

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144727.5 + phase: 0
(256 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
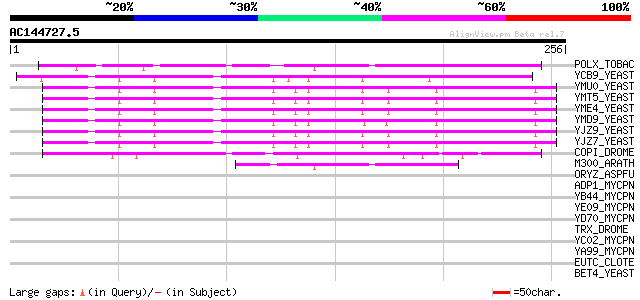
Score E
Sequences producing significant alignments: (bits) Value
POLX_TOBAC (P10978) Retrovirus-related Pol polyprotein from tran... 83 7e-16
YCB9_YEAST (P25384) Transposon Ty2 protein B (Ty1-17 protein B) 79 1e-14
YMU0_YEAST (Q04670) Transposon Ty1 protein B 74 3e-13
YMT5_YEAST (Q04214) Transposon Ty1 protein B 74 3e-13
YME4_YEAST (Q04711) Transposon Ty1 protein B 74 3e-13
YMD9_YEAST (Q03434) Transposon Ty1 protein B 74 3e-13
YJZ9_YEAST (P47100) Transposon Ty1 protein B 74 3e-13
YJZ7_YEAST (P47098) Transposon Ty1 protein B 72 1e-12
COPI_DROME (P04146) Copia protein (Gag-int-pol protein) [Contain... 68 2e-11
M300_ARATH (P93293) Hypothetical mitochondrial protein AtMg00300... 44 3e-04
ORYZ_ASPFU (P28296) Oryzin precursor (EC 3.4.21.63) (Alkaline pr... 35 0.23
ADP1_MYCPN (P11311) Adhesin P1 precursor (Cytadhesin P1) (Attach... 33 0.52
YB44_MYCPN (P75142) Very hypothetical adhesin P1-like protein MP... 33 0.88
YE09_MYCPN (P75375) Very hypothetical adhesin P1-like protein MP... 32 1.1
YD70_MYCPN (P75411) Very hypothetical adhesin P1-like protein MP... 32 1.5
TRX_DROME (P20659) Trithorax protein 32 2.0
YC02_MYCPN (Q50286) Very hypothetical adhesin P1-like protein MP... 31 2.6
YA99_MYCPN (P75593) Very hypothetical adhesin P1-like protein MP... 31 2.6
EUTC_CLOTE (Q892D0) Ethanolamine ammonia-lyase light chain (EC 4... 30 4.4
BET4_YEAST (Q00618) Geranylgeranyl transferase type II alpha sub... 30 4.4
>POLX_TOBAC (P10978) Retrovirus-related Pol polyprotein from
transposon TNT 1-94 [Contains: Protease (EC 3.4.23.-);
Reverse transcriptase (EC 2.7.7.49); Endonuclease]
Length = 1328
Score = 82.8 bits (203), Expect = 7e-16
Identities = 71/250 (28%), Positives = 111/250 (44%), Gaps = 33/250 (13%)
Query: 14 WFLDSGASNHMTGSSE-YLHNLASYHGNQQIQIADGNNLSITDVGDIN-----------S 61
W +D+ AS+H T + + +A G +++ + + I +GDI
Sbjct: 294 WVVDTAASHHATPVRDLFCRYVAGDFGT--VKMGNTSYSKIAGIGDICIKTNVGCTLVLK 351
Query: 62 DFRNVLVSPGLASNLLSVGQLVDNNCNVNFSRAGCVVQEQVSGKVIAKGPKVGRLFPLQF 121
D R+V P L NL+S L + F+ + + VIAKG G L+
