

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
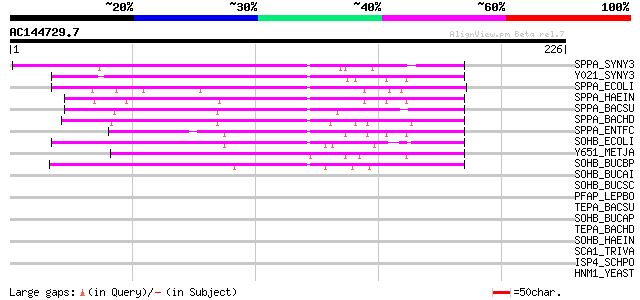
Query= AC144729.7 - phase: 0
(226 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
SPPA_SYNY3 (P73689) Protease IV homolog (EC 3.4.21.-) (Endopepti... 82 8e-16
Y021_SYNY3 (Q55682) Putative protease slr0021 (EC 3.4.21.-) 73 6e-13
SPPA_ECOLI (P08395) Protease IV (EC 3.4.21.-) (Endopeptidase IV)... 67 3e-11
SPPA_HAEIN (P45243) Protease IV (EC 3.4.21.-) (Endopeptidase IV)... 63 6e-10
SPPA_BACSU (O34525) Putative signal peptide peptidase sppA (EC 3... 60 4e-09
SPPA_BACHD (Q9K809) Putative signal peptide peptidase sppA (EC 3... 57 4e-08
SPPA_ENTFC (Q9X480) Putative signal peptide peptidase sppA (EC 3... 54 4e-07
SOHB_ECOLI (P24213) Possible protease sohB (EC 3.4.21.-) 50 4e-06
Y651_METJA (Q58067) Putative protease MJ0651 (EC 3.4.21.-) 49 1e-05
SOHB_BUCBP (Q89AL0) Putative protease sohB (EC 3.4.21.-) 45 2e-04
SOHB_BUCAI (P57370) Possible protease sohB (EC 3.4.21.-) 42 0.002
SOHB_BUCSC (Q44600) Possible protease sohB (EC 3.4.21.-) (Fragment) 40 0.004
PFAP_LEPBO (Q48513) Putative peptidase pfaP precursor (EC 3.4.21... 39 0.013
TEPA_BACSU (Q99171) Translocation-enhancing protein tepA 36 0.065
SOHB_BUCAP (Q8K9P8) Possible protease sohB (EC 3.4.21.-) 36 0.085
TEPA_BACHD (Q9KA93) Translocation-enhancing protein tepA 35 0.11
SOHB_HAEIN (P45315) Possible protease sohB (EC 3.4.21.-) 35 0.14
SCA1_TRIVA (P53399) Succinyl-CoA ligase [GDP-forming] alpha-chai... 34 0.25
ISP4_SCHPO (P40900) Sexual differentiation process protein isp4 33 0.42
HNM1_YEAST (P19807) Choline transport protein 33 0.72
>SPPA_SYNY3 (P73689) Protease IV homolog (EC 3.4.21.-)
(Endopeptidase IV) (Signal peptide peptidase)
Length = 610
Score = 82.4 bits (202), Expect = 8e-16
Identities = 68/223 (30%), Positives = 103/223 (45%), Gaps = 43/223 (19%)
Query: 2 ISFRKYSKVRKWTVGISEAKEQIAIIRASGTMDS------DIVTSNFIEKIGMVKDSKKF 55
IS +Y +++ W + +IAI+ G++ + +I + E + ++
Sbjct: 299 ISLAEYHRLQNWETENHDQDPKIAIVYLEGSIVNGRGTWENIGGDRYGELLRTIRQDDDI 358
Query: 56 KAVIVRIDSVGGDFHASQSTWEAIRSLASKKPVIASMSDAATSAGYYMAMGAGAIVAENL 115
KAV++RI+S GG A+ W + L ++KPVI SM + A S GY++A IVA+
Sbjct: 359 KAVVLRINSPGGSASAADIIWREVELLQAQKPVIISMGNVAASGGYWIATAGEKIVAQPN 418
Query: 116 TLTGSIRGTVSQNFNLDN---------------EL----PSFIPYDDAKV---------- 146
T+TGSI G S FN++N EL S P + ++
