

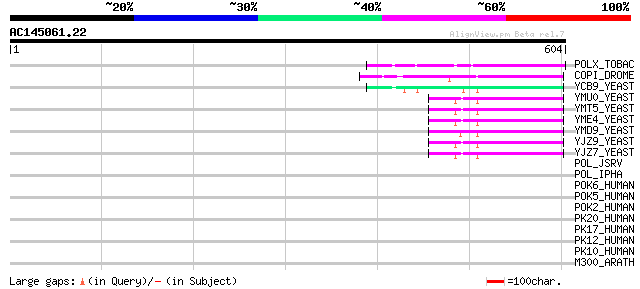
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC145061.22 - phase: 0 /pseudo
(604 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
POLX_TOBAC (P10978) Retrovirus-related Pol polyprotein from tran... 128 5e-29
COPI_DROME (P04146) Copia protein (Gag-int-pol protein) [Contain... 80 2e-14
YCB9_YEAST (P25384) Transposon Ty2 protein B (Ty1-17 protein B) 60 2e-08
YMU0_YEAST (Q04670) Transposon Ty1 protein B 57 1e-07
YMT5_YEAST (Q04214) Transposon Ty1 protein B 57 1e-07
YME4_YEAST (Q04711) Transposon Ty1 protein B 57 1e-07
YMD9_YEAST (Q03434) Transposon Ty1 protein B 57 1e-07
YJZ9_YEAST (P47100) Transposon Ty1 protein B 57 1e-07
YJZ7_YEAST (P47098) Transposon Ty1 protein B 57 1e-07
POL_JSRV (P31623) Pol polyprotein [Contains: Reverse transcripta... 35 0.57
POL_IPHA (P04026) Putative Pol polyprotein [Contains: Integrase ... 34 1.3
POK6_HUMAN (P63133) HERV-K_8p23.1 provirus ancestral Pol protein... 32 6.3
POK5_HUMAN (P63132) HERV-K_19p13.11 provirus ancestral Pol prote... 32 6.3
POK2_HUMAN (Q9BXR3) HERV-K_7p22.1 provirus ancestral Pol protein... 32 6.3
PK20_HUMAN (Q9UQG0) HERV-K_3q27.3 provirus ancestral Pol protein... 32 6.3
PK17_HUMAN (P63136) HERV-K_11q22.1 provirus ancestral Pol protei... 32 6.3
PK12_HUMAN (P63135) HERV-K_1q22 provirus ancestral Pol protein (... 32 6.3
PK10_HUMAN (P10266) HERV-K_5q33.3 provirus ancestral Pol protein... 32 6.3
M300_ARATH (P93293) Hypothetical mitochondrial protein AtMg00300... 32 6.3
>POLX_TOBAC (P10978) Retrovirus-related Pol polyprotein from
transposon TNT 1-94 [Contains: Protease (EC 3.4.23.-);
Reverse transcriptase (EC 2.7.7.49); Endonuclease]
Length = 1328
Score = 128 bits (321), Expect = 5e-29
Identities = 74/216 (34%), Positives = 121/216 (55%), Gaps = 6/216 (2%)
Query: 389 NLLSVGQLVQKGYSITMKDNSLKLFDKNQRLVMKTPLAKNKTF*TDMKVAKLNCLSAIVN 448
NL+S L + GY + +L + LV+ +A+ + T+ ++ + L+A +
Sbjct: 362 NLISGIALDRDGYESYFANQKWRLTKGS--LVIAKGVARGTLYRTNAEICQGE-LNAAQD 418
Query: 449 NEDSWLWHYMFGHLNYISLNHLVDKEMILVVPKLDIPHKFYDTRLIGKQPRTAFISTTTH 508
LWH GH++ L L K +I + K D L GKQ R +F T++
Sbjct: 419 EISVDLWHKRMGHMSEKGLQILAKKSLISYAKGTTV--KPCDYCLFGKQHRVSF-QTSSE 475
