

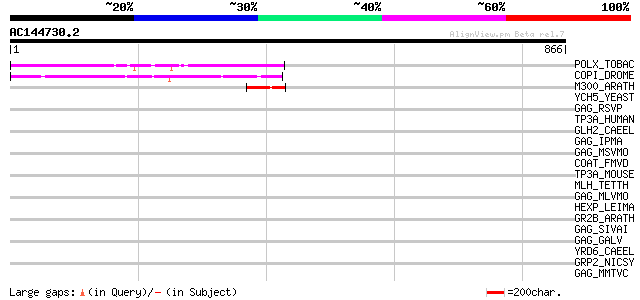
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144730.2 - phase: 0 /pseudo
(866 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
POLX_TOBAC (P10978) Retrovirus-related Pol polyprotein from tran... 230 1e-59
COPI_DROME (P04146) Copia protein (Gag-int-pol protein) [Contain... 110 1e-23
M300_ARATH (P93293) Hypothetical mitochondrial protein AtMg00300... 52 9e-06
YCH5_YEAST (P25601) Transposon Ty5-1 16.0 kDa hypothetical protein 45 0.001
GAG_RSVP (P03322) Gag polyprotein [Contains: Core protein p19; C... 39 0.059
TP3A_HUMAN (Q13472) DNA topoisomerase III alpha (EC 5.99.1.2) 38 0.10
GLH2_CAEEL (Q966L9) ATP-dependent RNA helicase glh-2 (EC 3.6.1.-... 38 0.13
GAG_IPMA (P11365) Retrovirus-related Gag polyprotein [Contains: ... 38 0.13
GAG_MSVMO (P03334) Gag polyprotein R65 [Contains: Core protein p... 37 0.23
COAT_FMVD (P09519) Probable coat protein 37 0.23
TP3A_MOUSE (O70157) DNA topoisomerase III alpha (EC 5.99.1.2) 37 0.29
MLH_TETTH (P40631) Micronuclear linker histone polyprotein (MIC ... 36 0.38
GAG_MLVMO (P03332) Gag polyprotein [Contains: Core protein p15; ... 36 0.50
HEXP_LEIMA (Q04832) DNA-binding protein HEXBP (Hexamer-binding p... 35 0.66
GR2B_ARATH (Q38896) Glycine-rich protein 2b (AtGRP2b) 35 0.66
GAG_SIVAI (Q02843) Gag polyprotein [Contains: Core protein p17; ... 35 0.66
GAG_GALV (P21416) Gag polyprotein [Contains: Core protein p15; C... 35 0.66
YRD6_CAEEL (Q09575) Hypothetical protein K02A2.6 in chromosome II 35 0.86
GRP2_NICSY (P27484) Glycine-rich protein 2 35 1.1
GAG_MMTVC (P11284) Gag polyprotein [Contains: Protein p10; Phosp... 35 1.1
>POLX_TOBAC (P10978) Retrovirus-related Pol polyprotein from
transposon TNT 1-94 [Contains: Protease (EC 3.4.23.-);
Reverse transcriptase (EC 2.7.7.49); Endonuclease]
Length = 1328
Score = 230 bits (586), Expect = 1e-59
Identities = 159/449 (35%), Positives = 234/449 (51%), Gaps = 37/449 (8%)
Query: 1 KWDIEKFTGSNDFGLWKVKMRAILIQQKCVEALKGEAQMDVHLTPAEKTEMNDKAVSAII 60
K+++ KF G N F W+ +MR +LIQQ + L +++ + + +++++A SAI
Sbjct: 5 KYEVAKFNGDNGFSTWQRRMRDLLIQQGLHKVLDVDSKKPDTMKAEDWADLDERAASAIR 64
Query: 61 LCLGDKVLREVSRESTAVSMRNKLDSLYMTKSLAHRQCLKQQLYFYRMVESKPIMEQLTE 120
L L D V+ + E TA + +L+SLYM+K+L ++ LK+QLY M E + L
Sbjct: 65 LHLSDDVVNNIIDEDTARGIWTRLESLYMSKTLTNKLYLKKQLYALHMSEGTNFLSHLNV 124
Query: 121 FNKIIDDLANIDVNLEDEDKALHLPCALPRSFENFKDTMLYGK*GTITLEEVQAALRTKE 180
