

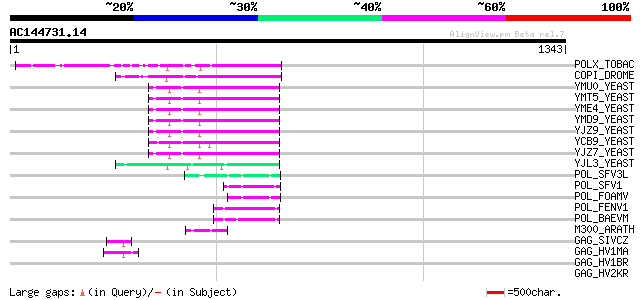
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144731.14 + phase: 0 /pseudo
(1343 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
POLX_TOBAC (P10978) Retrovirus-related Pol polyprotein from tran... 188 1e-46
COPI_DROME (P04146) Copia protein (Gag-int-pol protein) [Contain... 128 1e-28
YMU0_YEAST (Q04670) Transposon Ty1 protein B 127 2e-28
YMT5_YEAST (Q04214) Transposon Ty1 protein B 127 2e-28
YME4_YEAST (Q04711) Transposon Ty1 protein B 127 2e-28
YMD9_YEAST (Q03434) Transposon Ty1 protein B 127 2e-28
YJZ9_YEAST (P47100) Transposon Ty1 protein B 127 2e-28
YCB9_YEAST (P25384) Transposon Ty2 protein B (Ty1-17 protein B) 126 3e-28
YJZ7_YEAST (P47098) Transposon Ty1 protein B 125 6e-28
YJL3_YEAST (P47024) Transposon Ty4 207.7 kDa hypothetical protein 70 3e-11
POL_SFV3L (P27401) Pol polyprotein [Contains: Protease (EC 3.4.2... 54 3e-06
POL_SFV1 (P23074) Pol polyprotein [Contains: Protease (EC 3.4.23... 49 1e-04
POL_FOAMV (P14350) Pol polyprotein [Contains: Reverse transcript... 49 1e-04
POL_FENV1 (P31792) Pol polyprotein [Contains: Reverse transcript... 47 4e-04
POL_BAEVM (P10272) Pol polyprotein [Contains: Protease (EC 3.4.2... 46 8e-04
M300_ARATH (P93293) Hypothetical mitochondrial protein AtMg00300... 46 8e-04
GAG_SIVCZ (P17282) Gag polyprotein [Contains: Core protein p18; ... 46 8e-04
GAG_HV1MA (P04594) Gag polyprotein [Contains: Core protein p17 (... 46 8e-04
GAG_HV1BR (P03348) Gag polyprotein [Contains: Core protein p17 (... 45 0.001
GAG_HV2KR (Q74119) Gag polyprotein [Contains: Core protein p16; ... 45 0.001
>POLX_TOBAC (P10978) Retrovirus-related Pol polyprotein from
transposon TNT 1-94 [Contains: Protease (EC 3.4.23.-);
Reverse transcriptase (EC 2.7.7.49); Endonuclease]
Length = 1328
Score = 188 bits (477), Expect = 1e-46
Identities = 172/658 (26%), Positives = 279/658 (42%), Gaps = 64/658 (9%)
Query: 15 RFTGKN-YPAWEFQFRMYVKGNKLWSHLDDVSKAPTEKAALEEWEYKDAQIISWILSSID 73
+F G N + W+ + R + L LD SK P A E+W D + S I +
Sbjct: 10 KFNGDNGFSTWQRRMRDLLIQQGLHKVLDVDSKKPDTMKA-EDWADLDERAASAIRLHLS 68
Query: 74 PQMINNLRSFSTAQEMWNYLKRIYNQDNAAKRFQLELEIANYKQGNLYVQEFYSGFLNLW 133
++NN+ TA+ +W L+ +Y + L+ + LY G +
Sbjct: 69 DDVVNNIIDEDTARGIWTRLESLYMSKTLTNKLYLKKQ--------LYALHMSEG--TNF 118
Query: 134 TEHSAIIHADVPKASLAAVQEVYNTSRRDQFLMKLRPEFEVVRGALLNRNPVPSLDTCVG 193
H + + + + + V+ + + L L ++ + +L+ L
Sbjct: 119 LSHLNVFNGLITQLANLGVK-IEEEDKAILLLNSLPSSYDNLATTILHGKTTIELKDVTS 177
Query: 194 ELLREEQRLLTQGTMSHDAFISEPVPVAYAAQSRGKGRDMRQVQCFTCKQFGHVARSCTA 253
LL E ++ + A I+E +Y S GR + + +
Sbjct: 178 ALLLNE-KMRKKPENQGQALITEGRGRSYQRSSNNYGRSGARGKSKNRSK-------SRV 229
Query: 254 KFCKYCKQNGHVIFDCPIRPPRRTQYPTQALHATTSSAAPPTITSASDGGSLQPEMIQQM 313
+ C C Q GH DCP PR+ + T ++AA + +D L ++
Sbjct: 230 RNCYNCNQPGHFKRDCP--NPRKGKGETSGQKNDDNTAA---MVQNNDNVVLFINEEEEC 284
Query: 314 VLAALSNMGIHGKSSNVSRPWFLDSGASNHMTGSSE-YLHNLHSYHGNQQIQIADGNKLS 372
M + G S W +D+ AS+H T + + + G +++ + +
Sbjct: 285 -------MHLSGPESE----WVVDTAASHHATPVRDLFCRYVAGDFGT--VKMGNTSYSK 331
Query: 373 ITDVGDI--------NSDFQDVLVSPGLASNLLSVGQLVDNNCNVNFSRAGCLVQEQVSG 424
I +GDI +DV P L NL+S L + F+ + +
