

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144724.7 - phase: 0 /pseudo
(1419 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
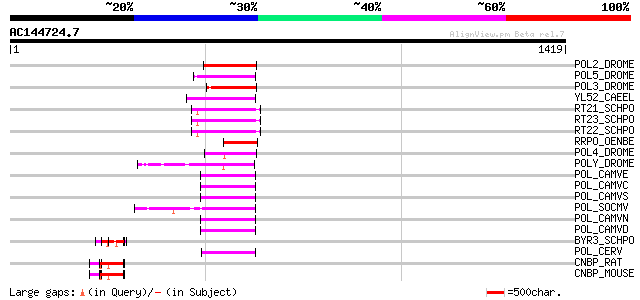
Score E
Sequences producing significant alignments: (bits) Value
POL2_DROME (P20825) Retrovirus-related Pol polyprotein from tran... 111 2e-23
POL5_DROME (Q8I7P9) Retrovirus-related Pol polyprotein from tran... 104 2e-21
POL3_DROME (P04323) Retrovirus-related Pol polyprotein from tran... 102 7e-21
YL52_CAEEL (P34431) Hypothetical protein F44E2.2 in chromosome III 100 2e-20
RT21_SCHPO (Q05654) Retrotransposable element Tf2 155 kDa protei... 97 2e-19
RT23_SCHPO (Q9UR07) Retrotransposable element Tf2 155 kDa protei... 96 5e-19
RT22_SCHPO (Q9C0R2) Retrotransposable element Tf2 155 kDa protei... 96 5e-19
RRPO_OENBE (P31843) RNA-directed DNA polymerase homolog (Reverse... 94 3e-18
POL4_DROME (P10394) Retrovirus-related Pol polyprotein from tran... 92 8e-18
POLY_DROME (P10401) Retrovirus-related Pol polyprotein from tran... 88 1e-16
POL_CAMVE (Q02964) Enzymatic polyprotein [Contains: Aspartic pro... 84 2e-15
POL_CAMVC (P03555) Enzymatic polyprotein [Contains: Aspartic pro... 84 2e-15
POL_CAMVS (P03554) Enzymatic polyprotein [Contains: Aspartic pro... 84 3e-15
POL_SOCMV (P15629) Enzymatic polyprotein [Contains: Aspartic pro... 84 4e-15
POL_CAMVN (Q00962) Enzymatic polyprotein [Contains: Aspartic pro... 82 1e-14
POL_CAMVD (P03556) Enzymatic polyprotein [Contains: Aspartic pro... 81 2e-14
BYR3_SCHPO (P36627) Cellular nucleic acid binding protein homolog 77 3e-13
POL_CERV (P05400) Enzymatic polyprotein [Contains: Aspartic prot... 74 3e-12
CNBP_RAT (P62634) Cellular nucleic acid binding protein (CNBP) (... 72 1e-11
CNBP_MOUSE (P53996) Cellular nucleic acid binding protein (CNBP)... 72 1e-11
>POL2_DROME (P20825) Retrovirus-related Pol polyprotein from
transposon 297 [Contains: Protease (EC 3.4.23.-);
Reverse transcriptase (EC 2.7.7.49); Endonuclease]
Length = 1059
Score = 111 bits (277), Expect = 2e-23
Identities = 54/139 (38%), Positives = 86/139 (61%), Gaps = 5/139 (3%)
Query: 497 SPIWITPYPMSASELGELKKQLEELLEKQFIRPSVSPWGAPVLLVKKKDGS-----VRLC 551
SPI+ YP++ + E++ Q++E+L + IR S SP+ +P +V KK + R+
Sbjct: 205 SPIYSKQYPLAQTHEIEVENQVQEMLNQGLIRESNSPYNSPTWVVPKKPDASGANKYRVV 264
