

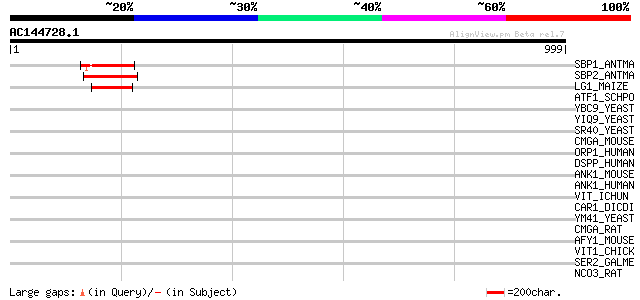
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144728.1 - phase: 0
(999 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
SBP1_ANTMA (Q38741) Squamosa-promoter binding protein 1 123 3e-27
SBP2_ANTMA (Q38740) Squamosa-promoter binding protein 2 119 3e-26
LG1_MAIZE (O04003) LIGULELESS1 protein 111 8e-24
ATF1_SCHPO (P52890) Transcription factor atf1 (Transcription fac... 44 0.002
YBC9_YEAST (P38201) Hypothetical 42.6 kDa protein in AAC2-RPL19B... 44 0.003
YIQ9_YEAST (P40442) Hypothetical 99.7 kDa protein in SDL1 5'regi... 43 0.004
SR40_YEAST (P32583) Suppressor protein SRP40 40 0.024
CMGA_MOUSE (P26339) Chromogranin A precursor (CgA) [Contains: Pa... 40 0.024
ORP1_HUMAN (Q9BXW6) Oxysterol binding protein-related protein 1 ... 39 0.091
DSPP_HUMAN (Q9NZW4) Dentin sialophosphoprotein precursor [Contai... 39 0.091
ANK1_MOUSE (Q02357) Ankyrin 1 (Erythrocyte ankyrin) 38 0.12
ANK1_HUMAN (P16157) Ankyrin 1 (Erythrocyte ankyrin) (Ankyrin R) 38 0.12
VIT_ICHUN (Q91062) Vitellogenin precursor (VTG) [Contains: Lipov... 37 0.26
CAR1_DICDI (P13773) Cyclic AMP receptor 1 37 0.26
YM41_YEAST (Q03213) Hypothetical 79.4 kDa protein in ALD2-DDR48 ... 37 0.34
CMGA_RAT (P10354) Chromogranin A precursor (CgA) [Contains: Panc... 37 0.34
AFY1_MOUSE (Q810B6) Ankyrin repeat and FYVE domain protein 1 (An... 37 0.34
VIT1_CHICK (P87498) Vitellogenin I precursor (Minor vitellogenin... 36 0.45
SER2_GALME (O96615) Sericin-2 (Silk gum protein 2) (Fragment) 36 0.59
NCO3_RAT (Q9EPU2) Nuclear receptor coactivator 3 (EC 2.3.1.48) (... 36 0.59
>SBP1_ANTMA (Q38741) Squamosa-promoter binding protein 1
Length = 131
Score = 123 bits (308), Expect = 3e-27
Identities = 59/104 (56%), Positives = 73/104 (69%), Gaps = 8/104 (7%)
Query: 128 VERDGKKSRG-------AGGTSNRAVCQVEDCGADLSRGKDYHRRHKVCEMHSKASRALV 180
+ D KK+R A G++ R+ CQVE+C A+++ K YHRRHKVCE H+KA L
Sbjct: 27 IGEDSKKTRALTPSGKRASGSTQRS-CQVENCAAEMTNAKPYHRRHKVCEFHAKAPVVLH 85
Query: 181 GNAMQRFCQQCSRFHILEEFDEGKRSCRRRLAGHNKRRRKTNQE 224
QRFCQQCSRFH L EFDE KRSCRRRLAGHN+RRRK++ +
Sbjct: 86 SGLQQRFCQQCSRFHELSEFDEAKRSCRRRLAGHNERRRKSSHD 129
>SBP2_ANTMA (Q38740) Squamosa-promoter binding protein 2
Length = 171
Score = 119 bits (299), Expect = 3e-26
Identities = 56/97 (57%), Positives = 69/97 (70%)
Query: 133 KKSRGAGGTSNRAVCQVEDCGADLSRGKDYHRRHKVCEMHSKASRALVGNAMQRFCQQCS 192
K + GG + C VE+CGADL K Y++RH+VCE+H+KA V MQRFCQQCS
Sbjct: 71 KHTASGGGVVAQPCCLVENCGADLRNCKKYYQRHRVCEVHAKAPVVSVEGLMQRFCQQCS 130
Query: 193 RFHILEEFDEGKRSCRRRLAGHNKRRRKTNQEAVPNG 229
RFH L EFD+ KRSCRRRLAGHN+RRRK++ E+ G
Sbjct: 131 RFHDLSEFDQTKRSCRRRLAGHNERRRKSSLESHKEG 167
>LG1_MAIZE (O04003) LIGULELESS1 protein
Length = 399
Score = 111 bits (278), Expect = 8e-24
Identities = 51/76 (67%), Positives = 59/76 (77%), Gaps = 1/76 (1%)
Query: 147 CQVEDCGADLSRGKDYHRRHKVCEMHSKASRALV-GNAMQRFCQQCSRFHILEEFDEGKR 205
CQ E C ADLS K YHRRHKVCE HSKA + G QRFCQQCSRFH+L+EFD+ K+
Sbjct: 185 CQAEGCKADLSSAKRYHRRHKVCEHHSKAPVVVTAGGLHQRFCQQCSRFHLLDEFDDAKK 244
Query: 206 SCRRRLAGHNKRRRKT 221
SCR+RLA HN+RRRK+
Sbjct: 245 SCRKRLADHNRRRRKS 260
>ATF1_SCHPO (P52890) Transcription factor atf1 (Transcription factor
mts1) (Protein sss1)
Length = 566
Score = 43.9 bits (102), Expect = 0.002
Identities = 65/269 (24%), Positives = 107/269 (39%), Gaps = 28/269 (10%)
Query: 226 VPNGSPTNDDQTSSYLLISLLKILSNMHYQPTDQDLLTHLLRSLASQNDEQGSKNLSNLL 285
V NG TN DQ+ + + + L + P Q L + ++S+ND+ S +NLL
Sbjct: 257 VVNG--TNGDQSDYFGANAAVHGLCLLSQVPDQQQKLQ---QPISSENDQAASTTANNLL 311
Query: 286 RE-QENLLREGGSSRNSGMVSALFSNGSQGSPTVITQHQPVSMNQMQQEMVHTHDVRTSD 344
