

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144727.4 - phase: 0
(762 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
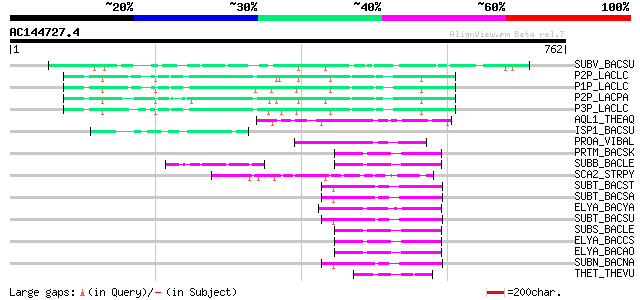
Score E
Sequences producing significant alignments: (bits) Value
SUBV_BACSU (P29141) Minor extracellular protease vpr precursor (... 132 4e-30
P2P_LACLC (P15293) PII-type proteinase precursor (EC 3.4.21.96) ... 87 2e-16
P1P_LACLC (P16271) PI-type proteinase precursor (EC 3.4.21.-) (W... 86 3e-16
P2P_LACPA (Q02470) PII-type proteinase precursor (EC 3.4.21.96) ... 80 2e-14
P3P_LACLC (P15292) PIII-type proteinase precursor (EC 3.4.21.96)... 77 1e-13
AQL1_THEAQ (P08594) Aqualysin I precursor (EC 3.4.21.-) 58 1e-07
ISP1_BACSU (P11018) Major intracellular serine protease precurso... 56 3e-07
PROA_VIBAL (P16588) Alkaline serine exoprotease A precursor (EC ... 56 4e-07
PRTM_BACSK (Q99405) M-protease (EC 3.4.21.-) 55 5e-07
SUBB_BACLE (P29599) Subtilisin BL (EC 3.4.21.62) (Alkaline prote... 54 1e-06
SCA2_STRPY (P58099) C5a peptidase precursor (EC 3.4.21.-) (SCP) 54 1e-06
SUBT_BACST (P29142) Subtilisin J precursor (EC 3.4.21.62) 54 2e-06
SUBT_BACSA (P00783) Subtilisin amylosacchariticus precursor (EC ... 54 2e-06
ELYA_BACYA (P20724) Alkaline elastase YaB precursor (EC 3.4.21.-) 53 3e-06
SUBT_BACSU (P04189) Subtilisin E precursor (EC 3.4.21.62) 53 3e-06
SUBS_BACLE (P29600) Subtilisin Savinase (EC 3.4.21.62) (Alkaline... 53 3e-06
ELYA_BACCS (P41362) Alkaline protease precursor (EC 3.4.21.-) 53 3e-06
ELYA_BACAO (P27693) Alkaline protease precursor (EC 3.4.21.-) 53 3e-06
SUBN_BACNA (P35835) Subtilisin NAT precursor (EC 3.4.21.62) 52 4e-06
THET_THEVU (P04072) Thermitase (EC 3.4.21.66) 52 6e-06
>SUBV_BACSU (P29141) Minor extracellular protease vpr precursor (EC
3.4.21.-)
Length = 806
Score = 132 bits (331), Expect = 4e-30
Identities = 177/710 (24%), Positives = 286/710 (39%), Gaps = 140/710 (19%)
Query: 54 AQVTTTNNNLNNSTSSKIFYTYTNVMNGFSANLSPEEHESLKTFSGFISSIPDLPLKLDT 113
A+ N + + K+ Y V +GFS L E L + P++ K D
Sbjct: 85 ARTKAKNKAIKAVKNGKVNREYEQVFSGFSMKLPANEIPKLLAVKDVKAVYPNVTYKTDN 144
Query: 114 TH-----------SPQFLGLNPYRGA---WPTSDFGKDIIVGVIDTGVWPESESFRDDGM 159
SPQ PY GA W GK I V +IDTGV + +
Sbjct: 145 MKDKDVTISEDAVSPQMDDSAPYIGANDAWDLGYTGKGIKVAIIDTGV-----EYNHPDL 199
Query: 160 TKIPSKWKGQLCQFENSNIQSINLSLCNKKLIGARFFNKGFLAKHSNISTTILNSTRDTN 219
K ++KG F + + K T + +
Sbjct: 200 KKNFGQYKGY------------------------DFVDNDYDPKE----TPTGDPRGEAT 231
Query: 220 GHGTHTSTTAAGSKVDGASFFGYANGTARGIASSSRVAIYKTAWGKDGDALSSDIIAAID 279
HGTH + T A ANGT +G+A + + Y+ G G + ++IA ++
Sbjct: 232 DHGTHVAGTVA------------ANGTIKGVAPDATLLAYRVL-GPGGSGTTENVIAGVE 278
Query: 280 AAISDGVDILSISLGSDDLLLYKDPVAIATFA--AMEKGIFVSTSAGNNGPSFKSIHNGI 337
A+ DG D++++SLG+ L A +T AM +G+ TS GN+GP NG
Sbjct: 279 RAVQDGADVMNLSLGNS---LNNPDWATSTALDWAMSEGVVAVTSNGNSGP------NG- 328
Query: 338 PWVITVAAGTLDREFLGTVTLGNGVSLTGLSFYLGNF-SANNFPIVFMGMCDNVKELNT- 395
W TV + RE + G + L+ Y F S ++ ++ D+VK LN
Sbjct: 329 -W--TVGSPGTSREAISV-----GATQLPLNEYAVTFGSYSSAKVMGYNKEDDVKALNNK 380
Query: 396 -------------------VKRKIVVCEGNNETLHEQMFNVYKAKVVGGVFISNI---LD 433
+ K+ V + + ++ N KA +G V +N+ ++
Sbjct: 381 EVELVEAGIGEAKDFEGKDLTGKVAVVKRGSIAFVDKADNAKKAGAIGMVVYNNLSGEIE 440
Query: 434 INDVDNSFPSIIINPVNGEIVKAYIKSHNSNASSIANMSFKKTAFGVKSTPSVDFYSSRG 493
N S P+I ++ +GE + + +K+ + + FK T DF SSRG
Sbjct: 441 ANVPGMSVPTIKLSLEDGEKLVSALKAGETKTT------FKLTVSKALGEQVADF-SSRG 493
Query: 494 PSNSCPYVLKPDITAPGTSILAAWPTNVPVSNFGTEVFNNFNLIDGTSMSCPHVAGVAAL 553
P +++KPDI+APG +I++ PT+ P +G + GTSM+ PH+AG A+
Sbjct: 494 PVMDT-WMIKPDISAPGVNIVSTIPTHDPDHPYG------YGSKQGTSMASPHIAGAVAV 546
Query: 554 LKGAHNGWSPSSIRSAIMTTSDILDNTKEHIKDIGNGNRAATPFALGAGHINPNRALDPG 613
+K A WS I++AIM T+ L ++ + N A + + A I + + PG
Sbjct: 547 IKQAKPKWSVEQIKAAIMNTAVTLKDSDGEVYP-HNAQGAGSARIMNA--IKADSLVSPG 603
Query: 614 -LVYDIGVQDYINLLCALNFTQKNISAITRSSFNDCSKPSLDLNYPSFIAFSNARNSSRT 672
Y +++ N FT +N S+I + S L Y + + +SR
Sbjct: 604 SYSYGTFLKENGNETKNETFTIENQSSI---------RKSYTLEYSFNGSGISTSGTSRV 654
Query: 673 TNEFHRT-----VTNVGEKKT---TYFASITPIKGFR-VTVIPNKLVFKK 713
H+T V KKT TY ++ +G + V +P L+ K+
Sbjct: 655 VIPAHQTGKATAKVKVNTKKTKAGTYEGTVIVREGGKTVAKVPTLLIVKE 704