Query: 552 VDYRQLNKVTIKNRYPLLRIDDLMDQLVGAEVFSKIDFRSGYHHNRVKAEDISKTVFRTR 611
+DYR+LN++TI +RYP+ +D+++ +L + F+ ID G+H + E ISKT F T+
Sbjct: 265 IDYRKLNEITIPDRYPIPNMDEILGKLGKCQYFTTIDLAKGFHQIEMDEESISKTAFSTK 324
Query: 612 YGHYEYFVMQFGVYNVPGT 630
GHYEY M FG+ N P T
Sbjct: 325 SGHYEYLRMPFGLRNAPAT 343
>POL5_DROME (Q8I7P9) Retrovirus-related Pol polyprotein from
transposon opus [Contains: Protease (EC 3.4.23.-);
Reverse transcriptase (EC 2.7.7.49); Endonuclease]
Length = 1003
Score = 104 bits (259), Expect = 2e-21
Identities = 62/169 (36%), Positives = 94/169 (54%), Gaps = 10/169 (5%)
Query: 469 IVQEFPEDITELPLE-REVEFAIDL---VPGMSPIWITPYPMSASELGELKKQLEELLEK 524
++ EFP I E PL VE A+ PI+ YP + GE+++Q++ELL+
Sbjct: 91 LLGEFPR-IFEPPLSGMSVETAVKAEIRTNTQDPIYAKSYPYPVNMRGEVERQIDELLQD 149
Query: 525 QFIRPSVSPWGAPVLLVKKK-----DGSVRLCVDYRQLNKVTIKNRYPLLRIDDLMDQLV 579
IRPS SP+ +P+ +V KK + R+ VD+++LN VTI + YP+ I+ + L
Sbjct: 150 GIIRPSNSPYNSPIWIVPKKPKPNGEKQYRMVVDFKRLNTVTIPDTYPIPDINATLASLG 209
Query: 580 GAEVFSKIDFRSGYHHNRVKAEDISKTVFRTRYGHYEYFVMQFGVYNVP 628
A+ F+ +D SG+H +K DI KT F T G YE+ + FG+ N P
Sbjct: 210 NAKYFTTLDLTSGFHQIHMKESDIPKTAFSTLNGKYEFLRLPFGLKNAP 258
>POL3_DROME (P04323) Retrovirus-related Pol polyprotein from
transposon 17.6 [Contains: Protease (EC 3.4.23.-);
Reverse transcriptase (EC 2.7.7.49); Endonuclease]
Length = 1058
Score = 102 bits (254), Expect = 7e-21
Identities = 49/132 (37%), Positives = 84/132 (63%), Gaps = 7/132 (5%)
Query: 504 YPMSASELGELKKQLEELLEKQFIRPSVSPWGAPVLLV-KKKDGS----VRLCVDYRQLN 558
YP + + E++ Q++++L + IR S SP+ +P+ +V KK+D S R+ +DYR+LN
Sbjct: 215 YPQAYEQ--EVESQIQDMLNQGIIRTSNSPYNSPIWVVPKKQDASGKQKFRIVIDYRKLN 272
Query: 559 KVTIKNRYPLLRIDDLMDQLVGAEVFSKIDFRSGYHHNRVKAEDISKTVFRTRYGHYEYF 618
++T+ +R+P+ +D+++ +L F+ ID G+H + E +SKT F T++GHYEY
Sbjct: 273 EITVGDRHPIPNMDEILGKLGRCNYFTTIDLAKGFHQIEMDPESVSKTAFSTKHGHYEYL 332
Query: 619 VMQFGVYNVPGT 630
M FG+ N P T
Sbjct: 333 RMPFGLKNAPAT 344
>YL52_CAEEL (P34431) Hypothetical protein F44E2.2 in chromosome III
Length = 2186
Score = 100 bits (250), Expect = 2e-20
Identities = 57/184 (30%), Positives = 100/184 (53%), Gaps = 8/184 (4%)
Query: 453 FASLKENSEKGVGD----LPIVQEFPEDIT----ELPLEREVEFAIDLVPGMSPIWITPY 504
F ++ E+ ++ GD ++++F + EL E I+L G PI P