++ Q+ + + + G+ TQ QP+ QQ + + +
Sbjct: 312 KQTQQQTFPDSIRPSFTQNTNPQAVTGTMNPQASRTQQQPMYFMGSQQ----FNGMPSVY 367
Query: 345 HQLISSIKPSISNSPPAYSETRDSSGQTKMNNFDLNDIYVDSDDGTEDLERLPVSTNLAT 404
++ PS++ +T D SGQ N N S+ +PV T
Sbjct: 368 GDTVNPADPSLT-----LRQTTDFSGQNAENG-STNLPQKTSNSDMPTANSMPVKLENGT 421
Query: 405 SSVDYPWTQQDS----HQSSPAQT-SGNSDSASAQSPSSSSGEAQ----SRTDRIVFKLF 455
DY +Q+ S +QSSP + +G + S SA S S G ++ + TD K F
Sbjct: 422 ---DYSTSQEPSSNANNQSSPTSSINGKASSESANGTSYSKGSSRRNSKNETDEEKRKSF 478
Query: 456 GKEPNEFPLVLRAQILDWLSQSPTDIESY 484
+ + L R + WLS +E Y
Sbjct: 479 LERNRQAALKCRQRKKQWLSNLQAKVEFY 507
>YBC9_YEAST (P38201) Hypothetical 42.6 kDa protein in AAC2-RPL19B
intergenic region
Length = 376
Score = 43.5 bits (101), Expect = 0.003
Identities = 56/227 (24%), Positives = 98/227 (42%), Gaps = 23/227 (10%)
Query: 230 SPTNDDQTSSYLLISLLKILSNMHYQP--TDQDLLTHLLRSLASQNDEQGSKNLSNLLRE 287
+P N D S +L I+ I N + T + L LA+ + S N SN
Sbjct: 25 APLNCDVNSPFLPINNNDINVNAYGDENLTYSNFLLSYNDKLATTTAKNNSINNSNSNNN 84
Query: 288 QENLLREGGSSRNSGMVSALFSNGSQGSPTVITQHQPVSMNQMQQEM----VHTHDVRTS 343
N + N+ ++ G+ S P ++N+ Q + ++ D+ T
Sbjct: 85 SNNNKNNNNNHNNNNLL------GNDISQMAFLLDYPSTLNEPQFAVNCKDIYRKDISTP 138
Query: 344 DHQLISSIKP-----SISNSP--PAYSETRDSSGQTKMNNF-DLNDIYVDSDDGTEDLER 395
L+SS+ P S+SNSP P S + G+ ++N + +DI+ D + +D
Sbjct: 139 S-SLVSSLPPAKFSLSLSNSPSPPPPSSSSLKHGEAIISNTSESSDIFADPNSFEKDTMP 197
Query: 396 LPVSTNLAT--SSVDYPWTQQDSHQSSPAQTSGNSDSASAQSPSSSS 440
L L + ++YP ++ + PA +S +S S+S++S SSS
Sbjct: 198 LTQELTLENLNNQLNYPDFTINAIEQDPAPSSFSSSSSSSESTVSSS 244
>YIQ9_YEAST (P40442) Hypothetical 99.7 kDa protein in SDL1 5'region
precursor
Length = 995
Score = 43.1 bits (100), Expect = 0.004
Identities = 53/240 (22%), Positives = 102/240 (42%), Gaps = 23/240 (9%)
Query: 221 TNQEAVPNGSPTNDDQTSSYLLISLLKILSNMHYQPTDQDLLTHLLRSLASQNDEQGSKN 280
+N ++ + S + +SS +S+ ++ TD +L+ + +S +S +
Sbjct: 27 SNSTSISSNSSSTSVVSSSSGSVSISSSIAETSSSATD--ILSSITQSASS------TSG 78
Query: 281 LSNLLREQENLLREGGSSRNSGMVSALFSNGSQGSPTVITQHQPVSMNQMQQEMVHTHDV 340
+S+ + + + S++S VS + S+ SQ S + VS Q T DV
Sbjct: 79 VSSSVGPSSSSVVSSSVSQSSSSVSDVSSSVSQSSSSASDVSSSVS-----QSASSTSDV 133
Query: 341 RTSDHQLISS---IKPSISNSPPAYSETRDSSGQTKMNNFDLNDIYVDSDDGTEDLERLP 397
+S Q SS + S+S S + S+ S Q+ + D++ S T D+
Sbjct: 134 SSSVSQSSSSASDVSSSVSQSSSSASDVSSSVSQSASSASDVSSSVSQSASSTSDVSSSV 193
Query: 398 VSTNLATSSVDYPWTQQDSHQ-------SSPAQTSGNSDSASAQSPSSSSGEAQSRTDRI 450
++ + S V +Q S S A ++ + S+ +QS SS+SG + S + +
Sbjct: 194 SQSSSSASDVSSSVSQSSSSASDVSSSVSQSASSTSDVSSSVSQSASSTSGVSSSGSQSV 253
Score = 38.1 bits (87), Expect = 0.12
Identities = 41/153 (26%), Positives = 63/153 (40%), Gaps = 12/153 (7%)
Query: 298 SRNSGMVSALFSNGSQGSPTVITQHQPVSMNQMQQEMVHTHDVRTSDHQLISS---IKPS 354
S++S S + S+ SQ S + VS Q DV +S Q SS + S
Sbjct: 138 SQSSSSASDVSSSVSQSSSSASDVSSSVS-----QSASSASDVSSSVSQSASSTSDVSSS 192
Query: 355 ISNSPPAYSETRDSSGQTKMNNFDLNDIYVDSDDGTEDLERLPVSTNLATSSVDYPWTQQ 414
+S S + S+ S Q+ + D++ S T D+ + +TS V +
Sbjct: 193 VSQSSSSASDVSSSVSQSSSSASDVSSSVSQSASSTSDVSSSVSQSASSTSGV----SSS 248
Query: 415 DSHQSSPAQTSGNSDSASAQSPSSSSGEAQSRT 447
S S A S +S S S S++SG A S +
Sbjct: 249 GSQSVSSASGSSSSFPQSTSSASTASGSATSNS 281
>SR40_YEAST (P32583) Suppressor protein SRP40
Length = 406
Score = 40.4 bits (93), Expect = 0.024
Identities = 40/179 (22%), Positives = 73/179 (40%), Gaps = 3/179 (1%)
Query: 270 ASQNDEQGSKNLSNLLREQENLLREGGSSRNSGMVSALFSNGSQGSPTVITQHQPVSMNQ 329
+S++D S + S+ + E S + A S+ T + +P S +
Sbjct: 95 SSESDSSSSGSSSSSSSSSDESSSES-ESEDETKKRARESDNEDAKETKKAKTEPESSSS 153
Query: 330 MQQEMVHTHDVRTSDHQLISSIKPSISNSPPAYSETRDSSGQTKMNNFDLNDIYVDSDDG 389
+ + S+ S S S+S + SE+ S ++ +D DSD