Sbjct: 352 DVRHV---PDLRMNLISGIALDRDGYESYFANQKWRLTK--GSLVIAKGVARGTLYRTN- 405
Query: 122 ISNHLSLPCNNVLNSYED------WHRKLGHPNSTVLSHLFKTGLLGNKQVVCTASISCL 175
+ C LN+ +D WH+++GH + L L K L+ + T C
Sbjct: 406 -----AEICQGELNAAQDEISVDLWHKRMGHMSEKGLQILAKKSLISYAKG--TTVKPCD 458
Query: 176 VCKLAKSKTLPFPSGAHRASNCFEMIHSDVWGMSPIASHARYKYFVTFIDDYSRFTWIYF 235
C K + F + + R N ++++SDV G I S KYFVTFIDD SR W+Y
Sbjct: 459 YCLFGKQHRVSFQTSSERKLNILDLVYSDVCGPMEIESMGGNKYFVTFIDDASRKLWVYI 518
Query: 236 LRSKSEVFSI 245
L++K +VF +
Sbjct: 519 LKTKDQVFQV 528
>YCB9_YEAST (P25384) Transposon Ty2 protein B (Ty1-17 protein B)
Length = 1770
Score = 78.6 bits (192), Expect = 1e-14
Identities = 73/269 (27%), Positives = 119/269 (44%), Gaps = 37/269 (13%)
Query: 4 HVKSSNVSRP--WFLDSGASNHMTGSSEYLHNLASYHGNQQIQIADGN--NLSITDVGDI 59
H SN P +DSGAS + S+ YLH+ N +I I D ++ I +G++
Sbjct: 442 HTIDSNDELPDHLLIDSGASQTLVRSAHYLHHATP---NSEINIVDAQKQDIPINAIGNL 498
Query: 60 NSDFRN-------VLVSPGLASNLLSVGQLVDNNCNVNFSRAGCVVQEQVSGKVIAKGPK 112
+ +F+N L +P +A +LLS+ +L + N F+R E+ G V+A K
Sbjct: 499 HFNFQNGTKTSIKALHTPNIAYDLLSLSELANQNITACFTRN---TLERSDGTVLAPIVK 555
Query: 113 VGRLFPLQ---FISNHLS-LPCNNVLNS-------YEDWHRKLGHPNSTVLSHLFKTGLL 161
G + L I +H+S L NNV S Y HR LGH N + K +
Sbjct: 556 HGDFYWLSKKYLIPSHISKLTINNVNKSKSVNKYPYPLIHRMLGHANFRSIQKSLKKNAV 615
Query: 162 -----GNKQVVCTASISCLVCKLAKSKTLPFPSGAH----RASNCFEMIHSDVWGMSPIA 212
+ + ++ C C + KS G+ + F+ +H+D++G
Sbjct: 616 TYLKESDIEWSNASTYQCPDCLIGKSTKHRHVKGSRLKYQESYEPFQYLHTDIFGPVHHL 675
Query: 213 SHARYKYFVTFIDDYSRFTWIYFLRSKSE 241
+ YF++F D+ +RF W+Y L + E
Sbjct: 676 PKSAPSYFISFTDEKTRFQWVYPLHDRRE 704
>YMU0_YEAST (Q04670) Transposon Ty1 protein B
Length = 1328
Score = 73.9 bits (180), Expect = 3e-13
Identities = 68/268 (25%), Positives = 125/268 (46%), Gaps = 37/268 (13%)
Query: 16 LDSGASNHMTGSSEYLHNLASYHGNQQIQIADGN--NLSITDVGDINSDFRN-------V 66
LDSGAS + S+ ++H+ +S N I + D N+ I +GD+ F++ V
Sbjct: 33 LDSGASRTLIRSAHHIHSASS---NPDINVVDAQKRNIPINAIGDLQFHFQDNTKTSIKV 89
Query: 67 LVSPGLASNLLSVGQLVDNNCNVNFSRAGCVVQEQVSGKVIAKGPKVGRLFPLQ---FIS 123
L +P +A +LLS+ +L + F++ V E+ G V+A K G + + +
Sbjct: 90 LHTPNIAYDLLSLNELAAVDITACFTKN---VLERSDGTVLAPIVKYGDFYWVSKKYLLP 146