Sbjct: 419 TVTGSI-GVFSILFNVENLGDRLGLNWDEVATGELANVGSSIKPKTELELAIFQRSVDQV 477
Query: 147 ----LDNVVRLRLMTLGKLDKMKKVAHGRIWTGKDAASHGLVD 185
LD V R R ++ LD VA GR+WTG A GLVD
Sbjct: 478 YEIFLDKVGRARNLSPTALD---SVAQGRVWTGLAAQKVGLVD 517
>Y021_SYNY3 (Q55682) Putative protease slr0021 (EC 3.4.21.-)
Length = 277
Score = 72.8 bits (177), Expect = 6e-13
Identities = 61/198 (30%), Positives = 96/198 (47%), Gaps = 33/198 (16%)
Query: 18 SEAKEQIAIIRASGTMDSDIVTSNFIEKIGMVKDSKKFKAVIVRIDSVGGDFHASQSTWE 77
+ +++IA I +G + S T + K + KK+ A++VRIDS GG SQ +
Sbjct: 7 TSTRKKIARIEVTGAIASG--TRKAVLKALKTVEEKKYPALLVRIDSPGGTVVDSQEIYT 64
Query: 78 AIRSLASKKPVIASMSDAATSAGYYMAMGAGAIVAENLTLTGSIRGTVSQNFNLDNELP- 136
++ L+ K V+AS + + S G Y+AMG I+A + T+TGSI G + + NL+ L
Sbjct: 65 KLKQLSEKIKVVASFGNISASGGVYIAMGCPHIMANSGTITGSI-GVILRGNNLERLLEK 123
Query: 137 ---SFI-----PYDDAKVLDNVVR------------------LRLMTLGK---LDKMKKV 167
SF PY D D + + + G+ ++K+K+
Sbjct: 124 VGVSFKVIKSGPYKDILSFDRELLPEEQSILQALIDDSYGQFVSTVAAGRNLAVEKVKEF 183
Query: 168 AHGRIWTGKDAASHGLVD 185
A GRI+TG+ A GLVD
Sbjct: 184 ADGRIFTGQQALELGLVD 201
>SPPA_ECOLI (P08395) Protease IV (EC 3.4.21.-) (Endopeptidase IV)
(Signal peptide peptidase)
Length = 618
Score = 67.0 bits (162), Expect = 3e-11
Identities = 59/205 (28%), Positives = 97/205 (46%), Gaps = 37/205 (18%)
Query: 18 SEAKEQIAIIRASGT-MDSDIVTSNF--IEKIGMVKDSK---KFKAVIVRIDSVGGDFHA 71
++ + I ++ A+G MD + N ++D++ K KA+++R++S GG A
Sbjct: 322 ADTGDSIGVVFANGAIMDGEETQGNVGGDTTAAQIRDARLDPKVKAIVLRVNSPGGSVTA 381
Query: 72 SQSTW-EAIRSLASKKPVIASMSDAATSAGYYMAMGAGAIVAENLTLTGSIRGTVSQNFN 130
S+ E + A+ KPV+ SM A S GY+++ A IVA TLTGSI G
Sbjct: 382 SEVIRAELAAARAAGKPVVVSMGGMAASGGYWISTPANYIVANPSTLTGSI-GIFGVITT 440
Query: 131 LDNELPSFIPYDD------------AKVLDNVVRL-----------RLMTL------GKL 161
++N L S + D + L +L R +TL
Sbjct: 441 VENSLDSIGVHTDGVSTSPLADVSITRALPPEAQLMMQLSIENGYKRFITLVADARHSTP 500
Query: 162 DKMKKVAHGRIWTGKDAASHGLVDA 186
+++ K+A G +WTG+DA ++GLVD+
Sbjct: 501 EQIDKIAQGHVWTGQDAKANGLVDS 525
>SPPA_HAEIN (P45243) Protease IV (EC 3.4.21.-) (Endopeptidase IV)
(Signal peptide peptidase)
Length = 615
Score = 62.8 bits (151), Expect = 6e-10
Identities = 55/199 (27%), Positives = 93/199 (46%), Gaps = 37/199 (18%)
Query: 23 QIAIIRASGTM-----DSDIVTSNFIEKI-GMVKDSKKFKAVIVRIDSVGGDFHASQSTW 76
+IA++ GT+ D + + I +I D KAVI+R++S GG AS+