FN +I LAN+ V +E+EDKA+ L +LP S++N T+L+GK TI L++V +AL E
Sbjct: 125 FNGLITQLANLGVKIEEEDKAILLLNSLPSSYDNLATTILHGK-TTIELKDVTSALLLNE 183
Query: 181 LTKFKELKVEDSG-----EGLNVSRERSQNRGKGKGKNSRSKSRSKGDGNKTQYKCFICH 235
+ K E+ G EG S +RS N G +SK+RSK C+ C+
Sbjct: 184 KMR---KKPENQGQALITEGRGRSYQRSSNNYGRSGARGKSKNRSK----SRVRNCYNCN 236
Query: 236 NPGHFKKDCPE-RKGNG---------------GGNPSVQIASNEEGYESAGALTVTSWEP 279
PGHFK+DCP RKG G N +V + NEE E L+ P
Sbjct: 237 QPGHFKRDCPNPRKGKGETSGQKNDDNTAAMVQNNDNVVLFINEE--EECMHLS----GP 290
Query: 280 EKGWVLDSGCSYHICPRKEYFEMLELEEGGVVCLGNNKACKIQVIGTIRLKMFDDRDFLL 339
E WV+D+ S+H P ++ F + G V +GN KI IG I +K +L
Sbjct: 291 ESEWVVDTAASHHATPVRDLFCRYVAGDFGTVKMGNTSYSKIAGIGDICIKTNVGCTLVL 350
Query: 340 KDVRYIPKLRRNLISISMFDGLGYCTRIERGVMRISHGALIIAKGSKIHGLYILEGSTVI 399
KDVR++P LR NLIS D GY + R++ G+L+IAKG LY
Sbjct: 351 KDVRHVPDLRMNLISGIALDRDGYESYFANQKWRLTKGSLVIAKGVARGTLYRTNAEICQ 410
Query: 400 ADASVASVDTLDVTKLWHLRLGHVSERGI 428
+ + A D + V LWH R+GH+SE+G+
Sbjct: 411 GELNAAQ-DEISV-DLWHKRMGHMSEKGL 437
>COPI_DROME (P04146) Copia protein (Gag-int-pol protein) [Contains:
Copia VLP protein; Copia protease (EC 3.4.23.-)]
Length = 1409
Score = 110 bits (276), Expect = 1e-23
Identities = 111/438 (25%), Positives = 193/438 (43%), Gaps = 28/438 (6%)
Query: 1 KWDIEKFTGSNDFGLWKVKMRAILIQQKCVEALKG--EAQMDVHLTPAEKTEMNDKAVSA 58
K +I+ F G + +WK ++RA+L +Q ++ + G ++D AE+ A S
Sbjct: 5 KRNIKPFDGEK-YAIWKFRIRALLAEQDVLKVVDGLMPNEVDDSWKKAERC-----AKST 58
Query: 59 IILCLGDKVLREVSRESTAVSMRNKLDSLYMTKSLAHRQCLKQQLYFYRMVESKPIMEQL 118
II L D L + + TA + LD++Y KSLA + L+++L ++ ++
Sbjct: 59 IIEYLSDSFLNFATSDITARQILENLDAVYERKSLASQLALRKRLLSLKLSSEMSLLSHF 118
Query: 119 TEFNKIIDDLANIDVNLEDEDKALHLPCALPRSFENFKDTMLYGK*GTITLEEVQAALRT 178
F+++I +L +E+ DK HL LP ++ + +TL V+ L
Sbjct: 119 HIFDELISELLAAGAKIEEMDKISHLLITLPSCYDGIITAIETLSEENLTLAFVKNRLLD 178
Query: 179 KELTKFKELKVEDSGEGLNVSRERSQNRGKG---KGKNSRSKSRSKGDGNKTQYKCFICH 235
+E+ K K + S + +N + N K K + ++ K KG+ +K + KC C
Sbjct: 179 QEI-KIKNDHNDTSKKVMNAIVHNNNNTYKNNLFKNRVTKPKKIFKGN-SKYKVKCHHCG 236
Query: 236 NPGHFKKDCPERK-----GNGGGNPSVQIASNEEGYESAGALTVTSWEPEKGWVLDSGCS 290
GH KKDC K N VQ A++ + TS G+VLDSG S
Sbjct: 237 REGHIKKDCFHYKRILNNKNKENEKQVQTATSHGIAFMVKEVNNTSVMDNCGFVLDSGAS 296
Query: 291 YHICPRKE-YFEMLELEEGGVVCLG-NNKACKIQVIGTIRLKMFDDRDFLLKDVRYIPKL 348
H+ + Y + +E+ + + + G +RL+ +D + L+DV + +
Sbjct: 297 DHLINDESLYTDSVEVVPPLKIAVAKQGEFIYATKRGIVRLR--NDHEITLEDVLFCKEA 354