>P2P_LACLC (P15293) PII-type proteinase precursor (EC 3.4.21.96)
(Lactocepin) (Cell wall-associated serine proteinase)
(LP151)
Length = 1902
Score = 86.7 bits (213), Expect = 2e-16
Identities = 139/601 (23%), Positives = 236/601 (39%), Gaps = 114/601 (18%)
Query: 74 TYTNVMNGFSANLSPEEHESLKTFSGFISSIPDLPLKLDTTHSPQFLGLNPY---RGAWP 130
+Y V+NGFS + + LK +G + + L + P N + W
Sbjct: 149 SYGYVVNGFSTKVRVVDIPKLKQIAGVKT------VTLAKVYYPTDAKANSMANVQAVWS 202
Query: 131 TSDF-GKDIIVGVIDTGVWPESESFR--DDGMTKIPSKWKGQLCQFENSNIQSINLSLCN 187
+ G+ +V VID+G+ P + R DD K+ K + +F ++
Sbjct: 203 NYKYKGEGTVVSVIDSGIDPTHKDMRLSDDKDVKLT---KSDVEKFTDTAKHG------- 252
Query: 188 KKLIGARFFNK----GFLAKHSNISTTILNSTRDTNGHGTHTSTT--AAGSKVDGASFFG 241
R+FN GF +++ + TI + T D HG H + A G+ D A
Sbjct: 253 ------RYFNSKVPYGF--NYADNNDTITDDTVDEQ-HGMHVAGIIGANGTGDDPAK--- 300
Query: 242 YANGTARGIASSSRVAIYKTAWGKDGDAL--SSDIIAAIDAAISDGVDILSISLGSDD-L 298
+ G+A +++ K D A S+ +++AI+ + G D+L++SLGSD
Sbjct: 301 ----SVVGVAPEAQLLAMKVFTNSDTSATTGSATLVSAIEDSAKIGADVLNMSLGSDSGN 356
Query: 299 LLYKDPVAIATFAAMEKGIFVSTSAGNNGPSFKSIHNGIPWVITVAAGTLDREFLGTVTL 358
+DP A A E G SAGN+G S + V G D E +GT
Sbjct: 357 QTLEDPELAAVQNANESGTAAVISAGNSGTSGSATEG----VNKDYYGLQDNEMVGTPGT 412
Query: 359 GNGVSL-------------------TGLSF-----------YLGNFSANNFPIV--FMGM 386
G + TGL + G+F F +V G
Sbjct: 413 SRGATTVASAENTDVITQAVTITDGTGLQLGPETIQLSSNDFTGSFDQKKFYVVKDASGN 472
Query: 387 CDNVKELN---TVKRKIVVCEGNNETLHEQMFNVYKAKVVGGVFISNILDINDVDN---- 439
K + K KI + + T ++ A G + ++N V +
Sbjct: 473 LSKGKVADYTADAKGKIAIVKRGELTFADKQKYAQAAGAAGLIIVNNDGTATPVTSMALT 532
Query: 440 -SFPSIIINPVNGEIVKAYIKSHNSNASSIANMSFKKTAFGVKSTPSVDFYSSRGPSNSC 498
+FP+ ++ V G+ + ++ +H ++ + ++ + + ++S GP ++
Sbjct: 533 TTFPTFGLSSVTGQKLVDWVAAHPDDSLGV-KIALTLVPNQKYTEDKMSDFTSYGPVSNL 591
Query: 499 PYVLKPDITAPGTSILAAWPTNVPVSNFGTEVFNNFNLIDGTSMSCPHVAGVAALLKGAH 558
+ KPDITAPG +I + T+ N + + GTSM+ P +AG ALLK A
Sbjct: 592 SF--KPDITAPGGNI------------WSTQNNNGYTNMSGTSMASPFIAGSQALLKQAL 637
Query: 559 NGWSP------SSIRSAIMT--TSDILDNTKEHIKDIGNGNRAATPFALGAGHINPNRAL 610
N + ++ +T + NT + I DI N +P GAG ++ A+
Sbjct: 638 NNKNNPFYAYYKQLKGTALTDFLKTVEMNTAQPINDINYNNVIVSPRRQGAGLVDVKAAI 697
Query: 611 D 611
D
Sbjct: 698 D 698
>P1P_LACLC (P16271) PI-type proteinase precursor (EC 3.4.21.-)
(Wall-associated serine proteinase)
Length = 1902
Score = 86.3 bits (212), Expect = 3e-16
Identities = 136/598 (22%), Positives = 234/598 (38%), Gaps = 108/598 (18%)
Query: 74 TYTNVMNGFSANLSPEEHESLKTFSGFISSIPDLPLKLDTTHSPQFLGLNPY---RGAWP 130
+Y V+NGFS + + LK +G + + L + P N + W
Sbjct: 149 SYGYVVNGFSTKVRVVDIPKLKQIAGVKT------VTLAKVYYPTDAKANSMANVQAVWS 202
Query: 131 TSDF-GKDIIVGVIDTGVWPESESFR--DDGMTKIPSKWKGQLCQFENSNIQSINLSLCN 187
+ G+ +V VID+G+ P + R DD K+ K + +F ++
Sbjct: 203 NYKYKGEGTVVSVIDSGIDPTHKDMRLSDDKDVKLT---KSDVEKFTDTAKHG------- 252
Query: 188 KKLIGARFFNK----GFLAKHSNISTTILNSTRDTNGHGTHTSTT--AAGSKVDGASFFG 241
R+FN GF +++ + TI + T D HG H + A G+ D A
Sbjct: 253 ------RYFNSKVPYGF--NYADNNDTITDDTVDEQ-HGMHVAGIIGANGTGDDPAK--- 300
Query: 242 YANGTARGIASSSRVAIYKTAWGKDGDAL--SSDIIAAIDAAISDGVDILSISLGSDD-L 298
+ G+A +++ K D A SS +++AI+ + G D+L++SLGSD
Sbjct: 301 ----SVVGVAPEAQLLAMKVFTNSDTSATTGSSTLVSAIEDSAKIGADVLNMSLGSDSGN 356
Query: 299 LLYKDPVAIATFAAMEKGIFVSTSAGNNGPSFKSIHN--------------GIPWVITVA 344
+DP A A E G SAGN+G S + G P A
Sbjct: 357 QTLEDPELAAVQNANESGTAAVISAGNSGTSGSATEGVNKDYYGLQDNEMVGTPGTSRGA 416
Query: 345 AGTLDREFLGTVT------------LGNGVSLTGLSFYLGNFSANNFPIVFMGMCDNVKE 392
E +T LG G + + G+F F +V N+ +
Sbjct: 417 TTVASAENTDVITQAVTITDGTGLQLGPGTIQLSSNDFTGSFDQKKFYVV-KDASGNLSK 475
Query: 393 ------LNTVKRKIVVCEGNNETLHEQMFNVYKAKVVGGVFISNILDINDVDN-----SF 441
K KI + + + ++ A G + ++N V + +F
Sbjct: 476 GALADYTADAKGKIAIVKRGELSFDDKQKYAQAAGAAGLIIVNNDGTATPVTSMALTTTF 535
Query: 442 PSIIINPVNGEIVKAYIKSHNSNASSIANMSFKKTAFGVKSTPSVDFYSSRGPSNSCPYV 501
P+ ++ V G+ + ++ +H ++ + ++ + + ++S GP ++ +
Sbjct: 536 PTFGLSSVTGQKLVDWVTAHPDDSLGV-KIALTLVPNQKYTEDKMSDFTSYGPVSNLSF- 593
Query: 502 LKPDITAPGTSILAAWPTNVPVSNFGTEVFNNFNLIDGTSMSCPHVAGVAALLKGAHNGW 561
KPDITAPG +I + T+ N + + GTSM+ P +AG ALLK A N
Sbjct: 594 -KPDITAPGGNI------------WSTQNNNGYTNMSGTSMASPFIAGSQALLKQALNNK 640