Sbjct: 332 IAGIGDICIKTNVGCTLVLKDVRHVPDLRMNLISGIALDRDGYESYFANQKWRLTK--GS 389
Query: 425 KVIAKGPKVGRLFPLQFISSHLSLACNNVLNSYED------WHRKLGHPNSTVLSHLFKT 478
VIAKG G L+ + C LN+ +D WH+++GH + L L K
Sbjct: 390 LVIAKGVARGTLYRTN------AEICQGELNAAQDEISVDLWHKRMGHMSEKGLQILAKK 443
Query: 479 GLLGNKQVVCTASISCPVCKLAKSKTLPFPSGAHRASNCFEMIHSDVWGMSPIASHAHYK 538
L+ + T C C K + F + + R N ++++SDV G I S K
Sbjct: 444 SLISYAKG--TTVKPCDYCLFGKQHRVSFQTSSERKLNILDLVYSDVCGPMEIESMGGNK 501
Query: 539 YFVTFIDDYSRFTWIYFLRSKSEVFSMFKKFLTYVETQFQASVKIFRSNSGGEYMSHEFQ 598
YFVTFIDD SR W+Y L++K +VF +F+KF VE + +K RS++GGEY S EF+
Sbjct: 502 YFVTFIDDASRKLWVYILKTKDQVFQVFQKFHALVERETGRKLKRLRSDNGGEYTSREFE 561
Query: 599 EYLQHKGILSQRSCPNTPQQNGLAERKNRHLLDVTRSLLLQASVPPRFWVEALSTVCF 656
EY GI +++ P TPQ NG+AER NR +++ RS+L A +P FW EA+ T C+
Sbjct: 562 EYCSSHGIRHEKTVPGTPQHNGVAERMNRTIVEKVRSMLRMAKLPKSFWGEAVQTACY 619
>COPI_DROME (P04146) Copia protein (Gag-int-pol protein) [Contains:
Copia VLP protein; Copia protease (EC 3.4.23.-)]
Length = 1409
Score = 128 bits (321), Expect = 1e-28
Identities = 101/414 (24%), Positives = 183/414 (43%), Gaps = 39/414 (9%)
Query: 256 CKYCKQNGHVIFDCPIRPPRRTQYPTQALHATTSSAAPPTITSASDGGSLQPEMIQQMVL 315
C +C + GH+ DC + + L+ T+ S G + + +
Sbjct: 232 CHHCGREGHIKKDC--------FHYKRILNNKNKENEKQVQTATSHGIAFMVKEVNNT-- 281
Query: 316 AALSNMGIHGKSSNVSRPWFLDSGASNHMTGSSEYLHNLHSYHGNQQIQIA-DGNKLSIT 374
+ + N G + LDSGAS+H+ + +I +A G + T
Sbjct: 282 SVMDNCG-----------FVLDSGASDHLINDESLYTDSVEVVPPLKIAVAKQGEFIYAT 330
Query: 375 DVG------DINSDFQDVLVSPGLASNLLSVGQLVDNNCNVNFSRAGCLVQEQVSGKVIA 428
G D +DVL A NL+SV +L + ++ F ++G + + +G ++
Sbjct: 331 KRGIVRLRNDHEITLEDVLFCKEAAGNLMSVKRLQEAGMSIEFDKSGVTISK--NGLMVV 388
Query: 429 KGPKVGRLFPLQFISSHLSLACNNVLNSYEDWHRKLGHPNSTVLSHLFKTGLLGNKQVVC 488
K + P+ ++ A + N++ WH + GH + L + + + ++ ++
Sbjct: 389 KNSGMLNNVPVINFQAYSINAKHK--NNFRLWHERFGHISDGKLLEIKRKNMFSDQSLLN 446
Query: 489 TASISCPVCKLA---KSKTLPFPS---GAHRASNCFEMIHSDVWGMSPIASHAHYKYFVT 542
+SC +C+ K LPF H F ++HSDV G + YFV
Sbjct: 447 NLELSCEICEPCLNGKQARLPFKQLKDKTHIKRPLF-VVHSDVCGPITPVTLDDKNYFVI 505
Query: 543 FIDDYSRFTWIYFLRSKSEVFSMFKKFLTYVETQFQASVKIFRSNSGGEYMSHEFQEYLQ 602
F+D ++ + Y ++ KS+VFSMF+ F+ E F V ++G EY+S+E +++
Sbjct: 506 FVDQFTHYCVTYLIKYKSDVFSMFQDFVAKSEAHFNLKVVYLYIDNGREYLSNEMRQFCV 565
Query: 603 HKGILSQRSCPNTPQQNGLAERKNRHLLDVTRSLLLQASVPPRFWVEALSTVCF 656
KGI + P+TPQ NG++ER R + + R+++ A + FW EA+ T +
Sbjct: 566 KKGISYHLTVPHTPQLNGVSERMIRTITEKARTMVSGAKLDKSFWGEAVLTATY 619
Score = 33.1 bits (74), Expect = 5.2
Identities = 22/110 (20%), Positives = 46/110 (41%), Gaps = 4/110 (3%)
Query: 7 EKAKDFCVRFTGKNYPAWEFQFRMYVKGNKLWSHLDDVSKAPTEKAALEEWEYKDAQIIS 66
+KAK F G+ Y W+F+ R + + +D + + + W+ + S
Sbjct: 2 DKAKRNIKPFDGEKYAIWKFRIRALLAEQDVLKVVDGLMPNEVD----DSWKKAERCAKS 57
Query: 67 WILSSIDPQMINNLRSFSTAQEMWNYLKRIYNQDNAAKRFQLELEIANYK 116
I+ + +N S TA+++ L +Y + + A + L + + K
Sbjct: 58 TIIEYLSDSFLNFATSDITARQILENLDAVYERKSLASQLALRKRLLSLK 107
>YMU0_YEAST (Q04670) Transposon Ty1 protein B
Length = 1328
Score = 127 bits (319), Expect = 2e-28
Identities = 98/347 (28%), Positives = 165/347 (47%), Gaps = 37/347 (10%)
Query: 336 LDSGASNHMTGSSEYLHNLHSYHGNQQIQIADGNK--LSITDVGDINSDFQD-------V 386
LDSGAS + S+ H++HS N I + D K + I +GD+ FQD V