Query: 349 RRNLISISMFDGLGYCTRIERGVMRISHGALIIAKGSKIHGLYILEGSTVI-ADASVASV 407
NL+S+ G ++ + IS L++ K S +L VI A +
Sbjct: 355 AGNLMSVKRLQEAGMSIEFDKSGVTISKNGLMVVKNSG-----MLNNVPVINFQAYSINA 409
Query: 408 DTLDVTKLWHLRLGHVSE 425
+ +LWH R GH+S+
Sbjct: 410 KHKNNFRLWHERFGHISD 427
>M300_ARATH (P93293) Hypothetical mitochondrial protein AtMg00300
(ORF145a) (ORF1451)
Length = 145
Score = 51.6 bits (122), Expect = 9e-06
Identities = 25/61 (40%), Positives = 39/61 (62%), Gaps = 1/61 (1%)
Query: 370 GVMRISHGALIIAKGSKIHGLYILEGSTVIADASVASVDTLDVTKLWHLRLGHVSERGIW 429
GV+++ G I KG++ LYIL+GS ++++A D T+LWH RL H+S+RG+
Sbjct: 27 GVLKVLKGCRTILKGNRHDSLYILQGSVETGESNLAET-AKDETRLWHSRLAHMSQRGME 85
Query: 430 L 430
L
Sbjct: 86 L 86
>YCH5_YEAST (P25601) Transposon Ty5-1 16.0 kDa hypothetical protein
Length = 146
Score = 44.7 bits (104), Expect = 0.001
Identities = 32/121 (26%), Positives = 47/121 (38%), Gaps = 15/121 (12%)
Query: 245 PERKGNGGGNPSVQIASNEEGYESAGALTVTSW----------EPEKGWVLDSGCSYHIC 294
P K + + SV I E ++AG +T SW W+ D+GC+ H+C
Sbjct: 31 PNDKTSRSSSASVAIPDYETQGQTAGQITPKSWLCMLSSTVPATKSSEWIFDTGCTSHMC 90
Query: 295 PRKEYFEMLELEEGGVVCLGNNKACKIQVIGTIRLKMFDDRDFLLKDVRYIPKLRRNLIS 354
+ F G + I GT+ + L DV Y+P L NLIS
Sbjct: 91 HDRSIFSSFTRSSRKDFVRGVGGSIPIMGSGTVNIGTVQ-----LHDVSYVPDLPVNLIS 145
Query: 355 I 355
+
Sbjct: 146 V 146
>GAG_RSVP (P03322) Gag polyprotein [Contains: Core protein p19; Core
protein p2A; Core protein p2B; Core protein p10; Capsid
protein p27; Inner coat protein p12; Protease p15 (EC
3.4.23.-)]
Length = 701
Score = 38.9 bits (89), Expect = 0.059
Identities = 12/48 (25%), Positives = 26/48 (54%)
Query: 214 SRSKSRSKGDGNKTQYKCFICHNPGHFKKDCPERKGNGGGNPSVQIAS 261
+R + G G + + C+ C +PGH++ CP+++ +G Q+ +
Sbjct: 492 NRERDGQTGSGGRARGLCYTCGSPGHYQAQCPKKRKSGNSRERCQLCN 539
Score = 33.5 bits (75), Expect = 2.5
Identities = 21/76 (27%), Positives = 31/76 (40%), Gaps = 13/76 (17%)
Query: 198 VSRERSQNRGKG------------KGKNSRSKSRSKGDGNKTQYKCFICHNPGHFKKDCP 245
V+RER G G G + + GN + +C +C+ GH K C
Sbjct: 491 VNRERDGQTGSGGRARGLCYTCGSPGHYQAQCPKKRKSGNSRE-RCQLCNGMGHNAKQCR 549
Query: 246 ERKGNGGGNPSVQIAS 261
+R GN G P ++S
Sbjct: 550 KRDGNQGQRPGKGLSS 565
>TP3A_HUMAN (Q13472) DNA topoisomerase III alpha (EC 5.99.1.2)
Length = 1001
Score = 38.1 bits (87), Expect = 0.10
Identities = 18/64 (28%), Positives = 31/64 (48%), Gaps = 5/64 (7%)
Query: 190 EDSGEGLNVSRERSQNRGK-----GKGKNSRSKSRSKGDGNKTQYKCFICHNPGHFKKDC 244