Sbjct: 889 FTTICEHLKRENGDDRKIWDVIEQFQDVFAISDDELGRNSGTECVIELKEGAEPIRQKPR 948
Query: 505 PMSASELGELKKQLEELLEKQFIRPSVSPWGAPVLLVKKKDGSVRLCVDYRQLNKVTIKN 564
P+ + E++K ++++L ++ IR S SPW +PV+LVKKKDGS+R+C+DYR++NKV N
Sbjct: 949 PIPLALKPEIRKMIQKMLNQKVIRESKSPWSSPVVLVKKKDGSIRMCIDYRKVNKVVKNN 1008
Query: 565 RYPLLRIDDLMDQLVGAEVFSKIDFRSGYHHNRVKAEDISKTVFRTRYGHYEYFVMQFGV 624
+PL I+ + L G ++++ D +G+ + + T F +E+ V+ FG+
Sbjct: 1009 AHPLPNIEATLQSLAGKKLYTVFDMIAGFWQIPLDEKSKEITAFAIGSELFEWNVLPFGL 1068
Query: 625 YNVP 628
P
Sbjct: 1069 VISP 1072
Score = 34.3 bits (77), Expect = 2.5
Identities = 22/83 (26%), Positives = 33/83 (39%), Gaps = 15/83 (18%)
Query: 234 KCYRCGTLGHYANDCKKDIS------CHKCGKAGHKAAECKDVARDVTCYNCGEKGHIST 287
+C C G + C K C +C ++G A C + ++ C+ C E GHI+
Sbjct: 544 RCSDCQQRGWHMFWCSKKSKDNASQKCDECQQSGWHMASCFKL-KNRACFRCNEMGHIAW 602
Query: 288 KCTKPKKAAGKVFALNAEEVEQP 310
C K + N E E P
Sbjct: 603 NCPKKNE--------NTSEKEAP 617
>RT21_SCHPO (Q05654) Retrotransposable element Tf2 155 kDa protein
type 1
Length = 1333
Score = 97.4 bits (241), Expect = 2e-19
Identities = 59/182 (32%), Positives = 98/182 (53%), Gaps = 12/182 (6%)
Query: 466 DLPIVQEFPEDIT------ELPLE-REVEFAIDLVPGMSPIWITPYPMSASELGELKKQL 518
+LP + + +DIT +LP + +EF ++L + I YP+ ++ + ++
Sbjct: 373 ELPDIYKEFKDITAETNTEKLPKPIKGLEFEVELTQENYRLPIRNYPLPPGKMQAMNDEI 432
Query: 519 EELLEKQFIRPSVSPWGAPVLLVKKKDGSVRLCVDYRQLNKVTIKNRYPLLRIDDLMDQL 578
+ L+ IR S + PV+ V KK+G++R+ VDY+ LNK N YPL I+ L+ ++
Sbjct: 433 NQGLKSGIIRESKAINACPVMFVPKKEGTLRMVVDYKPLNKYVKPNIYPLPLIEQLLAKI 492
Query: 579 VGAEVFSKIDFRSGYHHNRVKAEDISKTVFRTRYGHYEYFVMQFGVYNVPGTSWNI*IEY 638
G+ +F+K+D +S YH RV+ D K FR G +EY VM +G+ P +Y
Sbjct: 493 QGSTIFTKLDLKSAYHLIRVRKGDEHKLAFRCPRGVFEYLVMPYGISTAPAH-----FQY 547
Query: 639 FI 640
FI
Sbjct: 548 FI 549
>RT23_SCHPO (Q9UR07) Retrotransposable element Tf2 155 kDa protein
type 3
Length = 1333
Score = 96.3 bits (238), Expect = 5e-19
Identities = 59/182 (32%), Positives = 98/182 (53%), Gaps = 12/182 (6%)
Query: 466 DLPIVQEFPEDIT------ELPLE-REVEFAIDLVPGMSPIWITPYPMSASELGELKKQL 518
+LP + + +DIT +LP + +EF ++L + I YP+ ++ + ++
Sbjct: 373 ELPDIYKEFKDITAETNTEKLPKPIKGLEFEVELTQENYRLPIRNYPLPPGKMQAMNDEI 432
Query: 519 EELLEKQFIRPSVSPWGAPVLLVKKKDGSVRLCVDYRQLNKVTIKNRYPLLRIDDLMDQL 578
+ L+ IR S + PV+ V KK+G++R+ VDY+ LNK N YPL I+ L+ ++