Sbjct: 33 LDSGASRTLIRSA---HHIHSASSNPDINVVDAQKRNIPINAIGDLQFHFQDNTKTSIKV 89
Query: 387 LVSPGLASNLLSVGQLVDNNCNVNFSRAGCLVQEQVSGKVIAKGPKVGRLFPLQ---FIS 443
L +P +A +LLS+ +L + F++ V E+ G V+A K G + + +
Sbjct: 90 LHTPNIAYDLLSLNELAAVDITACFTKN---VLERSDGTVLAPIVKYGDFYWVSKKYLLP 146
Query: 444 SHLSL-ACNNVLNS-------YEDWHRKLGHPNSTVLSHLFKTGLL---GNKQVVCTASI 492
S++S+ NNV S Y HR L H N+ + + K + V +++I
Sbjct: 147 SNISVPTINNVHTSESTRKYPYPFIHRMLAHANAQTIRYSLKNNTITYFNESDVDWSSAI 206
Query: 493 S--CPVCKLAKSKTLPFPSGAH----RASNCFEMIHSDVWGMSPIASHAHYKYFVTFIDD 546
CP C + KS G+ + F+ +H+D++G + YF++F D+
Sbjct: 207 DYQCPDCLIGKSTKHRHIKGSRLKYQNSYEPFQYLHTDIFGPVHNLPKSAPSYFISFTDE 266
Query: 547 YSRFTWIYFLRSKSE--VFSMFKKFLTYVETQFQASVKIFRSNSGGEYMSHEFQEYLQHK 604
++F W+Y L + E + +F L +++ QFQASV + + + G EY + ++L+
Sbjct: 267 TTKFRWVYPLHDRREDSILDVFTTILAFIKNQFQASVLVIQMDRGSEYTNRTLHKFLEKN 326
Query: 605 GILSQRSCPNTPQQNGLAERKNRHLLDVTRSLLLQASVPPRFWVEAL 651
GI + + +G+AER NR LLD R+ L + +P W A+
Sbjct: 327 GITPCYTTTADSRAHGVAERLNRTLLDDCRTQLQCSGLPNHLWFSAI 373
>YMT5_YEAST (Q04214) Transposon Ty1 protein B
Length = 1328
Score = 127 bits (319), Expect = 2e-28
Identities = 98/347 (28%), Positives = 165/347 (47%), Gaps = 37/347 (10%)
Query: 336 LDSGASNHMTGSSEYLHNLHSYHGNQQIQIADGNK--LSITDVGDINSDFQD-------V 386
LDSGAS + S+ H++HS N I + D K + I +GD+ FQD V
Sbjct: 33 LDSGASRTLIRSA---HHIHSASSNPDINVVDAQKRNIPINAIGDLQFHFQDNTKTSIKV 89
Query: 387 LVSPGLASNLLSVGQLVDNNCNVNFSRAGCLVQEQVSGKVIAKGPKVGRLFPLQ---FIS 443
L +P +A +LLS+ +L + F++ V E+ G V+A K G + + +
Sbjct: 90 LHTPNIAYDLLSLNELAAVDITACFTKN---VLERSDGTVLAPIVKYGDFYWVSKKYLLP 146
Query: 444 SHLSL-ACNNVLNS-------YEDWHRKLGHPNSTVLSHLFKTGLL---GNKQVVCTASI 492
S++S+ NNV S Y HR L H N+ + + K + V +++I
Sbjct: 147 SNISVPTINNVHTSESTRKYPYPFIHRMLAHANAQTIRYSLKNNTITYFNESDVDWSSAI 206
Query: 493 S--CPVCKLAKSKTLPFPSGAH----RASNCFEMIHSDVWGMSPIASHAHYKYFVTFIDD 546
CP C + KS G+ + F+ +H+D++G + YF++F D+
Sbjct: 207 DYQCPDCLIGKSTKHRHIKGSRLKYQNSYEPFQYLHTDIFGPVHNLPKSAPSYFISFTDE 266
Query: 547 YSRFTWIYFLRSKSE--VFSMFKKFLTYVETQFQASVKIFRSNSGGEYMSHEFQEYLQHK 604
++F W+Y L + E + +F L +++ QFQASV + + + G EY + ++L+
Sbjct: 267 TTKFRWVYPLHDRREDSILDVFTTILAFIKNQFQASVLVIQMDRGSEYTNRTLHKFLEKN 326
Query: 605 GILSQRSCPNTPQQNGLAERKNRHLLDVTRSLLLQASVPPRFWVEAL 651
GI + + +G+AER NR LLD R+ L + +P W A+
Sbjct: 327 GITPCYTTTADSRAHGVAERLNRTLLDDCRTQLQCSGLPNHLWFSAI 373
>YME4_YEAST (Q04711) Transposon Ty1 protein B
Length = 1328
Score = 127 bits (319), Expect = 2e-28
Identities = 98/347 (28%), Positives = 165/347 (47%), Gaps = 37/347 (10%)
Query: 336 LDSGASNHMTGSSEYLHNLHSYHGNQQIQIADGNK--LSITDVGDINSDFQD-------V 386
LDSGAS + S+ H++HS N I + D K + I +GD+ FQD V
Sbjct: 33 LDSGASRTLIRSA---HHIHSASSNPDINVVDAQKRNIPINAIGDLQFHFQDNTKTSIKV 89
Query: 387 LVSPGLASNLLSVGQLVDNNCNVNFSRAGCLVQEQVSGKVIAKGPKVGRLFPLQ---FIS 443
L +P +A +LLS+ +L + F++ V E+ G V+A K G + + +
Sbjct: 90 LHTPNIAYDLLSLNELAAVDITACFTKN---VLERSDGTVLAPIVKYGDFYWVSKKYLLP 146
Query: 444 SHLSL-ACNNVLNS-------YEDWHRKLGHPNSTVLSHLFKTGLL---GNKQVVCTASI 492
S++S+ NNV S Y HR L H N+ + + K + V +++I
Sbjct: 147 SNISVPTINNVHTSESTRKYPYPFIHRMLAHANAQTIRYSLKNNTITYFNESDVDWSSAI 206
Query: 493 S--CPVCKLAKSKTLPFPSGAH----RASNCFEMIHSDVWGMSPIASHAHYKYFVTFIDD 546