E++ G + + + +RG+ + K R+ S G K KC +CH PGH + C
Sbjct: 938 ENTAPGTSGAPSWTGDRGRTLESEARSKRPRASSSDMGSTAKKPRKCSLCHQPGHTRPFC 997
Query: 245 PERK 248
P+ +
Sbjct: 998 PQNR 1001
>GLH2_CAEEL (Q966L9) ATP-dependent RNA helicase glh-2 (EC 3.6.1.-)
(Germline helicase-2)
Length = 974
Score = 37.7 bits (86), Expect = 0.13
Identities = 25/86 (29%), Positives = 33/86 (38%), Gaps = 11/86 (12%)
Query: 192 SGEGLNVSRERSQNRGKGKGKNSRSKSRSKGDGNKTQYKCFICHNPGHFKKDCPERK--- 248
SG G ER+ N + RS + + C+ C PGH +DCPE +
Sbjct: 359 SGGGGQDRGERNNNCFNCQQPGHRSNDCPEPKKEREPRVCYNCQQPGHNSRDCPEERKPR 418
Query: 249 --------GNGGGNPSVQIASNEEGY 266
G GGGN N EG+
Sbjct: 419 EGRNGFTSGFGGGNDGGFGGGNAEGF 444
Score = 36.2 bits (82), Expect = 0.38
Identities = 19/48 (39%), Positives = 20/48 (41%), Gaps = 2/48 (4%)
Query: 203 SQNRGKGKGKNSRSKSRSKG--DGNKTQYKCFICHNPGHFKKDCPERK 248
S G G G NS G D + CF C PGH DCPE K
Sbjct: 229 SGGSGFGSGGNSNGFGSGGGGQDRGERNNNCFNCQQPGHRSNDCPEPK 276
Score = 35.0 bits (79), Expect = 0.86
Identities = 16/42 (38%), Positives = 19/42 (45%), Gaps = 3/42 (7%)
Query: 207 GKGKGKNSRSKSRSKGDGNKTQYKCFICHNPGHFKKDCPERK 248
G G S + +G+ N CF C PGH DCPE K
Sbjct: 352 GNSNGFGSGGGGQDRGERNNN---CFNCQQPGHRSNDCPEPK 390
Score = 33.5 bits (75), Expect = 2.5
Identities = 21/64 (32%), Positives = 27/64 (41%), Gaps = 1/64 (1%)
Query: 192 SGEGLNVSRERSQNRGKGKGKNSRSKSRSKGDGNKTQYKCFICHNPGHFKKDCP-ERKGN 250
SG G ER+ N + RS + + C+ C PGH +DCP ERK
Sbjct: 245 SGGGGQDRGERNNNCFNCQQPGHRSNDCPEPKKEREPRVCYNCQQPGHNSRDCPEERKPR 304
Query: 251 GGGN 254
G N
Sbjct: 305 EGRN 308
>GAG_IPMA (P11365) Retrovirus-related Gag polyprotein [Contains:
Protease (EC 3.4.23.-)]
Length = 827
Score = 37.7 bits (86), Expect = 0.13
Identities = 18/38 (47%), Positives = 25/38 (65%), Gaps = 6/38 (15%)
Query: 214 SRSKSRSKGDGNKTQYKCFICHNPGHFKKDC--PERKG 249
S+++S S+ D Q CF C PGHFKKDC P+++G
Sbjct: 447 SQNRSMSRND----QRTCFNCGKPGHFKKDCRAPDKQG 480
>GAG_MSVMO (P03334) Gag polyprotein R65 [Contains: Core protein p15;
Inner coat protein p12; Core shell protein p30;
Nucleoprotein p10]
Length = 538
Score = 37.0 bits (84), Expect = 0.23
Identities = 24/103 (23%), Positives = 46/103 (44%), Gaps = 6/103 (5%)
Query: 168 TLEEVQAALRTKELTKFKELKVEDSGEGLNVSRERSQNRGK--GKGKNSRSKSRSKGDGN 225
T EE + +R + K + + ED + R R + + + + + R +G+
Sbjct: 436 TPEEREERIRREREEKEERRRTEDEQKEKERDRRRHREMSRLLATVVSGQRQDRQEGERR 495
Query: 226 KTQY---KCFICHNPGHFKKDCPER-KGNGGGNPSVQIASNEE 264
++Q +C C GH+ KDCP R +G G P + + ++
