSNILLGDDMRAK 732
Query: 192 ISDFGLAKIFGGKETGASTQRVMGTYGYMSPEYALDGFFSIKSDVFSFGVVLLEILSGKK 251
++DFGL ++ + G+ R+ GT+GY++PEYA+ G + K DV+SFGV+L+E+++G+K
Sbjct: 733 VADFGLVRL-APEGKGSIETRIAGTFGYLAPEYAVTGRVTTKVDVYSFGVILMELITGRK 791
Query: 252 NTGFFRSQQISSLLGYAWRLWTE-----NKLLDLMDSALSETCNENEFVKCAQIGLLCVQ 306
+ + ++ L+ + R++ K +D ET V A++ C
Sbjct: 792 SLDESQPEESIHLVSWFKRMYINKEASFKKAIDTTIDLDEETLASVHTV--AELAGHCCA 849
Query: 307 DEPGNRPTMSNILTML 322
EP RP M + + +L
Sbjct: 850 REPYQRPDMGHAVNIL 865
>APKA_ARATH (Q06548) Protein kinase APK1A, chloroplast precursor (EC
2.7.1.-)
Length = 410
Score = 199 bits (507), Expect = 7e-51
Identities = 119/304 (39%), Positives = 180/304 (59%), Gaps = 13/304 (4%)
Query: 32 ESIEVPYYTFRSIQAATNNFSDSNKLGQGGYGPVYKGRF---------PG-GQEIAIKRL 81
+S + ++F +++AT NF + LG+GG+G V+KG PG G IA+K+L
Sbjct: 49 QSPNLKSFSFAELKSATRNFRPDSVLGEGGFGCVFKGWIDEKSLTASRPGTGLVIAVKKL 108
Query: 82 SSVSTQGLQEFKNEIVLIAKLQHRNLVRLRGYCIKGDEKILLYEYMSNKSLDTFIFDRTR 141
+ QG QE+ E+ + + HR+LV+L GYC++ + ++L+YE+M SL+ +F R
Sbjct: 109 NQDGWQGHQEWLAEVNYLGQFSHRHLVKLIGYCLEDEHRLLVYEFMPRGSLENHLFRRGL 168
Query: 142 TVL-LGWKLRFDIIVGIARGMLYLHQDSRLRVIHRDLKTSNILLDDEMIPKISDFGLAKI 200
L WKLR + +G A+G+ +LH S RVI+RD KTSNILLD E K+SDFGLAK
Sbjct: 169 YFQPLSWKLRLKVALGAAKGLAFLHS-SETRVIYRDFKTSNILLDSEYNAKLSDFGLAKD 227
Query: 201 FGGKETGASTQRVMGTYGYMSPEYALDGFFSIKSDVFSFGVVLLEILSGKKNTGFFRSQQ 260
+ + RVMGT+GY +PEY G + KSDV+SFGVVLLE+LSG++ R
Sbjct: 228 GPIGDKSHVSTRVMGTHGYAAPEYLATGHLTTKSDVYSFGVVLLELLSGRRAVDKNRPSG 287
Query: 261 ISSLLGYAW-RLWTENKLLDLMDSALSETCNENEFVKCAQIGLLCVQDEPGNRPTMSNIL 319
+L+ +A L + K+ ++D+ L + + E K A + L C+ E RP MS ++
Sbjct: 288 ERNLVEWAKPYLVNKRKIFRVIDNRLQDQYSMEEACKVATLSLRCLTTEIKLRPNMSEVV 347
Query: 320 TMLD 323
+ L+
Sbjct: 348 SHLE 351
>APKB_ARATH (P46573) Protein kinase APK1B, chloroplast precursor (EC
2.7.1.-)
Length = 412
Score = 197 bits (502), Expect = 3e-50
Identities = 117/304 (38%), Positives = 177/304 (57%), Gaps = 13/304 (4%)
Query: 32 ESIEVPYYTFRSIQAATNNFSDSNKLGQGGYGPVYKGRF---------PG-GQEIAIKRL 81
