

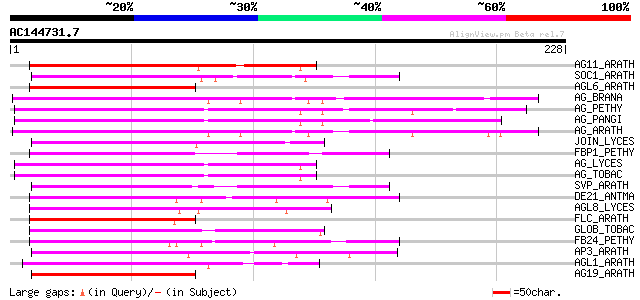
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144731.7 + phase: 0
(228 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
AG11_ARATH (Q38836) Agamous-like MADS box protein AGL11 84 4e-16
SOC1_ARATH (O64645) SUPPRESSOR OF CONSTANS OVEREXPRESSION 1 prot... 82 8e-16
AGL6_ARATH (P29386) Agamous-like MADS box protein AGL6 80 4e-15
AG_BRANA (Q01540) Floral homeotic protein AGAMOUS 80 5e-15
AG_PETHY (Q40885) Floral homeotic protein AGAMOUS (pMADS3) 79 7e-15
AG_PANGI (Q40872) Floral homeotic protein AGAMOUS (GAG2) 79 1e-14
AG_ARATH (P17839) Floral homeotic protein AGAMOUS 79 1e-14
JOIN_LYCES (Q9FUY6) MADS-box JOINTLESS protein (LeMADS) 78 2e-14
FBP1_PETHY (Q03488) Floral homeotic protein FBP1 (Floral binding... 78 2e-14
AG_LYCES (Q40168) Floral homeotic protein AGAMOUS (TAG1) 78 2e-14
AG_TOBAC (Q43585) Floral homeotic protein AGAMOUS (NAG1) 77 3e-14
SVP_ARATH (Q9FVC1) SHORT VEGETATIVE PHASE protein 75 1e-13
DE21_ANTMA (Q8RVL4) MADS box protein defh21 (DEFICIENS homolog 21) 75 2e-13
AGL8_LYCES (Q40170) Agamous-like MADS box protein AGL8 homolog (... 75 2e-13
FLC_ARATH (Q9S7Q7) FLOWERING LOCUS C protein (MADS box protein F... 74 3e-13
GLOB_TOBAC (Q03416) Floral homeotic protein GLOBOSA 74 4e-13
FB24_PETHY (Q9ATE5) MADS box protein FBP24 (Floral binding prote... 74 4e-13
AP3_ARATH (P35632) Floral homeotic protein APETALA3 74 4e-13
AGL1_ARATH (P29381) Agamous-like MADS box protein AGL1 (Protein ... 74 4e-13
AG19_ARATH (O82743) Agamous-like MADS box protein AGL19 74 4e-13
>AG11_ARATH (Q38836) Agamous-like MADS box protein AGL11
Length = 230
Score = 83.6 bits (205), Expect = 4e-16
Identities = 50/121 (41%), Positives = 75/121 (61%), Gaps = 6/121 (4%)
Query: 9 GRQKIEMKKISNESNLQVTFSKHHNGLFKKASELCTLCGADVALVVFSPSGKVFSFGHPN 68
GR KIE+K+I N +N QVTF K NGL KKA EL LC A+VAL+VFS G+++ + + N
Sbjct: 2 GRGKIEIKRIENSTNRQVTFCKRRNGLLKKAYELSVLCDAEVALIVFSTRGRLYEYANNN 61
Query: 69 LDTVIDRF-LSLIPTQNDGTMQFIEAHRNANVRELNAQLTQINNTLDAEKK--IGDELSN 125
+ + I+R+ + + N T+Q I A A ++ +A+L Q T+ + +GD LS+
Sbjct: 62 IRSTIERYKKACSDSTNTSTVQEINA---AYYQQESAKLRQQIQTIQNSNRNLMGDSLSS 118
Query: 126 L 126
L
Sbjct: 119 L 119
>SOC1_ARATH (O64645) SUPPRESSOR OF CONSTANS OVEREXPRESSION 1 protein
(Agamous-like MADS box protein AGL20)
Length = 214
Score = 82.4 bits (202), Expect = 8e-16