Sbjct: 323 KIAVVNVEGTIIDGESDEENAGGDTIARILRKAHDDNSVKAVILRVNSPGGSAFASEIIR 382
Query: 77 EAIRSLAS-KKPVIASMSDAATSAGYYMAMGAGAIVAENLTLTGSIRGTVSQNFNLDNEL 135
+ +L KPVI SM A S GY+++ A I+A++ T+TGSI G + +N +
Sbjct: 383 QETENLQKIGKPVIVSMGAMAASGGYWISSTADYIIADSNTITGSI-GIFTMFPTFENSI 441
Query: 136 PSFIPYDD----------------AKVLDNVVR----------LRLMTLGK---LDKMKK 166
+ D AK + ++ + L +++ G+ ++ K
Sbjct: 442 KKIGVHADGVSTTELANTSAFSPLAKPVQDIYQTEIEHGYDRFLEIVSKGRQLSKTQVDK 501
Query: 167 VAHGRIWTGKDAASHGLVD 185
+A G++W G DA +GLVD
Sbjct: 502 LAQGQVWLGSDAFQNGLVD 520
>SPPA_BACSU (O34525) Putative signal peptide peptidase sppA (EC
3.4.21.-)
Length = 335
Score = 60.1 bits (144), Expect = 4e-09
Identities = 50/209 (23%), Positives = 89/209 (41%), Gaps = 50/209 (23%)
Query: 23 QIAIIRASGTMDSDIVTSN-----------FIEKIGMVKDSKKFKAVIVRIDSVGGDFHA 71
+IA++ SGT+ + +S+ F++ + KD K K ++++++S GG +
Sbjct: 59 KIAVLEVSGTIQDNGDSSSLLGADGYNHRTFLKNLERAKDDKTVKGIVLKVNSPGGGVYE 118
Query: 72 SQSTWEAIRSLA--SKKPVIASMSDAATSAGYYMAMGAGAIVAENLTLTGSIRGTVSQNF 129
S + + + +KKP+ SM A S GYY++ A I A TLTGS+ G + ++
Sbjct: 119 SAEIHKKLEEIKKETKKPIYVSMGSMAASGGYYISTAADKIFATPETLTGSL-GVIMESV 177
Query: 130 NLD---------------------------------NELPSFIPYDDAKVLDNVVRLRLM 156
N N + S + +D + + R M
Sbjct: 178 NYSKLADKLGISFETIKSGAHKDIMSPSREMTKEEKNIMQSMVDNSYEGFVDVISKGRGM 237
Query: 157 TLGKLDKMKKVAHGRIWTGKDAASHGLVD 185
++KK+A GR++ G+ A LVD
Sbjct: 238 PKA---EVKKIADGRVYDGRQAKKLNLVD 263
>SPPA_BACHD (Q9K809) Putative signal peptide peptidase sppA (EC
3.4.21.-)
Length = 331
Score = 57.0 bits (136), Expect = 4e-08
Identities = 53/201 (26%), Positives = 93/201 (45%), Gaps = 38/201 (18%)
Query: 22 EQIAIIRASGTMDSDIVTS-----NFIEKIGMVKDSKKFKAVIVRIDSVGGDFHASQSTW 76
E +I+ +G+ S + T +F++++ + +I+++++ GG S
Sbjct: 62 ELSGVIQDTGSAPSLLNTGVYHHRDFLKQLEKAGEDPNIAGIILQVNTPGGGVLESAEIH 121
Query: 77 EAIRSLA--SKKPVIASMSDAATSAGYYMAMGAGAIVAENLTLTGSIRGTVSQNF----- 129
+ + + S+KPV SM + A S GYY++ A I A T+TGSI G + Q+
Sbjct: 122 KQVEEIVQDSEKPVYVSMGNMAASGGYYISAPATKIYAHPQTITGSI-GVIMQSIDISGL 180
Query: 130 --NLDNELPSFI--PYDDA--------------------KVLDNVVRLRLMTLGKLD-KM 164
NL E +F PY D ++ D VR+ + G + ++
Sbjct: 181 AENLGIEFNTFKSGPYKDILSQTREVTDEEEDILQTLVDEMYDEFVRVIVDGRGMSETEV 240
Query: 165 KKVAHGRIWTGKDAASHGLVD 185
+++A GRI+TG A + GLVD
Sbjct: 241 RELADGRIYTGSQAVATGLVD 261
>SPPA_ENTFC (Q9X480) Putative signal peptide peptidase sppA (EC
3.4.21.-)