CP C + KS G+ + F+ +H+D++G + YF++F D+
Sbjct: 207 DYQCPDCLIGKSTKHRHIKGSRLKYQNSYEPFQYLHTDIFGPVHNLPKSAPSYFISFTDE 266
Query: 547 YSRFTWIYFLRSKSE--VFSMFKKFLTYVETQFQASVKIFRSNSGGEYMSHEFQEYLQHK 604
++F W+Y L + E + +F L +++ QFQASV + + + G EY + ++L+
Sbjct: 267 TTKFRWVYPLHDRREDSILDVFTTILAFIKNQFQASVLVIQMDRGSEYTNRTLHKFLEKN 326
Query: 605 GILSQRSCPNTPQQNGLAERKNRHLLDVTRSLLLQASVPPRFWVEAL 651
GI + + +G+AER NR LLD R+ L + +P W A+
Sbjct: 327 GITPCYTTTADSRAHGVAERLNRTLLDDCRTQLQCSGLPNHLWFSAI 373
>YMD9_YEAST (Q03434) Transposon Ty1 protein B
Length = 1328
Score = 127 bits (319), Expect = 2e-28
Identities = 99/347 (28%), Positives = 165/347 (47%), Gaps = 37/347 (10%)
Query: 336 LDSGASNHMTGSSEYLHNLHSYHGNQQIQIADGNK--LSITDVGDINSDFQD-------V 386
LDSGAS + S+ H++HS N I + D K + I +GD+ FQD V
Sbjct: 33 LDSGASRTLIRSA---HHIHSASSNPDINVVDAQKRNIPINAIGDLQFHFQDNTKTSIKV 89
Query: 387 LVSPGLASNLLSVGQLVDNNCNVNFSRAGCLVQEQVSGKVIAKGPKVGRLFPLQ---FIS 443
L +P +A +LLS+ +L + F++ V E+ G V+A K G + + +
Sbjct: 90 LHTPNIAYDLLSLNELAAVDITACFTKN---VLERSDGTVLAPIVKYGDFYWVSKKYLLP 146
Query: 444 SHLSL-ACNNVLNS-------YEDWHRKLGHPNSTVLSHLFKTGLLG--NKQVVCTASI- 492
S++S+ NNV S Y HR L H N+ + + K + N+ V +S
Sbjct: 147 SNISVPTINNVHTSESTRKYPYPFIHRMLAHANAQTIRYSLKNNTITYFNESDVDRSSAI 206
Query: 493 --SCPVCKLAKSKTLPFPSGAH----RASNCFEMIHSDVWGMSPIASHAHYKYFVTFIDD 546
CP C + KS G+ + F+ +H+D++G + YF++F D+
Sbjct: 207 DYQCPDCLIGKSTKHRHIKGSRLKYQNSYEPFQYLHTDIFGPVHNLPKSAPSYFISFTDE 266
Query: 547 YSRFTWIYFLRSKSE--VFSMFKKFLTYVETQFQASVKIFRSNSGGEYMSHEFQEYLQHK 604
++F W+Y L + E + +F L +++ QFQASV + + + G EY + ++L+
Sbjct: 267 TTKFRWVYPLHDRREDSILDVFTTILAFIKNQFQASVLVIQMDRGSEYTNRTLHKFLEKN 326
Query: 605 GILSQRSCPNTPQQNGLAERKNRHLLDVTRSLLLQASVPPRFWVEAL 651
GI + + +G+AER NR LLD R+ L + +P W A+
Sbjct: 327 GITPCYTTTADSRAHGVAERLNRTLLDDCRTQLQCSGLPNHLWFSAI 373
>YJZ9_YEAST (P47100) Transposon Ty1 protein B
Length = 1755
Score = 127 bits (319), Expect = 2e-28
Identities = 98/347 (28%), Positives = 165/347 (47%), Gaps = 37/347 (10%)
Query: 336 LDSGASNHMTGSSEYLHNLHSYHGNQQIQIADGNK--LSITDVGDINSDFQD-------V 386
LDSGAS + S+ H++HS N I + D K + I +GD+ FQD V
Sbjct: 460 LDSGASRTLIRSA---HHIHSASSNPDINVVDAQKRNIPINAIGDLQFHFQDNTKTSIKV 516
Query: 387 LVSPGLASNLLSVGQLVDNNCNVNFSRAGCLVQEQVSGKVIAKGPKVGRLFPLQ---FIS 443
L +P +A +LLS+ +L + F++ V E+ G V+A K G + + +
Sbjct: 517 LHTPNIAYDLLSLNELAAVDITACFTKN---VLERSDGTVLAPIVKYGDFYWVSKKYLLP 573
Query: 444 SHLSL-ACNNVLNS-------YEDWHRKLGHPNSTVLSHLFKTGLL---GNKQVVCTASI 492
S++S+ NNV S Y HR L H N+ + + K + V +++I
Sbjct: 574 SNISVPTINNVHTSESTRKYPYPFIHRMLAHANAQTIRYSLKNNTITYFNESDVDWSSAI 633
Query: 493 S--CPVCKLAKSKTLPFPSGAH----RASNCFEMIHSDVWGMSPIASHAHYKYFVTFIDD 546
CP C + KS G+ + F+ +H+D++G + YF++F D+
Sbjct: 634 DYQCPDCLIGKSTKHRHIKGSRLKYQNSYEPFQYLHTDIFGPVHNLPKSAPSYFISFTDE 693
Query: 547 YSRFTWIYFLRSKSE--VFSMFKKFLTYVETQFQASVKIFRSNSGGEYMSHEFQEYLQHK 604
++F W+Y L + E + +F L +++ QFQASV + + + G EY + ++L+
Sbjct: 694 TTKFRWVYPLHDRREDSILDVFTTILAFIKNQFQASVLVIQMDRGSEYTNRTLHKFLEKN 753
Query: 605 GILSQRSCPNTPQQNGLAERKNRHLLDVTRSLLLQASVPPRFWVEAL 651
GI + + +G+AER NR LLD R+ L + +P W A+
Sbjct: 754 GITPCYTTTADSRAHGVAERLNRTLLDDCRTQLQCSGLPNHLWFSAI 800
>YCB9_YEAST (P25384) Transposon Ty2 protein B (Ty1-17 protein B)