Query: 562 SP------SSIRSAIMT--TSDILDNTKEHIKDIGNGNRAATPFALGAGHINPNRALD 611
+ ++ +T + NT + I DI N +P GAG ++ A+D
Sbjct: 641 NNPFYAYYKQLKGTALTDFLKTVEMNTAQPINDINYNNVIVSPRRQGAGLVDVKAAID 698
>P2P_LACPA (Q02470) PII-type proteinase precursor (EC 3.4.21.96)
(Lactocepin) (Cell wall-associated serine proteinase)
(LP151)
Length = 1902
Score = 80.1 bits (196), Expect = 2e-14
Identities = 135/604 (22%), Positives = 229/604 (37%), Gaps = 120/604 (19%)
Query: 74 TYTNVMNGFSANLSPEEHESLKTFSGFISSIPDLPLKLDTTHSPQFLGLNPY---RGAWP 130
+Y V+NGFS + + LK +G + + L + P N + W
Sbjct: 149 SYGYVVNGFSTKVRVVDIPKLKQIAGVKT------VTLAKVYYPTDAKANSMANVQAVWS 202
Query: 131 TSDF-GKDIIVGVIDTGVWPESESFRDDGMTKIPSKWKGQLCQFENSNIQSINLSLCNKK 189
+ G+ +V VIDTG+ P + R L ++ + ++
Sbjct: 203 NYKYKGEGTVVSVIDTGIDPTHKDMR--------------LSDDKDVKLTKYDVEKFTDT 248
Query: 190 LIGARFFNK----GFLAKHSNISTTILNSTRDTNGHGTHTSTTAAGSKVDGASFFGYANG 245
R+F GF +++ + TI + T D HG H A G ANG
Sbjct: 249 AKHGRYFTSKVPYGF--NYADNNDTITDDTVDEQ-HGMHV-----------AGIIG-ANG 293
Query: 246 TAR-------GIASSSRVAIYKTAWGKDGDAL--SSDIIAAIDAAISDGVDILSISLGSD 296
T G+A +++ K D A S+ +++AI+ + G D+L++SLGSD
Sbjct: 294 TGDDPTKSVVGVAPEAQLLAMKVFTNSDTSATTGSATLVSAIEDSAKIGADVLNMSLGSD 353
Query: 297 D-LLLYKDPVAIATFAAMEKGIFVSTSAGNNGPSFKSIHNGIPWVITVAAGTLDREFLGT 355
+DP A A E G SAGN+G S + V G D E +GT
Sbjct: 354 SGNQTLEDPEIAAVQNANESGTAAVISAGNSGTSGSATQG----VNKDYYGLQDNEMVGT 409
Query: 356 ----------------------VTLGNGVSLT--------GLSFYLGNFSANNFPIVFMG 385
VT+ +G L + + G+F F +V
Sbjct: 410 PGTSRGATTVASAENTDVISQAVTITDGKDLQLGPETIQLSSNDFTGSFDQKKFYVVKDA 469
Query: 386 MCDNVKELNT-----VKRKIVVCEGNNETLHEQMFNVYKAKVVGGVFISNI-----LDIN 435
D K K KI + + ++ A G + ++N L
Sbjct: 470 SGDLSKGAAADYTADAKGKIAIVKRGELNFADKQKYAQAAGAAGLIIVNNDGTATPLTSI 529
Query: 436 DVDNSFPSIIINPVNGEIVKAYIKSHNSNASSIANMSFKKTAFGVKSTPSVDFYSSRGPS 495
+ +FP+ ++ G+ + ++ +H ++ + ++ + + ++S GP
Sbjct: 530 RLTTTFPTFGLSSKTGQKLVDWVTAHPDDSLGV-KIALTLLPNQKYTEDKMSDFTSYGPV 588
Query: 496 NSCPYVLKPDITAPGTSILAAWPTNVPVSNFGTEVFNNFNLIDGTSMSCPHVAGVAALLK 555
++ + KPDITAPG +I + T+ N + + GTSM+ P +AG ALLK
Sbjct: 589 SNLSF--KPDITAPGGNI------------WSTQNNNGYTNMSGTSMASPFIAGSQALLK 634
Query: 556 GAHNGWSP------SSIRSAIMT--TSDILDNTKEHIKDIGNGNRAATPFALGAGHINPN 607
A N + ++ +T + NT + I DI N +P GAG ++
Sbjct: 635 QALNNKNNPFYADYKQLKGTALTDFLKTVEMNTAQPINDINYNNVIVSPRRQGAGLVDVK 694
Query: 608 RALD 611
A+D
Sbjct: 695 AAID 698
>P3P_LACLC (P15292) PIII-type proteinase precursor (EC 3.4.21.96)
(Lactocepin) (Cell wall-associated serine proteinase)
Length = 1902
Score = 77.4 bits (189), Expect = 1e-13
Identities = 134/602 (22%), Positives = 237/602 (39%), Gaps = 116/602 (19%)
Query: 74 TYTNVMNGFSANLSPEEHESLKTFSGFISSIPDLPLKLDTTHSPQFLGLNPY---RGAWP 130
+Y V+NGFS + + LK +G + + L + P N + W
Sbjct: 149 SYGYVVNGFSTKVRVVDIPKLKQIAGVKT------VTLAKVYYPTDAKANSMANVQAVWS 202
Query: 131 TSDF-GKDIIVGVIDTGVWPESESFR--DDGMTKIPSKWKGQLCQFENSNIQSINLSLCN 187
+ G+ +V VID+G+ P + R DD K+ S+++ ++ +
Sbjct: 203 NYKYKGEGTVVSVIDSGIDPTHKDMRLSDDKDVKLTK-----------SDVEKFTDTVKH 251
Query: 188 KKLIGARFFNK----GFLAKHSNISTTILNSTRDTNGHGTHTSTT--AAGSKVDGASFFG 241
R+FN GF +++ + TI + D HG H + A G+ D A
Sbjct: 252 -----GRYFNSKVPYGF--NYADNNDTITDDKVDEQ-HGMHVAGIIGANGTGDDPAK--- 300
Query: 242 YANGTARGIASSSRVAIYKTAWGKDGDAL--SSDIIAAIDAAISDGVDILSISLGSDD-L 298
+ G+A +++ K D A S+ +++AI+ + G D+L++SLGS+
Sbjct: 301 ----SVVGVAPEAQLLAMKVFSNSDTSAKTGSATVVSAIEDSAKIGADVLNMSLGSNSGN 356
Query: 299 LLYKDPVAIATFAAMEKGIFVSTSAGNNGPSFKSIHNGIPWVITVAAGTLDREFLG---- 354
+DP A A E G SAGN+G S + V G D E +G
Sbjct: 357 QTLEDPELAAVQNANESGTAAVISAGNSGTSGSATEG----VNKDYYGLQDNEMVGSPGT 412
Query: 355 --------------------TVTLGNGVSLTGLSFYL------GNFSANNFPIVFMGMCD 388
T+T G G+ L + L G+F F IV
Sbjct: 413 SRGATTVASAENTDVITQAVTITDGTGLQLGPETIQLSSHDFTGSFDQKKFYIV-KDASG 471
Query: 389 NVKE------LNTVKRKIVVCEGNNETLHEQMFNVYKAKVVGGVFISNILDINDVDN--- 439
N+ + K KI + + + ++ A G + ++ + +
Sbjct: 472 NLSKGALADYTADAKGKIAIVKRGEFSFDDKQKYAQAAGAAGLIIVNTDGTATPMTSIAL 531
Query: 440 --SFPSIIINPVNGEIVKAYIKSHNSNASSIANMSFKKTAFGVKSTPSVDFYSSRGPSNS 497
+FP+ ++ V G+ + ++ +H ++ + ++ + + ++S GP ++
Sbjct: 532 TTTFPTFGLSSVTGQKLVDWVTAHPDDSLGV-KITLAMLPNQKYTEDKMSDFTSYGPVSN 590
Query: 498 CPYVLKPDITAPGTSILAAWPTNVPVSNFGTEVFNNFNLIDGTSMSCPHVAGVAALLKGA 557
