

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144765.19 - phase: 0
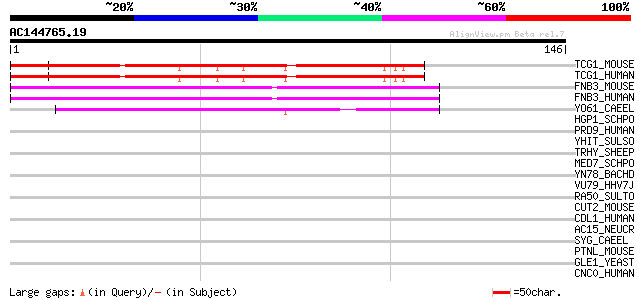
(146 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TCG1_MOUSE (Q8CGF7) Transcription elongation regulator 1 (TATA b... 91 1e-18
TCG1_HUMAN (O14776) Transcription elongation regulator 1 (TATA b... 91 1e-18
FNB3_MOUSE (Q9R1C7) Formin-binding protein 3 (Formin-binding pro... 52 4e-07
FNB3_HUMAN (O75400) Formin-binding protein 3 (Huntingtin yeast p... 52 4e-07
YO61_CAEEL (P34600) Hypothetical protein ZK1098.1 in chromosome III 45 7e-05
HGP1_SCHPO (Q09685) Hyphal growth protein 1 39 0.003
PRD9_HUMAN (Q9NQV7) PR-domain zinc finger protein 9 35 0.074
YHIT_SULSO (P95937) Hypothetical hit-like protein SSO2163 32 0.48
TRHY_SHEEP (P22793) Trichohyalin 30 1.8
MED7_SCHPO (O60104) RNA polymerase II mediator complex protein med7 30 1.8
YN78_BACHD (Q9KAB2) Hypothetical UPF0144 protein BH2378 30 2.4
VU79_HHV7J (P52531) Protein U79 30 2.4
RA50_SULTO (Q96YR5) DNA double-strand break repair rad50 ATPase 30 2.4
CUT2_MOUSE (P70298) Homeobox protein Cux-2 (Cut-like 2) 30 2.4
CDL1_HUMAN (P21127) PITSLRE serine/threonine-protein kinase CDC2... 30 2.4
AC15_NEUCR (P87000) Transcriptional activator protein acu-15 30 2.4
SYG_CAEEL (Q10039) Glycyl-tRNA synthetase (EC 6.1.1.14) (Glycine... 29 3.1
PTNL_MOUSE (Q62136) Protein tyrosine phosphatase, non-receptor t... 29 3.1
GLE1_YEAST (Q12315) Nucleoporin GLE1 (Nuclear pore protein GLE1)... 29 3.1
CNC0_HUMAN (Q8NEJ9) Protein C14orf120 29 3.1
>TCG1_MOUSE (Q8CGF7) Transcription elongation regulator 1 (TATA
box-binding protein-associated factor 2S) (Transcription
factor CA150) (p144) (Formin-binding protein 28) (FBP
28)
Length = 1100
Score = 90.5 bits (223), Expect = 1e-18
Identities = 51/110 (46%), Positives = 71/110 (64%), Gaps = 4/110 (3%)
Query: 1 MLKERGVAPFSKWEKELPKIVFDPRFKAIPSYSARRSLFEHYVKNRAEEERKEKRAAQKA 60
ML ERGV+ FS WEKEL KIVFDPR+ + + R+ +F+ YVK RAEEER+EK+
Sbjct: 671 MLLERGVSAFSTWEKELHKIVFDPRYLLL-NPKERKQVFDQYVKTRAEEERREKKNKIMQ 729
Query: 61 AIEGFKQLLDEASEDIDDKTDSHTFRKKWGNDPRFEALDR-KEREHLLNE 109
A E FK++++EA + + F K D RF+A+++ K+RE L NE
Sbjct: 730 AKEDFKKMMEEAK--FNPRATFSEFAAKHAKDSRFKAIEKMKDREALFNE 777
Score = 50.1 bits (118), Expect = 2e-06
Identities = 45/141 (31%), Positives = 61/141 (42%), Gaps = 46/141 (32%)
