

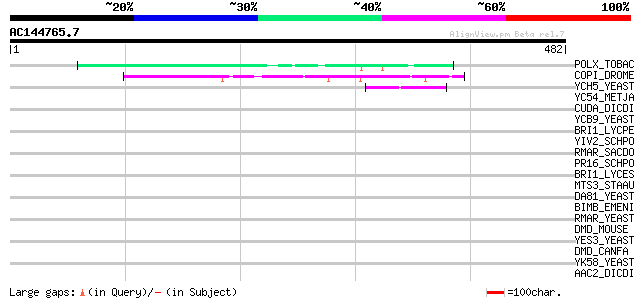
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144765.7 - phase: 0
(482 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
POLX_TOBAC (P10978) Retrovirus-related Pol polyprotein from tran... 66 2e-10
COPI_DROME (P04146) Copia protein (Gag-int-pol protein) [Contain... 65 5e-10
YCH5_YEAST (P25601) Transposon Ty5-1 16.0 kDa hypothetical protein 47 1e-04
YC54_METJA (Q58651) Hypothetical protein MJ1254 41 0.008
CUDA_DICDI (O00841) Putative transcriptional regulator cudA 38 0.066
YCB9_YEAST (P25384) Transposon Ty2 protein B (Ty1-17 protein B) 36 0.25
BRI1_LYCPE (Q8L899) Systemin receptor SR160 precursor (EC 2.7.1.... 35 0.33
YIV2_SCHPO (Q9UTM0) Hypothetical zinc finger protein C144.02 in ... 35 0.43
RMAR_SACDO (Q35905) Mitochondrial ribosomal protein VAR1 33 1.3
PR16_SCHPO (Q9P774) Pre-mRNA splicing factor RNA helicase prp16 ... 33 1.3
BRI1_LYCES (Q8GUQ5) Brassinosteroid LRR receptor kinase precurso... 33 1.3
MTS3_STAAU (P16668) Modification methylase Sau3AI (EC 2.1.1.37) ... 33 1.6
DA81_YEAST (P21657) Transcriptional activator protein DAL81 (Reg... 33 1.6
BIMB_EMENI (P33144) Separin (EC 3.4.22.49) (Separase) (Cell divi... 33 2.1
RMAR_YEAST (P02381) Mitochondrial ribosomal protein VAR1 32 2.8
DMD_MOUSE (P11531) Dystrophin 32 2.8
YES3_YEAST (P40061) Hypothetical 164.4 kDa protein in MET6-PUP3 ... 32 3.6
DMD_CANFA (O97592) Dystrophin 32 3.6
YK58_YEAST (P36158) Hypothetical 68.3 kDa protein in SIS2-MTD1 i... 32 4.8
AAC2_DICDI (P14196) AAC-rich mRNA clone AAC11 protein (Fragment) 32 4.8
>POLX_TOBAC (P10978) Retrovirus-related Pol polyprotein from
transposon TNT 1-94 [Contains: Protease (EC 3.4.23.-);
Reverse transcriptase (EC 2.7.7.49); Endonuclease]
Length = 1328
Score = 65.9 bits (159), Expect = 2e-10
Identities = 76/352 (21%), Positives = 134/352 (37%), Gaps = 45/352 (12%)
Query: 60 DSSKSNNPAFEEWQANDQRLLGWMLNSMATEMATQLLHCETSKQLWDEAQSLAGAHTRSQ 119
DS K + E+W D+R + ++ ++ ++ +T++ +W +SL + T +
Sbjct: 40 DSKKPDTMKAEDWADLDERAASAIRLHLSDDVVNNIIDEDTARGIWTRLESLYMSKTLTN 99
Query: 120 IIYLKSEFHSIRKGE-MKMEDYLIKMKNLADKLKLAGNPISNSDLIIQTLNGLDSEYNPI 178
+YLK + +++ E +L L +L G I D I LN L S Y+ +
Sbjct: 100 KLYLKKQLYALHMSEGTNFLSHLNVFNGLITQLANLGVKIEEEDKAILLLNSLPSSYDNL 159
Query: 179 VVK-LSDHTTLSWVDLQAQLLTFESRIEQLNNLTNLNLNATANVANKFDHRDNRFNSNNN 237