Query: 509 RSKEVLNVVYFDVNGPLEVPSMGRIKYFISFLDEFSKNIWVCLIKAKSGAFDIFQKFKFF 568
R +L++VY DV GP+E+ SMG KYF++F+D+ S+ +WV ++K K F +FQKF
Sbjct: 476 RKLNILDLVYSDVCGPMEIESMGGNKYFVTFIDDASRKLWVYILKTKDQVFQVFQKFHAL 535
Query: 569 VEKQSGKSITSLRTDGGGEYTIKVFEKFCEENGVIH 604
VE+++G+ + LR+D GGEYT + FE++C +G+ H
Sbjct: 536 VERETGRKLKRLRSDNGGEYTSREFEEYCSSHGIRH 571
>COPI_DROME (P04146) Copia protein (Gag-int-pol protein) [Contains:
Copia VLP protein; Copia protease (EC 3.4.23.-)]
Length = 1409
Score = 79.7 bits (195), Expect = 2e-14
Identities = 67/228 (29%), Positives = 103/228 (44%), Gaps = 13/228 (5%)
Query: 381 MFSMPPVCNLLSVGQLVQKGYSITMKDNSLKLFDKNQRLVMKTPLAKNKTF*TDMKVAKL 440
+F NL+SV +L + G SI D S KN +V+K N ++ V
Sbjct: 349 LFCKEAAGNLMSVKRLQEAGMSIEF-DKSGVTISKNGLMVVK-----NSGMLNNVPVINF 402
Query: 441 NCLSAIVNNEDSW-LWHYMFGHLNYISLNHLVDKEMIL---VVPKLDIPHKFYDTRLIGK 496
S +++++ LWH FGH++ L + K M ++ L++ + + L GK
Sbjct: 403 QAYSINAKHKNNFRLWHERFGHISDGKLLEIKRKNMFSDQSLLNNLELSCEICEPCLNGK 462
Query: 497 QPRTAF--ISTTTHRSKEVLNVVYFDVNGPLEVPSMGRIKYFISFLDEFSKNIWVCLIKA 554
Q R F + TH K L VV+ DV GP+ ++ YF+ F+D+F+ LIK
Sbjct: 463 QARLPFKQLKDKTH-IKRPLFVVHSDVCGPITPVTLDDKNYFVIFVDQFTHYCVTYLIKY 521
Query: 555 KSGAFDIFQKFKFFVEKQSGKSITSLRTDGGGEYTIKVFEKFCEENGV 602
KS F +FQ F E + L D G EY +FC + G+
Sbjct: 522 KSDVFSMFQDFVAKSEAHFNLKVVYLYIDNGREYLSNEMRQFCVKKGI 569
>YCB9_YEAST (P25384) Transposon Ty2 protein B (Ty1-17 protein B)
Length = 1770
Score = 59.7 bits (143), Expect = 2e-08
Identities = 56/235 (23%), Positives = 95/235 (39%), Gaps = 25/235 (10%)
Query: 389 NLLSVGQLVQKGYSITMKDNSLKLFDKNQRLVMKTPLAKN-------KTF*TDMKVAKLN 441
+LLS+ +L + + N+L+ D + P+ K+ K + ++KL
Sbjct: 521 DLLSLSELANQNITACFTRNTLERSDGT----VLAPIVKHGDFYWLSKKYLIPSHISKLT 576
Query: 442 C----LSAIVNNEDSWLWHYMFGHLNYISLNHLVDKEMILVVPKLDIPHKFYDTR----- 492
S VN L H M GH N+ S+ + K + + + DI T
Sbjct: 577 INNVNKSKSVNKYPYPLIHRMLGHANFRSIQKSLKKNAVTYLKESDIEWSNASTYQCPDC 636
Query: 493 LIGKQPRTAFISTTT---HRSKEVLNVVYFDVNGPLEVPSMGRIKYFISFLDEFSKNIWV 549
LIGK + + + S E ++ D+ GP+ YFISF DE ++ WV
Sbjct: 637 LIGKSTKHRHVKGSRLKYQESYEPFQYLHTDIFGPVHHLPKSAPSYFISFTDEKTRFQWV 696
Query: 550 CLI--KAKSGAFDIFQKFKFFVEKQSGKSITSLRTDGGGEYTIKVFEKFCEENGV 602