Identities = 56/161 (34%), Positives = 89/161 (54%), Gaps = 18/161 (11%)
Query: 10 RQKIEMKKISNESNLQVTFSKHHNGLFKKASELCTLCGADVALVVFSPSGKVFSFGHPNL 69
R K +MK+I N ++ QVTFSK NGL KKA EL LC A+V+L++FSP GK++ F N+
Sbjct: 3 RGKTQMKRIENATSRQVTFSKRRNGLLKKAFELSVLCDAEVSLIIFSPKGKLYEFASSNM 62
Query: 70 DTVIDRFL----SLIPTQ--NDGTMQFIEAHRNANVRELNAQLTQINNTLDAEKKIG--- 120
IDR+L + T+ ++ MQ ++ + AN+ + QL L E IG
Sbjct: 63 QDTIDRYLRHTKDRVSTKPVSEENMQHLK-YEAANMMKKIEQLEASKRKLLGE-GIGTCS 120
Query: 121 -DELSNLHKETEAKFWWACVVDGMNRDQLEIFKKALEELKK 160
+EL + ++ E V + + ++FK+ +E+LK+
Sbjct: 121 IEELQQIEQQLEKS------VKCIRARKTQVFKEQIEQLKQ 155
>AGL6_ARATH (P29386) Agamous-like MADS box protein AGL6
Length = 252
Score = 80.1 bits (196), Expect = 4e-15
Identities = 36/68 (52%), Positives = 50/68 (72%)
Query: 9 GRQKIEMKKISNESNLQVTFSKHHNGLFKKASELCTLCGADVALVVFSPSGKVFSFGHPN 68
GR ++EMK+I N+ N QVTFSK NGL KKA EL LC A+VAL++FS GK++ FG
Sbjct: 2 GRGRVEMKRIENKINRQVTFSKRRNGLLKKAYELSVLCDAEVALIIFSSRGKLYEFGSVG 61
Query: 69 LDTVIDRF 76
+++ I+R+
Sbjct: 62 IESTIERY 69
>AG_BRANA (Q01540) Floral homeotic protein AGAMOUS
Length = 252
Score = 79.7 bits (195), Expect = 5e-15
Identities = 69/237 (29%), Positives = 106/237 (44%), Gaps = 27/237 (11%)
Query: 2 SSVRKGRGRQKIEMKKISNESNLQVTFSKHHNGLFKKASELCTLCGADVALVVFSPSGKV 61
SS ++ GR KIE+K+I N +N QVTF K NGL KKA EL LC A+VAL+VFS G++
Sbjct: 11 SSPQRKAGRGKIEIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEVALIVFSSRGRL 70
Query: 62 FSFGHPNLDTVIDRFLSLI-PTQNDGTMQFIEA----HRNANVRELNAQLTQINNTLDAE 116
+ + + ++ I+R+ I N G++ I A +A +R+ + N L E
Sbjct: 71 YEYSNNSVKGTIERYKKAISDNSNTGSVAEINAQYYQQESAKLRQQIISIQNSNRQLMGE 130
Query: 117 KKIGD----ELSNLH------------KETEAKFWWACVVDGMNRDQLEIFKKALEELKK 160
IG EL NL K+ E F +D M + ++++ K
Sbjct: 131 -TIGSMSPKELRNLEGRLDRSVNRIRSKKNELLF---AEIDYMQKREVDLHNDNQLLRAK 186
Query: 161 LVIQHAATRTLPFFVGNTSSSNIYLHHQPNPQQAEMFPPQIFQNAMLQLQTHFFDGS 217
+ ++ G ++ I Q PQ + FQ A LQ H + +
Sbjct: 187 IAENERNNPSMSLMPGGSNYEQIMPPPQTQPQPFD--SRNYFQVAALQPNNHHYSSA 241
>AG_PETHY (Q40885) Floral homeotic protein AGAMOUS (pMADS3)
Length = 242
Score = 79.3 bits (194), Expect = 7e-15
Identities = 62/216 (28%), Positives = 105/216 (47%), Gaps = 10/216 (4%)
Query: 3 SVRKGRGRQKIEMKKISNESNLQVTFSKHHNGLFKKASELCTLCGADVALVVFSPSGKVF 62
S ++ GR KIE+K+I N +N QVTF K NGL KKA EL LC A+VAL+VFS G+++