L TT+ D+ + LL E ++ N + + + S+NN
Sbjct: 160 ATTILHGKTTIELKDVTSALLLNEKMRKKPENQGQALITEGRGRSYQ--------RSSNN 211
Query: 238 WRGSNFRGWRGGRGRGRSSKAPCQVCGKTNHTAINCFHRFDKNYSRSNYSADSDKQGSHN 297
+ S RG + R +S C C + H F R N + K +
Sbjct: 212 YGRSGARG--KSKNRSKSRVRNCYNCNQPGH-----FKRDCPNPRKGKGETSGQKNDDNT 264
Query: 298 AFIASQN----------------SVEDYDWYFDSGASNHVT--------HQTNKFQDLTE 333
A + N S + +W D+ AS+H T + F +
Sbjct: 265 AAMVQNNDNVVLFINEEEECMHLSGPESEWVVDTAASHHATPVRDLFCRYVAGDFGTVKM 324
Query: 334 HHGKNSLVVGNGDKLEIVATCSSKLKSLNLDDVLYVPNITKNLLSVSKLAAD 385
+ S + G GD + ++ +L L DV +VP++ NL+S L D
Sbjct: 325 GNTSYSKIAGIGD----ICIKTNVGCTLVLKDVRHVPDLRMNLISGIALDRD 372
>COPI_DROME (P04146) Copia protein (Gag-int-pol protein) [Contains:
Copia VLP protein; Copia protease (EC 3.4.23.-)]
Length = 1409
Score = 64.7 bits (156), Expect = 5e-10
Identities = 72/312 (23%), Positives = 136/312 (43%), Gaps = 27/312 (8%)
Query: 100 TSKQLWDEAQSLAGAHTRSQIIYLKSEFHSIR-KGEMKMEDYLIKMKNLADKLKLAGNPI 158
T++Q+ + ++ + + + L+ S++ EM + + L +L AG I
Sbjct: 76 TARQILENLDAVYERKSLASQLALRKRLLSLKLSSEMSLLSHFHIFDELISELLAAGAKI 135
Query: 159 SNSDLIIQTLNGLDSEYNPIVVKLS----DHTTLSWVDLQAQLLTFESRIEQLNNLTNLN 214
D I L L S Y+ I+ + ++ TL++V + +LL E +I+ +N T+
Sbjct: 136 EEMDKISHLLITLPSCYDGIITAIETLSEENLTLAFV--KNRLLDQEIKIKNDHNDTS-- 191
Query: 215 LNATANVANKFDHRDNRFNSNNNWRGSNFRGWRGGRGRGRSSKAPCQVCGKTNHTAINCF 274
V N H +N NN ++ + + +G + K C CG+ H +CF
Sbjct: 192 ----KKVMNAIVHNNNNTYKNNLFKNRVTKPKKIFKGNSKY-KVKCHHCGREGHIKKDCF 246
Query: 275 H--RFDKNYSRSNYSADSDKQGSHNAFIASQ----NSVEDYDWYFDSGASNHVTHQTNKF 328
H R N ++ N AF+ + + +++ + DSGAS+H+ + + +
Sbjct: 247 HYKRILNNKNKENEKQVQTATSHGIAFMVKEVNNTSVMDNCGFVLDSGASDHLINDESLY 306
Query: 329 QDLTEHHGKNSLVVGNGDKLEIVATCSSKLK-----SLNLDDVLYVPNITKNLLSVSKLA 383
D E + V + I AT ++ + L+DVL+ NL+SV +L
Sbjct: 307 TDSVEVVPPLKIAVAKQGEF-IYATKRGIVRLRNDHEITLEDVLFCKEAAGNLMSVKRL- 364
Query: 384 ADNNIFVEFDKN 395
+ + +EFDK+
Sbjct: 365 QEAGMSIEFDKS 376
>YCH5_YEAST (P25601) Transposon Ty5-1 16.0 kDa hypothetical protein
Length = 146
Score = 47.0 bits (110), Expect = 1e-04
Identities = 23/70 (32%), Positives = 40/70 (56%), Gaps = 1/70 (1%)
Query: 310 DWYFDSGASNHVTHQTNKFQDLTEHHGKNSLVVGNGDKLEIVATCSSKLKSLNLDDVLYV 369
+W FD+G ++H+ H + F T K+ V G G + I+ + + + ++ L DV YV
Sbjct: 78 EWIFDTGCTSHMCHDRSIFSSFTRSSRKD-FVRGVGGSIPIMGSGTVNIGTVQLHDVSYV 136