+S + +TF ++AAT NF + LG+GG+G V+KG PG G IA+K+L
Sbjct: 50 QSPNLKSFTFAELKAATRNFRPDSVLGEGGFGSVFKGWIDEQTLTASKPGTGVVIAVKKL 109
Query: 82 SSVSTQGLQEFKNEIVLIAKLQHRNLVRLRGYCIKGDEKILLYEYMSNKSLDTFIFDRTR 141
+ QG QE+ E+ + + H NLV+L GYC++ + ++L+YE+M SL+ +F R
Sbjct: 110 NQDGWQGHQEWLAEVNYLGQFSHPNLVKLIGYCLEDEHRLLVYEFMPRGSLENHLFRRGS 169
Query: 142 TVL-LGWKLRFDIIVGIARGMLYLHQDSRLRVIHRDLKTSNILLDDEMIPKISDFGLAKI 200
L W LR + +G A+G+ +LH ++ VI+RD KTSNILLD E K+SDFGLAK
Sbjct: 170 YFQPLSWTLRLKVALGAAKGLAFLH-NAETSVIYRDFKTSNILLDSEYNAKLSDFGLAKD 228
Query: 201 FGGKETGASTQRVMGTYGYMSPEYALDGFFSIKSDVFSFGVVLLEILSGKKNTGFFRSQQ 260
+ + R+MGTYGY +PEY G + KSDV+S+GVVLLE+LSG++ R
Sbjct: 229 GPTGDKSHVSTRIMGTYGYAAPEYLATGHLTTKSDVYSYGVVLLEVLSGRRAVDKNRPPG 288
Query: 261 ISSLLGYAWRLW-TENKLLDLMDSALSETCNENEFVKCAQIGLLCVQDEPGNRPTMSNIL 319
L+ +A L + KL ++D+ L + + E K A + L C+ E RP M+ ++
Sbjct: 289 EQKLVEWARPLLANKRKLFRVIDNRLQDQYSMEEACKVATLALRCLTFEIKLRPNMNEVV 348
Query: 320 TMLD 323
+ L+
Sbjct: 349 SHLE 352
>PSKR_DAUCA (Q8LPB4) Phytosulfokine receptor precursor (EC 2.7.1.37)
(Phytosulfokine LRR receptor kinase)
Length = 1021
Score = 195 bits (496), Expect = 1e-49
Identities = 102/281 (36%), Positives = 167/281 (59%), Gaps = 2/281 (0%)
Query: 44 IQAATNNFSDSNKLGQGGYGPVYKGRFPGGQEIAIKRLSSVSTQGLQEFKNEIVLIAKLQ 103
I +T++F+ +N +G GG+G VYK P G ++AIKRLS + Q +EF+ E+ +++ Q
Sbjct: 736 ILKSTSSFNQANIIGCGGFGLVYKATLPDGTKVAIKRLSGDTGQMDREFQAEVETLSRAQ 795
Query: 104 HRNLVRLRGYCIKGDEKILLYEYMSNKSLDTFIFDRT-RTVLLGWKLRFDIIVGIARGML 162
H NLV L GYC ++K+L+Y YM N SLD ++ ++ L WK R I G A G+
Sbjct: 796 HPNLVHLLGYCNYKNDKLLIYSYMDNGSLDYWLHEKVDGPPSLDWKTRLRIARGAAEGLA 855
Query: 163 YLHQDSRLRVIHRDLKTSNILLDDEMIPKISDFGLAKIFGGKETGASTQRVMGTYGYMSP 222
YLHQ ++HRD+K+SNILL D + ++DFGLA++ +T +T ++GT GY+ P
Sbjct: 856 YLHQSCEPHILHRDIKSSNILLSDTFVAHLADFGLARLILPYDTHVTTD-LVGTLGYIPP 914
Query: 223 EYALDGFFSIKSDVFSFGVVLLEILSGKKNTGFFRSQQISSLLGYAWRLWTENKLLDLMD 282
EY + K DV+SFGVVLLE+L+G++ + + L+ + ++ TE + ++ D
Sbjct: 915 EYGQASVATYKGDVYSFGVVLLELLTGRRPMDVCKPRGSRDLISWVLQMKTEKRESEIFD 974
Query: 283 SALSETCNENEFVKCAQIGLLCVQDEPGNRPTMSNILTMLD 323