Query: 11 SKWEKELPKIVFDPRFKAIPSYSARRSLFEHYV----KNRAEEERKE------------- 53
S+W K K+ DPR+KA+ S S R LF+ Y+ KN E+ KE
Sbjct: 814 SRWSKVKDKVESDPRYKAVDSSSMREDLFKQYIEKIAKNLDSEKEKELERQARIEASLRE 873
Query: 54 -KRAAQKA-------------------AIEGFKQLLDE--ASEDIDDKTDSHTFRKKWGN 91
+R QKA AI+ FK LL + S D+ T RK
Sbjct: 874 REREVQKARSEQTKEIDREREQHKREEAIQNFKALLSDMVRSSDVSWSDTRRTLRK---- 929
Query: 92 DPRFEA---LDRKEREHLLNE 109
D R+E+ L+R+E+E L NE
Sbjct: 930 DHRWESGSLLEREEKEKLFNE 950
Score = 41.6 bits (96), Expect = 6e-04
Identities = 28/110 (25%), Positives = 54/110 (48%), Gaps = 3/110 (2%)
Query: 1 MLKERGVAPFSKWEKELPKIVFDPRFKAIPSYSARRSLFEHYVKNRAEEERKEKRAAQKA 60
M++E P + + + K D RFKAI R +LF +V ++E+++ + +
Sbjct: 737 MMEEAKFNPRATFSEFAAKHAKDSRFKAIEKMKDREALFNEFVAAARKKEKEDSKTRGEK 796
Query: 61 AIEGFKQLLDEASEDIDDKTDSHTFRKKWGNDPRFEALDRKE-REHLLNE 109
F +LL ++ +D ++ + K +DPR++A+D RE L +
Sbjct: 797 IKSDFFELL--SNHHLDSQSRWSKVKDKVESDPRYKAVDSSSMREDLFKQ 844
>TCG1_HUMAN (O14776) Transcription elongation regulator 1 (TATA
box-binding protein-associated factor 2S) (Transcription
factor CA150)
Length = 1098
Score = 90.5 bits (223), Expect = 1e-18
Identities = 51/110 (46%), Positives = 71/110 (64%), Gaps = 4/110 (3%)
Query: 1 MLKERGVAPFSKWEKELPKIVFDPRFKAIPSYSARRSLFEHYVKNRAEEERKEKRAAQKA 60
ML ERGV+ FS WEKEL KIVFDPR+ + + R+ +F+ YVK RAEEER+EK+
Sbjct: 669 MLLERGVSAFSTWEKELHKIVFDPRYLLL-NPKERKQVFDQYVKTRAEEERREKKNKIMQ 727
Query: 61 AIEGFKQLLDEASEDIDDKTDSHTFRKKWGNDPRFEALDR-KEREHLLNE 109
A E FK++++EA + + F K D RF+A+++ K+RE L NE
Sbjct: 728 AKEDFKKMMEEAK--FNPRATFSEFAAKHAKDSRFKAIEKMKDREALFNE 775
Score = 50.1 bits (118), Expect = 2e-06
Identities = 45/141 (31%), Positives = 61/141 (42%), Gaps = 46/141 (32%)
Query: 11 SKWEKELPKIVFDPRFKAIPSYSARRSLFEHYV----KNRAEEERKE------------- 53
S+W K K+ DPR+KA+ S S R LF+ Y+ KN E+ KE
Sbjct: 812 SRWSKVKDKVESDPRYKAVDSSSMREDLFKQYIEKIAKNLDSEKEKELERQARIEASLRE 871
Query: 54 -KRAAQKA-------------------AIEGFKQLLDE--ASEDIDDKTDSHTFRKKWGN 91
+R QKA AI+ FK LL + S D+ T RK
Sbjct: 872 REREVQKARSEQTKEIDREREQHKREEAIQNFKALLSDMVRSSDVSWSDTRRTLRK---- 927
Query: 92 DPRFEA---LDRKEREHLLNE 109
D R+E+ L+R+E+E L NE
Sbjct: 928 DHRWESGSLLEREEKEKLFNE 948
Score = 41.6 bits (96), Expect = 6e-04
Identities = 28/110 (25%), Positives = 54/110 (48%), Gaps = 3/110 (2%)