+ KPDITAPG +I + T+ N + + GTSM+ P +AG ALLK A
Sbjct: 591 LSF--KPDITAPGGNI------------WSTQNNNGYTNMSGTSMASPFIAGSQALLKQA 636
Query: 558 HNGWSP------SSIRSAIMT--TSDILDNTKEHIKDIGNGNRAATPFALGAGHINPNRA 609
N + ++ +T + NT + I DI N +P GAG ++ A
Sbjct: 637 LNNKNNPFYAYYKQLKGTALTDFLKTVEMNTAQPINDINYNNVIVSPRRQGAGLVDVKAA 696
Query: 610 LD 611
+D
Sbjct: 697 ID 698
>AQL1_THEAQ (P08594) Aqualysin I precursor (EC 3.4.21.-)
Length = 513
Score = 57.8 bits (138), Expect = 1e-07
Identities = 79/289 (27%), Positives = 121/289 (41%), Gaps = 60/289 (20%)
Query: 339 WVITVAAGTLDREFLGTVTLG------NGVSLTGLSFYLGNFSANNFPIVFMGMCDNVKE 392
+VI T REF G +G NG G G A V G+ K
Sbjct: 163 YVIDTGIRTTHREFGGRARVGYDALGGNGQDCNGH----GTHVAGTIGGVTYGVA---KA 215
Query: 393 LNTVKRKIVVCEGNNETLHEQMFNVYKAKVVGGV----------FISNILDINDVDNSFP 442
+N +++ C G+ T + V+ GV ++N+ V +
Sbjct: 216 VNLYAVRVLDCNGSGST----------SGVIAGVDWVTRNHRRPAVANMSLGGGVSTALD 265
Query: 443 SIIINPVNGEIVKAYIKSHNSNASSIANMSFKKTA--FGVKSTPSVDFYSSRGPSNSCPY 500
+ + N + +V A + + N NA++ N S + A V +T S D +S SC
Sbjct: 266 NAVKNSIAAGVVYA-VAAGNDNANA-CNYSPARVAEALTVGATTSSDARASFSNYGSCV- 322
Query: 501 VLKPDITAPGTSILAAWPTNVPVSNFGTEVFNNFNLIDGTSMSCPHVAGVAALLKGAHNG 560
D+ APG SI +AW T S+ T+ N GTSM+ PHVAGVAAL +
Sbjct: 323 ----DLFAPGASIPSAWYT----SDTATQTLN------GTSMATPHVAGVAALYLEQNPS 368
Query: 561 WSPSSIRSAIMTTSDILDNTKEHIKDIGNGNRAATPFAL---GAGHINP 606
+P+S+ SAI+ + T + IG+G+ ++L G+G P
Sbjct: 369 ATPASVASAILNGA-----TTGRLSGIGSGSPNRLLYSLLSSGSGSTAP 412
Score = 31.6 bits (70), Expect = 8.2
Identities = 21/80 (26%), Positives = 37/80 (46%), Gaps = 7/80 (8%)
Query: 75 YTNVMNGFSANLSPEEHESLKTFSGFISSIPDLPLKLDTTHSPQFLGLN-------PYRG 127
YT + GF+A ++P+ E+ + D ++ T SP GL+ P
Sbjct: 90 YTGALQGFAAEMAPQALEAFRQSPDVEFIEADKVVRAWATQSPAPWGLDRIDQRDLPLSN 149
Query: 128 AWPTSDFGKDIIVGVIDTGV 147
++ + G+ + V VIDTG+
Sbjct: 150 SYTYTATGRGVNVYVIDTGI 169
>ISP1_BACSU (P11018) Major intracellular serine protease precursor
(EC 3.4.21.-) (ISP-1)
Length = 319
Score = 56.2 bits (134), Expect = 3e-07
Identities = 56/218 (25%), Positives = 88/218 (39%), Gaps = 54/218 (24%)
Query: 111 LDTTHSPQFLGLNPYRGAWPTSDFGKDIIVGVIDTGVWPESESFRDDGMTKIPSKWKGQL 170
+D P+ + + W GK+I V V+DTG
Sbjct: 17 MDVNELPEGIKVIKAPEMWAKGVKGKNIKVAVLDTG------------------------ 52
Query: 171 CQFENSNIQSINLSLCNKKLIGARFFNKGFLAKHSNISTTILNSTRDTNGHGTHTSTTAA 230
C + ++++ ++IG + F K IS D NGHGTH + T A
Sbjct: 53 CDTSHPDLKN--------QIIGGKNFTDDDGGKEDAIS--------DYNGHGTHVAGTIA 96
Query: 231 GSKVDGASFFGYANGTARGIASSSRVAIYKTAWGKDGDALSSDIIAAIDAAISDGVDILS 290
+ +NG G+A + + I K G++G II I+ A+ VDI+S
Sbjct: 97 AND---------SNGGIAGVAPEASLLIVKVLGGENGSGQYEWIINGINYAVEQKVDIIS 147
Query: 291 ISLGS-DDLLLYKDPVAIATFAAMEKGIFVSTSAGNNG 327
+SLG D+ K+ V A++ G+ V +AGN G
Sbjct: 148 MSLGGPSDVPELKEAVK----NAVKNGVLVVCAAGNEG 181
Score = 35.0 bits (79), Expect = 0.74
Identities = 18/51 (35%), Positives = 26/51 (50%), Gaps = 12/51 (23%)
Query: 505 DITAPGTSILAAWPTNVPVSNFGTEVFNNFNLIDGTSMSCPHVAGVAALLK 555
D+ APG +IL+ P + + GTSM+ PHV+G AL+K
Sbjct: 222 DLVAPGENILSTLPNK------------KYGKLTGTSMAAPHVSGALALIK 260
>PROA_VIBAL (P16588) Alkaline serine exoprotease A precursor (EC
3.4.21.-)
Length = 534
Score = 55.8 bits (133), Expect = 4e-07
Identities = 54/184 (29%), Positives = 81/184 (43%), Gaps = 21/184 (11%)
Query: 391 KELNTVKRKIVVCEGNNETLHEQMFNVYKAKVVGGVFISNILDINDVDNSFPSIIINPVN 450
K +N V +++ C G+ T + A G ++N+ + S + + V
Sbjct: 230 KNVNLVGVRVLSCSGSGSTSGVIAGVDWVAANASGPSVANMSLGGGQSVALDSAVQSAVQ 289
Query: 451 GEIVKAYIKSHNSNASSIANMSFKKTAFGVK--STPSVDFYSSRGPSNSCPYVLKPDITA 508
V + + NSNA + N S + A GV ST S D SS SC D+ A
Sbjct: 290 SG-VSFMLAAGNSNADA-CNYSPARVATGVTVGSTTSTDARSSFSNWGSCV-----DVFA 342
Query: 509 PGTSILAAWPTNVPVSNFGTEVFNNFNLIDGTSMSCPHVAGVAALLKGAHNGWSPSSIRS 568
PG+ I +AW + I GTSM+ PHVAGVAAL ++ SPS + +
Sbjct: 343 PGSQIKSAWYDG------------GYKTISGTSMATPHVAGVAALYLQENSSVSPSQVEA 390
Query: 569 AIMT 572
I++
Sbjct: 391 LIVS 394
Score = 36.6 bits (83), Expect = 0.25
Identities = 59/306 (19%), Positives = 115/306 (37%), Gaps = 58/306 (18%)
Query: 23 LALSDNYIIHMNLSDMPKSFSNQHSWYESTLAQVTTTNNNLNNSTSSKIFYTYTNVMNGF 82
+A++D YI+ + P+ ++ +E Q + L + +F + ++GF