+ + + E + +I C+ + P RPT +++ L+
Sbjct: 975 PFIYDKDHAEEMLLVLEIACRCLGENPKTRPTTQQLVSWLE 1015
>NAK_ARATH (P43293) Probable serine/threonine-protein kinase NAK (EC
2.7.1.37)
Length = 389
Score = 186 bits (473), Expect = 6e-47
Identities = 113/299 (37%), Positives = 175/299 (57%), Gaps = 17/299 (5%)
Query: 39 YTFRSIQAATNNFSDSNKLGQGGYGPVYKGRF---------PG-GQEIAIKRLSSVSTQG 88
++ +++AT NF + +G+GG+G V+KG PG G IA+KRL+ QG
Sbjct: 56 FSLSELKSATRNFRPDSVVGEGGFGCVFKGWIDESSLAPSKPGTGIVIAVKRLNQEGFQG 115
Query: 89 LQEFKNEIVLIAKLQHRNLVRLRGYCIKGDEKILLYEYMSNKSLDTFIFDR-TRTVLLGW 147
+E+ EI + +L H NLV+L GYC++ + ++L+YE+M+ SL+ +F R T L W
Sbjct: 116 HREWLAEINYLGQLDHPNLVKLIGYCLEEEHRLLVYEFMTRGSLENHLFRRGTFYQPLSW 175
Query: 148 KLRFDIIVGIARGMLYLHQDSRLRVIHRDLKTSNILLDDEMIPKISDFGLAKIFGGKETG 207
R + +G ARG+ +LH +++ +VI+RD K SNILLD K+SDFGLA+ +
Sbjct: 176 NTRVRMALGAARGLAFLH-NAQPQVIYRDFKASNILLDSNYNAKLSDFGLARDGPMGDNS 234
Query: 208 ASTQRVMGTYGYMSPEYALDGFFSIKSDVFSFGVVLLEILSGKKNTGFFRSQQISSLLGY 267
+ RVMGT GY +PEY G S+KSDV+SFGVVLLE+LSG++ ++Q +
Sbjct: 235 HVSTRVMGTQGYAAPEYLATGHLSVKSDVYSFGVVLLELLSGRR--AIDKNQPVGEHNLV 292
Query: 268 AWR---LWTENKLLDLMDSALSETCNENEFVKCAQIGLLCVQDEPGNRPTMSNILTMLD 323
W L + +LL +MD L + +K A + L C+ + +RPTM+ I+ ++
Sbjct: 293 DWARPYLTNKRRLLRVMDPRLQGQYSLTRALKIAVLALDCISIDAKSRPTMNEIVKTME 351
>SIRK_ARATH (O64483) Senescence-induced receptor-like
serine/threonine kinase precursor (FLG22-induced
receptor-like kinase 1)
Length = 876
Score = 181 bits (458), Expect = 3e-45
Identities = 99/300 (33%), Positives = 173/300 (57%), Gaps = 6/300 (2%)
Query: 24 GNIGE-NDSESIEVPYYTFRSIQAATNNFSDSNKLGQGGYGPVYKGRFPGGQEIAIKRLS 82
G +GE N Y+ + + TNNF +G+GG+G VY G G+++A+K LS
Sbjct: 548 GTLGERNGPLKTAKRYFKYSEVVNITNNFE--RVIGKGGFGKVYHGVI-NGEQVAVKVLS 604
Query: 83 SVSTQGLQEFKNEIVLIAKLQHRNLVRLRGYCIKGDEKILLYEYMSNKSLDTFIFDRTRT 142
S QG +EF+ E+ L+ ++ H NL L GYC + + +L+YEYM+N++L ++ + R+
Sbjct: 605 EESAQGYKEFRAEVDLLMRVHHTNLTSLVGYCNEINHMVLIYEYMANENLGDYLAGK-RS 663
Query: 143 VLLGWKLRFDIIVGIARGMLYLHQDSRLRVIHRDLKTSNILLDDEMIPKISDFGLAKIFG 202