Length = 344
Score = 53.5 bits (127), Expect = 4e-07
Identities = 46/179 (25%), Positives = 79/179 (43%), Gaps = 38/179 (21%)
Query: 41 NFIEKIGMVKDSKKFKAVIVRIDSVGGDFHASQSTWEAIRSLASKK----PVIASMSDAA 96
NF+ ++ +++ K K V++ ++S GG + S E + +A K P+ + + A
Sbjct: 92 NFLTQLKKIQEDKAVKGVLLEVNSPGGGIYESA---EIAKEMAKIKKLDIPIYTAFKNTA 148
Query: 97 TSAGYYMAMGAGAIVAENLTLTGSIRGTVSQNFNLDNEL--------------------P 136
S GYY++ G+ I A T TGSI G + N L P
Sbjct: 149 ASGGYYISAGSDKIFATEETTTGSI-GVIISGLNYSGLLEKLGVTDATYKSGALKDMMSP 207
Query: 137 SFIPYDDA-KVLDNVVR------LRLMTLGK---LDKMKKVAHGRIWTGKDAASHGLVD 185
P ++ KV+ V + ++ G+ + +K++A GRI+ G A +GLVD
Sbjct: 208 QHKPSEEENKVIQEFVMSAYDRFVNVVAKGRNMDTNAVKELADGRIYDGNQAVENGLVD 266
>SOHB_ECOLI (P24213) Possible protease sohB (EC 3.4.21.-)
Length = 349
Score = 50.1 bits (118), Expect = 4e-06
Identities = 56/202 (27%), Positives = 86/202 (41%), Gaps = 40/202 (19%)
Query: 18 SEAKEQIAIIRASGTMDSDIVTSNFIEKIGMVKDSKKFKAVIVRIDSVGGDFHASQSTWE 77
+++K ++ ++ G+MD+ V S E ++ K V++R++S GG H
Sbjct: 97 TDSKPRVWVLDFKGSMDAHEVNSLREEITAVLAAFKPQDQVVLRLESPGGMVHGYGLAAS 156
Query: 78 AIRSLASKK-PVIASMSDAATSAGYYMAMGAGAIVAENLTLTGSIRGTVSQ--NFN---- 130
++ L K P+ ++ A S GY MA A IV+ + GSI G V+Q NFN
Sbjct: 157 QLQRLRDKNIPLTVTVDKVAASGGYMMACVADKIVSAPFAIVGSI-GVVAQMPNFNRFLK 215
Query: 131 ---LDNELPSFIPYDDAKVL------------------------DNVVRLRLMTLGKLDK 163
+D EL + Y L D V R+R LD
Sbjct: 216 SKDIDIELHTAGQYKRTLTLLGENTEEGREKFREELNETHQLFKDFVKRMR----PSLD- 270
Query: 164 MKKVAHGRIWTGKDAASHGLVD 185
+++VA G W G+ A GLVD
Sbjct: 271 IEQVATGEHWYGQQAVEKGLVD 292
>Y651_METJA (Q58067) Putative protease MJ0651 (EC 3.4.21.-)
Length = 311
Score = 48.5 bits (114), Expect = 1e-05
Identities = 43/173 (24%), Positives = 75/173 (42%), Gaps = 29/173 (16%)
Query: 42 FIEKIGMVKDSKKFKAVIVRIDSVGGDFHASQSTWEAIRSLASKKPVIASMSDAATSAGY 101
+I + ++ K V++ ++S GG+ AS+ + LA KKPV+ + S Y
Sbjct: 72 YINLLDDLEKDDSVKGVLLVVNSPGGEVIASEKLARKVEELAKKKPVVVYVEGLDASGAY 131
Query: 102 YMAMGAGAIVAENLTLTGSI--RGTVSQNFNLDNEL----------------PSFIPY-- 141
++ A IVAE ++ GSI R + + L +L F P
Sbjct: 132 MVSAPADYIVAEKHSIVGSIGVRMDLMHYYGLMKKLGINVTTIKAGKYKDIGSPFRPMTK 191
Query: 142 DDAKVLDNVVRLRLMTLGK---------LDKMKKVAHGRIWTGKDAASHGLVD 185
++ + L ++ M K ++ K+A G+I++G+DA GLVD
Sbjct: 192 EEKEYLQKMINETYMDFVKWVAEHRHLSINYTLKIADGKIYSGEDAKKVGLVD 244
>SOHB_BUCBP (Q89AL0) Putative protease sohB (EC 3.4.21.-)