Sbjct: 496 RSQLDCDQCTYCEEQGHWAKDCPRRPRGPRGPRPQTSLLTLDD 538
>COAT_FMVD (P09519) Probable coat protein
Length = 489
Score = 37.0 bits (84), Expect = 0.23
Identities = 24/88 (27%), Positives = 41/88 (46%), Gaps = 4/88 (4%)
Query: 179 KELTKFKELKVEDSGEGLNVSRERSQNRGKGKGKNSRSKSRSKGDGNKTQYKCFICHNPG 238
K+ +K+K K + + + L ++R GK K K +G + + +C+IC G
Sbjct: 362 KKSSKYKAYKKKKTLKKLWKKKKRKFTPGKYFSKKKPEKFCPQG---RKKCRCWICTEEG 418
Query: 239 HFKKDCPERKGNGGGNPSVQIASNEEGY 266
H+ +CP RK + + I EGY
Sbjct: 419 HYANECPNRKSH-QEKVKILIHGMNEGY 445
>TP3A_MOUSE (O70157) DNA topoisomerase III alpha (EC 5.99.1.2)
Length = 1003
Score = 36.6 bits (83), Expect = 0.29
Identities = 15/43 (34%), Positives = 21/43 (47%)
Query: 206 RGKGKGKNSRSKSRSKGDGNKTQYKCFICHNPGHFKKDCPERK 248
R + K R+ S G K KC +CH PGH + CP+ +
Sbjct: 961 RPEAASKRPRAGSSDAGSTVKKPRKCSLCHQPGHTRTFCPQNR 1003
>MLH_TETTH (P40631) Micronuclear linker histone polyprotein (MIC LH)
[Contains: Micronuclear linker histone-alpha;
Micronuclear linker histone-beta; Micronuclear linker
histone-delta; Micronuclear linker histone-gamma]
Length = 633
Score = 36.2 bits (82), Expect = 0.38
Identities = 26/107 (24%), Positives = 51/107 (47%), Gaps = 4/107 (3%)
Query: 182 TKFKELKVEDSGEGLNVSRERSQNRGKGKGKNSRSKSRSKGDG-NKTQYKCFICHNPGHF 240
+K K S + + S++ + N G+ SRS+S+SK + NK K + P
Sbjct: 508 SKSKSKSASKSRKNASKSKKDTTNHGRQTRSKSRSESKSKSEAPNKPSNKMEVIEQP--- 564
Query: 241 KKDCPERKGNGGGNPSVQIASNEEGYESAGALTVTSWEPEKGWVLDS 287
K++ +RK + S + S+++ + + +T+ +P+K DS
Sbjct: 565 KEESSDRKRRESRSQSAKKTSDKKSKNRSDSKKMTAEDPKKNNAEDS 611
Score = 32.7 bits (73), Expect = 4.2
Identities = 19/53 (35%), Positives = 26/53 (48%), Gaps = 1/53 (1%)
Query: 177 RTKELTKFKELKVEDSG-EGLNVSRERSQNRGKGKGKNSRSKSRSKGDGNKTQ 228
RTK+ K SG + S +S+ + KGKNS+S+S SK N Q
Sbjct: 400 RTKKANNKSASKASKSGSKSKGKSASKSKGKSSSKGKNSKSRSASKPKSNAAQ 452
>GAG_MLVMO (P03332) Gag polyprotein [Contains: Core protein p15;
Inner coat protein p12; Core shell protein p30;
Nucleoprotein p10]
Length = 538
Score = 35.8 bits (81), Expect = 0.50
Identities = 24/103 (23%), Positives = 46/103 (44%), Gaps = 6/103 (5%)
Query: 168 TLEEVQAALRTKELTKFKELKVEDSGEGLNVSRERSQNRGK--GKGKNSRSKSRSKGDGN 225
T EE + +R + K + + ED + R R + K + + + R G+
Sbjct: 436 TPEEREERIRRETEEKEERRRTEDEQKEKERDRRRHREMSKLLATVVSGQKQDRQGGERR 495
Query: 226 KTQY---KCFICHNPGHFKKDCPER-KGNGGGNPSVQIASNEE 264
++Q +C C GH+ KDCP++ +G G P + + ++
Sbjct: 496 RSQLDRDQCAYCKEKGHWAKDCPKKPRGPRGPRPQTSLLTLDD 538