Sbjct: 433 NQGLKSGIIRESKAINACPVMFVPKKEGTLRMVVDYKPLNKYVKPNIYPLPLIEQLLAKI 492
Query: 579 VGAEVFSKIDFRSGYHHNRVKAEDISKTVFRTRYGHYEYFVMQFGVYNVPGTSWNI*IEY 638
G+ +F+K+D +S YH RV+ D K FR G +EY VM +G+ P +Y
Sbjct: 493 QGSTIFTKLDLKSAYHLIRVRKGDEHKLAFRCPRGVFEYLVMPYGISIAPAH-----FQY 547
Query: 639 FI 640
FI
Sbjct: 548 FI 549
>RT22_SCHPO (Q9C0R2) Retrotransposable element Tf2 155 kDa protein
type 2
Length = 1333
Score = 96.3 bits (238), Expect = 5e-19
Identities = 59/182 (32%), Positives = 98/182 (53%), Gaps = 12/182 (6%)
Query: 466 DLPIVQEFPEDIT------ELPLE-REVEFAIDLVPGMSPIWITPYPMSASELGELKKQL 518
+LP + + +DIT +LP + +EF ++L + I YP+ ++ + ++
Sbjct: 373 ELPDIYKEFKDITAETNTEKLPKPIKGLEFEVELTQENYRLPIRNYPLPPGKMQAMNDEI 432
Query: 519 EELLEKQFIRPSVSPWGAPVLLVKKKDGSVRLCVDYRQLNKVTIKNRYPLLRIDDLMDQL 578
+ L+ IR S + PV+ V KK+G++R+ VDY+ LNK N YPL I+ L+ ++
Sbjct: 433 NQGLKSGIIRESKAINACPVMFVPKKEGTLRMVVDYKPLNKYVKPNIYPLPLIEQLLAKI 492
Query: 579 VGAEVFSKIDFRSGYHHNRVKAEDISKTVFRTRYGHYEYFVMQFGVYNVPGTSWNI*IEY 638
G+ +F+K+D +S YH RV+ D K FR G +EY VM +G+ P +Y
Sbjct: 493 QGSTIFTKLDLKSAYHLIRVRKGDEHKLAFRCPRGVFEYLVMPYGISIAPAH-----FQY 547
Query: 639 FI 640
FI
Sbjct: 548 FI 549
>RRPO_OENBE (P31843) RNA-directed DNA polymerase homolog (Reverse
transcriptase homolog)
Length = 142
Score = 93.6 bits (231), Expect = 3e-18
Identities = 45/88 (51%), Positives = 59/88 (66%)
Query: 547 SVRLCVDYRQLNKVTIKNRYPLLRIDDLMDQLVGAEVFSKIDFRSGYHHNRVKAEDISKT 606
S+R+C+DYR L KVTIKN+YP+ R+DDL D+L A F+K+D RSGY R+ D KT
Sbjct: 5 SLRMCIDYRALTKVTIKNKYPIPRVDDLFDRLAQATWFTKLDLRSGYWQVRIAKGDEPKT 64
Query: 607 VFRTRYGHYEYFVMQFGVYNVPGTSWNI 634
TRYG +E+ VM FG+ N T N+
Sbjct: 65 TCVTRYGSFEFRVMPFGLTNALATFCNL 92
>POL4_DROME (P10394) Retrovirus-related Pol polyprotein from
transposon 412 [Contains: Protease (EC 3.4.23.-);
Reverse transcriptase (EC 2.7.7.49); Endonuclease]
Length = 1237
Score = 92.4 bits (228), Expect = 8e-18
Identities = 48/139 (34%), Positives = 80/139 (57%), Gaps = 6/139 (4%)
Query: 498 PIWITPYPMSASELGELKKQLEELLEKQFIRPSVSPWGAPVLLVKKKDG------SVRLC 551
P++ Y S++ E++ Q+++L++ + + PSVS + +P+LLV KK RL
Sbjct: 314 PVYTKNYRSPHSQVEEIQAQVQKLIKDKIVEPSVSQYNSPLLLVPKKSSPNSDKKKWRLV 373
Query: 552 VDYRQLNKVTIKNRYPLLRIDDLMDQLVGAEVFSKIDFRSGYHHNRVKAEDISKTVFRTR 611
+DYRQ+NK + +++PL RIDD++DQL A+ FS +D SG+H + T F T