Sbjct: 154 SESSSSGSSSSSESESGSESDSDSSSSSSSSSDSESDSESDSQSSSSSSSSDSSSDSDSS 213
Query: 390 TEDLERLPVSTNLATSSVDYPWTQQDSHQSSPAQTSGNSDSASAQSPSSSSGEAQSRTD 448
+ D S++ ++SS + DS SS + +SG+SDS+S+ SS + +D
Sbjct: 214 SSDSSSDSDSSSSSSSSSSD--SDSDSDSSSDSDSSGSSDSSSSSDSSSDESTSSDSSD 270
Score = 37.7 bits (86), Expect = 0.15
Identities = 42/183 (22%), Positives = 76/183 (40%), Gaps = 7/183 (3%)
Query: 268 SLASQNDEQGSKNLSNLLREQENLLREGGSSRNSGMVSALFSNGSQGSPTVITQHQPVSM 327
S +S + S + + + SS + S+ + S GS + + S
Sbjct: 58 SSSSSSSSDSSDSSDSESSSSSSSSSSSSSSSSDSESSSESDSSSSGSSSSSSSSSDESS 117
Query: 328 NQMQQEMVHTHDVRTSDHQLISSIK-----PSISNSPPAYSETRDSSGQTKMNNFDLNDI 382
++ + E R SD++ K P S+S + S SS +++ + +D
Sbjct: 118 SESESEDETKKRARESDNEDAKETKKAKTEPESSSSSESSSSGSSSSSESESGSESDSDS 177
Query: 383 YVDSDDGTEDLERLPVSTNLATSSVDYPWTQQDSHQSSPAQTSGNSDSASAQSPSSSSGE 442
S + D E S + ++SS + DS SS + +S +SDS+S+ S SSS +
Sbjct: 178 S-SSSSSSSDSESDSESDSQSSSSSSSSDSSSDSDSSS-SDSSSDSDSSSSSSSSSSDSD 235
Query: 443 AQS 445
+ S
Sbjct: 236 SDS 238
Score = 36.2 bits (82), Expect = 0.45
Identities = 27/102 (26%), Positives = 52/102 (50%), Gaps = 3/102 (2%)
Query: 349 SSIKPSISNSPPAYSETRDSSGQTKMNNFDLNDIYVDSDDGTEDLERLPVSTNLATSSVD 408
SS S S+S + S + SSG++ ++ + S D ++ + S++ ++SS
Sbjct: 29 SSSSSSSSSSSSSSSSSSSSSGESSSSSSSSSSS--SSSDSSDSSDSESSSSSSSSSSSS 86
Query: 409 YPWTQQDSHQSSPAQTSGNSDSASAQS-PSSSSGEAQSRTDR 449
+ +S S + +SG+S S+S+ S SSS E++ T +
Sbjct: 87 SSSSDSESSSESDSSSSGSSSSSSSSSDESSSESESEDETKK 128
>CMGA_MOUSE (P26339) Chromogranin A precursor (CgA) [Contains:
Pancreastatin; Beta-granin; WE-14]
Length = 463
Score = 40.4 bits (93), Expect = 0.024
Identities = 47/214 (21%), Positives = 88/214 (40%), Gaps = 26/214 (12%)
Query: 225 AVPNGSPTNDDQTSSYLLISLLKILSNMHYQPTDQDLLTHLLRSLASQNDEQGSKNLSNL 284
A+P SP T ++ +L+++S+ +P+ + L +L QG + + ++
Sbjct: 18 ALPVNSPMTKGDTK--VMKCVLEVISDSLSKPSPMPVSPECLETL------QGDERILSI 69
Query: 285 LREQENLLREGGSSRNSGMVSALFSNGSQGSPTVITQHQPVSMNQMQQEMVHTHDVRTSD 344
LR Q NLL+E + + + + Q QP Q QQ+ +S
Sbjct: 70 LRHQ-NLLKE--------LQDLALQGAKERAQQPLKQQQPPKQQQQQQQQQQQEQQHSSF 120
Query: 345 HQLISSIKPSISNSPPAYSETRDSSGQTKMNNFDLNDIYVDSDDGTEDLERLPVSTNLAT 404
+S + + SP A + RD++ + D + DSD G +D +T
Sbjct: 121 EDELSEVFEN--QSPDA--KHRDAAAEVPSR--DTMEKRKDSDKGQQDGFE---ATTEGP 171
Query: 405 SSVDYPWTQQDSHQSSPAQTSGNSDSASAQSPSS 438
+P Q+S +++ G + + QSP+S
Sbjct: 172 RPQAFPEPNQESPMMGDSESPGEDTATNTQSPTS 205
>ORP1_HUMAN (Q9BXW6) Oxysterol binding protein-related protein 1
(OSBP-related protein 1) (ORP-1)
Length = 950
Score = 38.5 bits (88), Expect = 0.091
Identities = 38/116 (32%), Positives = 50/116 (42%), Gaps = 8/116 (6%)
Query: 767 LSEMG--LLHRAVRRNSKQLVELLLRYVPDNT-SDELGPEDKALVGGKN-HSYLFRPDAV 822
L++MG LHRA K+LV LLL Y D T + G K + + S L +
Sbjct: 77 LNDMGDTPLHRAAFTGRKELVMLLLEYNADTTIVNGSGQTAKEVTHAEEIRSMLEAVERT 136
Query: 823 GPAGLTPLHIAAGKDGSEDVLDALTNDPCMVGIEAWKNARDSTGSTPEDYARLRGH 878
L L +AA ++G L AL N P + N D G+TP A R H
Sbjct: 137 QQRKLEELLLAAAREGKTTELTALLNRPNPPDV----NCSDQLGNTPLHCAAYRAH 188
>DSPP_HUMAN (Q9NZW4) Dentin sialophosphoprotein precursor [Contains:
Dentin phosphoprotein (Dentin phosphophoryn) (DPP);
Dentin sialoprotein (DSP)]
Length = 1253
Score = 38.5 bits (88), Expect = 0.091
Identities = 40/180 (22%), Positives = 73/180 (40%), Gaps = 11/180 (6%)
Query: 270 ASQNDEQGSKNLSNLLREQENLLREGGSSRNSGMVSALFSNGSQGSPTVITQHQPVSMNQ 329
+S +D + + SN +N S+ ++ S+ S+ S S + + S N
Sbjct: 633 SSDSDSKSDSSDSNSSDSSDNSDSSDSSNSSNSSDSSDSSDSSDSSSSSDSSSSSDSSNS 692
Query: 330 MQQEMVHTHDVRTSDHQLISSIKPSISNSPPAYSETRDSSGQTKMNNFDLNDI-YVDSDD 388
+SD S+ S +S + S++ DSS + N+ D + DS D