+ + + ++F F++ Q + ++ D G EYT K KF G+
Sbjct: 697 YPLHDRREESILNVFTSILAFIKNQFNARVLVIQMDRGSEYTNKTLHKFFTNRGI 751
>YMU0_YEAST (Q04670) Transposon Ty1 protein B
Length = 1328
Score = 57.0 bits (136), Expect = 1e-07
Identities = 43/158 (27%), Positives = 69/158 (43%), Gaps = 12/158 (7%)
Query: 456 HYMFGHLNYISLNHLVDKEMILVVPKLD------IPHKFYDTRLIGKQPRTAFISTTT-- 507
H M H N ++ + + I + D I ++ D LIGK + I +
Sbjct: 172 HRMLAHANAQTIRYSLKNNTITYFNESDVDWSSAIDYQCPDC-LIGKSTKHRHIKGSRLK 230
Query: 508 -HRSKEVLNVVYFDVNGPLEVPSMGRIKYFISFLDEFSKNIWVCLI--KAKSGAFDIFQK 564
S E ++ D+ GP+ YFISF DE +K WV + + + D+F
Sbjct: 231 YQNSYEPFQYLHTDIFGPVHNLPKSAPSYFISFTDETTKFRWVYPLHDRREDSILDVFTT 290
Query: 565 FKFFVEKQSGKSITSLRTDGGGEYTIKVFEKFCEENGV 602
F++ Q S+ ++ D G EYT + KF E+NG+
Sbjct: 291 ILAFIKNQFQASVLVIQMDRGSEYTNRTLHKFLEKNGI 328
>YMT5_YEAST (Q04214) Transposon Ty1 protein B
Length = 1328
Score = 57.0 bits (136), Expect = 1e-07
Identities = 43/158 (27%), Positives = 69/158 (43%), Gaps = 12/158 (7%)
Query: 456 HYMFGHLNYISLNHLVDKEMILVVPKLD------IPHKFYDTRLIGKQPRTAFISTTT-- 507
H M H N ++ + + I + D I ++ D LIGK + I +
Sbjct: 172 HRMLAHANAQTIRYSLKNNTITYFNESDVDWSSAIDYQCPDC-LIGKSTKHRHIKGSRLK 230
Query: 508 -HRSKEVLNVVYFDVNGPLEVPSMGRIKYFISFLDEFSKNIWVCLI--KAKSGAFDIFQK 564
S E ++ D+ GP+ YFISF DE +K WV + + + D+F
Sbjct: 231 YQNSYEPFQYLHTDIFGPVHNLPKSAPSYFISFTDETTKFRWVYPLHDRREDSILDVFTT 290
Query: 565 FKFFVEKQSGKSITSLRTDGGGEYTIKVFEKFCEENGV 602
F++ Q S+ ++ D G EYT + KF E+NG+
Sbjct: 291 ILAFIKNQFQASVLVIQMDRGSEYTNRTLHKFLEKNGI 328
>YME4_YEAST (Q04711) Transposon Ty1 protein B
Length = 1328
Score = 57.0 bits (136), Expect = 1e-07
Identities = 43/158 (27%), Positives = 69/158 (43%), Gaps = 12/158 (7%)
Query: 456 HYMFGHLNYISLNHLVDKEMILVVPKLD------IPHKFYDTRLIGKQPRTAFISTTT-- 507
H M H N ++ + + I + D I ++ D LIGK + I +
Sbjct: 172 HRMLAHANAQTIRYSLKNNTITYFNESDVDWSSAIDYQCPDC-LIGKSTKHRHIKGSRLK 230
Query: 508 -HRSKEVLNVVYFDVNGPLEVPSMGRIKYFISFLDEFSKNIWVCLI--KAKSGAFDIFQK 564
S E ++ D+ GP+ YFISF DE +K WV + + + D+F
Sbjct: 231 YQNSYEPFQYLHTDIFGPVHNLPKSAPSYFISFTDETTKFRWVYPLHDRREDSILDVFTT 290
Query: 565 FKFFVEKQSGKSITSLRTDGGGEYTIKVFEKFCEENGV 602
F++ Q S+ ++ D G EYT + KF E+NG+
Sbjct: 291 ILAFIKNQFQASVLVIQMDRGSEYTNRTLHKFLEKNGI 328