Query: 1 MLKERGVAPFSKWEKELPKIVFDPRFKAIPSYSARRSLFEHYVKNRAEEERKEKRAAQKA 60
M++E P + + + K D RFKAI R +LF +V ++E+++ + +
Sbjct: 735 MMEEAKFNPRATFSEFAAKHAKDSRFKAIEKMKDREALFNEFVAAARKKEKEDSKTRGEK 794
Query: 61 AIEGFKQLLDEASEDIDDKTDSHTFRKKWGNDPRFEALDRKE-REHLLNE 109
F +LL ++ +D ++ + K +DPR++A+D RE L +
Sbjct: 795 IKSDFFELL--SNHHLDSQSRWSKVKDKVESDPRYKAVDSSSMREDLFKQ 842
>FNB3_MOUSE (Q9R1C7) Formin-binding protein 3 (Formin-binding
protein 11) (FBP 11)
Length = 953
Score = 52.0 bits (123), Expect = 4e-07
Identities = 30/113 (26%), Positives = 55/113 (48%), Gaps = 1/113 (0%)
Query: 1 MLKERGVAPFSKWEKELPKIVFDPRFKAIPSYSARRSLFEHYVKNRAEEERKEKRAAQKA 60
+LKE+ V + WE+ + I+ DPR+ A+ S ++ F Y +EE++E R+ K
Sbjct: 399 LLKEKRVPSNASWEQAMKMIINDPRYSALAKLSEKKQAFNAYKVQTEKEEKEEARSKYKE 458
Query: 61 AIEGFKQLLDEASEDIDDKTDSHTFRKKWGNDPRFEALDRKEREHLLNERCHF 113
A E F++ L E E + T + +G + A+ ++R + + F
Sbjct: 459 AKESFQRFL-ENHEKMTSTTRYKKAEQMFGEMEVWNAISERDRLEIYEDVLFF 510
Score = 29.3 bits (64), Expect = 3.1
Identities = 17/79 (21%), Positives = 36/79 (45%), Gaps = 3/79 (3%)
Query: 13 WEKELPKIVFDPRFKAIPSYSARRSLFE---HYVKNRAEEERKEKRAAQKAAIEGFKQLL 69
WE + V +P F+ I S R+ +F+ H +++ + + + K + + ++
Sbjct: 763 WEDIRERFVKEPAFEDITLESERKRIFKDFMHVLEHECQHHHSKNKKHSKKSKKHHRKRS 822
Query: 70 DEASEDIDDKTDSHTFRKK 88
S D DSH+ +K+
Sbjct: 823 RSRSGSESDDDDSHSKKKR 841
Score = 28.9 bits (63), Expect = 4.1
Identities = 17/68 (25%), Positives = 30/68 (44%), Gaps = 1/68 (1%)
Query: 46 RAEEERKEKRAAQKAAIEGFKQLLDEASEDIDDKTDSHTFRKKWGNDPRFEALD-RKERE 104
R E KE+ K FK +L +A+ I+ R+++ +P FE + ER+
Sbjct: 727 REREREKEEARKMKRKESAFKSMLKQATPPIELDAVWEDIRERFVKEPAFEDITLESERK 786
Query: 105 HLLNERCH 112
+ + H
Sbjct: 787 RIFKDFMH 794
>FNB3_HUMAN (O75400) Formin-binding protein 3 (Huntingtin yeast
partner A) (Huntingtin-interacting protein HYPA/FBP11)
(Fas-ligand associated factor 1) (NY-REN-6 antigen)
(HSPC225)
Length = 957
Score = 52.0 bits (123), Expect = 4e-07
Identities = 30/113 (26%), Positives = 55/113 (48%), Gaps = 1/113 (0%)
Query: 1 MLKERGVAPFSKWEKELPKIVFDPRFKAIPSYSARRSLFEHYVKNRAEEERKEKRAAQKA 60
+LKE+ V + WE+ + I+ DPR+ A+ S ++ F Y +EE++E R+ K
Sbjct: 403 LLKEKRVPSNASWEQAMKMIINDPRYSALAKLSEKKQAFNAYKVQTEKEEKEEARSKYKE 462
Query: 61 AIEGFKQLLDEASEDIDDKTDSHTFRKKWGNDPRFEALDRKEREHLLNERCHF 113
A E F++ L E E + T + +G + A+ ++R + + F
Sbjct: 463 AKESFQRFL-ENHEKMTSTTRYKKAEQMFGEMEVWNAISERDRLEIYEDVLFF 514