>HEXP_LEIMA (Q04832) DNA-binding protein HEXBP (Hexamer-binding
protein)
Length = 271
Score = 35.4 bits (80), Expect = 0.66
Identities = 15/35 (42%), Positives = 21/35 (59%), Gaps = 3/35 (8%)
Query: 220 SKGDGNKTQYKCFICHNPGHFKKDCPERKGNGGGN 254
S G G++ YKC PGH ++CPE G+ GG+
Sbjct: 216 STGSGDRACYKC---GKPGHISRECPEAGGSYGGS 247
Score = 34.3 bits (77), Expect = 1.5
Identities = 20/81 (24%), Positives = 31/81 (37%), Gaps = 1/81 (1%)
Query: 188 KVEDSGEGLNVSRERSQNRGKGKGKNSRSKSRSKGDGNKTQYKCFICHNPGHFKKDCPER 247
+ ED S +N GK +G +R + G++ CF C GH ++CP
Sbjct: 3 ETEDVKRPRTESSTSCRNCGK-EGHYARECPEADSKGDERSTTCFRCGEEGHMSRECPNE 61
Query: 248 KGNGGGNPSVQIASNEEGYES 268
+G E G+ S
Sbjct: 62 ARSGAAGAMTCFRCGEAGHMS 82
Score = 33.5 bits (75), Expect = 2.5
Identities = 14/43 (32%), Positives = 19/43 (43%)
Query: 211 GKNSRSKSRSKGDGNKTQYKCFICHNPGHFKKDCPERKGNGGG 253
G SR S G ++C+ C GH +DCP +G G
Sbjct: 79 GHMSRDCPNSAKPGAAKGFECYKCGQEGHLSRDCPSSQGGSRG 121
>GR2B_ARATH (Q38896) Glycine-rich protein 2b (AtGRP2b)
Length = 201
Score = 35.4 bits (80), Expect = 0.66
Identities = 20/54 (37%), Positives = 25/54 (46%), Gaps = 2/54 (3%)
Query: 193 GEGLNVSRERSQNRG--KGKGKNSRSKSRSKGDGNKTQYKCFICHNPGHFKKDC 244
GE +++RE SQ G G G R S G G C+ C GHF +DC
Sbjct: 142 GEPGHMARECSQGGGGYSGGGGGGRYGSGGGGGGGGGGLSCYSCGESGHFARDC 195
Score = 32.0 bits (71), Expect = 7.2
Identities = 24/80 (30%), Positives = 32/80 (40%), Gaps = 12/80 (15%)
Query: 206 RGKGKGKNSR--------SKSRSKGDGNKTQYKCFICHNPGHFKKDCPERKGN-GGGNPS 256
RG G+G S S R G G+ + CF C PGH ++C + G GG
Sbjct: 108 RGGGRGGGSYGGGYGGRGSGGRGGGGGDNS---CFKCGEPGHMARECSQGGGGYSGGGGG 164
Query: 257 VQIASNEEGYESAGALTVTS 276
+ S G G L+ S
Sbjct: 165 GRYGSGGGGGGGGGGLSCYS 184
>GAG_SIVAI (Q02843) Gag polyprotein [Contains: Core protein p17;
Core protein p24; Core protein p15]
Length = 513
Score = 35.4 bits (80), Expect = 0.66
Identities = 37/131 (28%), Positives = 50/131 (37%), Gaps = 20/131 (15%)
Query: 168 TLEEVQAALRTKELTKFK-ELKVEDSGEGLNVSRERSQNRGK-------GKGKNSRSKSR 219
TLEE+ A + + K +L VE G N+ + Q +G GK +
Sbjct: 345 TLEEMLIACQGVGGPQHKAKLMVEMMSNGQNMVQVGPQKKGPRGPLKCFNCGKFGHMQRE 404
Query: 220 SKGDGNKTQYKCFICHNPGHFKKDCPERKGN-------GGGNPS--VQIASNEEGYESAG 270
K Q KCF C GH KDC + N GG P VQ + G E
Sbjct: 405 CKAP---RQIKCFKCGKIGHMAKDCKNGQANFLGYGHWGGAKPRNFVQYRGDTVGLEPTA 461
Query: 271 ALTVTSWEPEK 281
T+++P K
Sbjct: 462 PPMETAYDPAK 472
>GAG_GALV (P21416) Gag polyprotein [Contains: Core protein p15; Core