>YMD9_YEAST (Q03434) Transposon Ty1 protein B
Length = 1328
Score = 57.0 bits (136), Expect = 1e-07
Identities = 41/157 (26%), Positives = 66/157 (41%), Gaps = 10/157 (6%)
Query: 456 HYMFGHLNYISLNHLVDKEMILVVPKLDIPHKFY-----DTRLIGKQPRTAFISTTT--- 507
H M H N ++ + + I + D+ LIGK + I +
Sbjct: 172 HRMLAHANAQTIRYSLKNNTITYFNESDVDRSSAIDYQCPDCLIGKSTKHRHIKGSRLKY 231
Query: 508 HRSKEVLNVVYFDVNGPLEVPSMGRIKYFISFLDEFSKNIWVCLI--KAKSGAFDIFQKF 565
S E ++ D+ GP+ YFISF DE +K WV + + + D+F
Sbjct: 232 QNSYEPFQYLHTDIFGPVHNLPKSAPSYFISFTDETTKFRWVYPLHDRREDSILDVFTTI 291
Query: 566 KFFVEKQSGKSITSLRTDGGGEYTIKVFEKFCEENGV 602
F++ Q S+ ++ D G EYT + KF E+NG+
Sbjct: 292 LAFIKNQFQASVLVIQMDRGSEYTNRTLHKFLEKNGI 328
>YJZ9_YEAST (P47100) Transposon Ty1 protein B
Length = 1755
Score = 57.0 bits (136), Expect = 1e-07
Identities = 43/158 (27%), Positives = 69/158 (43%), Gaps = 12/158 (7%)
Query: 456 HYMFGHLNYISLNHLVDKEMILVVPKLD------IPHKFYDTRLIGKQPRTAFISTTT-- 507
H M H N ++ + + I + D I ++ D LIGK + I +
Sbjct: 599 HRMLAHANAQTIRYSLKNNTITYFNESDVDWSSAIDYQCPDC-LIGKSTKHRHIKGSRLK 657
Query: 508 -HRSKEVLNVVYFDVNGPLEVPSMGRIKYFISFLDEFSKNIWVCLI--KAKSGAFDIFQK 564
S E ++ D+ GP+ YFISF DE +K WV + + + D+F
Sbjct: 658 YQNSYEPFQYLHTDIFGPVHNLPKSAPSYFISFTDETTKFRWVYPLHDRREDSILDVFTT 717
Query: 565 FKFFVEKQSGKSITSLRTDGGGEYTIKVFEKFCEENGV 602
F++ Q S+ ++ D G EYT + KF E+NG+
Sbjct: 718 ILAFIKNQFQASVLVIQMDRGSEYTNRTLHKFLEKNGI 755
>YJZ7_YEAST (P47098) Transposon Ty1 protein B
Length = 1755
Score = 57.0 bits (136), Expect = 1e-07
Identities = 43/158 (27%), Positives = 69/158 (43%), Gaps = 12/158 (7%)
Query: 456 HYMFGHLNYISLNHLVDKEMILVVPKLD------IPHKFYDTRLIGKQPRTAFISTTT-- 507
H M H N ++ + + I + D I ++ D LIGK + I +
Sbjct: 599 HRMLAHANAQTIRYSLKNNTITYFNESDVDWSSAIDYQCPDC-LIGKSTKHRHIKGSRLK 657
Query: 508 -HRSKEVLNVVYFDVNGPLEVPSMGRIKYFISFLDEFSKNIWVCLI--KAKSGAFDIFQK 564
S E ++ D+ GP+ YFISF DE +K WV + + + D+F
Sbjct: 658 YQNSYEPFQYLHTDIFGPVHNLPKSAPSYFISFTDETTKFRWVYPLHDRREDSILDVFTT 717
Query: 565 FKFFVEKQSGKSITSLRTDGGGEYTIKVFEKFCEENGV 602
F++ Q S+ ++ D G EYT + KF E+NG+
Sbjct: 718 ILAFIKNQFQASVLVIQMDRGSEYTNRTLHKFLEKNGI 755
>POL_JSRV (P31623) Pol polyprotein [Contains: Reverse