Score = 32.0 bits (71), Expect = 0.48
Identities = 22/86 (25%), Positives = 43/86 (49%), Gaps = 5/86 (5%)
Query: 13 WEKELPKIVFDPRFKAIPSYSARRSLFE---HYVKNRAEEER-KEKRAAQKAAIEGFKQL 68
WE + V +P F+ I S R+ +F+ H +++ + K K+ ++K+ K+
Sbjct: 767 WEDIRERFVKEPAFEDITLESERKRIFKDFMHVLEHECQHHHSKNKKHSKKSKKHHRKRS 826
Query: 69 LDEASEDIDDKTDSHTFRKKWGNDPR 94
+ D DD DSH+ +K+ ++ R
Sbjct: 827 RSRSGSDSDD-DDSHSKKKRQRSESR 851
Score = 28.9 bits (63), Expect = 4.1
Identities = 17/68 (25%), Positives = 30/68 (44%), Gaps = 1/68 (1%)
Query: 46 RAEEERKEKRAAQKAAIEGFKQLLDEASEDIDDKTDSHTFRKKWGNDPRFEALD-RKERE 104
R E KE+ K FK +L +A+ I+ R+++ +P FE + ER+
Sbjct: 731 REREREKEEARKMKRKESAFKSMLKQAAPPIELDAVWEDIRERFVKEPAFEDITLESERK 790
Query: 105 HLLNERCH 112
+ + H
Sbjct: 791 RIFKDFMH 798
>YO61_CAEEL (P34600) Hypothetical protein ZK1098.1 in chromosome III
Length = 724
Score = 44.7 bits (104), Expect = 7e-05
Identities = 29/104 (27%), Positives = 52/104 (49%), Gaps = 7/104 (6%)
Query: 13 WEKELPKIVFDPRFKAIPSYSARRSLFEHYVKNRAEEERKEKRAAQKAAIEGFKQLLDE- 71
W++ + I DPRF+ + S ++ LF + R +EER EKR A K + E ++ L E
Sbjct: 250 WDQAVKWIQNDPRFRILNKVSEKKQLFNAWKVQRGKEERDEKRLAIKKSKEDLEKFLQEH 309
Query: 72 --ASEDIDDKTDSHTFRKKWGNDPRFEALDRKEREHLLNERCHF 113
E + + S F K +P + A++ ++R+ + + F
Sbjct: 310 PKMKESLKYQKASDIFSK----EPLWIAVNDEDRKEIFRDCIDF 349
Score = 33.9 bits (76), Expect = 0.13
Identities = 32/133 (24%), Positives = 50/133 (37%), Gaps = 29/133 (21%)
Query: 1 MLKERGVAPFSKWEKELPKIVFDPRFKAIPSYSARRSLFEHY----------------VK 44
+LKE V S+W PKI D + A+ + R + F HY K
Sbjct: 588 LLKEHNVDKDSEWTVIKPKIEKDKAYLAMENDDERETAFNHYKNGTSGTTAGSEILEKKK 647
Query: 45 NRAEEERKEKRAAQKAAIEG---FKQLLDEASEDIDDKTDSHTFRKK----------WGN 91
+ ++++K KR+ + EG K+ + +D+ D KK
Sbjct: 648 KKKDKKKKNKRSDNNSESEGEIREKEKKKKKKHSKEDRMDDEERGKKSKKSRKRSPSRSE 707
Query: 92 DPRFEALDRKERE 104
PR + RK RE
Sbjct: 708 SPRHSSEKRKRRE 720
>HGP1_SCHPO (Q09685) Hyphal growth protein 1
Length = 411
Score = 39.3 bits (90), Expect = 0.003
Identities = 16/55 (29%), Positives = 30/55 (54%)
Query: 1 MLKERGVAPFSKWEKELPKIVFDPRFKAIPSYSARRSLFEHYVKNRAEEERKEKR 55
+LK++ + + WE PK++ D RF + S R+ +FE Y K+ ++ +R
Sbjct: 249 LLKDKNIGAYQPWELVYPKLLDDDRFYVLDSGERRKEVFEEYCKSVVSTKKITRR 303
>PRD9_HUMAN (Q9NQV7) PR-domain zinc finger protein 9
Length = 689
Score = 34.7 bits (78), Expect = 0.074