Sbjct: 12 SPQRKLGRGKIEIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEVALIVFSSRGRLY 71
Query: 63 SFGHPNLDTVIDRFLSLIPTQNDGTMQFIEAHRNANVRELNAQLTQINNTLDAEKK-IGD 121
+ + ++ I+R+ + + T EA+ +E + QI N + + +G+
Sbjct: 72 EYANNSVKATIERYKKAC-SDSSNTGSIAEANAQYYQQEASKLRAQIGNLQNQNRNFLGE 130
Query: 122 ELSNLH----KETEAKFWWACVVDGMNRDQLEIFKKALEELKKLVIQ-HAATRTLPFFVG 176
L+ L+ + E K + + + E+ +E ++K I H + L +
Sbjct: 131 SLAALNLRDLRNLEQKIEKG--ISKIRAKKNELLFAEIEYMQKREIDLHNNNQYLRAKIA 188
Query: 177 NTSSSNIYLHHQPNPQQAEMFPPQIFQNAMLQLQTH 212
T S ++ P ++ PPQ +A LQ +
Sbjct: 189 ETERSQ-QMNLMPGSSSYDLVPPQQSFDARNYLQVN 223
>AG_PANGI (Q40872) Floral homeotic protein AGAMOUS (GAG2)
Length = 242
Score = 78.6 bits (192), Expect = 1e-14
Identities = 59/205 (28%), Positives = 99/205 (47%), Gaps = 7/205 (3%)
Query: 3 SVRKGRGRQKIEMKKISNESNLQVTFSKHHNGLFKKASELCTLCGADVALVVFSPSGKVF 62
S ++ GR KIE+K+I N +N QVTF K NGL KKA EL LC A+VAL+VFS G+++
Sbjct: 12 SPQRKLGRGKIEIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEVALIVFSTRGRLY 71
Query: 63 SFGHPNLDTVIDRFLSLIPTQNDGTMQFIEAHRNANVRELNAQLTQINNTLDAEKK-IGD 121
+ + ++ I+R+ T + T EA+ +E + +I++ + +G+
Sbjct: 72 EYANNSVKGTIERYKKAC-TDSPNTSSVSEANAQFYQQEASKLRQEISSIQKNNRNMMGE 130
Query: 122 ELSNLH----KETEAKFWWACVVDGMNRDQLEIFKKALEELKKLVIQHAATRTLPFFVGN 177
L +L K E K +++L +F + KK + H + L +
Sbjct: 131 SLGSLTVRDLKGLETKLEKGISRIRSKKNEL-LFAEIEYMQKKEIDLHNNNQYLRAKIAE 189
Query: 178 TSSSNIYLHHQPNPQQAEMFPPQIF 202
+ +++ P E+ PPQ F
Sbjct: 190 NERAQQHMNLMPGSSDYELAPPQSF 214
>AG_ARATH (P17839) Floral homeotic protein AGAMOUS
Length = 252
Score = 78.6 bits (192), Expect = 1e-14
Identities = 70/238 (29%), Positives = 106/238 (44%), Gaps = 29/238 (12%)
Query: 2 SSVRKGRGRQKIEMKKISNESNLQVTFSKHHNGLFKKASELCTLCGADVALVVFSPSGKV 61
SS + GR KIE+K+I N +N QVTF K NGL KKA EL LC A+VAL+VFS G++
Sbjct: 11 SSPLRKSGRGKIEIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEVALIVFSSRGRL 70
Query: 62 FSFGHPNLDTVIDRFLSLI-PTQNDGTMQFIEA----HRNANVRELNAQLTQINNTLDAE 116
+ + + ++ I+R+ I N G++ I A +A +R+ + N L E
Sbjct: 71 YEYSNNSVKGTIERYKKAISDNSNTGSVAEINAQYYQQESAKLRQQIISIQNSNRQLMGE 130
Query: 117 KKIGD----ELSNLHKETEAKFWWACVVDGMNRDQLEIFKKALEELKKLVIQ-HAATRTL 171
IG EL NL E + + + E+ ++ ++K + H + L
Sbjct: 131 -TIGSMSPKELRNLEGRLERS------ITRIRSKKNELLFSEIDYMQKREVDLHNDNQIL 183