Sbjct: 52 IAIADRYIV---VFQQPQMMASSSPEFEQFTQQSVDRMSGLYSIQVESVF---DHSISGF 105
Query: 83 SANLSPEEHESLKTFSGFISSIPDLPLKLDTTHSPQFLGLNPYRGAWPTSDFGKDIIVGV 142
ANLSPE+ + L++ ++D + L L+P A D + +
Sbjct: 106 VANLSPEQLKDLRSDP-----------RVDYIEQDRILSLDPIVSA----DANQTNAIWG 150
Query: 143 IDTGVWPESESFRDDGMTKIPSKWKGQLCQFENSNIQSINLSLCNKKLIGARFFNKGFLA 202
+D + + D G ++ + + ++ + + G F + A
Sbjct: 151 LDR---IDQRNLPLDNNYSANFDGTGVTAYVIDTGVNNAHVEFGGRSVSGYDFVDNDADA 207
Query: 203 KHSNISTTILNSTRDTNGHGTHTSTTAAGSKVDGASFFGYANGTARGIASSSRVAIYKTA 262
D NGHGTH + T GS G+A + + +
Sbjct: 208 S-------------DCNGHGTHVAGTIGGS--------------LYGVAKNVNLVGVRVL 240
Query: 263 WGKDGDALSSDIIAAID--AAISDGVDILSISLGSDDLLLYKDPVAIATFAAMEKGIFVS 320
G +S +IA +D AA + G + ++SLG + V +A++ G+
Sbjct: 241 -SCSGSGSTSGVIAGVDWVAANASGPSVANMSLGGGQSVALDSAVQ----SAVQSGVSFM 295
Query: 321 TSAGNN 326
+AGN+
Sbjct: 296 LAAGNS 301
>PRTM_BACSK (Q99405) M-protease (EC 3.4.21.-)
Length = 269
Score = 55.5 bits (132), Expect = 5e-07
Identities = 43/148 (29%), Positives = 65/148 (43%), Gaps = 17/148 (11%)
Query: 446 INPVNGEIVKAYIKSHNSNASSIANMSFKKTAFGVKSTPSVDFYSSRGPSNSCPYVLKPD 505
+N V S NS A SI+ + A V +T + ++ Y D
Sbjct: 137 VNSATSRGVLVVAASGNSGAGSISYPARYANAMAVGATDQ-----NNNRASFSQYGAGLD 191
Query: 506 ITAPGTSILAAWPTNVPVSNFGTEVFNNFNLIDGTSMSCPHVAGVAALLKGAHNGWSPSS 565
I APG ++ + +P + S ++GTSM+ PHVAGVAAL+K + WS
Sbjct: 192 IVAPGVNVQSTYPGSTYAS------------LNGTSMATPHVAGVAALVKQKNPSWSNVQ 239
Query: 566 IRSAIMTTSDILDNTKEHIKDIGNGNRA 593
IR+ + T+ L NT + + N A
Sbjct: 240 IRNHLKNTATGLGNTNLYGSGLVNAEAA 267
Score = 42.4 bits (98), Expect = 0.005
Identities = 38/137 (27%), Positives = 62/137 (44%), Gaps = 15/137 (10%)
Query: 214 STRDTNGHGTHTSTTAAGSKVDGASFFGYANGTARGIASSSRVAIYKTAWGKDGDALSSD 273
ST+D NGHGTH + T A + G G+A S+ + K G G S
Sbjct: 55 STQDGNGHGTHVAGTIAALN----NSIG-----VLGVAPSAELYAVKVL-GASGSGSVSS 104
Query: 274 IIAAIDAAISDGVDILSISLGSDDLLLYKDPVAIATFAAMEKGIFVSTSAGNNGPSFKSI 333
I ++ A ++G+ + ++SLGS + A +A +G+ V ++GN+G SI
Sbjct: 105 IAQGLEWAGNNGMHVANLSLGSPS---PSATLEQAVNSATSRGVLVVAASGNSGAG--SI 159
Query: 334 HNGIPWVITVAAGTLDR 350
+ +A G D+
Sbjct: 160 SYPARYANAMAVGATDQ 176
>SUBB_BACLE (P29599) Subtilisin BL (EC 3.4.21.62) (Alkaline
protease)
Length = 269
Score = 54.3 bits (129), Expect = 1e-06
Identities = 42/148 (28%), Positives = 65/148 (43%), Gaps = 17/148 (11%)
Query: 446 INPVNGEIVKAYIKSHNSNASSIANMSFKKTAFGVKSTPSVDFYSSRGPSNSCPYVLKPD 505
+N V S NS ASSI+ + A V +T + ++ Y D
Sbjct: 137 VNSATSRGVLVVAASGNSGASSISYPARYANAMAVGATDQ-----NNNRASFSQYGAGLD 191
Query: 506 ITAPGTSILAAWPTNVPVSNFGTEVFNNFNLIDGTSMSCPHVAGVAALLKGAHNGWSPSS 565
I APG ++ + +P + S ++GTSM+ PHVAG AAL+K + WS
Sbjct: 192 IVAPGVNVQSTYPGSTYAS------------LNGTSMATPHVAGAAALVKQKNPSWSNVQ 239
Query: 566 IRSAIMTTSDILDNTKEHIKDIGNGNRA 593
IR+ + T+ L +T + + N A
Sbjct: 240 IRNHLKNTATSLGSTNLYGSGLVNAEAA 267
Score = 45.8 bits (107), Expect = 4e-04
Identities = 40/137 (29%), Positives = 64/137 (46%), Gaps = 15/137 (10%)
Query: 214 STRDTNGHGTHTSTTAAGSKVDGASFFGYANGTARGIASSSRVAIYKTAWGKDGDALSSD 273
ST+D NGHGTH + T A + G G+A S+ + K G DG S
Sbjct: 55 STQDGNGHGTHVAGTIAALN----NSIG-----VLGVAPSAELYAVKVL-GADGRGAISS 104
Query: 274 IIAAIDAAISDGVDILSISLGSDDLLLYKDPVAIATFAAMEKGIFVSTSAGNNGPSFKSI 333
I ++ A ++G+ + ++SLGS + A +A +G+ V ++GN+G S SI
Sbjct: 105 IAQGLEWAGNNGMHVANLSLGSPS---PSATLEQAVNSATSRGVLVVAASGNSGAS--SI 159
Query: 334 HNGIPWVITVAAGTLDR 350
+ +A G D+
Sbjct: 160 SYPARYANAMAVGATDQ 176
>SCA2_STRPY (P58099) C5a peptidase precursor (EC 3.4.21.-) (SCP)
Length = 1181
Score = 54.3 bits (129), Expect = 1e-06
Identities = 85/336 (25%), Positives = 144/336 (42%), Gaps = 63/336 (18%)
Query: 277 AIDAAISDGVDILSISLGSDDLLLYK--DPVAIATFAAMEKGIFVSTSAGNNG------- 327
AI A++ G ++++S G+ L D A A KG+ + TSAGN+
Sbjct: 245 AIIDAVNLGAKVINMSFGNAALAYANLPDETKKAFDYAKSKGVSIVTSAGNDSSFGGKTR 304
Query: 328 ------PSFKSIHNGIPWV----ITVAAGTLDREFLGTVTLGNG------VSLTGLSFYL 371
P + + G P +TVA+ + D++ T T+ + + + +
Sbjct: 305 LPLADHPDYGVV--GTPAAADSTLTVASYSPDKQLTETATVKTADQQDKEMPVLSTNRFE 362
Query: 372 GNFSANNFPIVFMGMCDNVKELNTVKRKIVVCEGNNETLHEQMFNVYKAKVVGGVFISNI 431
N A ++ GM ++ + VK KI + E + +++ N KA VG + N
Sbjct: 363 PN-KAYDYAYANRGMKED--DFKDVKGKIALIERGDIDFKDKIANAKKAGAVGVLIYDNQ 419