Sbjct: 693 SD----------SSDSSDSSNSSESSDSSDSSDSDSSDSSDSSNSNSSDSDSSNSSDSSD 742
Query: 389 GTEDLERLPVSTNLATSSVDYPWTQQDSHQSSPAQTSGNSDSASAQSPSSSSGEAQSRTD 448
++ + S + +S DS SS + S NS ++ S SS S ++ + +D
Sbjct: 743 SSDSSDSSNSSDSSDSSDSSNSSDSSDSSDSSDSSDSSNSSDSNDSSNSSDSSDSSNSSD 802
Score = 37.7 bits (86), Expect = 0.15
Identities = 41/181 (22%), Positives = 70/181 (38%), Gaps = 4/181 (2%)
Query: 268 SLASQNDEQGSKNLSNLLREQENLLREGGSSRNSGMVSALFSNGSQGSPTVITQHQPVSM 327
S +S +D S N S+ + S+ + S+ SN S S + + + S
Sbjct: 901 SNSSDSDSSDSSNSSDSSDSSNSSDSSESSNSSDNSNSSDSSNSSDSSDSSDSSNSSDSS 960
Query: 328 NQMQQEMVHTHDVRTSDHQLISSIKPSISNSPPAYSETRDSSGQTKMNNFDLNDIYVDSD 387
N ++ +SD S S S+ S++ DSS + +N + DS
Sbjct: 961 NSGDS----SNSSDSSDSNSSDSSDSSNSSDSSDSSDSSDSSDSSDSSNSSDSSDSSDSS 1016
Query: 388 DGTEDLERLPVSTNLATSSVDYPWTQQDSHQSSPAQTSGNSDSASAQSPSSSSGEAQSRT 447
D + + S + +S DS SS + S +S +S S SS S ++ +
Sbjct: 1017 DSSNSSDSSNSSDSSDSSDSSDSSDSSDSSNSSDSSDSSDSSDSSDSSGSSDSSDSSDSS 1076
Query: 448 D 448
D
Sbjct: 1077 D 1077
Score = 36.6 bits (83), Expect = 0.34
Identities = 43/179 (24%), Positives = 73/179 (40%), Gaps = 9/179 (5%)
Query: 270 ASQNDEQGSKNLSNLLREQENLLREGGSSRNSGMVSALFSNGSQGSPTVITQHQPVSMNQ 329
+ +D S N S+ + N SS +S ++ S+ S S + + S N
Sbjct: 771 SDSSDSSDSSNSSDS-NDSSNSSDSSDSSNSSDSSNSSDSSDSSDSSDSDSSNSSDSSNS 829
Query: 330 MQQEMVHTHDVRTSDHQLISSIKPSISNSPPAYSETRDSSGQTKMNNFDLNDIYVDSDDG 389
++ +SD S S S++ S + DSS + +N + DS D
Sbjct: 830 SDSSD-SSNSSDSSDSSDSSDSSDSDSSNRSDSSNSSDSSDSSDSSNSSDSSDSSDSSDS 888
Query: 390 TEDLERLPVSTNLATSSVDYPWTQQDSHQSSPAQTSGNSDSASAQSPSSSSGEAQSRTD 448
E S+N + SS + DS SS + S +S ++S S SS+S + + +D
Sbjct: 889 NE-------SSNSSDSSDSSNSSDSDSSDSSNSSDSSDSSNSSDSSESSNSSDNSNSSD 940
Score = 36.2 bits (82), Expect = 0.45
Identities = 41/179 (22%), Positives = 68/179 (37%), Gaps = 14/179 (7%)
Query: 270 ASQNDEQGSKNLSNLLREQENLLREGGSSRNSGMVSALFSNGSQGSPTVITQHQPVSMNQ 329
+ +D S N S+ ++ S + S+ SN S + + + S N
Sbjct: 741 SDSSDSSDSSNSSDSSDSSDSSNSSDSSDSSDSSDSSDSSNSSDSNDSSNSSDSSDSSNS 800
Query: 330 MQQEMVHTHDVRTSDHQLISSIKPSISNSPPAYSETRDSSGQTKMNNFDLNDIYVDSDDG 389
+SD S S S++ S + DSS + N+ D +D SD
Sbjct: 801 SDSS-------NSSDSSDSSDSSDSDSSNSSDSSNSSDSSDSS--NSSDSSDSSDSSDSS 851
Query: 390 TEDLERLPVSTNLATSSVDYPWTQQDSHQSSPAQTSGNSDSASAQSPSSSSGEAQSRTD 448
D S+N + SS DS SS + S +S ++ S SS S ++ + +D
Sbjct: 852 DSDSSNRSDSSNSSDSS-----DSSDSSNSSDSSDSSDSSDSNESSNSSDSSDSSNSSD 905
Score = 35.4 bits (80), Expect = 0.77
Identities = 38/182 (20%), Positives = 67/182 (35%), Gaps = 6/182 (3%)
Query: 270 ASQNDEQGSKNLSNLLREQENLLREGGSSRNSGMVSALFSNGSQGSPTVITQHQPVSMNQ 329
+ +D S N SN ++ SS + S+ SN S S + + + S +
Sbjct: 651 SDNSDSSDSSNSSNSSDSSDSSDSSDSSSSSDSSSSSDSSNSSDSSDSSDSSNSSESSDS 710
Query: 330 MQQEMVHTHDV------RTSDHQLISSIKPSISNSPPAYSETRDSSGQTKMNNFDLNDIY 383
+ D +SD +S S S+ S + DSS + +N +
Sbjct: 711 SDSSDSDSSDSSDSSNSNSSDSDSSNSSDSSDSSDSSDSSNSSDSSDSSDSSNSSDSSDS 770
Query: 384 VDSDDGTEDLERLPVSTNLATSSVDYPWTQQDSHQSSPAQTSGNSDSASAQSPSSSSGEA 443
DS D ++ + + +S DS SS + S +S + + + S SS +
Sbjct: 771 SDSSDSSDSSNSSDSNDSSNSSDSSDSSNSSDSSNSSDSSDSSDSSDSDSSNSSDSSNSS 830
Query: 444 QS 445
S
Sbjct: 831 DS 832
Score = 35.0 bits (79), Expect = 1.0
Identities = 37/176 (21%), Positives = 70/176 (39%), Gaps = 6/176 (3%)
Query: 270 ASQNDEQGSKNLSNLLREQENLLREGGSSRNSGMVSALFSNGSQGSPTVITQHQPVSMNQ 329
+ +D S + S +N S+ + S+ SN S S + + + S +
Sbjct: 915 SDSSDSSNSSDSSESSNSSDNSNSSDSSNSSDSSDSSDSSNSSDSSNSGDSSNSSDSSDS 974
Query: 330 MQQEMVHTHDVRTSDHQLISSIKPSISNSPPAYSETRDSSGQTKMNNFDLNDIYVDSDDG 389
+ + + S SS S+S + S++ DSS + +N + DS D
Sbjct: 975 NSSDSSDSSNSSDSSDSSDSSDSSDSSDSSNS-SDSSDSSDSSDSSNSSDSSNSSDSSDS 1033
Query: 390 TEDLERLPVSTNLATSSVDYPWTQQDSHQSSPAQTSGNSDSASAQSPSSSSGEAQS 445