Query: 124 NHLSLPC-NNVLNS-------YEDWHRKLGHPNSTVLSHLFKTGLL---GNKQVVCTASI 172
+++S+P NNV S Y HR L H N+ + + K + V +++I
Sbjct: 147 SNISVPTINNVHTSESTRKYPYPFIHRMLAHANAQTIRYSLKNNTITYFNESDVDWSSAI 206
Query: 173 S-----CLVCKLAKSKTLPFPSGAHRAS-NCFEMIHSDVWGMSPIASHARYKYFVTFIDD 226
CL+ K K + + ++ S F+ +H+D++G + YF++F D+
Sbjct: 207 DYQCPDCLIGKSTKHRHIKGSRLKYQNSYEPFQYLHTDIFGPVHNLPKSAPSYFISFTDE 266
Query: 227 YSRFTWIYFLRSKSE--VFSIVITISGY 252
++F W+Y L + E + + TI +
Sbjct: 267 TTKFRWVYPLHDRREDSILDVFTTILAF 294
>YMT5_YEAST (Q04214) Transposon Ty1 protein B
Length = 1328
Score = 73.9 bits (180), Expect = 3e-13
Identities = 68/268 (25%), Positives = 125/268 (46%), Gaps = 37/268 (13%)
Query: 16 LDSGASNHMTGSSEYLHNLASYHGNQQIQIADGN--NLSITDVGDINSDFRN-------V 66
LDSGAS + S+ ++H+ +S N I + D N+ I +GD+ F++ V
Sbjct: 33 LDSGASRTLIRSAHHIHSASS---NPDINVVDAQKRNIPINAIGDLQFHFQDNTKTSIKV 89
Query: 67 LVSPGLASNLLSVGQLVDNNCNVNFSRAGCVVQEQVSGKVIAKGPKVGRLFPLQ---FIS 123
L +P +A +LLS+ +L + F++ V E+ G V+A K G + + +
Sbjct: 90 LHTPNIAYDLLSLNELAAVDITACFTKN---VLERSDGTVLAPIVKYGDFYWVSKKYLLP 146
Query: 124 NHLSLPC-NNVLNS-------YEDWHRKLGHPNSTVLSHLFKTGLL---GNKQVVCTASI 172
+++S+P NNV S Y HR L H N+ + + K + V +++I
Sbjct: 147 SNISVPTINNVHTSESTRKYPYPFIHRMLAHANAQTIRYSLKNNTITYFNESDVDWSSAI 206
Query: 173 S-----CLVCKLAKSKTLPFPSGAHRAS-NCFEMIHSDVWGMSPIASHARYKYFVTFIDD 226
CL+ K K + + ++ S F+ +H+D++G + YF++F D+
Sbjct: 207 DYQCPDCLIGKSTKHRHIKGSRLKYQNSYEPFQYLHTDIFGPVHNLPKSAPSYFISFTDE 266
Query: 227 YSRFTWIYFLRSKSE--VFSIVITISGY 252
++F W+Y L + E + + TI +
Sbjct: 267 TTKFRWVYPLHDRREDSILDVFTTILAF 294
>YME4_YEAST (Q04711) Transposon Ty1 protein B
Length = 1328
Score = 73.9 bits (180), Expect = 3e-13
Identities = 68/268 (25%), Positives = 125/268 (46%), Gaps = 37/268 (13%)
Query: 16 LDSGASNHMTGSSEYLHNLASYHGNQQIQIADGN--NLSITDVGDINSDFRN-------V 66
LDSGAS + S+ ++H+ +S N I + D N+ I +GD+ F++ V
Sbjct: 33 LDSGASRTLIRSAHHIHSASS---NPDINVVDAQKRNIPINAIGDLQFHFQDNTKTSIKV 89
Query: 67 LVSPGLASNLLSVGQLVDNNCNVNFSRAGCVVQEQVSGKVIAKGPKVGRLFPLQ---FIS 123
L +P +A +LLS+ +L + F++ V E+ G V+A K G + + +
Sbjct: 90 LHTPNIAYDLLSLNELAAVDITACFTKN---VLERSDGTVLAPIVKYGDFYWVSKKYLLP 146
Query: 124 NHLSLPC-NNVLNS-------YEDWHRKLGHPNSTVLSHLFKTGLL---GNKQVVCTASI 172