Query: 432 -----LDINDVDNSFPSIIINPVNGEIVKAYIKSHNSNASSIANMSFKKTAFGVKSTPSV 486
+++ +VD P+ I+ +G ++K ++ + A TA G K +
Sbjct: 420 DKGFPIELPNVDQ-MPAAFISRKDGLLLK---ENPQKTITFNATPKVLPTASGTK----L 471
Query: 487 DFYSSRGPSNSCPYVLKPDITAPGTSILAAWPTNVPVSNFGTEVFNNFNLIDGTSMSCPH 546
+SS G + +KPDI APG IL++ V+N N + + GTSMS P
Sbjct: 472 SRFSSWGLTADGN--IKPDIAAPGQDILSS------VAN------NKYAKLSGTSMSAPL 517
Query: 547 VAGVAALLKGAHNGWSPSSIRSAIMTTSDILDNTKE 582
VAG+ LL+ + P MT S+ LD K+
Sbjct: 518 VAGIMGLLQKQYETQYPD------MTPSERLDLAKK 547
>SUBT_BACST (P29142) Subtilisin J precursor (EC 3.4.21.62)
Length = 381
Score = 53.5 bits (127), Expect = 2e-06
Identities = 44/174 (25%), Positives = 77/174 (43%), Gaps = 21/174 (12%)
Query: 428 ISNILDINDVDNSFPS-------IIINPVNGEIVKAYIKSHNSNASSIANMSFKKTAFGV 480
ISN +D+ ++ PS ++ V+ IV A + ++ S + + +
Sbjct: 221 ISNNMDVINMSLGGPSGSTALKTVVDKAVSSGIVVAAAAGNEGSSGSSSTVGYPAKYPST 280
Query: 481 KSTPSVDFYSSRGPSNSCPYVLKPDITAPGTSILAAWPTNVPVSNFGTEVFNNFNLIDGT 540
+ +V+ + R +S L D+ APG SI + P + +GT
Sbjct: 281 IAVGAVNSSNQRASFSSAGSEL--DVMAPGVSIQSTLPGGT------------YGAYNGT 326
Query: 541 SMSCPHVAGVAALLKGAHNGWSPSSIRSAIMTTSDILDNTKEHIKDIGNGNRAA 594
SM+ PHVAG AAL+ H W+ + +R + +T+ L N+ + K + N AA
Sbjct: 327 SMATPHVAGAAALILSKHPTWTNAQVRDRLESTATYLGNSFYYGKGLINVQAAA 380
Score = 43.5 bits (101), Expect = 0.002
Identities = 40/142 (28%), Positives = 62/142 (43%), Gaps = 21/142 (14%)
Query: 213 NSTRDTNGHGTHTSTTAAGSKVDGASFFGYANGTARGIASSSRVAIYKT-AWGKDGDALS 271
N +D + HGTH + T A N + + S ++Y G
Sbjct: 162 NPYQDGSSHGTHVAGTIAA-----------LNNSIGVLGVSPSASLYAVKVLDSTGSGQY 210
Query: 272 SDIIAAIDAAISDGVDILSISLGSDDLLLYKDPVAIATFA--AMEKGIFVSTSAGNNGPS 329
S II I+ AIS+ +D++++SLG A+ T A+ GI V+ +AGN G S
Sbjct: 211 SWIINGIEWAISNNMDVINMSLGGP-----SGSTALKTVVDKAVSSGIVVAAAAGNEGSS 265
Query: 330 FKSIHNGIP--WVITVAAGTLD 349
S G P + T+A G ++
Sbjct: 266 GSSSTVGYPAKYPSTIAVGAVN 287
>SUBT_BACSA (P00783) Subtilisin amylosacchariticus precursor (EC
3.4.21.62)
Length = 381
Score = 53.5 bits (127), Expect = 2e-06
Identities = 44/174 (25%), Positives = 77/174 (43%), Gaps = 21/174 (12%)
Query: 428 ISNILDINDVDNSFPS-------IIINPVNGEIVKAYIKSHNSNASSIANMSFKKTAFGV 480
ISN +D+ ++ PS ++ V+ IV A + ++ S + + +
Sbjct: 221 ISNNMDVINMSLGGPSGSTALKTVVDKAVSSGIVVAAAAGNEGSSGSSSTVGYPAKYPST 280
Query: 481 KSTPSVDFYSSRGPSNSCPYVLKPDITAPGTSILAAWPTNVPVSNFGTEVFNNFNLIDGT 540
+ +V+ + R +S L D+ APG SI + P + +GT
Sbjct: 281 IAVGAVNSSNQRASFSSAGSEL--DVMAPGVSIQSTLPGGT------------YGAYNGT 326
Query: 541 SMSCPHVAGVAALLKGAHNGWSPSSIRSAIMTTSDILDNTKEHIKDIGNGNRAA 594
SM+ PHVAG AAL+ H W+ + +R + +T+ L N+ + K + N AA
Sbjct: 327 SMATPHVAGAAALILSKHPTWTNAQVRDRLESTATYLGNSFYYGKGLINVQAAA 380
Score = 43.5 bits (101), Expect = 0.002
Identities = 40/142 (28%), Positives = 62/142 (43%), Gaps = 21/142 (14%)
Query: 213 NSTRDTNGHGTHTSTTAAGSKVDGASFFGYANGTARGIASSSRVAIYKT-AWGKDGDALS 271
N +D + HGTH + T A N + + S ++Y G
Sbjct: 162 NPYQDGSSHGTHVAGTIAA-----------LNNSIGVLGVSPSASLYAVKVLDSTGSGQY 210
Query: 272 SDIIAAIDAAISDGVDILSISLGSDDLLLYKDPVAIATFA--AMEKGIFVSTSAGNNGPS 329
S II I+ AIS+ +D++++SLG A+ T A+ GI V+ +AGN G S
Sbjct: 211 SWIINGIEWAISNNMDVINMSLGGP-----SGSTALKTVVDKAVSSGIVVAAAAGNEGSS 265
Query: 330 FKSIHNGIP--WVITVAAGTLD 349
S G P + T+A G ++
Sbjct: 266 GSSSTVGYPAKYPSTIAVGAVN 287
>ELYA_BACYA (P20724) Alkaline elastase YaB precursor (EC 3.4.21.-)
Length = 378
Score = 53.1 bits (126), Expect = 3e-06
Identities = 47/169 (27%), Positives = 70/169 (40%), Gaps = 17/169 (10%)
Query: 425 GVFISNILDINDVDNSFPSIIINPVNGEIVKAYIKSHNSNASSIANMSFKKTAFGVKSTP 484
G+ I+N+ + ++ +N V S NS A ++ + A V +T
Sbjct: 225 GMHIANMSLGSSAGSATMEQAVNQATASGVLVVAASGNSGAGNVGFPARYANAMAVGATD 284
Query: 485 SVDFYSSRGPSNSCPYVLKPDITAPGTSILAAWPTNVPVSNFGTEVFNNFNLIDGTSMSC 544
+ + Y DI APG + + P N G FN GTSM+
Sbjct: 285 Q-----NNNRATFSQYGAGLDIVAPGVGVQSTVPGN------GYASFN------GTSMAT 327
Query: 545 PHVAGVAALLKGAHNGWSPSSIRSAIMTTSDILDNTKEHIKDIGNGNRA 593
PHVAGVAAL+K + WS IR+ + T+ L NT + + N A
Sbjct: 328 PHVAGVAALVKQKNPSWSNVQIRNHLKNTATNLGNTTQFGSGLVNAEAA 376
Score = 33.5 bits (75), Expect = 2.2
Identities = 43/169 (25%), Positives = 68/169 (39%), Gaps = 40/169 (23%)
Query: 217 DTNGHGTHTSTTAA------------------GSKVDGASFFGYANGTARGIASSSRVAI 258