Query: 172 PFFVGNTSSSNIYLHHQPNPQQAE--MFPPQ----------IFQNAMLQLQTHFFDGS 217
+ +N + P E M PPQ FQ A LQ H + +
Sbjct: 184 RAKIAENERNNPSISLMPGGSNYEQLMPPPQTQSQPFDSRNYFQVAALQPNNHHYSSA 241
>JOIN_LYCES (Q9FUY6) MADS-box JOINTLESS protein (LeMADS)
Length = 265
Score = 78.2 bits (191), Expect = 2e-14
Identities = 45/122 (36%), Positives = 73/122 (58%), Gaps = 4/122 (3%)
Query: 10 RQKIEMKKISNESNLQVTFSKHHNGLFKKASELCTLCGADVALVVFSPSGKVFSFGHPNL 69
R+KI++KKI N + QVTFSK GLFKKA EL LC ADVAL++FS +GK+F + ++
Sbjct: 3 REKIQIKKIDNSTARQVTFSKRRRGLFKKAEELSVLCDADVALIIFSSTGKLFDYSSSSM 62
Query: 70 DTVIDR--FLSLIPTQNDGTMQFIEAHRNANVRELNAQLTQINNTLDAEKKIGDELSNLH 127
+++R S + D ++ N+N L+ ++++ ++ L + G+EL L+
Sbjct: 63 KQILERRDLHSKNLEKLDQPSLELQLVENSNYSRLSKEISEKSHRL--RQMRGEELQGLN 120
Query: 128 KE 129
E
Sbjct: 121 IE 122
>FBP1_PETHY (Q03488) Floral homeotic protein FBP1 (Floral binding
protein 1)
Length = 210
Score = 77.8 bits (190), Expect = 2e-14
Identities = 47/148 (31%), Positives = 77/148 (51%), Gaps = 22/148 (14%)
Query: 9 GRQKIEMKKISNESNLQVTFSKHHNGLFKKASELCTLCGADVALVVFSPSGKVFSFGHPN 68
GR KIE+K+I N SN QVT+SK NG+ KKA E+ LC A V++++F+ SGK+ F +
Sbjct: 2 GRGKIEIKRIENSSNRQVTYSKRRNGILKKAKEISVLCDARVSVIIFASSGKMHEFSSTS 61
Query: 69 LDTVIDRFLSLIPTQNDGTMQFIEAHRNANVRELNAQLTQINNTLDAEKKIGDELSNLHK 128
L ++D++ H+ R L+A+ ++N ++ KK D +
Sbjct: 62 LVDILDQY-----------------HKLTGRRLLDAKHENLDNEINKVKKDNDNM----- 99
Query: 129 ETEAKFWWACVVDGMNRDQLEIFKKALE 156
+ E + + +N +L I + ALE
Sbjct: 100 QIELRHLKGEDITSLNHRELMILEDALE 127
>AG_LYCES (Q40168) Floral homeotic protein AGAMOUS (TAG1)
Length = 248
Score = 77.8 bits (190), Expect = 2e-14
Identities = 45/125 (36%), Positives = 72/125 (57%), Gaps = 2/125 (1%)
Query: 3 SVRKGRGRQKIEMKKISNESNLQVTFSKHHNGLFKKASELCTLCGADVALVVFSPSGKVF 62
S ++ GR KIE+K+I N +N QVTF K NGL KKA EL LC A+VALVVFS G+++
Sbjct: 12 SPQRKLGRGKIEIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEVALVVFSNRGRLY 71
Query: 63 SFGHPNLDTVIDRFLSLIPTQNDGTMQFIEAHRNANVRELNAQLTQINNTLDAEKK-IGD 121
+ + ++ I+R+ + + T EA+ +E + QI N ++ + +G+
Sbjct: 72 EYANNSVKATIERYKKAC-SDSSNTGSVSEANAQYYQQEASKLRAQIGNLMNQNRNMMGE 130
Query: 122 ELSNL 126
L+ +
Sbjct: 131 ALAGM 135
>AG_TOBAC (Q43585) Floral homeotic protein AGAMOUS (NAG1)
Length = 248
Score = 77.4 bits (189), Expect = 3e-14
Identities = 45/125 (36%), Positives = 71/125 (56%), Gaps = 2/125 (1%)