+++S+P NNV S Y HR L H N+ + + K + V +++I
Sbjct: 147 SNISVPTINNVHTSESTRKYPYPFIHRMLAHANAQTIRYSLKNNTITYFNESDVDWSSAI 206
Query: 173 S-----CLVCKLAKSKTLPFPSGAHRAS-NCFEMIHSDVWGMSPIASHARYKYFVTFIDD 226
CL+ K K + + ++ S F+ +H+D++G + YF++F D+
Sbjct: 207 DYQCPDCLIGKSTKHRHIKGSRLKYQNSYEPFQYLHTDIFGPVHNLPKSAPSYFISFTDE 266
Query: 227 YSRFTWIYFLRSKSE--VFSIVITISGY 252
++F W+Y L + E + + TI +
Sbjct: 267 TTKFRWVYPLHDRREDSILDVFTTILAF 294
>YMD9_YEAST (Q03434) Transposon Ty1 protein B
Length = 1328
Score = 73.9 bits (180), Expect = 3e-13
Identities = 69/268 (25%), Positives = 125/268 (45%), Gaps = 37/268 (13%)
Query: 16 LDSGASNHMTGSSEYLHNLASYHGNQQIQIADGN--NLSITDVGDINSDFRN-------V 66
LDSGAS + S+ ++H+ +S N I + D N+ I +GD+ F++ V
Sbjct: 33 LDSGASRTLIRSAHHIHSASS---NPDINVVDAQKRNIPINAIGDLQFHFQDNTKTSIKV 89
Query: 67 LVSPGLASNLLSVGQLVDNNCNVNFSRAGCVVQEQVSGKVIAKGPKVGRLFPLQ---FIS 123
L +P +A +LLS+ +L + F++ V E+ G V+A K G + + +
Sbjct: 90 LHTPNIAYDLLSLNELAAVDITACFTKN---VLERSDGTVLAPIVKYGDFYWVSKKYLLP 146
Query: 124 NHLSLPC-NNVLNS-------YEDWHRKLGHPNSTVLSHLFKTGLLG--NKQVVCTASI- 172
+++S+P NNV S Y HR L H N+ + + K + N+ V +S
Sbjct: 147 SNISVPTINNVHTSESTRKYPYPFIHRMLAHANAQTIRYSLKNNTITYFNESDVDRSSAI 206
Query: 173 -----SCLVCKLAKSKTLPFPSGAHRAS-NCFEMIHSDVWGMSPIASHARYKYFVTFIDD 226
CL+ K K + + ++ S F+ +H+D++G + YF++F D+
Sbjct: 207 DYQCPDCLIGKSTKHRHIKGSRLKYQNSYEPFQYLHTDIFGPVHNLPKSAPSYFISFTDE 266
Query: 227 YSRFTWIYFLRSKSE--VFSIVITISGY 252
++F W+Y L + E + + TI +
Sbjct: 267 TTKFRWVYPLHDRREDSILDVFTTILAF 294
>YJZ9_YEAST (P47100) Transposon Ty1 protein B
Length = 1755
Score = 73.9 bits (180), Expect = 3e-13
Identities = 68/268 (25%), Positives = 125/268 (46%), Gaps = 37/268 (13%)
Query: 16 LDSGASNHMTGSSEYLHNLASYHGNQQIQIADGN--NLSITDVGDINSDFRN-------V 66
LDSGAS + S+ ++H+ +S N I + D N+ I +GD+ F++ V
Sbjct: 460 LDSGASRTLIRSAHHIHSASS---NPDINVVDAQKRNIPINAIGDLQFHFQDNTKTSIKV 516
Query: 67 LVSPGLASNLLSVGQLVDNNCNVNFSRAGCVVQEQVSGKVIAKGPKVGRLFPLQ---FIS 123
L +P +A +LLS+ +L + F++ V E+ G V+A K G + + +
Sbjct: 517 LHTPNIAYDLLSLNELAAVDITACFTKN---VLERSDGTVLAPIVKYGDFYWVSKKYLLP 573
Query: 124 NHLSLPC-NNVLNS-------YEDWHRKLGHPNSTVLSHLFKTGLL---GNKQVVCTASI 172
+++S+P NNV S Y HR L H N+ + + K + V +++I