Sbjct: 374 IDYRQINKKLLADKFPLPRIDDILDQLGRAKYFSCLDLMSGFHQIELDEGSRDITSFSTS 433
Query: 612 YGHYEYFVMQFGVYNVPGT 630
G Y + + FG+ P +
Sbjct: 434 NGSYRFTRLPFGLKIAPNS 452
>POLY_DROME (P10401) Retrovirus-related Pol polyprotein from
transposon gypsy [Contains: Reverse transcriptase (EC
2.7.7.49); Endonuclease]
Length = 1035
Score = 88.2 bits (217), Expect = 1e-16
Identities = 85/307 (27%), Positives = 135/307 (43%), Gaps = 26/307 (8%)
Query: 328 IIDIGATHSFISVSCVKRLKLVVTPLLRGMVIDTPARGSVTTSFMCAKCPVNFGNVDFEL 387
+ID A ++I VK LK V+ P+ + + GS C + F L
Sbjct: 27 LIDTDAAKNYIRP--VKELKNVM-PVASPFSVSS-IHGSTEIKHKCLMKVFKHISPFFLL 82
Query: 388 DLVCLPLKHMDVIFGMDWLLFFGVSINCLTKSVTFSKPVEKLDRKFLTAEQVKKSLDGEA 447
D L D I G+D L GV +N S+ + EKL + + V + +
Sbjct: 83 D----SLNAFDAIIGLDLLTQAGVKLNLAEDSLEYQGIAEKLH--YFSCPSVNFTDVNDI 136
Query: 448 CVFMMFASLKENSEKGVGDLPIVQE--FPEDITELPLEREVEFAIDLVPGMSPIWITPYP 505
V ++ +K D I ++ F LP V I V P++ YP
Sbjct: 137 VV-------PDSVKKEFKDTIIRRKKAFSTTNEALPFNTAVTATIRTVDN-EPVYSRAYP 188
Query: 506 MSASELGELKKQLEELLEKQFIRPSVSPWGAPVLLVKKK------DGSVRLCVDYRQLNK 559
+ ++++LL+ IRPS SP+ +P +V KK + + RL +D+R+LN+
Sbjct: 189 TLMGVSDFVNNEVKQLLKDGIIRPSRSPYNSPTWVVDKKGTDAFGNPNKRLVIDFRKLNE 248
Query: 560 VTIKNRYPLLRIDDLMDQLVGAEVFSKIDFRSGYHHNRVKAEDISKTVFRTRYGHYEYFV 619
TI +RYP+ I ++ L A+ F+ +D +SGYH + D KT F G YE+
Sbjct: 249 KTIPDRYPMPSIPMILANLGKAKFFTTLDLKSGYHQIYLAEHDREKTSFSVNGGKYEFCR 308
Query: 620 MQFGVYN 626
+ FG+ N
Sbjct: 309 LPFGLRN 315
>POL_CAMVE (Q02964) Enzymatic polyprotein [Contains: Aspartic
protease (EC 3.4.23.-); Endonuclease; Reverse
transcriptase (EC 2.7.7.49)]
Length = 679
Score = 84.3 bits (207), Expect = 2e-15
Identities = 50/144 (34%), Positives = 76/144 (52%), Gaps = 4/144 (2%)
Query: 489 AIDLVPGMSPIWITPYPMSASELGELKKQLEELLEKQFIRPSVSPWGAPVLLV----KKK 544
+I L I + P S + E KQ++ELL+ + I+PS SP AP LV +K+
Sbjct: 237 SIKLSDPSKAIKVKPMKYSPMDREEFDKQIKELLDLKVIKPSKSPHMAPAFLVNNEAEKR 296
Query: 545 DGSVRLCVDYRQLNKVTIKNRYPLLRIDDLMDQLVGAEVFSKIDFRSGYHHNRVKAEDIS 604
G R+ V+Y+ +NK TI + Y L D+L+ + G ++FS D +SG+ + E
Sbjct: 297 RGKKRMVVNYKAMNKATIGDAYNLPNKDELLTLIRGKKIFSSFDCKSGFWQVLLDQESRP 356
Query: 605 KTVFRTRYGHYEYFVMQFGVYNVP 628
T F GHYE+ V+ FG+ P
Sbjct: 357 LTAFTCPQGHYEWNVVPFGLKQAP 380
>POL_CAMVC (P03555) Enzymatic polyprotein [Contains: Aspartic