++ + S + +S + S S + +SG+SDS+ + S SS + S
Sbjct: 1034 SDSSDSSDSSDSSNSSD-----SSDSSDSSDSSDSSGSSDSSDSSDSSDSSDSSDS 1084
Score = 34.3 bits (77), Expect = 1.7
Identities = 42/155 (27%), Positives = 60/155 (38%), Gaps = 12/155 (7%)
Query: 294 EGGSSRNSGMVSALFSNGSQGSPTVITQHQPVSMNQMQQEMVHTHDVRTSDHQLISSIKP 353
E SS + + SN S S + S N + D S SS
Sbjct: 630 ESDSSDSDSKSDSSDSNSSDSSDNSDSSDSSNSSNSSDSS--DSSDSSDSSSSSDSSSSS 687
Query: 354 SISNSPPAYSETRDSSGQTKMNNFDLNDIYVDSDDGTEDLERLPVSTNLATSSVDYPWTQ 413
SNS S++ DSS + N+ + +D SD D S+N +S D
Sbjct: 688 DSSNS----SDSSDSSDSS--NSSESSD---SSDSSDSDSSDSSDSSNSNSSDSDSS-NS 737
Query: 414 QDSHQSSPAQTSGNSDSASAQSPSSSSGEAQSRTD 448
DS SS + S NS +S S SS+S ++ +D
Sbjct: 738 SDSSDSSDSSDSSNSSDSSDSSDSSNSSDSSDSSD 772
Score = 34.3 bits (77), Expect = 1.7
Identities = 36/178 (20%), Positives = 72/178 (40%), Gaps = 1/178 (0%)
Query: 268 SLASQNDEQGSKNLSNLLREQENLLREGGSSRNSGMVSALFSNGSQGSPTVITQHQPVSM 327
S +S +D S + S+ ++ S + S+ S+ S S + + + S
Sbjct: 727 SNSSDSDSSNSSDSSDSSDSSDSSNSSDSSDSSDSSNSSDSSDSSDSSDSSDSSNSSDS- 785
Query: 328 NQMQQEMVHTHDVRTSDHQLISSIKPSISNSPPAYSETRDSSGQTKMNNFDLNDIYVDSD 387
N + +SD S S +S S + DSS + ++ + DS
Sbjct: 786 NDSSNSSDSSDSSNSSDSSNSSDSSDSSDSSDSDSSNSSDSSNSSDSSDSSNSSDSSDSS 845
Query: 388 DGTEDLERLPVSTNLATSSVDYPWTQQDSHQSSPAQTSGNSDSASAQSPSSSSGEAQS 445
D ++ + + + +++S D + S+ S + +S +SDS + + S SS + S
Sbjct: 846 DSSDSSDSDSSNRSDSSNSSDSSDSSDSSNSSDSSDSSDSSDSNESSNSSDSSDSSNS 903
Score = 32.7 bits (73), Expect = 5.0
Identities = 41/187 (21%), Positives = 76/187 (39%), Gaps = 8/187 (4%)
Query: 268 SLASQNDEQGSKNLSNLLREQENLLREGGSSRNSGMVSALFSNGSQGSPTVITQHQPVSM 327
S +S + + + S+ ++ + SS +S S+ SN S S + + S
Sbjct: 558 SNSSDSSDSSDSDSSDSNSSSDSDSSDSDSSDSSDSDSSDSSNSSDSSDSSDSSDSSDSS 617
Query: 328 NQMQQEMVHTH-DVRTSDHQLISSIKPSISNSPPAYSETRDSSGQTKMNNFDLNDIYVDS 386
+ + + + +SD S S S+ S++ DSS + N+ D +D S
Sbjct: 618 DSSDSKSDSSKSESDSSDSDSKSDSSDSNSSDSSDNSDSSDSSNSS--NSSDSSDSSDSS 675
Query: 387 D-----DGTEDLERLPVSTNLATSSVDYPWTQQDSHQSSPAQTSGNSDSASAQSPSSSSG 441
D D + + S + +S DS SS + +S +SDS+++ S S S
Sbjct: 676 DSSSSSDSSSSSDSSNSSDSSDSSDSSNSSESSDSSDSSDSDSSDSSDSSNSNSSDSDSS 735
Query: 442 EAQSRTD 448
+ +D
Sbjct: 736 NSSDSSD 742
Score = 32.3 bits (72), Expect = 6.5
Identities = 39/176 (22%), Positives = 71/176 (40%), Gaps = 13/176 (7%)
Query: 270 ASQNDEQGSKNLSNLLREQENLLREGGSSRNSGMVSALFSNGSQGSPTVITQHQPVSMNQ 329
+S + + + S+ + ++ R S+ + S+ SN S S + + S N
Sbjct: 835 SSNSSDSSDSSDSSDSSDSDSSNRSDSSNSSDSSDSSDSSNSSDSSDSSDSSDSNESSNS 894
Query: 330 MQQEMVHTHDVRTSDHQLISSIKPSISNSPPAYSETRDSSGQTKMNNFDLNDIYVDSDDG 389
+ +SD S S S+ S++ +SS + +N + DS D
Sbjct: 895 SDS----SDSSNSSDSDSSDSSNSSDSSDSSNSSDSSESSNSSDNSNSSDSSNSSDSSDS 950
Query: 390 TEDLERLPVSTNLATSSVDYPWTQQDSHQSSPAQTSGNSDSASAQSPSSSSGEAQS 445
+ D S+N SS +S SS + +S +SDS+++ S SS + S
Sbjct: 951 S-DSSNSSDSSNSGDSS--------NSSDSSDSNSSDSSDSSNSSDSSDSSDSSDS 997
>ANK1_MOUSE (Q02357) Ankyrin 1 (Erythrocyte ankyrin)
Length = 1862
Score = 38.1 bits (87), Expect = 0.12
Identities = 45/160 (28%), Positives = 70/160 (43%), Gaps = 21/160 (13%)
Query: 759 DHPTLYQALSEMGL--LHRAVRRNSKQLVELLLRY---VPDNTSDELGPEDKALVGGKNH 813
DH QA ++ GL +H A + + V LLL+Y + D T D L P A G +
Sbjct: 289 DHGAPIQAKTKNGLSPIHMAAQGDHLDCVRLLLQYNAEIDDITLDHLTPLHVAAHCGHHR 348
Query: 814 SYLF------RPDAVGPAGLTPLHIAAGKDGSEDVLDALTNDPCMVGIEAWKNARDSTGS 867
+P++ G TPLHIA K+ V++ L + +A +G
Sbjct: 349 VAKVLLDKGAKPNSRALNGFTPLHIACKKNHIR-VMELLLKTGASI------DAVTESGL 401
Query: 868 TPEDYARLRGHYTYI-HLVQKKINKTQGAAHVVVEIPSNM 906
TP A GH + +L+Q+ + ++V VE P +M
Sbjct: 402 TPLHVASFMGHLPIVKNLLQR--GASPNVSNVKVETPLHM 439
Score = 34.3 bits (77), Expect = 1.7