D NGHGT + T A G KV GAS G +G A+G+ ++ +
Sbjct: 167 DGNGHGTQVAGTIAALNNSIGVLGVAPNVDLYGVKVLGASGSGSISGIAQGLQWAANNGM 226
Query: 259 YKTAWGKDGDALSSDIIAAIDAAISDGVDILSIS--LGSDDL---LLYKDPVAI------ 307
+ A S+ + A++ A + GV +++ S G+ ++ Y + +A+
Sbjct: 227 HIANMSLGSSAGSATMEQAVNQATASGVLVVAASGNSGAGNVGFPARYANAMAVGATDQN 286
Query: 308 ---ATFAAMEKGIFV--------STSAGNNGPSFKSIHNGIPWVITVAA 345
ATF+ G+ + ST GN SF P V VAA
Sbjct: 287 NNRATFSQYGAGLDIVAPGVGVQSTVPGNGYASFNGTSMATPHVAGVAA 335
>SUBT_BACSU (P04189) Subtilisin E precursor (EC 3.4.21.62)
Length = 381
Score = 52.8 bits (125), Expect = 3e-06
Identities = 43/174 (24%), Positives = 77/174 (43%), Gaps = 21/174 (12%)
Query: 428 ISNILDINDVDNSFPS-------IIINPVNGEIVKAYIKSHNSNASSIANMSFKKTAFGV 480
ISN +D+ ++ P+ ++ V+ IV A + ++ S + + +
Sbjct: 221 ISNNMDVINMSLGGPTGSTALKTVVDKAVSSGIVVAAAAGNEGSSGSTSTVGYPAKYPST 280
Query: 481 KSTPSVDFYSSRGPSNSCPYVLKPDITAPGTSILAAWPTNVPVSNFGTEVFNNFNLIDGT 540
+ +V+ + R +S L D+ APG SI + P + +GT
Sbjct: 281 IAVGAVNSSNQRASFSSAGSEL--DVMAPGVSIQSTLPGGT------------YGAYNGT 326
Query: 541 SMSCPHVAGVAALLKGAHNGWSPSSIRSAIMTTSDILDNTKEHIKDIGNGNRAA 594
SM+ PHVAG AAL+ H W+ + +R + +T+ L N+ + K + N AA
Sbjct: 327 SMATPHVAGAAALILSKHPTWTNAQVRDRLESTATYLGNSFYYGKGLINVQAAA 380
Score = 42.0 bits (97), Expect = 0.006
Identities = 39/142 (27%), Positives = 62/142 (43%), Gaps = 21/142 (14%)
Query: 213 NSTRDTNGHGTHTSTTAAGSKVDGASFFGYANGTARGIASSSRVAIYKT-AWGKDGDALS 271
N +D + HGTH + T A N + + S ++Y G
Sbjct: 162 NPYQDGSSHGTHVAGTIAA-----------LNNSIGVLGVSPSASLYAVKVLDSTGSGQY 210
Query: 272 SDIIAAIDAAISDGVDILSISLGSDDLLLYKDPVAIATFA--AMEKGIFVSTSAGNNGPS 329
S II I+ AIS+ +D++++SLG A+ T A+ GI V+ +AGN G S
Sbjct: 211 SWIINGIEWAISNNMDVINMSLGGP-----TGSTALKTVVDKAVSSGIVVAAAAGNEGSS 265
Query: 330 FKSIHNGIP--WVITVAAGTLD 349
+ G P + T+A G ++
Sbjct: 266 GSTSTVGYPAKYPSTIAVGAVN 287
>SUBS_BACLE (P29600) Subtilisin Savinase (EC 3.4.21.62) (Alkaline
protease)
Length = 269
Score = 52.8 bits (125), Expect = 3e-06
Identities = 41/148 (27%), Positives = 64/148 (42%), Gaps = 17/148 (11%)
Query: 446 INPVNGEIVKAYIKSHNSNASSIANMSFKKTAFGVKSTPSVDFYSSRGPSNSCPYVLKPD 505
+N V S NS A SI+ + A V +T + ++ Y D
Sbjct: 137 VNSATSRGVLVVAASGNSGAGSISYPARYANAMAVGATDQ-----NNNRASFSQYGAGLD 191
Query: 506 ITAPGTSILAAWPTNVPVSNFGTEVFNNFNLIDGTSMSCPHVAGVAALLKGAHNGWSPSS 565
I APG ++ + +P + S ++GTSM+ PHVAG AAL+K + WS
Sbjct: 192 IVAPGVNVQSTYPGSTYAS------------LNGTSMATPHVAGAAALVKQKNPSWSNVQ 239
Query: 566 IRSAIMTTSDILDNTKEHIKDIGNGNRA 593
IR+ + T+ L +T + + N A
Sbjct: 240 IRNHLKNTATSLGSTNLYGSGLVNAEAA 267
Score = 42.4 bits (98), Expect = 0.005
Identities = 38/137 (27%), Positives = 62/137 (44%), Gaps = 15/137 (10%)
Query: 214 STRDTNGHGTHTSTTAAGSKVDGASFFGYANGTARGIASSSRVAIYKTAWGKDGDALSSD 273
ST+D NGHGTH + T A + G G+A S+ + K G G S
Sbjct: 55 STQDGNGHGTHVAGTIAALN----NSIG-----VLGVAPSAELYAVKVL-GASGSGSVSS 104
Query: 274 IIAAIDAAISDGVDILSISLGSDDLLLYKDPVAIATFAAMEKGIFVSTSAGNNGPSFKSI 333
I ++ A ++G+ + ++SLGS + A +A +G+ V ++GN+G SI
Sbjct: 105 IAQGLEWAGNNGMHVANLSLGSPS---PSATLEQAVNSATSRGVLVVAASGNSGAG--SI 159
Query: 334 HNGIPWVITVAAGTLDR 350
+ +A G D+
Sbjct: 160 SYPARYANAMAVGATDQ 176
>ELYA_BACCS (P41362) Alkaline protease precursor (EC 3.4.21.-)
Length = 380
Score = 52.8 bits (125), Expect = 3e-06
Identities = 41/148 (27%), Positives = 64/148 (42%), Gaps = 17/148 (11%)
Query: 446 INPVNGEIVKAYIKSHNSNASSIANMSFKKTAFGVKSTPSVDFYSSRGPSNSCPYVLKPD 505
+N V S NS A SI+ + A V +T + ++ Y D
Sbjct: 248 VNSATSRGVLVVAASGNSGAGSISYPARYANAMAVGATDQ-----NNNRASFSQYGAGLD 302
Query: 506 ITAPGTSILAAWPTNVPVSNFGTEVFNNFNLIDGTSMSCPHVAGVAALLKGAHNGWSPSS 565
I APG ++ + +P + S ++GTSM+ PHVAG AAL+K + WS
Sbjct: 303 IVAPGVNVQSTYPGSTYAS------------LNGTSMATPHVAGAAALVKQKNPSWSNVQ 350
Query: 566 IRSAIMTTSDILDNTKEHIKDIGNGNRA 593
IR+ + T+ L +T + + N A
Sbjct: 351 IRNHLKNTATSLGSTNLYGSGLVNAEAA 378
Score = 42.4 bits (98), Expect = 0.005
Identities = 38/137 (27%), Positives = 62/137 (44%), Gaps = 15/137 (10%)
Query: 214 STRDTNGHGTHTSTTAAGSKVDGASFFGYANGTARGIASSSRVAIYKTAWGKDGDALSSD 273
ST+D NGHGTH + T A + G G+A S+ + K G G S
Sbjct: 166 STQDGNGHGTHVAGTIAALN----NSIG-----VLGVAPSAELYAVKVL-GASGSGSVSS 215
Query: 274 IIAAIDAAISDGVDILSISLGSDDLLLYKDPVAIATFAAMEKGIFVSTSAGNNGPSFKSI 333