Sbjct: 574 SNISVPTINNVHTSESTRKYPYPFIHRMLAHANAQTIRYSLKNNTITYFNESDVDWSSAI 633
Query: 173 S-----CLVCKLAKSKTLPFPSGAHRAS-NCFEMIHSDVWGMSPIASHARYKYFVTFIDD 226
CL+ K K + + ++ S F+ +H+D++G + YF++F D+
Sbjct: 634 DYQCPDCLIGKSTKHRHIKGSRLKYQNSYEPFQYLHTDIFGPVHNLPKSAPSYFISFTDE 693
Query: 227 YSRFTWIYFLRSKSE--VFSIVITISGY 252
++F W+Y L + E + + TI +
Sbjct: 694 TTKFRWVYPLHDRREDSILDVFTTILAF 721
>YJZ7_YEAST (P47098) Transposon Ty1 protein B
Length = 1755
Score = 72.4 bits (176), Expect = 1e-12
Identities = 67/268 (25%), Positives = 125/268 (46%), Gaps = 37/268 (13%)
Query: 16 LDSGASNHMTGSSEYLHNLASYHGNQQIQIADGN--NLSITDVGDINSDFRN-------V 66
LDSGAS + S+ ++H+ +S N I + D N+ I +GD+ F++ V
Sbjct: 460 LDSGASRTLIRSAHHIHSASS---NPDINVVDAQKRNIPINAIGDLQFHFQDNTKTSIKV 516
Query: 67 LVSPGLASNLLSVGQLVDNNCNVNFSRAGCVVQEQVSGKVIAKGPKVGRLFPLQ---FIS 123
L +P +A +LLS+ +L + F++ V E+ G V+A + G + + +
Sbjct: 517 LHTPNIAYDLLSLNELAAVDITACFTKN---VLERSDGTVLAPIVQYGDFYWVSKRYLLP 573
Query: 124 NHLSLPC-NNVLNS-------YEDWHRKLGHPNSTVLSHLFKTGLL---GNKQVVCTASI 172
+++S+P NNV S Y HR L H N+ + + K + V +++I
Sbjct: 574 SNISVPTINNVHTSESTRKYPYPFIHRMLAHANAQTIRYSLKNNTITYFNESDVDWSSAI 633
Query: 173 S-----CLVCKLAKSKTLPFPSGAHRAS-NCFEMIHSDVWGMSPIASHARYKYFVTFIDD 226
CL+ K K + + ++ S F+ +H+D++G + YF++F D+
Sbjct: 634 DYQCPDCLIGKSTKHRHIKGSRLKYQNSYEPFQYLHTDIFGPVHNLPKSAPSYFISFTDE 693
Query: 227 YSRFTWIYFLRSKSE--VFSIVITISGY 252
++F W+Y L + E + + TI +
Sbjct: 694 TTKFRWVYPLHDRREDSILDVFTTILAF 721
>COPI_DROME (P04146) Copia protein (Gag-int-pol protein) [Contains:
Copia VLP protein; Copia protease (EC 3.4.23.-)]
Length = 1409
Score = 68.2 bits (165), Expect = 2e-11
Identities = 60/245 (24%), Positives = 110/245 (44%), Gaps = 22/245 (8%)
Query: 16 LDSGASNHMTGSSEYLHNLASYHGNQQIQIA-DGNNLSITDVG------DINSDFRNVLV 68
LDSGAS+H+ + +I +A G + T G D +VL
Sbjct: 291 LDSGASDHLINDESLYTDSVEVVPPLKIAVAKQGEFIYATKRGIVRLRNDHEITLEDVLF 350
Query: 69 SPGLASNLLSVGQLVDNNCNVNFSRAGCVVQEQVSGKVIAKGPKVGRLFPLQFISNHLSL 128
A NL+SV +L + ++ F ++G + + +G ++ K + P + N +
Sbjct: 351 CKEAAGNLMSVKRLQEAGMSIEFDKSGVTISK--NGLMVVKNSGMLNNVP---VINFQAY 405
Query: 129 PCN-NVLNSYEDWHRKLGHPNSTVLSHLFKTGLLGNKQVVCTASISCLVCKLA---KSKT 184
N N++ WH + GH + L + + + ++ ++ +SC +C+ K