Query: 3 SVRKGRGRQKIEMKKISNESNLQVTFSKHHNGLFKKASELCTLCGADVALVVFSPSGKVF 62
S ++ GR KIE+K+I N +N QVTF K NGL KKA EL LC A+VAL+VFS G+++
Sbjct: 12 SPQRKLGRGKIEIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEVALIVFSSRGRLY 71
Query: 63 SFGHPNLDTVIDRFLSLIPTQNDGTMQFIEAHRNANVRELNAQLTQINNTLDAEKK-IGD 121
+ + ++ I+R+ + + T EA+ +E + QI N + + +G+
Sbjct: 72 EYANNSVKATIERYKKAC-SDSSNTGSISEANAQYYQQEASKLRAQIGNLQNQNRNMLGE 130
Query: 122 ELSNL 126
L+ L
Sbjct: 131 SLAAL 135
>SVP_ARATH (Q9FVC1) SHORT VEGETATIVE PHASE protein
Length = 240
Score = 75.5 bits (184), Expect = 1e-13
Identities = 49/147 (33%), Positives = 80/147 (54%), Gaps = 17/147 (11%)
Query: 10 RQKIEMKKISNESNLQVTFSKHHNGLFKKASELCTLCGADVALVVFSPSGKVFSFGHPNL 69
R+KI+++KI N + QVTFSK GLFKKA EL LC ADVAL++FS +GK+F F ++
Sbjct: 3 REKIQIRKIDNATARQVTFSKRRRGLFKKAEELSVLCDADVALIIFSSTGKLFEFCSSSM 62
Query: 70 DTVIDRFLSLIPTQNDGTMQFIEAHRNANVRELNAQLTQINNTLDAEKKIGDELSNLHKE 129
V++R + ++N + L QL + ++ K+I D+ L +
Sbjct: 63 KEVLER--HNLQSKN---------LEKLDQPSLELQLVENSDHARMSKEIADKSHRLRQM 111
Query: 130 TEAKFWWACVVDGMNRDQLEIFKKALE 156
+ + G++ ++L+ +KALE
Sbjct: 112 RGEE------LQGLDIEELQQLEKALE 132
>DE21_ANTMA (Q8RVL4) MADS box protein defh21 (DEFICIENS homolog 21)
Length = 247
Score = 74.7 bits (182), Expect = 2e-13
Identities = 51/160 (31%), Positives = 87/160 (53%), Gaps = 10/160 (6%)
Query: 9 GRQKIEMKKISNESNLQVTFSKHHNGLFKKASELCTLCGADVALVVFSPSGKVFSFGHP- 67
GR KIE+K+I N ++ QVTFSK +GL KK EL LC A + L+VFS GK+ + P
Sbjct: 2 GRGKIEVKRIENNTSRQVTFSKRRSGLMKKTHELSVLCDAQIGLIVFSTKGKLTEYCTPP 61
Query: 68 -NLDTVIDRFL---SLIPTQNDGTMQFIEAHRNANVRELNAQLTQ-INNTLDAEKKIGDE 122
++ +IDR++ ++P + A + ++EL + +N L+ ++ GD+
Sbjct: 62 FSMKQIIDRYVKAKGILPEMENRAGP--HADNDQVIKELTRMKEETLNLQLNLQRYKGDD 119
Query: 123 LSNLHKE--TEAKFWWACVVDGMNRDQLEIFKKALEELKK 160
LS + E TE + ++ + +LE+ + +E LK+
Sbjct: 120 LSTVRFEELTELEKLLDQSLNKVRARKLELLHEQMENLKR 159
>AGL8_LYCES (Q40170) Agamous-like MADS box protein AGL8 homolog
(TM4)
Length = 227
Score = 74.7 bits (182), Expect = 2e-13
Identities = 44/133 (33%), Positives = 72/133 (54%), Gaps = 9/133 (6%)
Query: 9 GRQKIEMKKISNESNLQVTFSKHHNGLFKKASELCTLCGADVALVVFSPSGKVFSFGHPN 68
GR ++++K+I N+ N QVTFSK +GL KKA E+ LC A+V L+VFS GK+F + + +
Sbjct: 2 GRGRVQLKRIENKINRQVTFSKRRSGLLKKAHEISVLCDAEVGLIVFSTKGKLFEYANDS 61
Query: 69 -LDTVIDRF-------LSLIPTQNDGTMQFIEAHRNANVRELNAQLTQINNT-LDAEKKI 119