protein p12; Core protein p30; Core protein p10]
Length = 520
Score = 35.4 bits (80), Expect = 0.66
Identities = 17/51 (33%), Positives = 24/51 (46%)
Query: 198 VSRERSQNRGKGKGKNSRSKSRSKGDGNKTQYKCFICHNPGHFKKDCPERK 248
VSRE S R G N K+ G + +C C GH+ ++CP +K
Sbjct: 458 VSREGSTGRQTGNLSNQAKKTPRDGRPPLDKDQCAYCKEKGHWARECPRKK 508
>YRD6_CAEEL (Q09575) Hypothetical protein K02A2.6 in chromosome II
Length = 1268
Score = 35.0 bits (79), Expect = 0.86
Identities = 22/89 (24%), Positives = 39/89 (43%), Gaps = 4/89 (4%)
Query: 170 EEVQAALRTKELTKFKELKVEDSGEGLNVSRERSQNRGKGKGKNSRSKSRSKGDGNKTQY 229
+E ++ + +K +K S + ++V++ + K K R +S D K
Sbjct: 178 DEWMKFIQMHQQSKIVSVKPSKSSQQVDVNKVDTNRSKKKKKPIPRKPEKSSQDSKKKGE 237
Query: 230 --KCFICHNPGHFKKDCPE--RKGNGGGN 254
CF C+ GH+ +C + GN GGN
Sbjct: 238 IPTCFYCNKKGHYATNCRSNPKTGNQGGN 266
>GRP2_NICSY (P27484) Glycine-rich protein 2
Length = 214
Score = 34.7 bits (78), Expect = 1.1
Identities = 14/32 (43%), Positives = 17/32 (52%)
Query: 222 GDGNKTQYKCFICHNPGHFKKDCPERKGNGGG 253
G G+ CF C GHF +DC + G GGG
Sbjct: 150 GGGSGGGSGCFKCGESGHFARDCSQSGGGGGG 181
>GAG_MMTVC (P11284) Gag polyprotein [Contains: Protein p10;
Phosphorylated protein pp21; Protein p3; Protein p8;
Major core protein p27; Nucleic acid binding protein
p14]
Length = 591
Score = 34.7 bits (78), Expect = 1.1
Identities = 20/54 (37%), Positives = 24/54 (44%), Gaps = 11/54 (20%)
Query: 204 QNRGKGKGKNSRSKSRSKGDGNKTQYKCFICHNPGHFKKDCPERKGNGGGNPSV 257
Q G GKG G G+K CF C GH K+DC E KG+ P +
Sbjct: 511 QTYGGGKG----------GQGSKGPV-CFSCGKTGHIKRDCKEEKGSKRAPPGL 553
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.344 0.151 0.505
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 88,546,118
Number of Sequences: 164201
Number of extensions: 3436305
Number of successful extensions: 15421
Number of sequences better than 10.0: 57
Number of HSP's better than 10.0 without gapping: 22
Number of HSP's successfully gapped in prelim test: 35
Number of HSP's that attempted gapping in prelim test: 15330
Number of HSP's gapped (non-prelim): 97
length of query: 866
length of database: 59,974,054
effective HSP length: 119
effective length of query: 747
effective length of database: 40,434,135
effective search space: 30204298845
effective search space used: 30204298845
T: 11
A: 40
X1: 15 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 38 (21.6 bits)
S2: 70 (31.6 bits)
Medicago: description of AC144730.2