Length = 349
Score = 44.7 bits (104), Expect = 2e-04
Identities = 50/198 (25%), Positives = 85/198 (42%), Gaps = 30/198 (15%)
Query: 17 ISEAKEQIAIIRASGTMDSDIVTSNFIEKIGMVKDSKKFKAVIVRIDSVGGDFHASQSTW 76
+ A+ + +I G + + V S E ++ ++K V++R++S GG H
Sbjct: 99 VVRAQPTLYVIDFKGGIAAHEVKSLREEISSIISVAQKHDEVLLRLESSGGTIHGYGLAA 158
Query: 77 EAIRSLASKKPVIA-SMSDAATSAGYYMAMGAGAIVAENLTLTGSIRGTVSQ-------- 127
++ L S+K + S+ ATS GY MA A I+A ++ GSI G V+Q
Sbjct: 159 VQLQRLRSRKIFLTISIDKIATSGGYMMACVANYIIATPFSIIGSI-GVVAQFPNIHKFL 217
Query: 128 -NFNLDNELPSF-------------IPYDDAK------VLDNVVRLRLMTLGKLDKMKKV 167
N+D EL + P D K V ++ + + T+ ++K+
Sbjct: 218 KKNNIDVELHTAGVHKRTLTIFGENTPEDRKKFVEELNVAHDLFKKFVKTMRPSLNIEKL 277
Query: 168 AHGRIWTGKDAASHGLVD 185
++G W G A LVD
Sbjct: 278 SNGECWFGSIALKKKLVD 295
>SOHB_BUCAI (P57370) Possible protease sohB (EC 3.4.21.-)
Length = 336
Score = 41.6 bits (96), Expect = 0.002
Identities = 37/132 (28%), Positives = 61/132 (46%), Gaps = 4/132 (3%)
Query: 2 ISFRKYSKVRKWTVGISEAKEQIAIIRASGTMDSDIVTSNFIEKIGMVKDSKKFKAVIVR 61
I F+K + K + + K+++ ++ G + ++ V E ++ + K V++R
Sbjct: 70 IWFKKQKEKNKKELLLKNNKKKLFVLDFKGDVYANEVVGLREEISAILLVANKHDEVLLR 129
Query: 62 IDSVGGDFHASQSTWEAIRSLASKK-PVIASMSDAATSAGYYMAMGAGAIVAENLTLTGS 120
++S GG H + L K +I S+ A S GY MA A IV+ + GS
Sbjct: 130 LESSGGVIHGYGLAASQLNRLRQKGIRLIVSVDKIAASGGYMMACVADYIVSAPFAIIGS 189
Query: 121 IRGTVSQ--NFN 130
I G V Q NFN
Sbjct: 190 I-GVVGQIPNFN 200
>SOHB_BUCSC (Q44600) Possible protease sohB (EC 3.4.21.-) (Fragment)
Length = 305
Score = 40.0 bits (92), Expect = 0.004
Identities = 33/108 (30%), Positives = 52/108 (47%), Gaps = 4/108 (3%)
Query: 26 IIRASGTMDSDIVTSNFIEKIGMVKDSKKFKAVIVRIDSVGGDFHASQSTWEAIRSLASK 85
++ G + + VTS E ++ +K+ V++R++S GG H + L K
Sbjct: 106 VLDFKGNVSASEVTSLREEISAIILAAKENDEVLLRLESGGGVIHGYGLASSQLSRLREK 165
Query: 86 K-PVIASMSDAATSAGYYMAMGAGAIVAENLTLTGSIRGTVSQ--NFN 130
+ S+ A S GY MA A I+A ++ GSI G V+Q NFN
Sbjct: 166 NIRLTVSVDKIAASGGYMMACVANYIIAAPFSVIGSI-GVVAQIPNFN 212
>PFAP_LEPBO (Q48513) Putative peptidase pfaP precursor (EC 3.4.21.-)
(Periplasmic flagella-associated protein) (PF-associated
peptidase)
Length = 204
Score = 38.5 bits (88), Expect = 0.013
Identities = 34/132 (25%), Positives = 56/132 (41%), Gaps = 28/132 (21%)
Query: 82 LASKKPVIASMSDAATSAGYYMAMGAGAIVAENLTLTGSI--------------RGTVSQ 127
L + ++ SM D A S GYY+A A I A + T+TGSI G +