+L W+ R I + A+G+ YLH + ++HRD+K +NILL++++ K++DFGL++ F
Sbjct: 664 FILSWEERLKISLDAAQGLEYLHNGCKPPIVHRDVKPTNILLNEKLQAKMADFGLSRSFS 723
Query: 203 GKETGASTQRVMGTYGYMSPEYALDGFFSIKSDVFSFGVVLLEILSGKKNTGFFRSQQIS 262
+ +G + V G+ GY+ PEY + KSDV+S GVVLLE+++G+ +++++
Sbjct: 724 VEGSGQISTVVAGSIGYLDPEYYSTRQMNEKSDVYSLGVVLLEVITGQPAIASSKTEKV- 782
Query: 263 SLLGYAWRLWTENKLLDLMDSALSETCNENEFVKCAQIGLLCVQDEPGNRPTMSNILTML 322
+ + + + ++D L E + K ++I L C + RPTMS ++ L
Sbjct: 783 HISDHVRSILANGDIRGIVDQRLRERYDVGSAWKMSEIALACTEHTSAQRPTMSQVVMEL 842
>KPRO_MAIZE (P17801) Putative receptor protein kinase ZmPK1
precursor (EC 2.7.1.37)
Length = 817
Score = 176 bits (445), Expect = 1e-43
Identities = 101/292 (34%), Positives = 167/292 (56%), Gaps = 11/292 (3%)
Query: 39 YTFRSIQAATNNFSDSNKLGQGGYGPVYKGRFPGGQEIAIKRLSSVSTQGLQEFKNEIVL 98
Y++R + AT F +LG+G G VYKG + +A+K+L +V QG + F+ E+ +
Sbjct: 524 YSYRELVKATRKFKV--ELGRGESGTVYKGVLEDDRHVAVKKLENVR-QGKEVFQAELSV 580
Query: 99 IAKLQHRNLVRLRGYCIKGDEKILLYEYMSNKSLDTFIFDRTRTVLLGWKLRFDIIVGIA 158
I ++ H NLVR+ G+C +G ++L+ EY+ N SL +F +LL W+ RF+I +G+A
Sbjct: 581 IGRINHMNLVRIWGFCSEGSHRLLVSEYVENGSLANILFSEGGNILLDWEGRFNIALGVA 640
Query: 159 RGMLYLHQDSRLRVIHRDLKTSNILLDDEMIPKISDFGLAKIFGGKETGASTQRVMGTYG 218
+G+ YLH + VIH D+K NILLD PKI+DFGL K+ + + V GT G
Sbjct: 641 KGLAYLHHECLEWVIHCDVKPENILLDQAFEPKITDFGLVKLLNRGGSTQNVSHVRGTLG 700
Query: 219 YMSPEYALDGFFSIKSDVFSFGVVLLEILSGKKNTGFF-RSQQISSLLGYAWRLWT---- 273
Y++PE+ + K DV+S+GVVLLE+L+G + + + ++ S+L R+ +
Sbjct: 701 YIAPEWVSSLPITAKVDVYSYGVVLLELLTGTRVSELVGGTDEVHSMLRKLVRMLSAKLE 760
Query: 274 --ENKLLD-LMDSALSETCNENEFVKCAQIGLLCVQDEPGNRPTMSNILTML 322
E +D +DS L+ N + ++ + C++++ RPTM + + L
Sbjct: 761 GEEQSWIDGYLDSKLNRPVNYVQARTLIKLAVSCLEEDRSKRPTMEHAVQTL 812
>CRI4_MAIZE (O24585) Putative receptor protein kinase CRINKLY4
precursor (EC 2.7.1.-)
Length = 901
Score = 173 bits (438), Expect = 7e-43
Identities = 105/305 (34%), Positives = 171/305 (55%), Gaps = 11/305 (3%)
Query: 39 YTFRSIQAATNNFSDSNKLGQGGYGPVYKGRFPGGQEIAIKRLSSVST--QGLQEFKNEI 96
+++ ++ AT FS+ +++G+G + V+KG G +A+KR S + +EF NE+
