

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC145109.3 - phase: 0
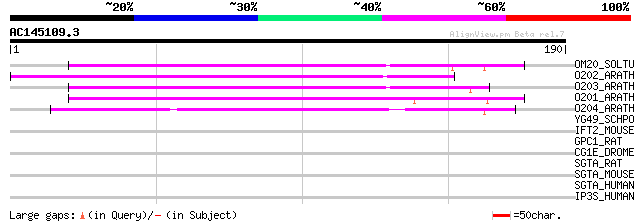
(190 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
OM20_SOLTU (P92792) Mitochondrial import receptor subunit TOM20 ... 111 1e-24
O202_ARATH (P82873) Mitochondrial import receptor subunit TOM20-... 107 1e-23
O203_ARATH (P82874) Mitochondrial import receptor subunit TOM20-... 106 3e-23
O201_ARATH (P82872) Mitochondrial import receptor subunit TOM20-... 100 2e-21
O204_ARATH (P82805) Mitochondrial import receptor subunit TOM20-... 80 2e-15
YG49_SCHPO (O60184) Protein C23E6.09 in chromosome II 35 0.080
IFT2_MOUSE (Q64112) Interferon-induced protein with tetratricope... 32 0.68
GPC1_RAT (P35053) Glypican-1 precursor (HSPG M12) 30 2.6
CG1E_DROME (P54733) G1/S-specific cyclin E (DmCycE) 30 4.4
SGTA_RAT (O70593) Small glutamine-rich tetratricopeptide repeat-... 29 5.7
SGTA_MOUSE (Q8BJU0) Small glutamine-rich tetratricopeptide repea... 29 5.7
SGTA_HUMAN (O43765) Small glutamine-rich tetratricopeptide repea... 28 9.8
IP3S_HUMAN (Q14571) Inositol 1,4,5-trisphosphate receptor type 2... 28 9.8
>OM20_SOLTU (P92792) Mitochondrial import receptor subunit TOM20
(Translocase of outer membrane 20 kDa subunit)
Length = 204
Score = 111 bits (277), Expect = 1e-24
Identities = 69/165 (41%), Positives = 94/165 (56%), Gaps = 10/165 (6%)
Query: 21 AEQRYNKNPHDAENLTRWGVALLRLSQAHSFPDSLHTIQVSISKLEEAFSLNPNNPDVHW 80
AE Y +NP DA+NLTRWG ALL LSQ +S I + SKLEEA ++NP D W
Sbjct: 20 AETTYAQNPLDADNLTRWGGALLELSQFQPVAESKQMISDATSKLEEALTVNPEKHDALW 79
Query: 81 LLGMALTMQALLTPDSHDAKLHFDSADVYFKRAFRQDPSDPTYQISLELPDTKDHEQHPK 140
LG A T LTPD +AK++F+ A F++AF DPS+ Y+ SLE+ K E H +
Sbjct: 80 CLGNAHTSHVFLTPDMDEAKVYFEKATQCFQQAFDADPSNDLYRKSLEV-TAKAPELHME 138
Query: 141 IVNHGLGQQS----KGSSSATKVIK-----HYKFLVFLFTLLVIG 176
I HG QQ+ +S++TK K K+ +F + +L +G
Sbjct: 139 IHRHGPMQQTMAAEPSTSTSTKSSKKTKSSDLKYDIFGWVILAVG 183
>O202_ARATH (P82873) Mitochondrial import receptor subunit TOM20-2
(Translocase of outer membrane 20 kDa subunit 2)
Length = 210
Score = 107 bits (268), Expect = 1e-23
Identities = 58/152 (38%), Positives = 84/152 (55%), Gaps = 1/152 (0%)
Query: 1 MDYPLRKFKFHLPMKHECILAEQRYNKNPHDAENLTRWGVALLRLSQAHSFPDSLHTIQV 60
M++ F+ + +H +E +Y +P D+ENL +WG ALL LSQ P++ +
Sbjct: 1 MEFSTADFERFIMFEHARKNSEAQYKNDPLDSENLLKWGGALLELSQFQPIPEAKLMLND 60
Query: 61 SISKLEEAFSLNPNNPDVHWLLGMALTMQALLTPDSHDAKLHFDSADVYFKRAFRQDPSD 120
+ISKLEEA ++NP W + A T A D +AK HFD A YF+RA +DP +