Query: 370 PNITKNLLSV 379
P++ NL+SV
Sbjct: 137 PDLPVNLISV 146
>YC54_METJA (Q58651) Hypothetical protein MJ1254
Length = 469
Score = 40.8 bits (94), Expect = 0.008
Identities = 27/86 (31%), Positives = 43/86 (49%), Gaps = 11/86 (12%)
Query: 123 LKSEFHSIRKGEMKMEDYLIKMKNLADKLKLAGNPISNSDLIIQTLNGLDSEYNPIVVKL 182
L S + +++ E ++++ IK+K+L DKL A + N D II LN SEY + KL
Sbjct: 289 LTSLKNKLKEKEEEIKNLAIKIKDLEDKLSKANKNLLNKDEIISVLNERISEYESQIQKL 348
Query: 183 SDHTTLSWVDLQAQLLTFESRIEQLN 208
D + ++ +IE LN
Sbjct: 349 LDEN-----------IIYKEKIESLN 363
>CUDA_DICDI (O00841) Putative transcriptional regulator cudA
Length = 791
Score = 37.7 bits (86), Expect = 0.066
Identities = 59/245 (24%), Positives = 107/245 (43%), Gaps = 39/245 (15%)
Query: 8 NKNDLPSSVSVKLDRNNYPLWKSLVLPVVRGCKLDGYMLGTKKCPE-EFITSSDSSKSNN 66
+KN + +V V + N + L SL LD + + PE + + S+S K +
Sbjct: 194 SKNGVQKNVHVVVKNNPFLLTLSL---------LDSSLNFHQLTPEVQLVYDSESLKEVD 244
Query: 67 -----PAFEEWQANDQRLLGWMLNSMATEMATQLLHCETSKQLWDEAQSLAGAHTRSQII 121
P + +AN++ + + E+ ++L + L+ + TR +
Sbjct: 245 SATVKPLEYKTRANEEG------DQLTIELRIKVLSSQLEDMLFRAKVKIVDPRTRKETH 298
Query: 122 YLKSEFHSIRKGEMKMEDYLIKMKNLADKLKLAGNPISNSDLIIQTLNGLD---SEYNPI 178
L H IR + D ++K A K K A +D ++ TLN ++ E +
Sbjct: 299 GLSVITHPIRV--VSKPD---QVKKKAKKRKRAP-----TDSLMDTLNRIEHQQKEQQRL 348
Query: 179 VVKLSDHTTLSWVDLQAQLLTFESRIEQL-NNLTNLNLNATANVANKFDHRDNRFNSNNN 237
+ KL H + + QL+ + + +QL NN+TN N+N N+ N ++ +N N+NNN
Sbjct: 349 LKKLCYHDKENNI---IQLIQQQQQQQQLLNNVTN-NINNNNNINNNNNNNNNNNNNNNN 404
Query: 238 WRGSN 242
+N
Sbjct: 405 NNNNN 409
>YCB9_YEAST (P25384) Transposon Ty2 protein B (Ty1-17 protein B)
Length = 1770
Score = 35.8 bits (81), Expect = 0.25
Identities = 64/283 (22%), Positives = 117/283 (40%), Gaps = 37/283 (13%)
Query: 133 GEMKMEDYLIKMKNLADKLKLAGNPISNSD-----LIIQTLNGLDSEYNPIVVKLSDHTT 187
G + + I + + +LK N I+ SD LI++ L+G D +Y + +
Sbjct: 275 GSTSADTFEITVSTIIQRLK--ENNINVSDRLACQLILKGLSG-DFKYLRNQYRTKTNMK 331
Query: 188 LSWVDLQAQLLTFESRIEQLNNLTNLNLNAT-ANVANKFDHRDNRFNSNNNWRGSNFRGW 246
LS + + QL+ E++I LN + ++ NV+ + N + N++ +N
Sbjct: 332 LSQLFAEIQLIYDENKIMNLNKPSQYKQHSEYKNVSRTSPNTTNTKVTTRNYQRTN---- 387
Query: 247 RGGRGRGRSSKAPCQVCGKTNHTAINCFHRFDKNYSRSNYSADSD-------KQGSHNAF 299
+ R++KA + + + +N H + S S D++ K+
Sbjct: 388 ---SSKPRAAKAH-NIATSSKFSRVNNDHINESTVSSQYLSDDNELSLGQQQKESKPTHT 443