Length = 1770
Score = 126 bits (317), Expect = 3e-28
Identities = 97/347 (27%), Positives = 165/347 (46%), Gaps = 37/347 (10%)
Query: 336 LDSGASNHMTGSSEYLHNLHSYHGNQQIQIADGNK--LSITDVGDINSDFQD-------V 386
+DSGAS + S+ YLH+ N +I I D K + I +G+++ +FQ+
Sbjct: 456 IDSGASQTLVRSAHYLHHATP---NSEINIVDAQKQDIPINAIGNLHFNFQNGTKTSIKA 512
Query: 387 LVSPGLASNLLSVGQLVDNNCNVNFSRAGCLVQEQVSGKVIAKGPKVGRLFPLQ---FIS 443
L +P +A +LLS+ +L + N F+R E+ G V+A K G + L I
Sbjct: 513 LHTPNIAYDLLSLSELANQNITACFTRN---TLERSDGTVLAPIVKHGDFYWLSKKYLIP 569
Query: 444 SHLS-LACNNVLNS-------YEDWHRKLGHPNSTVLSHLFKTGLL-----GNKQVVCTA 490
SH+S L NNV S Y HR LGH N + K + + + +
Sbjct: 570 SHISKLTINNVNKSKSVNKYPYPLIHRMLGHANFRSIQKSLKKNAVTYLKESDIEWSNAS 629
Query: 491 SISCPVCKLAKSKTLPFPSGAH----RASNCFEMIHSDVWGMSPIASHAHYKYFVTFIDD 546
+ CP C + KS G+ + F+ +H+D++G + YF++F D+
Sbjct: 630 TYQCPDCLIGKSTKHRHVKGSRLKYQESYEPFQYLHTDIFGPVHHLPKSAPSYFISFTDE 689
Query: 547 YSRFTWIYFLRSKSE--VFSMFKKFLTYVETQFQASVKIFRSNSGGEYMSHEFQEYLQHK 604
+RF W+Y L + E + ++F L +++ QF A V + + + G EY + ++ ++
Sbjct: 690 KTRFQWVYPLHDRREESILNVFTSILAFIKNQFNARVLVIQMDRGSEYTNKTLHKFFTNR 749
Query: 605 GILSQRSCPNTPQQNGLAERKNRHLLDVTRSLLLQASVPPRFWVEAL 651
GI + + + +G+AER NR LL+ R+LL + +P W A+
Sbjct: 750 GITACYTTTADSRAHGVAERLNRTLLNDCRTLLHCSGLPNHLWFSAV 796
>YJZ7_YEAST (P47098) Transposon Ty1 protein B
Length = 1755
Score = 125 bits (315), Expect = 6e-28
Identities = 97/347 (27%), Positives = 165/347 (46%), Gaps = 37/347 (10%)
Query: 336 LDSGASNHMTGSSEYLHNLHSYHGNQQIQIADGNK--LSITDVGDINSDFQD-------V 386
LDSGAS + S+ H++HS N I + D K + I +GD+ FQD V
Sbjct: 460 LDSGASRTLIRSA---HHIHSASSNPDINVVDAQKRNIPINAIGDLQFHFQDNTKTSIKV 516
Query: 387 LVSPGLASNLLSVGQLVDNNCNVNFSRAGCLVQEQVSGKVIAKGPKVGRLFPLQ---FIS 443
L +P +A +LLS+ +L + F++ V E+ G V+A + G + + +
Sbjct: 517 LHTPNIAYDLLSLNELAAVDITACFTKN---VLERSDGTVLAPIVQYGDFYWVSKRYLLP 573
Query: 444 SHLSL-ACNNVLNS-------YEDWHRKLGHPNSTVLSHLFKTGLL---GNKQVVCTASI 492
S++S+ NNV S Y HR L H N+ + + K + V +++I
Sbjct: 574 SNISVPTINNVHTSESTRKYPYPFIHRMLAHANAQTIRYSLKNNTITYFNESDVDWSSAI 633
Query: 493 S--CPVCKLAKSKTLPFPSGAH----RASNCFEMIHSDVWGMSPIASHAHYKYFVTFIDD 546
CP C + KS G+ + F+ +H+D++G + YF++F D+
Sbjct: 634 DYQCPDCLIGKSTKHRHIKGSRLKYQNSYEPFQYLHTDIFGPVHNLPKSAPSYFISFTDE 693
Query: 547 YSRFTWIYFLRSKSE--VFSMFKKFLTYVETQFQASVKIFRSNSGGEYMSHEFQEYLQHK 604
++F W+Y L + E + +F L +++ QFQASV + + + G EY + ++L+
Sbjct: 694 TTKFRWVYPLHDRREDSILDVFTTILAFIKNQFQASVLVIQMDRGSEYTNRTLHKFLEKN 753
Query: 605 GILSQRSCPNTPQQNGLAERKNRHLLDVTRSLLLQASVPPRFWVEAL 651
GI + + +G+AER NR LLD R+ L + +P W A+
Sbjct: 754 GITPCYTTTADSRAHGVAERLNRTLLDDCRTQLQCSGLPNHLWFSAI 800
>YJL3_YEAST (P47024) Transposon Ty4 207.7 kDa hypothetical protein
Length = 1803
Score = 70.5 bits (171), Expect = 3e-11
Identities = 94/437 (21%), Positives = 170/437 (38%), Gaps = 55/437 (12%)
Query: 256 CKYCKQNGHVIFDCPIRPPRRTQYPTQALHATTSSAAPPTITSAS-DGGSLQPEMIQQMV 314
C YCK H +C +P R L T + P I D L P +Q
Sbjct: 342 CMYCKSVFHCSINCKKKPNRN-------LGLTRPISQKPIIYKVHRDNNHLSPVQNEQKS 394
Query: 315 LAALSNMGIHGKSSNVSRPWFLDSGASNHMTGSSEYLHNLH-SYHGNQQIQIADGNKLSI 373
K N + +D+G+ ++T LHN S + I + +S+
Sbjct: 395 WNKTQKRS--NKVYNSKKLVIIDTGSGVNITNDKTLLHNYEDSNRSTRFFGIGKNSSVSV 452