Sbjct: 493 FSYEELEQATGGFSEDSQVGKGSFSCVFKGILRDGTVVAVKRAIKASDVKKSSKEFHNEL 552
Query: 97 VLIAKLQHRNLVRLRGYCIKGDEKILLYEYMSNKSLDTFIFDRTRTVL--LGWKLRFDII 154
L+++L H +L+ L GYC G E++L+YE+M++ SL + + + L W R I
Sbjct: 553 DLLSRLNHAHLLNLLGYCEDGSERLLVYEFMAHGSLYQHLHGKDPNLKKRLNWARRVTIA 612
Query: 155 VGIARGMLYLHQDSRLRVIHRDLKTSNILLDDEMIPKISDFGLAKIFGGKETGASTQRV- 213
V ARG+ YLH + VIHRD+K+SNIL+D++ +++DFGL+ I G ++G +
Sbjct: 613 VQAARGIEYLHGYACPPVIHRDIKSSNILIDEDHNARVADFGLS-ILGPADSGTPLSELP 671
Query: 214 MGTYGYMSPEYALDGFFSIKSDVFSFGVVLLEILSGKKNTGFFRSQQISSLLGYAWRLWT 273
GT GY+ PEY + + KSDV+SFGVVLLEILSG+K + +++ +A L
Sbjct: 672 AGTLGYLDPEYYRLHYLTTKSDVYSFGVVLLEILSGRK--AIDMQFEEGNIVEWAVPLIK 729
Query: 274 ENKLLDLMDSALSETCNENEFVKCAQIGLLCVQDEPGNRPTMSNILTMLDGETATI---P 330
+ ++D LS + K A + CV+ +RP+M + T L+ A + P
Sbjct: 730 AGDIFAILDPVLSPPSDLEALKKIASVACKCVRMRGKDRPSMDKVTTALEHALALLMGSP 789
Query: 331 IPSQP 335
QP
Sbjct: 790 CIEQP 794
>RPK1_IPONI (P93194) Receptor-like protein kinase precursor (EC
2.7.1.37)
Length = 1109
Score = 169 bits (429), Expect = 8e-42
Identities = 98/284 (34%), Positives = 155/284 (54%), Gaps = 11/284 (3%)
Query: 47 ATNNFSDSNKLGQGGYGPVYKGRFPGGQEIAIKRLSSVSTQ-GLQEFKNEIVLIAKLQHR 105
AT N +D +G+G +G +YK + A+K+L + G EI I K++HR
Sbjct: 812 ATENLNDKYVIGKGAHGTIYKATLSPDKVYAVKKLVFTGIKNGSVSMVREIETIGKVRHR 871
Query: 106 NLVRLRGYCIKGDEKILLYEYMSNKSLDTFIFDRTRTVLLGWKLRFDIIVGIARGMLYLH 165
NL++L + ++ + ++LY YM N SL + + L W R +I VG A G+ YLH
Sbjct: 872 NLIKLEEFWLRKEYGLILYTYMENGSLHDILHETNPPKPLDWSTRHNIAVGTAHGLAYLH 931
Query: 166 QDSRLRVIHRDLKTSNILLDDEMIPKISDFGLAKIFGGKETGASTQRVMGTYGYMSPEYA 225
D ++HRD+K NILLD ++ P ISDFG+AK+ T + V GT GYM+PE A
Sbjct: 932 FDCDPAIVHRDIKPMNILLDSDLEPHISDFGIAKLLDQSATSIPSNTVQGTIGYMAPENA 991
Query: 226 LDGFFSIKSDVFSFGVVLLEILSGKKNTG-FFRSQQISSLLGYAWRLWTE----NKLLD- 279
S +SDV+S+GVVLLE+++ KK F + + ++G+ +WT+ K++D
Sbjct: 992 FTTVKSRESDVYSYGVVLLELITRKKALDPSFNGE--TDIVGWVRSVWTQTGEIQKIVDP 1049
Query: 280 -LMDSALSETCNENEFVKCAQIGLLCVQDEPGNRPTMSNILTML 322
L+D + + E + + + L C + E RPTM +++ L
Sbjct: 1050 SLLDELIDSSVME-QVTEALSLALRCAEKEVDKRPTMRDVVKQL 1092