Query: 300 IASQNSVEDYDWYFDSGASNHVTHQTNKFQDLTEHHGKNSLVVGNGDKLEIVATCSSKLK 359
I S + + D+ DSGAS + + T + N + + K +I L
Sbjct: 444 IDSNDELPDH-LLIDSGASQTLVRSAHYLHHATPNSEIN---IVDAQKQDIPINAIGNLH 499
Query: 360 SLNLDD-------VLYVPNITKNLLSVSKLAADNNIFVEFDKN 395
N + L+ PNI +LLS+S+L A+ NI F +N
Sbjct: 500 -FNFQNGTKTSIKALHTPNIAYDLLSLSEL-ANQNITACFTRN 540
>BRI1_LYCPE (Q8L899) Systemin receptor SR160 precursor (EC 2.7.1.37)
(Brassinosteroid LRR receptor kinase)
Length = 1207
Score = 35.4 bits (80), Expect = 0.33
Identities = 41/170 (24%), Positives = 70/170 (41%), Gaps = 11/170 (6%)
Query: 54 EFITSSDSSKSNNPAFEEWQANDQRLLGWMLNSMATEMATQL--LHCETSKQLWDEAQSL 111
+F+ S SN P E + L G + + + + L L+ + + SL
Sbjct: 388 KFVGGLPDSFSNLPKLETLDMSSNNLTGIIPSGICKDPMNNLKVLYLQNNLFKGPIPDSL 447
Query: 112 AGAHTRSQIIYLKSEFHSIRKGEMKMEDYLIKMKNLADKLKLAGNPISNSDLIIQTLNGL 171
+ SQ++ L F+ + L K+K+L L I + +Q L L
Sbjct: 448 SNC---SQLVSLDLSFNYLTGSIPSSLGSLSKLKDLILWLNQLSGEIPQELMYLQALENL 504
Query: 172 DSEYN----PIVVKLSDHTTLSWVDLQAQLLTFE--SRIEQLNNLTNLNL 215
++N PI LS+ T L+W+ L L+ E + + +L+NL L L
Sbjct: 505 ILDFNDLTGPIPASLSNCTKLNWISLSNNQLSGEIPASLGRLSNLAILKL 554
>YIV2_SCHPO (Q9UTM0) Hypothetical zinc finger protein C144.02 in
chromosome I
Length = 249
Score = 35.0 bits (79), Expect = 0.43
Identities = 16/56 (28%), Positives = 28/56 (49%)
Query: 176 NPIVVKLSDHTTLSWVDLQAQLLTFESRIEQLNNLTNLNLNATANVANKFDHRDNR 231
+PI ++ T W + LLT ++ + ++N T NL +N+ + DH NR
Sbjct: 14 SPIDLEPESETICHWQSCEQDLLTLDNLVHHIHNGTTSNLRLISNINSILDHIGNR 69
>RMAR_SACDO (Q35905) Mitochondrial ribosomal protein VAR1
Length = 373
Score = 33.5 bits (75), Expect = 1.3
Identities = 21/73 (28%), Positives = 34/73 (45%), Gaps = 19/73 (26%)
Query: 169 NGLDSEY----NPIVVKLSDHTTLSWVDLQAQLLTFESRIEQLNNLTNLNLNATANVANK 224
NG+ + Y N I+ KL+DH + +NN+ N+N N N+ N
Sbjct: 211 NGILTNYQRMLNNIMPKLNDHNI---------------SMNYINNINNINNNKYNNMINL 255
Query: 225 FDHRDNRFNSNNN 237
++ +N N+NNN
Sbjct: 256 LNNNNNNNNNNNN 268
>PR16_SCHPO (Q9P774) Pre-mRNA splicing factor RNA helicase prp16 (EC
3.6.1.-)
Length = 1173
Score = 33.5 bits (75), Expect = 1.3
Identities = 32/131 (24%), Positives = 54/131 (40%), Gaps = 27/131 (20%)
Query: 224 KFDHRDNRF--NSNNNWRGSNFRGWRGGRGRGRSSKAPCQVCGKTNHTAINCFHRFDKNY 281
+F+ D+++ NS ++W GSNF G R ++ + K + N R +
Sbjct: 182 EFESEDDKYRRNSYSSWSGSNFESSNGSNRRRYRTQMEEPLSSKRRNRFGNGSRR---DV 238