++ +++R+ L+PT + + + HR R Q Q + D E
Sbjct: 62 CMERILERYERYSFAEKQLVPTDHTSPVSWTLEHRKLKARLEVLQRNQKHYVGEDLESLS 121
Query: 120 GDELSNLHKETEA 132
EL NL + ++
Sbjct: 122 MKELQNLEHQLDS 134
>FLC_ARATH (Q9S7Q7) FLOWERING LOCUS C protein (MADS box protein
FLOWERING LOCUS F)
Length = 196
Score = 73.9 bits (180), Expect = 3e-13
Identities = 37/69 (53%), Positives = 51/69 (73%), Gaps = 1/69 (1%)
Query: 9 GRQKIEMKKISNESNLQVTFSKHHNGLFKKASELCTLCGADVALVVFSPSGKVFSFGH-P 67
GR+K+E+K+I N+S+ QVTFSK NGL +KA +L LC A VAL+V S SGK++SF
Sbjct: 2 GRKKLEIKRIENKSSRQVTFSKRRNGLIEKARQLSVLCDASVALLVVSASGKLYSFSSGD 61
Query: 68 NLDTVIDRF 76
NL ++DR+
Sbjct: 62 NLVKILDRY 70
>GLOB_TOBAC (Q03416) Floral homeotic protein GLOBOSA
Length = 209
Score = 73.6 bits (179), Expect = 4e-13
Identities = 41/122 (33%), Positives = 68/122 (55%), Gaps = 6/122 (4%)
Query: 9 GRQKIEMKKISNESNLQVTFSKHHNGLFKKASELCTLCGADVALVVFSPSGKVFSFGHPN 68
GR KIE+K+I N SN QVT+SK NG+ KKA E+ LC A V++++F+ SGK+ F +
Sbjct: 2 GRGKIEIKRIENSSNRQVTYSKRRNGILKKAKEISVLCDARVSVIIFASSGKMHEFSSTS 61
Query: 69 LDTVIDRFLSLIPTQNDGTMQFIEAHRNANVRELNAQLTQINNTLDAEKKIGDELSNL-H 127
L ++D++ L G + H N + + N ++ G+++++L H
Sbjct: 62 LVDILDQYHKL-----TGRRLWDAKHENLDNEINKVKKDNDNMQIELRHLKGEDITSLNH 116
Query: 128 KE 129
+E
Sbjct: 117 RE 118
>FB24_PETHY (Q9ATE5) MADS box protein FBP24 (Floral binding protein
24)
Length = 268
Score = 73.6 bits (179), Expect = 4e-13
Identities = 53/159 (33%), Positives = 84/159 (52%), Gaps = 14/159 (8%)
Query: 9 GRQKIEMKKISNESNLQVTFSKHHNGLFKKASELCTLCGADVALVVFSPSGKVFSF-GHP 67
GR KIE+K+I N+++ QVTFSK GL KK EL LC A + L++FS GK+F + P
Sbjct: 5 GRGKIEVKRIENKTSRQVTFSKRRAGLLKKTHELSVLCDAQIGLIIFSSKGKLFEYCSQP 64
Query: 68 -NLDTVIDRFL----SLIPTQNDGTMQFIEAHRNANVRELNAQLT-QINNTLDAEKKIGD 121
++ +I R+L + +P + D +Q + LN QL+ Q D +
Sbjct: 65 HSMSQIISRYLQTTGASLPVE-DNRVQLYDEVAKMRRDTLNLQLSLQRYKGDDLSLAQYE 123
Query: 122 ELSNLHKETEAKFWWACVVDGMNRDQLEIFKKALEELKK 160
EL+ L K+ E ++ + +LE+ ++ +E LKK
Sbjct: 124 ELNELEKQLEH------ALNKIRARKLELMQQQMENLKK 156
>AP3_ARATH (P35632) Floral homeotic protein APETALA3
Length = 232
Score = 73.6 bits (179), Expect = 4e-13
Identities = 49/168 (29%), Positives = 85/168 (50%), Gaps = 19/168 (11%)
Query: 10 RQKIEMKKISNESNLQVTFSKHHNGLFKKASELCTLCGADVALVVFSPSGKVFSFGHPNL 69
R KI++K+I N++N QVT+SK NGLFKKA EL LC A V++++FS S K+ + PN