transcriptase/ribonuclease H (EC 2.7.7.49) (EC 3.1.26.4)
(RT); Integrase (IN)]
Length = 870
Score = 35.0 bits (79), Expect = 0.57
Identities = 24/71 (33%), Positives = 34/71 (47%), Gaps = 3/71 (4%)
Query: 527 VPSMGRIKYFISFLDEFSKNIWVCLIKAKSGAFDIFQKFKFFVEKQSGKSITSLRTDGGG 586
+P GR+KY +D FS + L +S I + F SG T L+TD G
Sbjct: 659 IPQFGRLKYVHVSIDTFSNFLMASLHTGESTRHCI--QHLLFCFSTSGIPQT-LKTDNGP 715
Query: 587 EYTIKVFEKFC 597
YT + F++FC
Sbjct: 716 GYTSRSFQRFC 726
>POL_IPHA (P04026) Putative Pol polyprotein [Contains: Integrase
(IN); Reverse transcriptase/ribonuclease H (EC 2.7.7.49)
(EC 3.1.26.4) (RT)]
Length = 863
Score = 33.9 bits (76), Expect = 1.3
Identities = 20/78 (25%), Positives = 37/78 (46%), Gaps = 4/78 (5%)
Query: 527 VPSMGRIKYFISFLDEFSKNIWVCLIKAKSGAFDIFQKFKFFVEKQSGKSITSLRTDGGG 586
+PS GR++Y +D S ++ + + ++ I + +E S ++TD G
Sbjct: 611 IPSFGRLQYVHVSVDTCSGVMFATPLTGEKASYVI----QHCLEAWSAWGKPRIKTDNGP 666
Query: 587 EYTIKVFEKFCEENGVIH 604
YT + F +FC + V H
Sbjct: 667 AYTSQKFRQFCRQMDVTH 684
>POK6_HUMAN (P63133) HERV-K_8p23.1 provirus ancestral Pol protein
(HERV-K115 Pol protein) [Includes: Reverse transcriptase
(RT) (EC 2.7.7.49); Ribonuclease H (EC 3.1.26.4) (RNase
H); Integrase (IN)]
Length = 956
Score = 31.6 bits (70), Expect = 6.3
Identities = 22/78 (28%), Positives = 32/78 (40%), Gaps = 3/78 (3%)
Query: 527 VPSMGRIKYFISFLDEFSKNIWVCLIKAKSGAFDIFQKFKFFVEKQSGKSITSLRTDGGG 586
VPS GR+ Y +D +S IW +S + F + I +TD G
Sbjct: 657 VPSFGRLSYVHVTVDTYSHFIWATCQTGESTSHVKKHLLSCFAVMGVPEKI---KTDNGP 713
Query: 587 EYTIKVFEKFCEENGVIH 604
Y K F+KF + + H
Sbjct: 714 GYCSKAFQKFLSQWKISH 731
>POK5_HUMAN (P63132) HERV-K_19p13.11 provirus ancestral Pol protein
(HERV-K113 Pol protein) [Includes: Reverse transcriptase
(RT) (EC 2.7.7.49); Ribonuclease H (EC 3.1.26.4) (RNase
H); Integrase (IN)]
Length = 956
Score = 31.6 bits (70), Expect = 6.3
Identities = 22/78 (28%), Positives = 32/78 (40%), Gaps = 3/78 (3%)
Query: 527 VPSMGRIKYFISFLDEFSKNIWVCLIKAKSGAFDIFQKFKFFVEKQSGKSITSLRTDGGG 586
VPS GR+ Y +D +S IW +S + F + I +TD G
Sbjct: 657 VPSFGRLSYVHVTVDTYSHFIWATCQTGESTSHVKKHLLSCFAVMGVPEKI---KTDNGP 713
Query: 587 EYTIKVFEKFCEENGVIH 604
Y K F+KF + + H
Sbjct: 714 GYCSKAFQKFLSQWKISH 731