Sbjct: 61 AISKLEEALTINPGKHQALWCIANAYTAHAFYVHDPEEAKEHFDKATEYFQRAENEDPGN 120
Query: 121 PTYQISLELPDTKDHEQHPKIVNHGLGQQSKG 152
TY+ SL+ K E H + +N G+GQQ G
Sbjct: 121 DTYRKSLD-SSLKAPELHMQFMNQGMGQQILG 151
>O203_ARATH (P82874) Mitochondrial import receptor subunit TOM20-3
(Translocase of outer membrane 20 kDa subunit 3)
Length = 202
Score = 106 bits (265), Expect = 3e-23
Identities = 61/149 (40%), Positives = 82/149 (54%), Gaps = 6/149 (4%)
Query: 21 AEQRYNKNPHDAENLTRWGVALLRLSQAHSFPDSLHTIQVSISKLEEAFSLNPNNPDVHW 80
AE Y NP DA+NLTRWG LL LSQ HS D+ IQ +I+K EEA ++P + W
Sbjct: 20 AENTYKSNPLDADNLTRWGGVLLELSQFHSISDAKQMIQEAITKFEEALLIDPKKDEAVW 79
Query: 81 LLGMALTMQALLTPDSHDAKLHFDSADVYFKRAFRQDPSDPTYQISLELPDTKDHEQHPK 140
+G A T A LTPD +AK +FD A +F++A + P + Y SLE+ K + H +
Sbjct: 80 CIGNAYTSFAFLTPDETEAKHNFDLATQFFQQAVDEQPDNTHYLKSLEM-TAKAPQLHAE 138
Query: 141 IVNHGLGQQSKGSSSA-----TKVIKHYK 164
GLG Q G A +K +K+ K
Sbjct: 139 AYKQGLGSQPMGRVEAPAPPSSKAVKNKK 167
>O201_ARATH (P82872) Mitochondrial import receptor subunit TOM20-1
(Translocase of outer membrane 20 kDa subunit 1)
Length = 188
Score = 100 bits (250), Expect = 2e-21
Identities = 65/164 (39%), Positives = 87/164 (52%), Gaps = 8/164 (4%)
Query: 21 AEQRYNKNPHDAENLTRWGVALLRLSQAHSFPDSLHTIQVSISKLEEAFSLNPNNPDVHW 80
AE+ Y NP DA+NL RWG ALL LSQ + DSL IQ +ISKLE+A ++P D W
Sbjct: 14 AEETYKLNPEDADNLMRWGEALLELSQFQNVIDSLKMIQDAISKLEDAILIDPMKHDAVW 73
Query: 81 LLGMALTMQALLTPDSHDAKLHFDSADVYFKRAFRQDPSDPTYQISLELPDTKDHEQ--- 137
LG A T A LTPD A+L+F A ++F A Q P + Y SLE+ D
Sbjct: 74 CLGNAYTSYARLTPDDTQARLNFGLAYLFFGIAVAQQPDNQVYHKSLEMADKAPQLHTGF 133
Query: 138 HPKIVNHGLGQQSKGSSSATKVIKH-----YKFLVFLFTLLVIG 176
H + LG + + KV+K+ K++V + +L IG
Sbjct: 134 HKNRLLSLLGGVETLAIPSPKVVKNKKSSDEKYIVMGWVILAIG 177
>O204_ARATH (P82805) Mitochondrial import receptor subunit TOM20-4
(Translocase of outer membrane 20 kDa subunit 4)
Length = 187
Score = 80.5 bits (197), Expect = 2e-15
Identities = 56/161 (34%), Positives = 80/161 (48%), Gaps = 9/161 (5%)
Query: 15 KHECILAEQRYNKNPHDAENLTRWGVALLRLSQAHSFPDSLHTIQVSISKLEEAFSLNPN 74
+H +AE Y KNP DAENLTRW ALL LSQ + P + I +I KL EA ++P
Sbjct: 14 EHARKVAEATYVKNPLDAENLTRWAGALLELSQFQTEPKQM--ILEAILKLGEALVIDPK 71
Query: 75 NPDVHWLLGMALTMQALLTPDSHDAKLHFDSADVYFKRAFRQDPSDPTYQISLELPDTKD 134
D WL+G A L+ D +A +F+ A +F+ A + P Y+ SL L
Sbjct: 72 KHDALWLIGNAHLSFGFLSSDQTEASDNFEKASQFFQLAVEEQPESELYRKSLTLA---- 127
Query: 135 HEQHPKIVNHGLGQQSKGSSSATKVIK--HYKFLVFLFTLL 173
+ P++ G S S+ K K +K+ VF + +L
Sbjct: 128 -SKAPELHTGGTAGPSSNSAKTMKQKKTSEFKYDVFGWVIL 167