Identities = 40/146 (27%), Positives = 59/146 (40%), Gaps = 20/146 (13%)
Query: 753 ETVNKGDHPTLYQALSEMGL--LHRAVRRNSKQLVELLLRYVPDN---TSDELGPEDKAL 807
E VN G + A S+ G L+ A + N ++V+ LL + T D P AL
Sbjct: 92 ELVNYGAN---VNAQSQKGFTPLYMAAQENHLEVVKFLLENGANQNVATEDGFTPLAVAL 148
Query: 808 VGGKNH--SYLFRPDAVGPAGLTPLHIAAGKDGSEDVLDALTNDPCMVGIEAWKNARDST 865
G + ++L G L LHIAA D + L NDP + T
Sbjct: 149 QQGHENVVAHLINYGTKGKVRLPALHIAARNDDTRTAAVLLQNDPN-------PDVLSKT 201
Query: 866 GSTPEDYARLRGHYTYIHLVQKKINK 891
G TP + HY +++ Q +N+
Sbjct: 202 GFTP---LHIAAHYENLNVAQLLLNR 224
>ANK1_HUMAN (P16157) Ankyrin 1 (Erythrocyte ankyrin) (Ankyrin R)
Length = 1880
Score = 38.1 bits (87), Expect = 0.12
Identities = 45/160 (28%), Positives = 70/160 (43%), Gaps = 21/160 (13%)
Query: 759 DHPTLYQALSEMGL--LHRAVRRNSKQLVELLLRY---VPDNTSDELGPEDKALVGGKNH 813
DH QA ++ GL +H A + + V LLL+Y + D T D L P A G +
Sbjct: 292 DHGAPIQAKTKNGLSPIHMAAQGDHLDCVRLLLQYDAEIDDITLDHLTPLHVAAHCGHHR 351
Query: 814 SYLF------RPDAVGPAGLTPLHIAAGKDGSEDVLDALTNDPCMVGIEAWKNARDSTGS 867
+P++ G TPLHIA K+ V++ L + +A +G
Sbjct: 352 VAKVLLDKGAKPNSRALNGFTPLHIACKKNHVR-VMELLLKTGASI------DAVTESGL 404
Query: 868 TPEDYARLRGHYTYI-HLVQKKINKTQGAAHVVVEIPSNM 906
TP A GH + +L+Q+ + ++V VE P +M
Sbjct: 405 TPLHVASFMGHLPIVKNLLQR--GASPNVSNVKVETPLHM 442
Score = 34.3 bits (77), Expect = 1.7
Identities = 40/146 (27%), Positives = 59/146 (40%), Gaps = 20/146 (13%)
Query: 753 ETVNKGDHPTLYQALSEMGL--LHRAVRRNSKQLVELLLRYVPDN---TSDELGPEDKAL 807
E VN G + A S+ G L+ A + N ++V+ LL + T D P AL
Sbjct: 95 ELVNYGAN---VNAQSQKGFTPLYMAAQENHLEVVKFLLENGANQNVATEDGFTPLAVAL 151
Query: 808 VGGKNH--SYLFRPDAVGPAGLTPLHIAAGKDGSEDVLDALTNDPCMVGIEAWKNARDST 865
G + ++L G L LHIAA D + L NDP + T
Sbjct: 152 QQGHENVVAHLINYGTKGKVRLPALHIAARNDDTRTAAVLLQNDPN-------PDVLSKT 204
Query: 866 GSTPEDYARLRGHYTYIHLVQKKINK 891
G TP + HY +++ Q +N+
Sbjct: 205 GFTP---LHIAAHYENLNVAQLLLNR 227
>VIT_ICHUN (Q91062) Vitellogenin precursor (VTG) [Contains:
Lipovitellin LV-1N; Lipovitellin LV-1C; Lipovitellin
LV-2]
Length = 1823
Score = 37.0 bits (84), Expect = 0.26
Identities = 54/240 (22%), Positives = 97/240 (39%), Gaps = 39/240 (16%)
Query: 268 SLASQNDEQGSK-----NLSNLLREQENLLREGGSSRNSGMVSALFSNGSQ--------- 313
S +S +D+ G K + NL ++ + + G S +S S+ S+ S+
Sbjct: 1143 SSSSSSDKSGKKTPRQGSTVNLAAKRASKKQRGKDSSSSSSSSSSSSDSSKSPHKHGGAK 1202
Query: 314 ------GSPTVITQHQPVSMNQMQQEMVHTHDVRTSDHQLISSIKPSISNSP-PAYSETR 366
G+P + Q S + +S + S++KP ++ P PA S +
Sbjct: 1203 RQHAGHGAPHLGPQSHSSSSSSSSSSS------SSSASKSFSTVKPPMTRKPRPARSSSS 1256
Query: 367 DSSGQTKMNNFDLNDIYVDSDDGT------EDLERLPVSTNLATSSVDYPWTQQDSHQSS 420
SS + ++ + S + + LE L V ++ ++ + Q Q+S
Sbjct: 1257 SSSSDSSSSSSSSSSSSSSSSSSSSSSSESKSLEWLAVKDVNQSAFYNFKYVPQRKPQTS 1316
Query: 421 ----PAQTSGNSDSASAQSPSSSSGEAQSRTDRIVFKLFGKEPNEFPLVLRAQILDWLSQ 476
PA +S +S S+S+ S SSSS ++ F+ K + +VLRA D Q
Sbjct: 1317 RRHTPASSSSSSSSSSSSSSSSSSSDSDMTVSAESFEKHSKP--KVVIVLRAVRADGKQQ 1374
>CAR1_DICDI (P13773) Cyclic AMP receptor 1
Length = 392
Score = 37.0 bits (84), Expect = 0.26
Identities = 27/90 (30%), Positives = 43/90 (47%), Gaps = 10/90 (11%)
Query: 356 SNSPPAYSETRDSSGQTKMNNFDLNDIYVDSDDGTEDLERLPVSTNLATSSVDYPWTQQD 415
+N+P YS +R +SG+T + +D+ SD LER P N QQ+
Sbjct: 295 NNNPSPYSSSRGTSGKTMGGHPTGDDVQCSSDMEQCSLERHPNMVN----------NQQN 344
Query: 416 SHQSSPAQTSGNSDSASAQSPSSSSGEAQS 445
+ + Q + N + +S+ S SSS E Q+
Sbjct: 345 LNNNYGLQQNYNDEGSSSSSLSSSDEEKQT 374
>YM41_YEAST (Q03213) Hypothetical 79.4 kDa protein in ALD2-DDR48
intergenic region
Length = 719
Score = 36.6 bits (83), Expect = 0.34
Identities = 44/182 (24%), Positives = 72/182 (39%), Gaps = 18/182 (9%)