Query: 374 TDVGDI-------NSDFQDVLVS--PGLASNLLSVGQLVDNNCNV---NFSRAGCLV--- 418
G I N+D + +L P S ++S L V ++R G +
Sbjct: 453 KGYGYIKIKNGHNNTDNKCLLTYYVPEEESTIISCYDLAKKTKMVLSRKYTRLGNKIIKI 512
Query: 419 -QEQVSGKVIAK----------GPKVGRLFPLQFISSHLSLACNNVLNSYEDWHRKLGHP 467
+ V+G + K K+ + P +S N + ED H+++GH
Sbjct: 513 KTKIVNGVIHVKMNELIERPSDDSKINAIKP----TSSPGFKLNKRSITLEDAHKRMGHT 568
Query: 468 NSTVLSHLFKTGLLGNKQVVCTA--SISCPVCKLAKSKTLPFPSGA-------HRASNCF 518
+ + K + C CK++K+ +G+ H + +
Sbjct: 569 GIQQIENSIKHNHYEESLDLIKEPNEFWCQTCKISKATKRNHYTGSMNNHSTDHEPGSSW 628
Query: 519 EMIHSDVWGMSPIASHAHYKYFVTFIDDYSRF--TWIYFLRSKSEVFSMFKKFLTYVETQ 576
M D++G ++ +Y + +D+ +R+ T +F ++ + + +K + YVETQ
Sbjct: 629 CM---DIFGPVSSSNADTKRYMLIMVDNNTRYCMTSTHFNKNAETILAQVRKNIQYVETQ 685
Query: 577 FQASVKIFRSNSGGEYMSHEFQEYLQHKGILSQRSCPNTPQQNGLAERKNRHLLDVTRSL 636
F V+ S+ G E+ + + +EY KGI + NG AER R ++ +L
Sbjct: 686 FDRKVREINSDRGTEFTNDQIEEYFISKGIHHILTSTQDHAANGRAERYIRTIITDATTL 745
Query: 637 LLQASVPPRFWVEALST 653
L Q+++ +FW A+++
Sbjct: 746 LRQSNLRVKFWEYAVTS 762
>POL_SFV3L (P27401) Pol polyprotein [Contains: Protease (EC 3.4.23.-);
Reverse transcriptase/ribonuclease H (EC 2.7.7.49) (EC
3.1.26.4) (RT); Integrase (IN)]
Length = 1157
Score = 53.9 bits (128), Expect = 3e-06
Identities = 57/234 (24%), Positives = 93/234 (39%), Gaps = 20/234 (8%)
Query: 423 SGKVIAKGPKVGRLFPLQFISSHLSLACNNVLNSYEDWHRKLGHPNSTVLSHLFKTGLLG 482
+G+V+ P R+ P + + L +N+ ++ D ST L K
Sbjct: 798 NGQVMVTRPNGKRIIPPKSDRPQIILQAHNIAHTGRD---------STFLKVSSKYWWPN 848
Query: 483 NKQVVCTASISCPVCKLAKSKTLPFPS--GAHRASNCFEMIHSDVWGMSPIASHAHYKYF 540
++ V C C + + TL P R F+ D G P+ Y +
Sbjct: 849 LRKDVVKVIRQCKQCLVTNAATLAAPPILRPERPVKPFDKFFIDYIG--PLPPSNGYLHV 906
Query: 541 VTFIDDYSRFTWIYFLRSKSEVFSMFKKFLTYVETQFQASVKIFRSNSGGEYMSHEFQEY 600
+ +D + F W+Y ++ S S K L + + A K+ S+ G + S F ++
Sbjct: 907 LVVVDSMTGFVWLYPTKAPST--SATVKALNMLTSI--AVPKVIHSDQGAAFTSATFADW 962
Query: 601 LQHKGILSQRSCPNTPQQNGLAERKNRHLLDVTRSLLLQASVPPRFWVEALSTV 654
++KGI + S P PQ +G ERKN D+ R L P W + L V
Sbjct: 963 AKNKGIQLEFSTPYHPQSSGKVERKNS---DIKRLLTKLLVGRPAKWYDLLPVV 1013
>POL_SFV1 (P23074) Pol polyprotein [Contains: Protease (EC 3.4.23.-);
Reverse transcriptase/ribonuclease H (EC 2.7.7.49) (EC
3.1.26.4) (RT); Integrase (IN)]
Length = 1161
Score = 48.5 bits (114), Expect = 1e-04
Identities = 37/137 (27%), Positives = 61/137 (44%), Gaps = 9/137 (6%)
Query: 518 FEMIHSDVWGMSPIASHAHYKYFVTFIDDYSRFTWIYFLRSKSEVFSMFKKFLTYVETQF 577
F+ + D G P+ Y + + +D + F W+Y ++ S S K L + +
Sbjct: 884 FDKFYIDYIG--PLPPSNGYLHVLVVVDSMTGFVWLYPTKAPST--SATVKALNMLTSI- 938
Query: 578 QASVKIFRSNSGGEYMSHEFQEYLQHKGILSQRSCPNTPQQNGLAERKNRHLLDVTRSLL 637
A K+ S+ G + S F ++ + KGI + S P PQ +G ERKN + + LL
Sbjct: 939 -AIPKVLHSDQGAAFTSSTFADWAKEKGIQLEFSTPYHPQSSGKVERKNSDIKRLLTKLL 997
Query: 638 LQASVPPRFWVEALSTV 654
+ P W + L V
Sbjct: 998 IGR---PAKWYDLLPVV 1011
>POL_FOAMV (P14350) Pol polyprotein [Contains: Reverse
transcriptase/ribonuclease H (EC 2.7.7.49) (EC 3.1.26.4)
(RT); Integrase (IN)]
Length = 886
Score = 48.5 bits (114), Expect = 1e-04
Identities = 36/127 (28%), Positives = 55/127 (42%), Gaps = 7/127 (5%)
Query: 528 MSPIASHAHYKYFVTFIDDYSRFTWIYFLRSKSEVFSMFKKFLTYVETQFQASVKIFRSN 587