protease (EC 3.4.23.-); Endonuclease; Reverse
transcriptase (EC 2.7.7.49)]
Length = 679
Score = 84.3 bits (207), Expect = 2e-15
Identities = 50/144 (34%), Positives = 76/144 (52%), Gaps = 4/144 (2%)
Query: 489 AIDLVPGMSPIWITPYPMSASELGELKKQLEELLEKQFIRPSVSPWGAPVLLV----KKK 544
+I L I + P S + E KQ++ELL+ + I+PS SP AP LV +K+
Sbjct: 237 SIKLSDPSKAIKVKPMKYSPMDREEFDKQIKELLDLKVIKPSKSPHMAPAFLVNNEAEKR 296
Query: 545 DGSVRLCVDYRQLNKVTIKNRYPLLRIDDLMDQLVGAEVFSKIDFRSGYHHNRVKAEDIS 604
G R+ V+Y+ +NK TI + Y L D+L+ + G ++FS D +SG+ + E
Sbjct: 297 RGKKRMVVNYKAMNKATIGDAYNLPNKDELLTLIRGKKIFSSFDCKSGFWQVLLDQESRP 356
Query: 605 KTVFRTRYGHYEYFVMQFGVYNVP 628
T F GHYE+ V+ FG+ P
Sbjct: 357 LTAFTCPQGHYEWNVVPFGLKQAP 380
>POL_CAMVS (P03554) Enzymatic polyprotein [Contains: Aspartic
protease (EC 3.4.23.-); Endonuclease; Reverse
transcriptase (EC 2.7.7.49)]
Length = 679
Score = 84.0 bits (206), Expect = 3e-15
Identities = 49/144 (34%), Positives = 76/144 (52%), Gaps = 4/144 (2%)
Query: 489 AIDLVPGMSPIWITPYPMSASELGELKKQLEELLEKQFIRPSVSPWGAPVLLV----KKK 544
+I L I + P S + E KQ++ELL+ + I+PS SP AP LV +K+
Sbjct: 237 SIKLSDPSKAIKVKPMKYSPMDREEFDKQIKELLDLKVIKPSKSPHMAPAFLVNNEAEKR 296
Query: 545 DGSVRLCVDYRQLNKVTIKNRYPLLRIDDLMDQLVGAEVFSKIDFRSGYHHNRVKAEDIS 604
G R+ V+Y+ +NK T+ + Y L D+L+ + G ++FS D +SG+ + E
Sbjct: 297 RGKKRMVVNYKAMNKATVGDAYNLPNKDELLTLIRGKKIFSSFDCKSGFWQVLLDQESRP 356
Query: 605 KTVFRTRYGHYEYFVMQFGVYNVP 628
T F GHYE+ V+ FG+ P
Sbjct: 357 LTAFTCPQGHYEWNVVPFGLKQAP 380
>POL_SOCMV (P15629) Enzymatic polyprotein [Contains: Aspartic
protease (EC 3.4.23.-); Endonuclease; Reverse
transcriptase (EC 2.7.7.49)]
Length = 692
Score = 83.6 bits (205), Expect = 4e-15
Identities = 80/325 (24%), Positives = 150/325 (45%), Gaps = 28/325 (8%)
Query: 320 INSTHLIGIIDIGATHSFISVSCVKRLKLVVTPLLRGMVIDTPARGSVTTSFMCAKCPVN 379
I + + ID GAT F +++ P + ++I ++ + + + +
Sbjct: 26 IGKRNFLAYIDTGATLCFGKRKISNNWEILKQP--KEIIIADKSKHYIREAI--SNVFLK 81
Query: 380 FGNVDFELDLVCLPLKHMDVIFGMDWLLFFGVSINCLT------KSVTFSKPVEKLDRKF 433
N +F + ++ L +D+I G ++L + I L K++ K + + K
Sbjct: 82 IENKEFLIPIIYLHDSGLDLIIGNNFLKLYQPFIQRLETIELRWKNLNNPKESQMISTKI 141
Query: 434 LTAEQVKKSLDGEACV----FMMFASLKENSEKGVGDLPIVQEFPEDITELPLEREVEFA 489
LT +V K + + ++ F +++E E+ + E P D T+ +E