Identities = 25/108 (23%), Positives = 47/108 (43%), Gaps = 9/108 (8%)
Query: 3 KERGVAPFSKWEKELPKIVFDPRFKAIPSYSARRSLFEHYVKNRAEEERKEKRAAQKAAI 62
+E G+ SKW+KE PK P ++S+++ L +H +N + + A
Sbjct: 163 QELGIKWGSKWKKEEPKPEIHPCPSCCLAFSSQKFLSQHVERNHSSQNFPGPSA------ 216
Query: 63 EGFKQLLDEASEDIDDKTDSHTFRKKWGNDPRFEALDRKEREHLLNER 110
++LL + D+ + + + + + KER LLN+R
Sbjct: 217 ---RKLLQPENPCPGDQNQEQQYPDPHSRNDKTKGQEIKERSKLLNKR 261
>YHIT_SULSO (P95937) Hypothetical hit-like protein SSO2163
Length = 139
Score = 32.0 bits (71), Expect = 0.48
Identities = 19/67 (28%), Positives = 32/67 (47%), Gaps = 1/67 (1%)
Query: 2 LKERGVAPFSKWEKELPKIVFDPRFKAIPSYSARRSLFEHYVKNRAEEERKEKRAAQKAA 61
LK G+ + K ++VF F +P++S + + +V R E+ R+ QKA
Sbjct: 74 LKADGIRILTNIGKSAGQVVFHSHFHIVPTWSQDPDIMKDFVP-RKEQSREYYEYVQKAI 132
Query: 62 IEGFKQL 68
IE K +
Sbjct: 133 IETLKNI 139
>TRHY_SHEEP (P22793) Trichohyalin
Length = 1549
Score = 30.0 bits (66), Expect = 1.8
Identities = 18/89 (20%), Positives = 41/89 (45%), Gaps = 3/89 (3%)
Query: 22 FDPRFKAIPSYSARRSLFEHYVKNRAEEERKEKRAAQKAAIEGFKQLLDEASEDIDDKTD 81
F+P+ + + +R E + EEE +EK+ ++ ++ +Q E++ +
Sbjct: 120 FEPQDRQLEERRLKRQELEELAE---EEELREKQVRREQRLQRREQEEYGGEEELQQRPK 176
Query: 82 SHTFRKKWGNDPRFEALDRKEREHLLNER 110
+ + RFE +++ER+ L E+
Sbjct: 177 GRELEELLNREQRFERQEQRERQRLQVEQ 205
>MED7_SCHPO (O60104) RNA polymerase II mediator complex protein med7
Length = 376
Score = 30.0 bits (66), Expect = 1.8
Identities = 24/84 (28%), Positives = 37/84 (43%), Gaps = 6/84 (7%)
Query: 2 LKERGVAPFSK----WEKELPKIVFDPRFKAIPSYSARRSLFEHYVKNRAEEERKEKRAA 57
LKE G+ K E E+ + +DP+ AI S R Y KN E K
Sbjct: 90 LKEFGIPELYKDIKDGEDEIEVVEYDPKSNAIVGTSFTR--VHDYDKNSPIENEKILEDD 147
Query: 58 QKAAIEGFKQLLDEASEDIDDKTD 81
Q A++ D+ +D+++KT+
Sbjct: 148 QHTAMKEKDNESDKTMKDVEEKTE 171
>YN78_BACHD (Q9KAB2) Hypothetical UPF0144 protein BH2378
Length = 521
Score = 29.6 bits (65), Expect = 2.4
Identities = 20/67 (29%), Positives = 35/67 (51%), Gaps = 11/67 (16%)
Query: 41 HYVKNRAEEERKEKR---AAQKAAIEGFKQLLDEASEDIDDKTDSHTFRKKWGNDPRFEA 97
H ++ AE E +E+R Q+ + +++LD SE +D K +S D R E+
Sbjct: 68 HKLRTEAEREIRERRNEIQKQENRLVQKEEILDRKSETLDKKEES--------LDKREES 119
Query: 98 LDRKERE 104
L +K+R+
Sbjct: 120 LTKKQRQ 126
>VU79_HHV7J (P52531) Protein U79
Length = 233
Score = 29.6 bits (65), Expect = 2.4