Sbjct: 3 LRKTRKIVVSMKDMAASGGYYIASSADKIFALSGTITGSIGVLQWLRYQRAFGSLGVKMR 62
Query: 128 NFNLDNELPSFIPYDDA---------KVLDNVVRLRLMTLGK-----LDKMKKVAHGRIW 173
+ S + D+ K+L + + + K + ++ +A GRI+
Sbjct: 63 TYKEGKYKDSLSLFRDSTPEEDEMIQKMLSDTYNEFVQDVAKGRNQTVKSVQNLAEGRIY 122
Query: 174 TGKDAASHGLVD 185
+G+DA + LVD
Sbjct: 123 SGQDAFRNKLVD 134
>TEPA_BACSU (Q99171) Translocation-enhancing protein tepA
Length = 223
Score = 36.2 bits (82), Expect = 0.065
Identities = 38/154 (24%), Positives = 71/154 (45%), Gaps = 18/154 (11%)
Query: 43 IEKIGMVKDSKKFKAVIVRIDSVGGDFHASQSTWEAIRSLASKKPVIASMSDAATSAGYY 102
I +I ++ + K + +++ +++VGGD A + E + SL+ KP ++ + S G
Sbjct: 49 IPQIVAIEQNPKIEGLLIILNTVGGDVEAGLAIAEMLASLS--KPTVSIVLGGGHSIGVP 106
Query: 103 MAMGAG-AIVAENLT-------LTGSIRGTVSQNFN-LDNELPSFIPY--DDAKVLDNVV 151
+A+ + +AE T LTG + G V Q F LD + + + + +
Sbjct: 107 IAVSCDYSYIAETSTVTIHPVRLTGLVIG-VPQTFEYLDKMQERVVKFVTSHSNITEEKF 165
Query: 152 RLRLMTLGKLDKMKKVAHGRIWTGKDAASHGLVD 185
+ + + G L + G GKDA +GL+D
Sbjct: 166 KELMFSKGNLTR----DIGTNVVGKDAVKYGLID 195
>SOHB_BUCAP (Q8K9P8) Possible protease sohB (EC 3.4.21.-)
Length = 342
Score = 35.8 bits (81), Expect = 0.085
Identities = 28/82 (34%), Positives = 39/82 (47%), Gaps = 4/82 (4%)
Query: 52 SKKFKAVIVRIDSVGGDFHASQSTWEAIRSLASKK-PVIASMSDAATSAGYYMAMGAGAI 110
+ K V++R++S GG H + L K +I S+ A S GY MA A I
Sbjct: 125 ANKDDEVLLRLESSGGVIHGYGLAAAQLERLRQNKIRLIISIDKIAASGGYMMACVADYI 184
Query: 111 VAENLTLTGSIRGTVSQ--NFN 130
++ + GSI G V Q NFN
Sbjct: 185 ISAPFAIIGSI-GVVGQLPNFN 205
>TEPA_BACHD (Q9KA93) Translocation-enhancing protein tepA
Length = 256
Score = 35.4 bits (80), Expect = 0.11
Identities = 36/157 (22%), Positives = 73/157 (45%), Gaps = 20/157 (12%)
Query: 41 NFIEKIGMVKDSKKFKAVIVRIDSVGGDFHASQSTWEAIRSLASKKPVIASMSDAATSAG 100
+ I ++ + +KK + +++ +++VGGD A + E + +++ KP + + S G
Sbjct: 78 HIIPQLVAAEQNKKIEGLLIILNTVGGDVEAGLAIAEMVATMS--KPTVTLVLGGGHSIG 135
Query: 101 YYMAMGAG-AIVAENLT-------LTGSIRGTVSQNF----NLDNELPSFIPYDDAKVLD 148
+A+ A + +AE T LTG + G V Q F + + SF+ + + +
Sbjct: 136 VPIAVAANYSFIAETATMTIHPVRLTGLVIG-VPQTFEYLDKMQERVVSFVT-KHSHISE 193
Query: 149 NVVRLRLMTLGKLDKMKKVAHGRIWTGKDAASHGLVD 185
+ + + G L + G G DA ++GL+D
Sbjct: 194 EKFKELMFSKGNLTR----DIGTNVIGTDAVTYGLID 226
>SOHB_HAEIN (P45315) Possible protease sohB (EC 3.4.21.-)
Length = 353
Score = 35.0 bits (79), Expect = 0.14