>YG49_SCHPO (O60184) Protein C23E6.09 in chromosome II
Length = 1102
Score = 35.4 bits (80), Expect = 0.080
Identities = 37/134 (27%), Positives = 55/134 (40%), Gaps = 26/134 (19%)
Query: 18 CILAEQRYNKNPH--------DAENLTRWGVALLRLSQAHSFPDSLHTIQVSISKLEEAF 69
C +A+Q+YNK D N T W + Q + + D+L A
Sbjct: 593 CYVAQQKYNKAYEAYQQAVYRDGRNPTFWCSIGVLYYQINQYQDALDAYS-------RAI 645
Query: 70 SLNPNNPDVHWLLGMALTMQALLTPDSHDAKLHFDSADVYFKRAFRQDPSDPTYQISLEL 129
LNP +V + LG D+ DA ++RA DP++P + L+L
Sbjct: 646 RLNPYISEVWYDLGTLYESCHNQISDALDA----------YQRAAELDPTNPHIKARLQL 695
Query: 130 PDTKDHEQHPKIVN 143
++EQH KIVN
Sbjct: 696 LRGPNNEQH-KIVN 708
>IFT2_MOUSE (Q64112) Interferon-induced protein with
tetratricopeptide repeats 2 (IFIT-2) (Interferon-induced
54 kDa protein) (IFI-54K) (Glucocorticoid-attenuated
response gene 39 protein) (GARG-39)
Length = 472
Score = 32.3 bits (72), Expect = 0.68
Identities = 25/98 (25%), Positives = 40/98 (40%), Gaps = 17/98 (17%)
Query: 22 EQRYNKNPHDAENLTRWGVALLRLSQAHSFPDSLHTIQVSISKLEEAFSLNPNNPDVHWL 81
++ K+P + E + W +A RL D I LE+A L+P+N V L
Sbjct: 162 QKALEKDPKNPEFTSGWAIAFYRL-------DDWPARNYCIDSLEQAIQLSPDNTYVKVL 214
Query: 82 LGMALTMQALLTPDSHDAKLHFDSADVYFKRAFRQDPS 119
L + L +H + A + A ++DPS
Sbjct: 215 LALKLDA----------VHVHKNQAMALVEEALKKDPS 242
>GPC1_RAT (P35053) Glypican-1 precursor (HSPG M12)
Length = 558
Score = 30.4 bits (67), Expect = 2.6
Identities = 13/27 (48%), Positives = 21/27 (77%)
Query: 148 QQSKGSSSATKVIKHYKFLVFLFTLLV 174
Q+ + +S+AT+ HY FL+FLFTL++
Sbjct: 524 QEGQKTSAATRPEPHYFFLLFLFTLVL 550
>CG1E_DROME (P54733) G1/S-specific cyclin E (DmCycE)
Length = 709
Score = 29.6 bits (65), Expect = 4.4
Identities = 17/56 (30%), Positives = 25/56 (44%)
Query: 62 ISKLEEAFSLNPNNPDVHWLLGMALTMQALLTPDSHDAKLHFDSADVYFKRAFRQD 117
IS+ LN N V G+ L ++T DSH + H + D+Y + QD
Sbjct: 571 ISQKAPYLQLNEQNEQVSNKFGLGLICPNIVTDDSHIIQTHTTTMDMYDEVLMAQD 626
>SGTA_RAT (O70593) Small glutamine-rich tetratricopeptide
repeat-containing protein A
Length = 314
Score = 29.3 bits (64), Expect = 5.7
Identities = 17/76 (22%), Positives = 35/76 (45%), Gaps = 11/76 (14%)
Query: 61 SISKLEEAFSLNPNNPDVHWLLGMALTMQALLTPDSHDAKLHFDSADVYFKRAFRQDPSD 120
++ E A ++P + +G+AL+ H ++ Y+K+A DP +
Sbjct: 144 AVQDCERAIGIDPGYSKAYGRMGLALS----------SLNKHAEAV-AYYKKALELDPDN 192
Query: 121 PTYQISLELPDTKDHE 136
TY+ +L++ + K E
Sbjct: 193 DTYKSNLKIAELKLRE 208
>SGTA_MOUSE (Q8BJU0) Small glutamine-rich tetratricopeptide
repeat-containing protein A
Length = 315
Score = 29.3 bits (64), Expect = 5.7