>RLK5_ARATH (P47735) Receptor-like protein kinase 5 precursor (EC
2.7.1.37)
Length = 999
Score = 169 bits (428), Expect = 1e-41
Identities = 107/299 (35%), Positives = 160/299 (52%), Gaps = 20/299 (6%)
Query: 53 DSNKLGQGGYGPVYKGRFPGGQEIAIKRLSSVSTQGLQEFKN----------EIVLIAKL 102
+ N +G G G VYK GG+ +A+K+L+ G E+ + E+ + +
Sbjct: 685 EKNVIGFGSSGKVYKVELRGGEVVAVKKLNKSVKGGDDEYSSDSLNRDVFAAEVETLGTI 744
Query: 103 QHRNLVRLRGYCIKGDEKILLYEYMSNKSL-DTFIFDRTRTVLLGWKLRFDIIVGIARGM 161
+H+++VRL C GD K+L+YEYM N SL D DR V+LGW R I + A G+
Sbjct: 745 RHKSIVRLWCCCSSGDCKLLVYEYMPNGSLADVLHGDRKGGVVLGWPERLRIALDAAEGL 804
Query: 162 LYLHQDSRLRVIHRDLKTSNILLDDEMIPKISDFGLAKI--FGGKETGASTQRVMGTYGY 219
YLH D ++HRD+K+SNILLD + K++DFG+AK+ G +T + + G+ GY
Sbjct: 805 SYLHHDCVPPIVHRDVKSSNILLDSDYGAKVADFGIAKVGQMSGSKTPEAMSGIAGSCGY 864
Query: 220 MSPEYALDGFFSIKSDVFSFGVVLLEILSGKKNTGFFRSQQISSLLGYAWRLWTENK--L 277
++PEY + KSD++SFGVVLLE+++GK+ T ++ W +K L
Sbjct: 865 IAPEYVYTLRVNEKSDIYSFGVVLLELVTGKQPT----DSELGDKDMAKWVCTALDKCGL 920
Query: 278 LDLMDSALSETCNENEFVKCAQIGLLCVQDEPGNRPTMSNILTMLDGETATIPIPSQPT 336
++D L E E K IGLLC P NRP+M ++ ML + +P S T
Sbjct: 921 EPVIDPKLDLKFKE-EISKVIHIGLLCTSPLPLNRPSMRKVVIMLQEVSGAVPCSSPNT 978
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.319 0.137 0.393
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 42,331,012
Number of Sequences: 164201
Number of extensions: 1817722
Number of successful extensions: 7865
Number of sequences better than 10.0: 1629
Number of HSP's better than 10.0 without gapping: 1183
Number of HSP's successfully gapped in prelim test: 446
Number of HSP's that attempted gapping in prelim test: 4271
Number of HSP's gapped (non-prelim): 1818
length of query: 365
length of database: 59,974,054
effective HSP length: 112
effective length of query: 253
effective length of database: 41,583,542
effective search space: 10520636126
effective search space used: 10520636126
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 66 (30.0 bits)
Medicago: description of AC145218.2