Sbjct: 406 SINAKHKNNFRLWHERFGHISDGKLLEIKRKNMFSDQSLLNNLELSCEICEPCLNGKQAR 465
Query: 185 LPFPS---GAHRASNCFEMIHSDVWG-MSPIASHARYKYFVTFIDDYSRFTWIYFLRSKS 240
LPF H F ++HSDV G ++P+ + YFV F+D ++ + Y ++ KS
Sbjct: 466 LPFKQLKDKTHIKRPLF-VVHSDVCGPITPVTLDDK-NYFVIFVDQFTHYCVTYLIKYKS 523
Query: 241 EVFSI 245
+VFS+
Sbjct: 524 DVFSM 528
>M300_ARATH (P93293) Hypothetical mitochondrial protein AtMg00300
(ORF145a) (ORF1451)
Length = 145
Score = 44.3 bits (103), Expect = 3e-04
Identities = 29/107 (27%), Positives = 50/107 (46%), Gaps = 9/107 (8%)
Query: 105 KVIAKGPKVGRLFPLQFISNHLSLPCNNVLNSYED----WHRKLGHPNSTVLSHLFKTGL 160
+ I KG + L+ LQ + +N+ + +D WH +L H + + L K G
Sbjct: 36 RTILKGNRHDSLYILQ---GSVETGESNLAETAKDETRLWHSRLAHMSQRGMELLVKKGF 92
Query: 161 LGNKQVVCTASISCLVCKLAKSKTLPFPSGAHRASNCFEMIHSDVWG 207
L + +V ++ C C K+ + F +G H N + +HSD+WG
Sbjct: 93 LDSSKV--SSLKFCEDCIYGKTHRVNFSTGQHTTKNPLDYVHSDLWG 137
>ORYZ_ASPFU (P28296) Oryzin precursor (EC 3.4.21.63) (Alkaline
proteinase) (ALP) (Elastase) (Elastinolytic serine
proteinase)
Length = 403
Score = 34.7 bits (78), Expect = 0.23
Identities = 26/73 (35%), Positives = 35/73 (47%), Gaps = 3/73 (4%)
Query: 36 SYHGNQQIQIA-DGNNLSITDVGDINSDFRNVLVSPGLASNLLSVGQLVDNNCNVNFSRA 94
SY N ++ A D LS+ G+ NSD N SP A N L+V + +N +FS
Sbjct: 261 SYAFNNAVENAFDEGVLSVVAAGNENSDASNT--SPASAPNALTVAAINKSNARASFSNY 318
Query: 95 GCVVQEQVSGKVI 107
G VV G+ I
Sbjct: 319 GSVVDIFAPGQDI 331
>ADP1_MYCPN (P11311) Adhesin P1 precursor (Cytadhesin P1) (Attachment
protein)
Length = 1627
Score = 33.5 bits (75), Expect = 0.52
Identities = 21/67 (31%), Positives = 33/67 (48%), Gaps = 4/67 (5%)
Query: 36 SYHGNQQIQIADGNNLSITDVG----DINSDFRNVLVSPGLASNLLSVGQLVDNNCNVNF 91
+Y GN + + N S +G + N+D + L++PGLA VG LV + V+F
Sbjct: 1056 TYFGNTRAGGSGSNTTSSPGIGFKIPEQNNDSKATLITPGLAWTPQDVGNLVVSGTTVSF 1115
Query: 92 SRAGCVV 98
G +V
Sbjct: 1116 QLGGWLV 1122
>YB44_MYCPN (P75142) Very hypothetical adhesin P1-like protein
MPN144 (E07_orf413)
Length = 413
Score = 32.7 bits (73), Expect = 0.88
Identities = 20/67 (29%), Positives = 34/67 (49%), Gaps = 4/67 (5%)
Query: 36 SYHGNQQIQIADGNNLSITDVG----DINSDFRNVLVSPGLASNLLSVGQLVDNNCNVNF 91
+Y GN + + N S +G + N+D + L++PGLA VG LV + +++F