Query: 216 KRRRKTNQEAVPNGSPTNDDQTSSYLLISLLKILSNMHYQPTDQDLLTHLLRSLASQNDE 275
KRRR+ Q N + TND +++ IS L L+N H T + + S
Sbjct: 381 KRRRRNTQSN--NNASTNDHASAAQKPISALSPLTNSHNSTTSMNYTNSSIHS------- 431
Query: 276 QGSKNLSNLLREQENLLREGGSSRNS----GMVSALFSNGSQGSPTVITQ-HQPVSMNQM 330
G + SN + +L G ++ S + +A F P +I QP+S +Q+
Sbjct: 432 -GVTSASNSFHDLNSLNNFGTTTALSLPSLALDNASFPPNQNVIPPIINNTQQPLSFSQL 490
Query: 331 QQEMVHTHDVRTSDHQLISSIKPSISNSPPAYSETRDSSGQTKMNNFDLNDIYVDSDDGT 390
+ T ++ S S + +I N A + T +N D + D DD
Sbjct: 491 INQDSTTSELLPSGK---SGVNTNIVNRNRASTLPSYPKPMTVKSNVDDDGYQEDDDDDG 547
Query: 391 ED 392
+D
Sbjct: 548 DD 549
>CMGA_RAT (P10354) Chromogranin A precursor (CgA) [Contains:
Pancreastatin; Beta-granin; WE-14]
Length = 466
Score = 36.6 bits (83), Expect = 0.34
Identities = 49/214 (22%), Positives = 84/214 (38%), Gaps = 31/214 (14%)
Query: 225 AVPNGSPTNDDQTSSYLLISLLKILSNMHYQPTDQDLLTHLLRSLASQNDEQGSKNLSNL 284
A+P SP T ++ +L+++S+ +P+ + L +L QG + + ++
Sbjct: 18 ALPVNSPMTKGDTK--VMKCVLEVISDSLSKPSPMPVSPECLETL------QGDERVLSI 69
Query: 285 LREQENLLREGGSSRNSGMVSALFSNGSQGSPTVITQHQPVSMNQMQQEMVHTHDVRTSD 344
LR Q NLL+E L QG+ Q Q Q QQ+ + +
Sbjct: 70 LRHQ-NLLKE------------LQDLALQGAKERAQQQQQQQQQQQQQQQQQQQQHSSFE 116
Query: 345 HQLISSIKPSISNSPPAYSETRDSSGQTKMNNFDLNDIYVDSDDGTEDLERLPVSTNLAT 404
+L + + SP A S +K D + DSD G +D T
Sbjct: 117 DELSEVFE---NQSPAAKHGDAASEAPSK----DTVEKREDSDKGQQDAFE---GTTEGP 166
Query: 405 SSVDYPWTQQDSHQSSPAQTSGNSDSASAQSPSS 438
+P +Q+S +Q+ G + + QSP+S
Sbjct: 167 RPQAFPEPKQESSMMGNSQSPGEDTANNTQSPTS 200
>AFY1_MOUSE (Q810B6) Ankyrin repeat and FYVE domain protein 1
(Ankyrin repeats hooked to a zinc finger motif)
Length = 1169
Score = 36.6 bits (83), Expect = 0.34
Identities = 45/163 (27%), Positives = 59/163 (35%), Gaps = 19/163 (11%)
Query: 772 LLHRAVRRNSKQLVELLLRYVPDNTSDELGPEDKALVGGKNHSYLFRPDAVGPAGLTPLH 831
LLHRAV N++ L+R D S P G + G TPLH
Sbjct: 728 LLHRAVDENNESTACFLIRSGCDVNS----PRQPGTNG--------EGEEEARDGQTPLH 775
Query: 832 IAAGKDGSEDVLDALTNDPCMVGIEAWKNARDSTGSTPEDYARLRGHYTYIHLVQKKINK 891
+AA E V C++ A NA+D+ G TP A H I L+ N
Sbjct: 776 LAASWGLEETV-------QCLLEFGANVNAQDAEGRTPVHVAISNQHSVIIQLLISHPNI 828
Query: 892 TQGAAHVVVEIPSNMTESNKNPKQNESFTSLEIGKAEVRRSQG 934
P + KN K E+ E G AE ++G
Sbjct: 829 ELSVRDRQGLTPFACAMTYKNNKAAEAILKRESGAAEQVDNKG 871
>VIT1_CHICK (P87498) Vitellogenin I precursor (Minor vitellogenin)
[Contains: Lipovitellin I (LVI); Phosvitin (PV);
Lipovitellin II (LVII); YGP42]
Length = 1912
Score = 36.2 bits (82), Expect = 0.45
Identities = 31/124 (25%), Positives = 59/124 (47%), Gaps = 3/124 (2%)
Query: 327 MNQMQQEMVHTHDVRTSDHQLISSIKPSISNSPPAYSET---RDSSGQTKMNNFDLNDIY 383
M ++++ + T + T + + SS SIS S + + T DS + + +N
Sbjct: 1075 MKRVKKILDDTDNQATRNSRSSSSSASSISESSESTTSTPSSSDSDNRASQGDPQINLKS 1134
Query: 384 VDSDDGTEDLERLPVSTNLATSSVDYPWTQQDSHQSSPAQTSGNSDSASAQSPSSSSGEA 443
S + S++ +SS + S SS +++S +SDS+S+ S SSSS +
Sbjct: 1135 RQSKANEKKFYPFGDSSSSGSSSSSSSSSSSSSDSSSSSRSSSSSDSSSSSSSSSSSSSS 1194
Query: 444 QSRT 447
+S++
Sbjct: 1195 KSKS 1198
Score = 32.7 bits (73), Expect = 5.0
Identities = 36/183 (19%), Positives = 73/183 (39%), Gaps = 12/183 (6%)
Query: 266 LRSLASQNDEQGSKNLSNLLREQENLLREGGSSRNSGMVSALFSNGSQGSPTVITQHQPV 325
++ + D Q ++N + ++ S+ ++ S + SQG P + + +
Sbjct: 1078 VKKILDDTDNQATRNSRSSSSSASSISESSESTTSTPSSSDSDNRASQGDPQINLKSRQS 1137
Query: 326 SMNQMQ-QEMVHTHDVRTSDHQLISSIKPSISNSPPAYSETRDSSGQTKMNNFDLNDIYV 384
N+ + + +S SS S S+S S + DSS + ++
Sbjct: 1138 KANEKKFYPFGDSSSSGSSSSSSSSSSSSSDSSSSSRSSSSSDSSSSSSSSSSS------ 1191
Query: 385 DSDDGTEDLERLPVSTNLATSSVDYPWTQQDSHQSSPAQTSGNSDSASAQSPSSSSGEAQ 444
S ++ R S ++SS ++ S SS + + G+S S+S S + + Q