>POK2_HUMAN (Q9BXR3) HERV-K_7p22.1 provirus ancestral Pol protein
(HERV-K(HML-2.HOM) Pol protein) (HERV-K108 Pol protein)
(HERV-K(C7) Pol protein) [Includes: Reverse
transcriptase (RT) (EC 2.7.7.49); Ribonuclease H (EC
3.1.26.4) (RNase H); Integrase (IN)]
Length = 956
Score = 31.6 bits (70), Expect = 6.3
Identities = 22/78 (28%), Positives = 32/78 (40%), Gaps = 3/78 (3%)
Query: 527 VPSMGRIKYFISFLDEFSKNIWVCLIKAKSGAFDIFQKFKFFVEKQSGKSITSLRTDGGG 586
VPS GR+ Y +D +S IW +S + F + I +TD G
Sbjct: 657 VPSFGRLSYVHVTVDTYSHFIWATCQTGESTSHVKKHLLSCFAVMGVPEKI---KTDNGP 713
Query: 587 EYTIKVFEKFCEENGVIH 604
Y K F+KF + + H
Sbjct: 714 GYCSKAFQKFLSQWKISH 731
>PK20_HUMAN (Q9UQG0) HERV-K_3q27.3 provirus ancestral Pol protein
[Includes: Reverse transcriptase (RT) (EC 2.7.7.49);
Ribonuclease H (EC 3.1.26.4) (RNase H); Integrase (IN)]
Length = 969
Score = 31.6 bits (70), Expect = 6.3
Identities = 22/78 (28%), Positives = 32/78 (40%), Gaps = 3/78 (3%)
Query: 527 VPSMGRIKYFISFLDEFSKNIWVCLIKAKSGAFDIFQKFKFFVEKQSGKSITSLRTDGGG 586
VPS GR+ Y +D +S IW +S + F + I +TD G
Sbjct: 657 VPSFGRLSYVHVTVDTYSHFIWATCQTGESTSHVKKHLLSCFAVMGVPEKI---KTDNGP 713
Query: 587 EYTIKVFEKFCEENGVIH 604
Y K F+KF + + H
Sbjct: 714 GYCSKAFQKFLSQWKISH 731
>PK17_HUMAN (P63136) HERV-K_11q22.1 provirus ancestral Pol protein
[Includes: Reverse transcriptase (RT) (EC 2.7.7.49);
Ribonuclease H (EC 3.1.26.4) (RNase H); Integrase (IN)]
Length = 954
Score = 31.6 bits (70), Expect = 6.3
Identities = 22/78 (28%), Positives = 32/78 (40%), Gaps = 3/78 (3%)
Query: 527 VPSMGRIKYFISFLDEFSKNIWVCLIKAKSGAFDIFQKFKFFVEKQSGKSITSLRTDGGG 586
VPS GR+ Y +D +S IW +S + F + I +TD G
Sbjct: 655 VPSFGRLSYVHVTVDTYSHFIWATCHTGESTSHVKKHLLSCFAVMGVPEKI---KTDNGP 711
Query: 587 EYTIKVFEKFCEENGVIH 604
Y K F+KF + + H
Sbjct: 712 GYCSKAFQKFLSQWKISH 729
>PK12_HUMAN (P63135) HERV-K_1q22 provirus ancestral Pol protein
(HERV-K102 Pol protein) (HERV-K(III) Pol protein)
[Includes: Reverse transcriptase (RT) (EC 2.7.7.49);
Ribonuclease H (EC 3.1.26.4) (RNase H); Integrase (IN)]
Length = 1459
Score = 31.6 bits (70), Expect = 6.3
Identities = 22/78 (28%), Positives = 32/78 (40%), Gaps = 3/78 (3%)
Query: 527 VPSMGRIKYFISFLDEFSKNIWVCLIKAKSGAFDIFQKFKFFVEKQSGKSITSLRTDGGG 586