Sbjct: 136 TYFGNTRAGGSGSNTTSSPGIGFKIPEQNNDSKATLITPGLAWTPQDVGNLVVSGTSLSF 195
Query: 92 SRAGCVV 98
G +V
Sbjct: 196 QLGGWLV 202
>YE09_MYCPN (P75375) Very hypothetical adhesin P1-like protein
MPN409 (F11_orf533L)
Length = 533
Score = 32.3 bits (72), Expect = 1.1
Identities = 21/60 (35%), Positives = 29/60 (48%), Gaps = 3/60 (5%)
Query: 39 GNQQIQIADGNNLSITDVGDINSDFRNVLVSPGLASNLLSVGQLVDNNCNVNFSRAGCVV 98
GNQ G I + N+D + VL++PGLA VG LV + V+F G +V
Sbjct: 260 GNQNRTTVPGIGFKIPEQ---NNDSKAVLITPGLAWTPQDVGNLVVSGTTVSFQLGGWLV 316
>YD70_MYCPN (P75411) Very hypothetical adhesin P1-like protein
MPN370 (A19_orf737V)
Length = 737
Score = 32.0 bits (71), Expect = 1.5
Identities = 20/67 (29%), Positives = 34/67 (49%), Gaps = 4/67 (5%)
Query: 36 SYHGNQQIQIADGNNLSITDVG----DINSDFRNVLVSPGLASNLLSVGQLVDNNCNVNF 91
+Y G + + N S +G + N+D + VL++PGLA VG LV + +++F
Sbjct: 291 TYFGTTRAGGSGSNTTSSPGIGFKIPEQNNDSKAVLITPGLAWTPQDVGNLVVSGTSLSF 350
Query: 92 SRAGCVV 98
G +V
Sbjct: 351 QLGGWLV 357
>TRX_DROME (P20659) Trithorax protein
Length = 3726
Score = 31.6 bits (70), Expect = 2.0
Identities = 28/127 (22%), Positives = 55/127 (43%), Gaps = 5/127 (3%)
Query: 7 SSNVSRPWFLDSGASNHMTGSSEYLHNLASYHGNQQIQIADGNNLSITDVGDINSDFRNV 66
S++ P SG + ++ + +S Q+ + N L +D + F N
Sbjct: 395 SASNGPPAMASSGDGSSPKSGADTGPSTSSTTAKQKKTVTFRNVLETSDDKSVVKRFYN- 453
Query: 67 LVSPGLASNLLSVGQLVDNNCNVNFSRAG-CVVQEQVSGKVIAKGPKVGRLFPLQFISNH 125
P + ++S+ + N +N+SR G C+V+ + K++ K + +L L+F S
Sbjct: 454 ---PDIRIPIVSIMKKDSLNRPLNYSRGGECIVRPSILSKILNKNSNIDKLNSLKFRSAG 510
Query: 126 LSLPCNN 132
S +N
Sbjct: 511 ASSSSSN 517
>YC02_MYCPN (Q50286) Very hypothetical adhesin P1-like protein
MPN202 (GT9_orf313)
Length = 313
Score = 31.2 bits (69), Expect = 2.6
Identities = 18/57 (31%), Positives = 32/57 (55%), Gaps = 4/57 (7%)
Query: 46 ADGNNLSITDVG----DINSDFRNVLVSPGLASNLLSVGQLVDNNCNVNFSRAGCVV 98
A+ N+ ++ +G + N+D + VL++PGLA VG LV + + +F G +V
Sbjct: 145 ANQNSTTVPGIGFKIPEQNNDSKAVLITPGLAWTPQDVGNLVVSGTSFSFQLGGWLV 201
>YA99_MYCPN (P75593) Very hypothetical adhesin P1-like protein
MPN099 (R02_orf347L)
Length = 347
Score = 31.2 bits (69), Expect = 2.6
Identities = 18/57 (31%), Positives = 31/57 (53%), Gaps = 4/57 (7%)
Query: 46 ADGNNLSITDVG----DINSDFRNVLVSPGLASNLLSVGQLVDNNCNVNFSRAGCVV 98