Query: 282 SRSNYSADSD-KQGSHNAFIA--------------------SQNSVEDYDWYFDSGASNH 320
S YS DSD G+H+++ A + + D DWY +S + N
Sbjct: 239 DSSKYSHDSDYSYGAHSSWDARDVEYPEEDPESKADRQRWEEEQAHLDRDWYMNSESQNL 298
Query: 321 VTHQT-NKFQD 330
+ + N F D
Sbjct: 299 LGDEVHNPFSD 309
>BRI1_LYCES (Q8GUQ5) Brassinosteroid LRR receptor kinase precursor
(EC 2.7.1.37) (tBRI1) (Altered brassinolide sensitivity
1) (Systemin receptor SR160)
Length = 1207
Score = 33.5 bits (75), Expect = 1.3
Identities = 29/104 (27%), Positives = 47/104 (44%), Gaps = 6/104 (5%)
Query: 118 SQIIYLKSEFHSIRKGEMKMEDYLIKMKNLADKLKLAGNPISNSDLIIQTLNGLDSEYN- 176
SQ++ L F+ + L K+K+L L I + +Q L L ++N
Sbjct: 451 SQLVSLDLSFNYLTGSIPSSLGSLSKLKDLILWLNQLSGEIPQELMYLQALENLILDFND 510
Query: 177 ---PIVVKLSDHTTLSWVDLQAQLLTFE--SRIEQLNNLTNLNL 215
PI LS+ T L+W+ L L+ E + + +L+NL L L
Sbjct: 511 LTGPIPASLSNCTKLNWISLSNNQLSGEIPASLGRLSNLAILKL 554
>MTS3_STAAU (P16668) Modification methylase Sau3AI (EC 2.1.1.37)
(Cytosine-specific methyltransferase Sau3AI) (M.Sau3AI)
Length = 412
Score = 33.1 bits (74), Expect = 1.6
Identities = 26/93 (27%), Positives = 41/93 (43%), Gaps = 2/93 (2%)
Query: 275 HRFDKNYSRSNYSADSDKQGSHNAFIASQNSVEDYDWYFDSGASNHVTHQTNKFQDLTEH 334
++ D NYS++ + DK HN A +EDY + H+TH D
Sbjct: 181 YKQDLNYSKAMEESPLDKIIYHNGLFAEAFPIEDYA-NKNRVNRTHITHDIVDISDNFSF 239
Query: 335 HGKNSLVVGNGDKLEIVATCSSKLKSLNLDDVL 367
NS ++ NG+ L I T KS+ L +++
Sbjct: 240 QFYNSGIMKNGEILTI-DTIPKYEKSVTLGEII 271
>DA81_YEAST (P21657) Transcriptional activator protein DAL81
(Regulatory protein UGA35)
Length = 970
Score = 33.1 bits (74), Expect = 1.6
Identities = 43/185 (23%), Positives = 72/185 (38%), Gaps = 19/185 (10%)
Query: 194 QAQLLTFESRIEQLNNLTNLNLNATA---NVANKFDHRDNRFNSNNNWRGSNFRG-WRGG 249
Q Q + ++ + +L + + NA A NV+N DH + N+NNN + G G
Sbjct: 90 QQQQQSLDTLLHHYQSLLSKSDNAIAFDDNVSNSADHNGSNSNNNNNNNDISSPGNLMGS 149
Query: 250 RGRGRSSKAPCQV------CGKTNHTAINCFHRFDKNY-SRSNYSADSDKQGSHNAFIAS 302
+ R K C C + + C NY R++ A+++ S N+
Sbjct: 150 CNQCRLKKTKCNYFPDLGNCLECETSRTKCTFSIAPNYLKRTSSGANNNMPTSSNS--KR 207
Query: 303 QNSVEDYDWYFDSG-ASNHVTHQTNKFQDLTEHHGKNSLVVGNGDKLEIVATCSSKLKSL 361
+ EDY S H Q + Q + ++S VG ++ T K +
Sbjct: 208 MKNFEDYSNRLPSSMLYRHQQQQQQQQQQQRIQYPRSSFFVGPASVFDLNLT-----KHV 262
Query: 362 NLDDV 366
LD+V
Sbjct: 263 RLDNV 267
>BIMB_EMENI (P33144) Separin (EC 3.4.22.49) (Separase) (Cell