Sbjct: 3 RGKIQIKRIENQTNRQVTYSKRRNGLFKKAHELTVLCDARVSIIMFSSSNKLHEYISPNT 62
Query: 70 DTV-----------IDRFLSLIPTQNDGTMQFIEAHRNANVRELNAQLTQINNTLDAE-- 116
T +D + + + + +E +RN ++ +L + + LD +
Sbjct: 63 TTKEIVDLYQTISDVDVWATQYERMQETKRKLLETNRNLRT-QIKQRLGECLDELDIQEL 121
Query: 117 KKIGDELSNLHKETEAKFWWAC-----VVDGMNRDQLEIFKKALEELK 159
+++ DE+ N K + + + N+ Q +I K + EL+
Sbjct: 122 RRLEDEMENTFKLVRERKFKSLGNQIETTKKKNKSQQDIQKNLIHELE 169
>AGL1_ARATH (P29381) Agamous-like MADS box protein AGL1 (Protein
Shatterproof 1)
Length = 248
Score = 73.6 bits (179), Expect = 4e-13
Identities = 44/128 (34%), Positives = 70/128 (54%), Gaps = 12/128 (9%)
Query: 6 KGRGRQKIEMKKISNESNLQVTFSKHHNGLFKKASELCTLCGADVALVVFSPSGKVFSFG 65
K GR KIE+K+I N +N QVTF K NGL KKA EL LC A+VALV+FS G+++ +
Sbjct: 14 KKLGRGKIEIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEVALVIFSTRGRLYEYA 73
Query: 66 HPNLDTVIDRFLSLI------PTQNDGTMQFIEAHRNANVRELNAQLTQINNTLDAEKKI 119
+ ++ I+R+ P+ + Q+ + + +L Q+ I N+ +
Sbjct: 74 NNSVRGTIERYKKACSDAVNPPSVTEANTQYYQQEAS----KLRRQIRDIQNS--NRHIV 127
Query: 120 GDELSNLH 127
G+ L +L+
Sbjct: 128 GESLGSLN 135
>AG19_ARATH (O82743) Agamous-like MADS box protein AGL19
Length = 219
Score = 73.6 bits (179), Expect = 4e-13
Identities = 35/67 (52%), Positives = 46/67 (68%)
Query: 10 RQKIEMKKISNESNLQVTFSKHHNGLFKKASELCTLCGADVALVVFSPSGKVFSFGHPNL 69
R K EMK+I N ++ QVTFSK NGL KKA EL LC A+VALV+FSP K++ F ++
Sbjct: 3 RGKTEMKRIENATSRQVTFSKRRNGLLKKAFELSVLCDAEVALVIFSPRSKLYEFSSSSI 62
Query: 70 DTVIDRF 76
I+R+
Sbjct: 63 AATIERY 69
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.320 0.133 0.393
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 25,759,997
Number of Sequences: 164201
Number of extensions: 1035078
Number of successful extensions: 3331
Number of sequences better than 10.0: 104
Number of HSP's better than 10.0 without gapping: 79
Number of HSP's successfully gapped in prelim test: 25
Number of HSP's that attempted gapping in prelim test: 3239
Number of HSP's gapped (non-prelim): 115
length of query: 228
length of database: 59,974,054
effective HSP length: 106
effective length of query: 122
effective length of database: 42,568,748
effective search space: 5193387256
effective search space used: 5193387256
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 64 (29.3 bits)
Medicago: description of AC144731.7