A+ N+ ++ +G + N+D + L++PGLA VG LV + V+F G +V
Sbjct: 68 ANQNSTTVPGIGFKIPEQNNDSKATLITPGLAWTPQDVGNLVVSGTTVSFQLGGWLV 124
>EUTC_CLOTE (Q892D0) Ethanolamine ammonia-lyase light chain (EC
4.3.1.7) (Ethanolamine ammonia-lyase small subunit)
Length = 294
Score = 30.4 bits (67), Expect = 4.4
Identities = 37/137 (27%), Positives = 54/137 (39%), Gaps = 20/137 (14%)
Query: 27 SSEYLHNLASYHGNQQIQIADGNNLSITDVGDINSDFRNVLVSPGLASNLLSVGQLVDNN 86
S E L L+SY N Q+QI + LS V ++ +N+L P L L G V
Sbjct: 145 SKEELSKLSSYKKNAQVQIYVSDGLSSKAV---EANVKNIL--PALIQGLEGYGISVGKP 199
Query: 87 CNVNFSRAGC--VVQEQVSGKVIAKGPKVGRLFPLQFISNHLSLPCNNVLNSYEDWHRKL 144
V R G V+ E+ V I L +++Y + +
Sbjct: 200 FFVKLGRVGAMDVISEEFGADVTC-----------VLIGERPGLVTAESMSAYIAYKGTV 248
Query: 145 GHPNS--TVLSHLFKTG 159
G P S TV+S++ K G
Sbjct: 249 GMPESRRTVVSNIHKGG 265
>BET4_YEAST (Q00618) Geranylgeranyl transferase type II alpha
subunit (EC 2.5.1.60) (Type II protein
geranyl-geranyltransferase alpha subunit)
(GGTase-II-alpha) (PGGT) (YPT1/SEC4 proteins
geranylgeranyltransferase alpha subunit)
Length = 290
Score = 30.4 bits (67), Expect = 4.4
Identities = 12/52 (23%), Positives = 30/52 (57%), Gaps = 5/52 (9%)
Query: 125 HLSLPCNNVLNSYEDWHRKLGHPNSTVLSHLFKTGLLGNKQVVCTASISCLV 176
+ ++ NN +++Y WH+++ ++S +F+ G +GN++ IS ++
Sbjct: 133 YTTIKINNNISNYSAWHQRV-----QIISRMFQKGEVGNQKEYIRTEISYII 179
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.320 0.135 0.413
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 29,473,720
Number of Sequences: 164201
Number of extensions: 1173356
Number of successful extensions: 2530
Number of sequences better than 10.0: 22
Number of HSP's better than 10.0 without gapping: 15
Number of HSP's successfully gapped in prelim test: 7
Number of HSP's that attempted gapping in prelim test: 2504
Number of HSP's gapped (non-prelim): 22
length of query: 256
length of database: 59,974,054
effective HSP length: 108
effective length of query: 148
effective length of database: 42,240,346
effective search space: 6251571208
effective search space used: 6251571208
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 64 (29.3 bits)
Medicago: description of AC144727.5