division-associated protein bimB)
Length = 2067
Score = 32.7 bits (73), Expect = 2.1
Identities = 19/73 (26%), Positives = 33/73 (45%), Gaps = 9/73 (12%)
Query: 78 RLLGWMLNSMATEMATQLLHCETSKQLWDEAQSLAGAHTRSQIIYLKSEFHSIRKGEMKM 137
R + W L+ A CE +K++WD GA ++ K+EF I K +++
Sbjct: 377 RCMSWKLSGHA---------CEDNKEMWDPLARYIGAFAQATKSIEKAEFAVIYKNIVRL 427
Query: 138 EDYLIKMKNLADK 150
+ K +N A +
Sbjct: 428 QSAFSKTQNCATR 440
>RMAR_YEAST (P02381) Mitochondrial ribosomal protein VAR1
Length = 404
Score = 32.3 bits (72), Expect = 2.8
Identities = 21/79 (26%), Positives = 36/79 (44%), Gaps = 19/79 (24%)
Query: 169 NGLDSEY----NPIVVKLSDHTTLSWVDLQAQLLTFESRIEQLNNLTNLNLNATANVANK 224
NG+ + Y N I+ KL+DH + +NN+ N+N N N+ N
Sbjct: 234 NGILTNYQRMLNNIMPKLNDHNI---------------SMNYINNINNINNNKYNNMINL 278
Query: 225 FDHRDNRFNSNNNWRGSNF 243
++ +N N+NN +N+
Sbjct: 279 LNNNNNNNNNNNINNNNNY 297
>DMD_MOUSE (P11531) Dystrophin
Length = 3678
Score = 32.3 bits (72), Expect = 2.8
Identities = 31/131 (23%), Positives = 57/131 (42%), Gaps = 21/131 (16%)
Query: 99 ETSKQLWDEAQSLAGAHTRSQIIYLKSEFHSIRKGEMKMEDYLIKMKNLADK-------- 150
E K+L + L TRS+ + SEF RK E + D K+ +A +
Sbjct: 721 ELRKRLDVDITELHSWITRSEAVLQSSEFAVYRK-EGNISDLQEKVNAIAREKAEKFRKL 779
Query: 151 ----------LKLAGNPISNSDLIIQTLNGLDSEYNPIVVKLSDHTTLSWVDLQAQLLTF 200
++ N N++ I Q L+S + LS+ ++W++ Q ++TF
Sbjct: 780 QDASRSAQALVEQMANEGVNAESIRQASEQLNSRWTEFCQLLSER--VNWLEYQTNIITF 837
Query: 201 ESRIEQLNNLT 211
++++QL +T
Sbjct: 838 YNQLQQLEQMT 848
>YES3_YEAST (P40061) Hypothetical 164.4 kDa protein in MET6-PUP3
intergenic region
Length = 1430
Score = 32.0 bits (71), Expect = 3.6
Identities = 31/134 (23%), Positives = 56/134 (41%), Gaps = 3/134 (2%)
Query: 96 LHCETSKQLWDEAQSLAGAHTRSQIIYLKSEFHSIRKGEMKMEDYLIKMKNLADKLKLAG 155
L T ++ + S S I ++ I ++E ++ +K DK +
Sbjct: 129 LRDSTISDIYSGSYSTNHLQKHSMRIRANTQLREIDNSIKRVEKHIFDLKQQFDKKRQRS 188
Query: 156 NPISNS-DLIIQTLNGLDSEYNPIVVKLSDHTTLS-WVDLQAQLLTFESRIEQLNNLTNL 213
S+S + ++ D + N +L DH +L+ V L + LT + + N+ NL
Sbjct: 189 LTTSSSIKADVGSIRNDDGQNNDSE-ELGDHDSLTDQVTLDDEYLTTPTSGTERNSQQNL 247
Query: 214 NLNATANVANKFDH 227
N N+T N N +H
Sbjct: 248 NRNSTVNSRNNENH 261
>DMD_CANFA (O97592) Dystrophin
Length = 3680
Score = 32.0 bits (71), Expect = 3.6
Identities = 34/134 (25%), Positives = 59/134 (43%), Gaps = 19/134 (14%)
Query: 95 LLHCETSKQLWDEAQSLAGAHTRSQIIYLKSEFHSIRK----GEMKMEDYLI------KM 144