+ P+ Y Y + +D + FTW+Y ++ S S K L + + A K+ S+
Sbjct: 684 IGPLPPSQGYLYVLVVVDGMTGFTWLYPTKAPST--SATVKSLNVLTSI--AIPKVIHSD 739
Query: 588 SGGEYMSHEFQEYLQHKGILSQRSCPNTPQQNGLAERKNRHLLDVTRSLLLQASVPPRFW 647
G + S F E+ + +GI + S P PQ ERKN D+ R L P W
Sbjct: 740 QGAAFTSSTFAEWAKERGIHLEFSTPYHPQSGSKVERKNS---DIKRLLTKLLVGRPTKW 796
Query: 648 VEALSTV 654
+ L V
Sbjct: 797 YDLLPVV 803
>POL_FENV1 (P31792) Pol polyprotein [Contains: Reverse
transcriptase/ribonuclease H (EC 2.7.7.49) (EC 3.1.26.4)
(RT); Integrase (IN)] (Fragment)
Length = 1046
Score = 47.0 bits (110), Expect = 4e-04
Identities = 39/160 (24%), Positives = 69/160 (42%), Gaps = 5/160 (3%)
Query: 493 SCPVCKLAKSKTLPFPSGAHRASNCFEMIHSDVWGMSPIASHAHYKYFVTFIDDYSRFTW 552
+C VC+ + P G N ++ ++ +A YKY + F+D +S +
Sbjct: 736 ACKVCQQVNAGATRVPEGKRTRGNR-PGVYWEIDFTEVKPHYAGYKYLLVFVDTFSGWVE 794
Query: 553 IYFLRSKSEVFSMFKKFLTYVETQFQASVKIFRSNSGGEYMSHEFQEYLQHKGILSQRSC 612
Y R ++ + KK L + +F K+ S++G ++S Q + GI + C
Sbjct: 795 AYPTRQET-AHMVAKKILEEIFPRF-GLPKVIGSDNGPAFVSQVSQGLARTLGINWKLHC 852
Query: 613 PNTPQQNGLAERKNRHLLDVTRSLLLQASVPPRFWVEALS 652
PQ +G ER NR + + L L+ + + W LS
Sbjct: 853 AYRPQSSGQVERMNRTIKETLTKLTLETGL--KDWRRLLS 890
>POL_BAEVM (P10272) Pol polyprotein [Contains: Protease (EC 3.4.23.-);
Reverse transcriptase/ribonuclease H (EC 2.7.7.49) (EC
3.1.26.4) (RT); Integrase (IN)]
Length = 1189
Score = 45.8 bits (107), Expect = 8e-04
Identities = 39/161 (24%), Positives = 70/161 (43%), Gaps = 7/161 (4%)
Query: 493 SCPVCKLAKSKTLPFPSGAHRASNCFEMIHSDVWGMSPIASHAHYKYFVTFIDDYSRFTW 552
+C VC+ + P+G N ++ ++ +A YKY + F+D +S W
Sbjct: 879 ACKVCQQVNAGATRVPAGKRTRGNR-PGVYWEIDFTEVKPHYAGYKYLLVFVDTFS--GW 935
Query: 553 IYFLRSKSEVFSMF-KKFLTYVETQFQASVKIFRSNSGGEYMSHEFQEYLQHKGILSQRS 611
+ ++ E + KK L + +F K+ S++G ++S Q + GI +
Sbjct: 936 VEAFPTRQETAHIVAKKILEEIFPRF-GLPKVIGSDNGPAFVSQVSQGLARILGINWKLH 994
Query: 612 CPNTPQQNGLAERKNRHLLDVTRSLLLQASVPPRFWVEALS 652
C PQ +G ER NR + + L L+ + + W LS
Sbjct: 995 CAYRPQSSGQVERMNRTIKETLTKLTLETGL--KDWRRLLS 1033
>M300_ARATH (P93293) Hypothetical mitochondrial protein AtMg00300
(ORF145a) (ORF1451)
Length = 145
Score = 45.8 bits (107), Expect = 8e-04
Identities = 29/107 (27%), Positives = 50/107 (46%), Gaps = 9/107 (8%)
Query: 425 KVIAKGPKVGRLFPLQFISSHLSLACNNVLNSYED----WHRKLGHPNSTVLSHLFKTGL 480
+ I KG + L+ LQ + +N+ + +D WH +L H + + L K G
Sbjct: 36 RTILKGNRHDSLYILQ---GSVETGESNLAETAKDETRLWHSRLAHMSQRGMELLVKKGF 92
Query: 481 LGNKQVVCTASISCPVCKLAKSKTLPFPSGAHRASNCFEMIHSDVWG 527
L + +V ++ C C K+ + F +G H N + +HSD+WG
Sbjct: 93 LDSSKV--SSLKFCEDCIYGKTHRVNFSTGQHTTKNPLDYVHSDLWG 137
>GAG_SIVCZ (P17282) Gag polyprotein [Contains: Core protein p18;
Core protein p25; Core protein p16]
Length = 508
Score = 45.8 bits (107), Expect = 8e-04
Identities = 26/72 (36%), Positives = 35/72 (48%), Gaps = 11/72 (15%)
Query: 234 RQVQCFTCKQFGHVARSCTA---KFCKYCKQNGHVIFDCPIRP--------PRRTQYPTQ 282
R+++CF C + GH+AR+C A K C C Q GH + DC R P R+ P
Sbjct: 398 RKIKCFNCGKEGHLARNCKAPRRKGCWRCGQEGHQMKDCTGRQVNFLGKGWPSRSGRPGN 457
Query: 283 ALHATTSSAAPP 294
+ T APP
Sbjct: 458 FVQNRTEPTAPP 469
>GAG_HV1MA (P04594) Gag polyprotein [Contains: Core protein p17
(Matrix protein); Core protein p24 (Core antigen); Core