Identities = 16/64 (25%), Positives = 32/64 (50%)
Query: 40 EHYVKNRAEEERKEKRAAQKAAIEGFKQLLDEASEDIDDKTDSHTFRKKWGNDPRFEALD 99
EH + + +++ K+K QK +++L++ + + K T ++K D R E D
Sbjct: 148 EHAERKKQDDDYKKKALKQKDKRRSEQKILEDCDKKDEKKRMDDTEKRKLQEDRRNEKQD 207
Query: 100 RKER 103
K+R
Sbjct: 208 LKKR 211
>RA50_SULTO (Q96YR5) DNA double-strand break repair rad50 ATPase
Length = 879
Score = 29.6 bits (65), Expect = 2.4
Identities = 23/75 (30%), Positives = 37/75 (48%), Gaps = 3/75 (4%)
Query: 14 EKELPKIVFDPRFKAIPSYSARRSLFEHYVKNRAEEERKEKRAAQKAAIEGFKQLLDEAS 73
EKEL I + I R+ +E VK EEE+KEKR + ++ +L D+ S
Sbjct: 204 EKELENIKRELEDLNIKEEKERKK-YEDIVKLNEEEEKKEKRYVELISL--LNKLKDDIS 260
Query: 74 EDIDDKTDSHTFRKK 88
E ++ D + R++
Sbjct: 261 ELREEVKDENRLREE 275
>CUT2_MOUSE (P70298) Homeobox protein Cux-2 (Cut-like 2)
Length = 1426
Score = 29.6 bits (65), Expect = 2.4
Identities = 15/40 (37%), Positives = 24/40 (59%)
Query: 24 PRFKAIPSYSARRSLFEHYVKNRAEEERKEKRAAQKAAIE 63
P +IP+ +A S E +KN E+ R+E +A Q+A +E
Sbjct: 612 PASVSIPNGTASSSTSEDAIKNILEQARREMQAQQQALLE 651
>CDL1_HUMAN (P21127) PITSLRE serine/threonine-protein kinase CDC2L1
(EC 2.7.1.37) (Galactosyltransferase associated protein
kinase p58/GTA) (Cell division cycle 2-like 1) (CLK-1)
(CDK11) (p58 CLK-1)
Length = 795
Score = 29.6 bits (65), Expect = 2.4
Identities = 23/77 (29%), Positives = 33/77 (41%), Gaps = 9/77 (11%)
Query: 43 VKNRAEEERKEKRAAQKAAIEGFKQLLDEASEDIDDKTDSH-----TFRKKWGNDPRFEA 97
V +R +E+RKEKR + + EG K + E ++ H R++W R E
Sbjct: 97 VHHRKDEKRKEKRRHRSHSAEGGKHARVKEKEREHERRKRHREEQDKARREWERQKRRE- 155
Query: 98 LDRKEREHLLNERCHFE 114
REH ER E
Sbjct: 156 ---MAREHSRRERDRLE 169
>AC15_NEUCR (P87000) Transcriptional activator protein acu-15
Length = 926
Score = 29.6 bits (65), Expect = 2.4
Identities = 16/46 (34%), Positives = 23/46 (49%)
Query: 49 EERKEKRAAQKAAIEGFKQLLDEASEDIDDKTDSHTFRKKWGNDPR 94
E +E+ A +A I K LLDE E +D + H+ R + PR
Sbjct: 68 ESLEERVRALEAEIRELKDLLDEKDEKLDMLSKMHSNRSRSAEPPR 113
>SYG_CAEEL (Q10039) Glycyl-tRNA synthetase (EC 6.1.1.14)
(Glycine--tRNA ligase) (GlyRS)
Length = 742
Score = 29.3 bits (64), Expect = 3.1
Identities = 28/122 (22%), Positives = 49/122 (39%), Gaps = 17/122 (13%)
Query: 18 PKIVFDPRFKAIPSYSARRSLFEHYVKNRAEEERKEKRAAQKAAIEGFKQLLDEASE--- 74
P+ V PRF+ R Y+K A E + K A +AA++ + L+ +