Sbjct: 1192 -SSSKSKSSSRSSKSNRSSSSSN----SKDSSSSSSKSNSKGSSSSSSKASGTRQKAKKQ 1246
Query: 445 SRT 447
S+T
Sbjct: 1247 SKT 1249
>SER2_GALME (O96615) Sericin-2 (Silk gum protein 2) (Fragment)
Length = 220
Score = 35.8 bits (81), Expect = 0.59
Identities = 41/173 (23%), Positives = 70/173 (39%), Gaps = 19/173 (10%)
Query: 270 ASQNDEQGSKNLSNLLREQENLLREGGSSRNSGMVSALFSNGSQGSPTVITQHQPVSMNQ 329
+S N+ GS ++ N G S+ NS S+ S+GS G ++ S +Q
Sbjct: 38 SSSNNTSGSSRNNSSGSSSSNNSSSGSSAGNSSGSSSSNSSGSSGIGSLSRNSSSSSSSQ 97
Query: 330 MQQEMVHTHDVRTSDHQLISSIKPSISNSPPAYSETRDSSGQTKMNNFDLNDIYVDSDDG 389
+S + +S+ S S+S + ++S+ ++ N D G
Sbjct: 98 A---------AGSSSSRRVSADGSSSSSSASNSAAAQNSASSSETINAD----------G 138
Query: 390 TEDLERLPVSTNLATSSVDYPWTQQDSHQSSPAQTSGNSDSASAQSPSSSSGE 442
+E+ S+ S+ D QSS + +S S+ ASA S SS S +
Sbjct: 139 SENESSSSSSSAAQNSATRSQVINADGSQSSSSSSSSASNQASATSSSSVSAD 191
Score = 35.4 bits (80), Expect = 0.77
Identities = 25/88 (28%), Positives = 38/88 (42%), Gaps = 7/88 (7%)
Query: 354 SISNSPPAYSETRDSSGQTKMNNFDLNDIYVDSDDGTEDLERLPVSTNLATSSVDYPWTQ 413
S +N+ S T +SSG + NN T + S N + SS +
Sbjct: 2 SSTNNSSGSSSTNNSSGSSSTNNSS-------GSSSTNNASGSSSSNNTSGSSRNNSSGS 54
Query: 414 QDSHQSSPAQTSGNSDSASAQSPSSSSG 441
S+ SS ++GNS +S+ + S SSG
Sbjct: 55 SSSNNSSSGSSAGNSSGSSSSNSSGSSG 82
Score = 33.9 bits (76), Expect = 2.2
Identities = 41/157 (26%), Positives = 65/157 (41%), Gaps = 10/157 (6%)
Query: 295 GGSSRNSGMVSALFSNGSQGSPTVITQHQPVSMNQMQQEMVHTHDVRTSDHQLISSIKPS 354
G SS N+ S+ +N S S T S N ++ +S++ SS S
Sbjct: 9 GSSSTNNSSGSSSTNNSSGSSSTNNASGSSSSNNTSGSSRNNSSGSSSSNN---SSSGSS 65
Query: 355 ISNSPPAYSETRDSSGQTKMNNFDLNDIYVDSDD--GTEDLERLPVSTNLATSSVDYPWT 412
NS + S + +SSG + + + N S G+ R+ + ++SS
Sbjct: 66 AGNS--SGSSSSNSSGSSGIGSLSRNSSSSSSSQAAGSSSSRRVSADGSSSSSSASNSAA 123
Query: 413 QQDSHQSSPAQTSGNSDSASAQSPSSSSGEAQSRTDR 449
Q+S SS N+D + +S SSSS AQ+ R
Sbjct: 124 AQNSASSSETI---NADGSENESSSSSSSAAQNSATR 157
>NCO3_RAT (Q9EPU2) Nuclear receptor coactivator 3 (EC 2.3.1.48)
(NCoA-3) (Amplified in breast cancer-1 protein homolog)
(AIB-1) (Fragment)
Length = 1082
Score = 35.8 bits (81), Expect = 0.59
Identities = 46/181 (25%), Positives = 75/181 (41%), Gaps = 32/181 (17%)
Query: 300 NSGMVSALFSNGSQGSPTVITQHQPVSMNQMQQEMVHTHDVRT------SDHQLISSIKP 353
+ G+ ++L S S P + + P +MN Q + D ++ + + SS+ P
Sbjct: 230 SEGVGTSLLSTLSSPGPKL--DNSP-NMNINQPSKASSQDSKSPLGLYCEQNPVESSVCP 286
Query: 354 SISNSPPAYSETRDSSG----------QTKMNNFDLNDIYVDSDD-GTEDLERLPVSTNL 402
S S PP E +++SG ++K + L + SDD G L P+ +N
Sbjct: 287 SNSRDPPVTKENKENSGEASETPRGPLESKGHKKLLQLLTCSSDDRGHSSLTNSPLDSNC 346
Query: 403 ATSSVDYPWTQQDSHQSSPAQTSGNSDSASAQSPSSSSGEAQSRTDRIVFKLF--GKEPN 460
SS+ +SP+ S +S S + S S+ G RI+ KL G P
Sbjct: 347 KDSSISV---------TSPSGVS-SSTSGAVSSTSNMHGSLLQEKHRILHKLLQNGNSPA 396
Query: 461 E 461
E
Sbjct: 397 E 397
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.316 0.132 0.388
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 120,454,373
Number of Sequences: 164201
Number of extensions: 5289702
Number of successful extensions: 15582
Number of sequences better than 10.0: 74
Number of HSP's better than 10.0 without gapping: 17
Number of HSP's successfully gapped in prelim test: 61
Number of HSP's that attempted gapping in prelim test: 15310
Number of HSP's gapped (non-prelim): 242
length of query: 999
length of database: 59,974,054
effective HSP length: 120
effective length of query: 879
effective length of database: 40,269,934
effective search space: 35397271986
effective search space used: 35397271986
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 71 (32.0 bits)
Medicago: description of AC144728.1