protein p2; Core protein p7 (Nucleocapsid protein); Core
protein p1; Core protein p6]
Length = 504
Score = 45.8 bits (107), Expect = 8e-04
Identities = 29/97 (29%), Positives = 43/97 (43%), Gaps = 12/97 (12%)
Query: 227 RGKGRDMRQVQCFTCKQFGHVARSCTA---KFCKYCKQNGHVIFDCPIRP--------PR 275
RG + ++++CF C + GH+AR+C A K C C + GH + DC R P
Sbjct: 386 RGNFKGQKRIKCFNCGKEGHLARNCRAPRKKGCWKCGKEGHQMKDCTERQANFLGKIWPS 445
Query: 276 RTQYPTQALHATTSSAAPPTITSASDGGSLQPEMIQQ 312
P L + APP S G ++P Q+
Sbjct: 446 HKGRPGNFLQSRPEPTAPPA-ESFGFGEEIKPSQKQE 481
>GAG_HV1BR (P03348) Gag polyprotein [Contains: Core protein p17
(Matrix protein); Core protein p24 (Core antigen); Core
protein p2; Core protein p7 (Nucleocapsid protein); Core
protein p1; Core protein p6]
Length = 511
Score = 45.4 bits (106), Expect = 0.001
Identities = 48/170 (28%), Positives = 67/170 (39%), Gaps = 26/170 (15%)
Query: 152 VQEVYNTSRRDQFLMKLRPEFEVVRGALLNRNPVPSLDTCVGEL----LREEQRLLTQGT 207
V Y T R +Q +++ + LL +N P T + L EE QG
Sbjct: 296 VDRFYKTLRAEQASQEVK---NWMTETLLVQNANPDCKTILKALGPAATLEEMMTACQGV 352
Query: 208 --MSHDAFI-----SEPVPVAYAAQSRGKGRDMRQ-VQCFTCKQFGHVARSCTA---KFC 256
H A + S+ A RG R+ R+ V+CF C + GH+AR+C A K C
Sbjct: 353 GGPGHKARVLAEAMSQVTNSATIMMQRGNFRNQRKIVKCFNCGKEGHIARNCRAPRKKGC 412
Query: 257 KYCKQNGHVIFDCPIRP--------PRRTQYPTQALHATTSSAAPPTITS 298
C + GH + DC R P P L + APP + S
Sbjct: 413 WKCGKEGHQMKDCTERQANFLGKIWPSYKGRPGNFLQSRPEPTAPPFLQS 462
>GAG_HV2KR (Q74119) Gag polyprotein [Contains: Core protein p16;
Core protein p26]
Length = 521
Score = 45.1 bits (105), Expect = 0.001
Identities = 53/210 (25%), Positives = 82/210 (38%), Gaps = 44/210 (20%)
Query: 174 VVRGALLNRNPVPSLDTCVGELLREEQRLLTQGTMSHDAFISEPVPVAYAAQSRGKGRDM 233
V++G +N L C G + Q+ +A P+P A AAQ R
Sbjct: 335 VLKGLGMNPTLEEMLTACQG-IGGPGQKARLMAEALKEALAPAPIPFA-AAQQR------ 386
Query: 234 RQVQCFTCKQFGHVARSCTA---KFCKYCKQNGHVIFDCPIRP----------PRRTQYP 280
R ++C+ C + GH AR C A + C C ++GHV+ +CP R + +P
Sbjct: 387 RTIKCWNCGKDGHSARQCRAPRRQGCWKCGKSGHVMANCPERQAGFLGIGPWGKKPRNFP 446
Query: 281 TQALHATTSSAAPPTITSASDGGSLQPEMIQQMVLAALSNMGIHGKSSNVSRPW------ 334
+ + APP A L + +QQ G K + RP+
Sbjct: 447 VTRVPQGLTPTAPP----ADPAADLLEKYLQQ---------GRKQKEQKM-RPYKEVTED 492
Query: 335 --FLDSGASNHMTGSSEYLHNLHSYHGNQQ 362
L+ G + H + + LH L+S G Q
Sbjct: 493 LLHLEQGETPHKEATEDLLH-LNSLFGKDQ 521
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.338 0.147 0.468
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 140,063,334
Number of Sequences: 164201
Number of extensions: 5456256
Number of successful extensions: 20771
Number of sequences better than 10.0: 129
Number of HSP's better than 10.0 without gapping: 31
Number of HSP's successfully gapped in prelim test: 98
Number of HSP's that attempted gapping in prelim test: 20515
Number of HSP's gapped (non-prelim): 223
length of query: 1343
length of database: 59,974,054
effective HSP length: 122
effective length of query: 1221
effective length of database: 39,941,532
effective search space: 48768610572
effective search space used: 48768610572
T: 11
A: 40
X1: 15 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (21.8 bits)
S2: 72 (32.3 bits)
Medicago: description of AC144731.14