VPS GR+ Y +D +S IW +S + F + I +TD G
Sbjct: 657 VPSFGRLSYVHVTVDTYSHFIWATCQTGESTSHVKKHLLSCFAVMGVPEKI---KTDNGP 713
Query: 587 EYTIKVFEKFCEENGVIH 604
Y K F+KF + + H
Sbjct: 714 GYCSKAFQKFLSQWKISH 731
>PK10_HUMAN (P10266) HERV-K_5q33.3 provirus ancestral Pol protein
(HERV-K10 Pol protein) (HERV-K107 Pol protein)
[Includes: Reverse transcriptase (RT) (EC 2.7.7.49);
Ribonuclease H (EC 3.1.26.4) (RNase H); Integrase (IN)]
Length = 1014
Score = 31.6 bits (70), Expect = 6.3
Identities = 22/78 (28%), Positives = 32/78 (40%), Gaps = 3/78 (3%)
Query: 527 VPSMGRIKYFISFLDEFSKNIWVCLIKAKSGAFDIFQKFKFFVEKQSGKSITSLRTDGGG 586
VPS GR+ Y +D +S IW +S + F + I +TD G
Sbjct: 657 VPSFGRLSYVHVTVDTYSHFIWATCQTGESTSHVKKHLLSCFAVMGVPEKI---KTDNGP 713
Query: 587 EYTIKVFEKFCEENGVIH 604
Y K F+KF + + H
Sbjct: 714 GYCSKAFQKFLSQWKISH 731
>M300_ARATH (P93293) Hypothetical mitochondrial protein AtMg00300
(ORF145a) (ORF1451)
Length = 145
Score = 31.6 bits (70), Expect = 6.3
Identities = 23/79 (29%), Positives = 37/79 (46%), Gaps = 3/79 (3%)
Query: 450 EDSWLWHYMFGHLNYISLNHLVDKEMILVVPKLDIPHKFYDTRLIGKQPRTAFISTTTHR 509
+++ LWH H++ + LV K + + KF + + GK R F ST H
Sbjct: 67 DETRLWHSRLAHMSQRGMELLVKKGFLDSSKVSSL--KFCEDCIYGKTHRVNF-STGQHT 123
Query: 510 SKEVLNVVYFDVNGPLEVP 528
+K L+ V+ D+ G VP
Sbjct: 124 TKNPLDYVHSDLWGAPSVP 142
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.349 0.155 0.516
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 60,103,888
Number of Sequences: 164201
Number of extensions: 2244818
Number of successful extensions: 8995
Number of sequences better than 10.0: 19
Number of HSP's better than 10.0 without gapping: 8
Number of HSP's successfully gapped in prelim test: 11
Number of HSP's that attempted gapping in prelim test: 8966
Number of HSP's gapped (non-prelim): 22
length of query: 604
length of database: 59,974,054
effective HSP length: 116
effective length of query: 488
effective length of database: 40,926,738
effective search space: 19972248144
effective search space used: 19972248144
T: 11
A: 40
X1: 14 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 38 (21.8 bits)
S2: 69 (31.2 bits)
Medicago: description of AC145061.22