Identities = 31/124 (25%), Positives = 55/124 (44%), Gaps = 2/124 (1%)
Query: 5 RKYSKVRKWTVGISEAKEQIAIIRASGTMDSDIVTSNFIEKIGMVKDSKKFKAVIVRIDS 64
++ K++K E K + ++ G + + T+ E ++ +K V++R++S
Sbjct: 87 KRKEKLKKGETLEDEKKACVYVLDFCGDISASETTALREEISAILNVAKSEDEVLLRLES 146
Query: 65 VGGDFHASQSTWEAIRSLASKK-PVIASMSDAATSAGYYMAMGAGAIVAENLTLTGSIRG 123
GG H + L K + ++ A S GY MA A IV+ + GSI G
Sbjct: 147 PGGIVHNYGFAASQLSRLKQKGIKLTVAVDKVAASGGYMMACVADKIVSAPFAVIGSI-G 205
Query: 124 TVSQ 127
V+Q
Sbjct: 206 VVAQ 209
>SCA1_TRIVA (P53399) Succinyl-CoA ligase [GDP-forming] alpha-chain 1
precursor (EC 6.2.1.4) (Succinyl-CoA synthetase, alpha
chain 1)
Length = 309
Score = 34.3 bits (77), Expect = 0.25
Identities = 20/54 (37%), Positives = 28/54 (51%), Gaps = 1/54 (1%)
Query: 59 IVRIDSVGGDFHASQSTWEAIRSLASKKPVIASMSDAATSAGYYMAMGAGAIVA 112
I+ I +GG + W A L +KPV+AS++ A G M AGAIV+
Sbjct: 216 IILIGEIGGTSEEDAAEWIAKTKLTQEKPVVASIAGATAPPGKRMG-HAGAIVS 268
>ISP4_SCHPO (P40900) Sexual differentiation process protein isp4
Length = 785
Score = 33.5 bits (75), Expect = 0.42
Identities = 13/32 (40%), Positives = 19/32 (58%)
Query: 189 PVISYGFAGMPPATPTNFARWPFTRMIFNYFV 220
P+I G +PPATP N+ W + FNY++
Sbjct: 676 PLIFGGTGYIPPATPVNYLAWSGIGLFFNYYL 707
>HNM1_YEAST (P19807) Choline transport protein
Length = 563
Score = 32.7 bits (73), Expect = 0.72
Identities = 16/43 (37%), Positives = 19/43 (43%)
Query: 161 LDKMKKVAHGRIWTGKDAASHGLVDAGMPVISYGFAGMPPATP 203
L K + +AHG W GK +V G V S F PP P
Sbjct: 444 LAKKRNIAHGPFWLGKFGFFSNIVLLGWTVFSVVFFSFPPVLP 486
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.321 0.134 0.393
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 24,898,568
Number of Sequences: 164201
Number of extensions: 962014
Number of successful extensions: 2516
Number of sequences better than 10.0: 35
Number of HSP's better than 10.0 without gapping: 18
Number of HSP's successfully gapped in prelim test: 17
Number of HSP's that attempted gapping in prelim test: 2479
Number of HSP's gapped (non-prelim): 45
length of query: 226
length of database: 59,974,054
effective HSP length: 106
effective length of query: 120
effective length of database: 42,568,748
effective search space: 5108249760
effective search space used: 5108249760
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 64 (29.3 bits)
Medicago: description of AC144729.7