++ E K+L + L TRS+ + EF RK ++K + I K
Sbjct: 716 IVDSEIRKRLDVDITELHSWITRSEAVLQSPEFAIYRKEGNFSDLKEKVNAIEREKAEKF 775
Query: 145 KNLADKLKLAG-------NPISNSDLIIQTLNGLDSEYNPIVVKLSDHTTLSWVDLQAQL 197
+ L D + A N N+D I Q L+S + LS+ L+W++ Q +
Sbjct: 776 RKLQDASRSAQALVEQMVNEGVNADSIKQASEQLNSRWIEFCQLLSER--LNWLEYQNNI 833
Query: 198 LTFESRIEQLNNLT 211
+TF ++++QL +T
Sbjct: 834 ITFYNQLQQLEQMT 847
>YK58_YEAST (P36158) Hypothetical 68.3 kDa protein in SIS2-MTD1
intergenic region
Length = 585
Score = 31.6 bits (70), Expect = 4.8
Identities = 22/66 (33%), Positives = 30/66 (45%), Gaps = 4/66 (6%)
Query: 141 LIKMKNLADKLKLAGNPISNSDLIIQTLNGLDSEYNPIVVKLSDHTTLSWVDLQAQLLTF 200
L K KN DKL N +N DL I+T + EY IV+K + SW + +L
Sbjct: 485 LDKKKNFLDKLSPGNNNSNNEDLKIRT---AEDEYR-IVLKRYNRVKQSWEKIMEDILNE 540
Query: 201 ESRIEQ 206
E+
Sbjct: 541 RKEFEK 546
>AAC2_DICDI (P14196) AAC-rich mRNA clone AAC11 protein (Fragment)
Length = 448
Score = 31.6 bits (70), Expect = 4.8
Identities = 29/139 (20%), Positives = 55/139 (38%), Gaps = 20/139 (14%)
Query: 202 SRIEQLNNLTNLNLNATANVANKFDHRDNRFNSNNNWRGSNFRGWRGGRGRGRSSKAPCQ 261
S I +NN N N N N +N ++ +N N+N N +N
Sbjct: 280 SNINNINNNNNNNSNNNNNSSNNNNNNNNSTNNNTNNNNNNTN----------------- 322
Query: 262 VCGKTNHTAINCFHRFDKNYSRSNYSADSDKQGSHNAFIASQNSVEDYDWYFDSGASNHV 321
TN+ N + + N + +N +A++ ++N S N+ + + +N+
Sbjct: 323 --NNTNNNN-NNINNNNNNTNNNNNNANNQNTNNNNMGNNSNNNNNPNNNNHQNNNNNNT 379
Query: 322 THQTNKFQDLTEHHGKNSL 340
++ +N T G N+L
Sbjct: 380 SNNSNTTTATTTAPGGNNL 398
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.316 0.130 0.391
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 51,842,758
Number of Sequences: 164201
Number of extensions: 2026844
Number of successful extensions: 6631
Number of sequences better than 10.0: 24
Number of HSP's better than 10.0 without gapping: 6
Number of HSP's successfully gapped in prelim test: 18
Number of HSP's that attempted gapping in prelim test: 6559
Number of HSP's gapped (non-prelim): 53
length of query: 482
length of database: 59,974,054
effective HSP length: 114
effective length of query: 368
effective length of database: 41,255,140
effective search space: 15181891520
effective search space used: 15181891520
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 68 (30.8 bits)
Medicago: description of AC144765.7