Sbjct: 42 PRKVVAPRFE-----KKTRQKISDYLKQMATPEIEAKLAPLRAAVKEYGDLIRDLKAKGA 96
Query: 75 ---DIDDKTDSHTFRKKWGND------PRFEALDRKEREHLLNERCHFEFNWTFLNSLNF 125
DID RK+ D P+ + DR + E LL R ++ ++ +
Sbjct: 97 PKIDIDKAVVELKARKRLLEDTEIALAPKEASFDRLKLEDLLKRRFFYDQSFAIYGGVTG 156
Query: 126 IF 127
++
Sbjct: 157 LY 158
>PTNL_MOUSE (Q62136) Protein tyrosine phosphatase, non-receptor type
21 (EC 3.1.3.48) (Protein-tyrosine phosphatase PTP-RL10)
Length = 1176
Score = 29.3 bits (64), Expect = 3.1
Identities = 15/52 (28%), Positives = 27/52 (51%)
Query: 31 SYSARRSLFEHYVKNRAEEERKEKRAAQKAAIEGFKQLLDEASEDIDDKTDS 82
S ++ S+F +K EE++ R + K ++ L+D + ED D + DS
Sbjct: 670 SAGSQSSVFSDKMKQEGTEEQEGGRYSHKKSLSDATMLIDSSEEDEDLEEDS 721
>GLE1_YEAST (Q12315) Nucleoporin GLE1 (Nuclear pore protein GLE1)
(RNA export factor GLE1)
Length = 538
Score = 29.3 bits (64), Expect = 3.1
Identities = 23/80 (28%), Positives = 33/80 (40%), Gaps = 2/80 (2%)
Query: 27 KAIPSYSARRSLFEHYVKNRAEEERKEKRAAQKAAIEG--FKQLLDEASEDIDDKTDSHT 84
K I S + E K + EEERK K A +KA E +Q DE +
Sbjct: 144 KEIQSIRENKRRVEEQRKRKEEEERKRKEAEEKAKREQELLRQKKDEEERKRKEAEAKLA 203
Query: 85 FRKKWGNDPRFEALDRKERE 104
+K+ + E + KER+
Sbjct: 204 QQKQEEERKKIEEQNEKERQ 223
>CNC0_HUMAN (Q8NEJ9) Protein C14orf120
Length = 315
Score = 29.3 bits (64), Expect = 3.1
Identities = 16/52 (30%), Positives = 25/52 (47%), Gaps = 5/52 (9%)
Query: 41 HYVKNRAEEERK-----EKRAAQKAAIEGFKQLLDEASEDIDDKTDSHTFRK 87
HY + AE E+K ++RA + I K+ +A E+I D H R+
Sbjct: 180 HYDETEAEREKKRLERAKRRALSSSVIRELKEQYSDAPEEIRDARHPHVTRQ 231
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.322 0.137 0.437
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 18,128,575
Number of Sequences: 164201
Number of extensions: 714099
Number of successful extensions: 2827
Number of sequences better than 10.0: 48
Number of HSP's better than 10.0 without gapping: 16
Number of HSP's successfully gapped in prelim test: 34
Number of HSP's that attempted gapping in prelim test: 2785
Number of HSP's gapped (non-prelim): 69
length of query: 146
length of database: 59,974,054
effective HSP length: 100
effective length of query: 46
effective length of database: 43,553,954
effective search space: 2003481884
effective search space used: 2003481884
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 60 (27.7 bits)
Medicago: description of AC144765.19