I ++ A ++G+ + ++SLGS + A +A +G+ V ++GN+G SI
Sbjct: 216 IAQGLEWAGNNGMHVANLSLGSPS---PSATLEQAVNSATSRGVLVVAASGNSGAG--SI 270
Query: 334 HNGIPWVITVAAGTLDR 350
+ +A G D+
Sbjct: 271 SYPARYANAMAVGATDQ 287
>ELYA_BACAO (P27693) Alkaline protease precursor (EC 3.4.21.-)
Length = 380
Score = 52.8 bits (125), Expect = 3e-06
Identities = 41/148 (27%), Positives = 64/148 (42%), Gaps = 17/148 (11%)
Query: 446 INPVNGEIVKAYIKSHNSNASSIANMSFKKTAFGVKSTPSVDFYSSRGPSNSCPYVLKPD 505
+N V S NS A SI+ + A V +T + ++ Y D
Sbjct: 248 VNSATSRGVLVVAASGNSGAGSISYPARYANAMAVGATDQ-----NNNRASFSQYGAGLD 302
Query: 506 ITAPGTSILAAWPTNVPVSNFGTEVFNNFNLIDGTSMSCPHVAGVAALLKGAHNGWSPSS 565
I APG ++ + +P + S ++GTSM+ PHVAG AAL+K + WS
Sbjct: 303 IVAPGVNVQSTYPGSTYAS------------LNGTSMATPHVAGAAALVKQKNPSWSNVQ 350
Query: 566 IRSAIMTTSDILDNTKEHIKDIGNGNRA 593
IR+ + T+ L +T + + N A
Sbjct: 351 IRNHLKNTATSLGSTNLYGSGLVNAEAA 378
Score = 41.2 bits (95), Expect = 0.010
Identities = 37/137 (27%), Positives = 62/137 (45%), Gaps = 15/137 (10%)
Query: 214 STRDTNGHGTHTSTTAAGSKVDGASFFGYANGTARGIASSSRVAIYKTAWGKDGDALSSD 273
ST+D NGHGTH + T A + G G+A ++ + K G G S
Sbjct: 166 STQDGNGHGTHVAGTIAALN----NSIG-----VLGVAPNAELYAVKVL-GASGSGSVSS 215
Query: 274 IIAAIDAAISDGVDILSISLGSDDLLLYKDPVAIATFAAMEKGIFVSTSAGNNGPSFKSI 333
I ++ A ++G+ + ++SLGS + A +A +G+ V ++GN+G SI
Sbjct: 216 IAQGLEWAGNNGMHVANLSLGSPS---PSATLEQAVNSATSRGVLVVAASGNSGAG--SI 270
Query: 334 HNGIPWVITVAAGTLDR 350
+ +A G D+
Sbjct: 271 SYPARYANAMAVGATDQ 287
>SUBN_BACNA (P35835) Subtilisin NAT precursor (EC 3.4.21.62)
Length = 381
Score = 52.4 bits (124), Expect = 4e-06
Identities = 43/174 (24%), Positives = 77/174 (43%), Gaps = 21/174 (12%)
Query: 428 ISNILDINDVDNSFPS-------IIINPVNGEIVKAYIKSHNSNASSIANMSFKKTAFGV 480
ISN +D+ ++ P+ ++ V+ IV A + ++ S + + +
Sbjct: 221 ISNNMDVINMSLGGPTGSTALKTVVDKAVSSGIVVAAAAGNEGSSGSTSTVGYPAKYPST 280
Query: 481 KSTPSVDFYSSRGPSNSCPYVLKPDITAPGTSILAAWPTNVPVSNFGTEVFNNFNLIDGT 540
+ +V+ + R +S L D+ APG SI + P + +GT
Sbjct: 281 IAVGAVNSSNQRASFSSVGSEL--DVMAPGVSIQSTLPGGT------------YGAYNGT 326
Query: 541 SMSCPHVAGVAALLKGAHNGWSPSSIRSAIMTTSDILDNTKEHIKDIGNGNRAA 594
SM+ PHVAG AAL+ H W+ + +R + +T+ L N+ + K + N AA
Sbjct: 327 SMATPHVAGAAALILSKHPTWTNAQVRDRLESTATYLGNSFYYGKGLINVQAAA 380
Score = 42.0 bits (97), Expect = 0.006
Identities = 41/141 (29%), Positives = 64/141 (45%), Gaps = 19/141 (13%)
Query: 213 NSTRDTNGHGTHTSTTAAGSKVDGASFFGYANGTARGIASSSRVAIYKTAWGKDGDALSS 272
N +D + HGTH + T A + G G+A S+ + K G S
Sbjct: 162 NPYQDGSSHGTHVAGTIAALN----NSIGVL-----GVAPSASLYAVKVL-DSTGSGQYS 211
Query: 273 DIIAAIDAAISDGVDILSISLGSDDLLLYKDPVAIATFA--AMEKGIFVSTSAGNNGPSF 330
II I+ AIS+ +D++++SLG A+ T A+ GI V+ +AGN G S
Sbjct: 212 WIINGIEWAISNNMDVINMSLGGP-----TGSTALKTVVDKAVSSGIVVAAAAGNEGSSG 266
Query: 331 KSIHNGIP--WVITVAAGTLD 349
+ G P + T+A G ++
Sbjct: 267 STSTVGYPAKYPSTIAVGAVN 287
>THET_THEVU (P04072) Thermitase (EC 3.4.21.66)
Length = 279
Score = 52.0 bits (123), Expect = 6e-06
Identities = 40/109 (36%), Positives = 53/109 (47%), Gaps = 19/109 (17%)
Query: 472 SFKKTAFGVKSTPSVDFYSSRGPSNSCPYVLKPDITAPGTSILAAWPTNVPVSNFGTEVF 531
++ A V ST D SS S D+ APG+ I + +PT+ S
Sbjct: 173 AYYSNAIAVASTDQNDNKSSFSTYGSVV-----DVAAPGSWIYSTYPTSTYAS------- 220
Query: 532 NNFNLIDGTSMSCPHVAGVAALLKGAHNGWSPSSIRSAIMTTSDILDNT 580
+ GTSM+ PHVAGVA LL A G S S+IR+AI T+D + T
Sbjct: 221 -----LSGTSMATPHVAGVAGLL--ASQGRSASNIRAAIENTADKISGT 262
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.317 0.133 0.394
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 89,793,314
Number of Sequences: 164201
Number of extensions: 3913807
Number of successful extensions: 10597
Number of sequences better than 10.0: 85
Number of HSP's better than 10.0 without gapping: 39
Number of HSP's successfully gapped in prelim test: 47
Number of HSP's that attempted gapping in prelim test: 10361
Number of HSP's gapped (non-prelim): 197
length of query: 762
length of database: 59,974,054
effective HSP length: 118
effective length of query: 644
effective length of database: 40,598,336
effective search space: 26145328384
effective search space used: 26145328384
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 70 (31.6 bits)
Medicago: description of AC144727.4