Identities = 17/76 (22%), Positives = 35/76 (45%), Gaps = 11/76 (14%)
Query: 61 SISKLEEAFSLNPNNPDVHWLLGMALTMQALLTPDSHDAKLHFDSADVYFKRAFRQDPSD 120
++ E A ++P + +G+AL+ H ++ Y+K+A DP +
Sbjct: 145 AVQDCERAIGIDPGYSKAYGRMGLALS----------SLNKHAEAV-AYYKKALELDPDN 193
Query: 121 PTYQISLELPDTKDHE 136
TY+ +L++ + K E
Sbjct: 194 DTYKSNLKIAELKLRE 209
>SGTA_HUMAN (O43765) Small glutamine-rich tetratricopeptide
repeat-containing protein A (Vpu-binding protein) (UBP)
Length = 313
Score = 28.5 bits (62), Expect = 9.8
Identities = 17/76 (22%), Positives = 35/76 (45%), Gaps = 11/76 (14%)
Query: 61 SISKLEEAFSLNPNNPDVHWLLGMALTMQALLTPDSHDAKLHFDSADVYFKRAFRQDPSD 120
++ E A ++P + +G+AL+ H ++ Y+K+A DP +
Sbjct: 144 AVQDCERAICIDPAYSKAYGRMGLALS----------SLNKHVEAV-AYYKKALELDPDN 192
Query: 121 PTYQISLELPDTKDHE 136
TY+ +L++ + K E
Sbjct: 193 ETYKSNLKIAELKLRE 208
>IP3S_HUMAN (Q14571) Inositol 1,4,5-trisphosphate receptor type 2
(Type 2 inositol 1,4,5-trisphosphate receptor) (Type 2
InsP3 receptor) (IP3 receptor isoform 2) (InsP3R2)
Length = 2701
Score = 28.5 bits (62), Expect = 9.8
Identities = 38/158 (24%), Positives = 66/158 (41%), Gaps = 27/158 (17%)
Query: 40 VALLRLSQAHSFPDSLHTIQVS-------ISKLEEAFSLNPNNPDVHWLLGMALTMQALL 92
V+LLR ++ F D L + VS +L F L+P N D+ + ++MQA
Sbjct: 619 VSLLRRNREPRFLDYLSDLCVSNTTAIPVTQELICKFMLSPGNADI-LIQTKVVSMQADN 677
Query: 93 TPDSHDAKLHFDSADVYFKRAFRQDPSDPTYQISLELPDTKDHEQHPKIVNHGLGQQSKG 152
+S D +V+ Y I + E H K + H + +G
Sbjct: 678 PMESSILSDDIDDEEVWL------------YWID------SNKEPHGKAIRHLAQEAKEG 719
Query: 153 SSSATKVIKHYKFLVFLFTLLVIGYQ-LPNSQFSCILS 189
+ + +V+ +Y++ + LF + + Q L +Q S LS
Sbjct: 720 TKADLEVLTYYRYQLNLFARMCLDRQYLAINQISTQLS 757
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.321 0.136 0.417
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 22,829,005
Number of Sequences: 164201
Number of extensions: 860508
Number of successful extensions: 1847
Number of sequences better than 10.0: 13
Number of HSP's better than 10.0 without gapping: 7
Number of HSP's successfully gapped in prelim test: 6
Number of HSP's that attempted gapping in prelim test: 1838
Number of HSP's gapped (non-prelim): 14
length of query: 190
length of database: 59,974,054
effective HSP length: 104
effective length of query: 86
effective length of database: 42,897,150
effective search space: 3689154900
effective search space used: 3689154900
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 62 (28.5 bits)
Medicago: description of AC145109.3