Sbjct: 142 LTKNEVLKLSFEKIHICLEKYLFFKTIEEQLEE------VCSEHPLDETKNKNGLLIE-- 193
Query: 490 IDLVPGMSPIWITP-YPMSASELGELKKQLEELLEKQFIRPSVSPWGAPVLLVKK----K 544
I L + I +T P + ++ E K++ E+LL+K IR S SP AP V+ K
Sbjct: 194 IRLKDPLQEINVTNRIPYTIRDVQEFKEECEDLLKKGLIRESQSPHSAPAFYVENHNEIK 253
Query: 545 DGSVRLCVDYRQLNKVTIKNRYPLLRIDDLMDQLVGAEVFSKIDFRSGYHHNRVKAEDIS 604
G R+ ++Y+++N+ TI + Y L R D +++++ G+ FS +D +SGY+ R+
Sbjct: 254 RGKRRMVINYKKMNEATIGDSYKLPRKDFILEKIKGSLWFSSLDAKSGYYQLRLHENTKP 313
Query: 605 KTVFR-TRYGHYEYFVMQFGVYNVP 628
T F HYE+ V+ FG+ P
Sbjct: 314 LTAFSCPPQKHYEWNVLSFGLKQAP 338
>POL_CAMVN (Q00962) Enzymatic polyprotein [Contains: Aspartic
protease (EC 3.4.23.-); Endonuclease; Reverse
transcriptase (EC 2.7.7.49)]
Length = 680
Score = 81.6 bits (200), Expect = 1e-14
Identities = 48/144 (33%), Positives = 75/144 (51%), Gaps = 4/144 (2%)
Query: 489 AIDLVPGMSPIWITPYPMSASELGELKKQLEELLEKQFIRPSVSPWGAPVLLVKKKD--- 545
+I L I + P S + E KQ++ELL+ + I+PS SP AP LV +
Sbjct: 238 SIKLSDPSKAIKVKPMKYSPMDREEFDKQIKELLDLKVIKPSKSPHMAPAFLVNNEAENG 297
Query: 546 -GSVRLCVDYRQLNKVTIKNRYPLLRIDDLMDQLVGAEVFSKIDFRSGYHHNRVKAEDIS 604
G+ R+ V+Y+ +NK T+ + Y L D+L+ + G ++FS D +SG+ + E
Sbjct: 298 RGNKRMVVNYKAMNKATVGDAYNLPNKDELLTLIRGKKIFSSFDCKSGFWQVLLDQESRP 357
Query: 605 KTVFRTRYGHYEYFVMQFGVYNVP 628
T F GHYE+ V+ FG+ P
Sbjct: 358 LTAFTCPQGHYEWNVVPFGLKQAP 381
>POL_CAMVD (P03556) Enzymatic polyprotein [Contains: Aspartic
protease (EC 3.4.23.-); Endonuclease; Reverse
transcriptase (EC 2.7.7.49)]
Length = 674
Score = 81.3 bits (199), Expect = 2e-14
Identities = 48/144 (33%), Positives = 75/144 (51%), Gaps = 4/144 (2%)
Query: 489 AIDLVPGMSPIWITPYPMSASELGELKKQLEELLEKQFIRPSVSPWGAPVLLV----KKK 544
+I L I + P S + E KQ++ELL+ + I+PS SP AP LV +K+
Sbjct: 232 SIKLSDPSKAIKVKPMKYSPMDREEFDKQIKELLDLKVIKPSKSPHMAPAFLVNNEAEKR 291
Query: 545 DGSVRLCVDYRQLNKVTIKNRYPLLRIDDLMDQLVGAEVFSKIDFRSGYHHNRVKAEDIS 604
G R+ V+Y+ +NK T+ + Y D+L+ + G ++FS D +SG+ + E
Sbjct: 292 RGKKRMVVNYKAMNKATVGDAYNPPNKDELLTLIRGKKIFSSFDCKSGFWQVLLDQESRP 351
Query: 605 KTVFRTRYGHYEYFVMQFGVYNVP 628
T F GHYE+ V+ FG+ P
Sbjct: 352 LTAFTCPQGHYEWNVVPFGLKQAP 375
>BYR3_SCHPO (P36627) Cellular nucleic acid binding protein homolog
Length = 179
Score = 77.0 bits (188), Expect = 3e-13
Identities = 28/73 (38%), Positives = 42/73 (57%)
Query: 220 REVGGGSKPSLADIKCYRCGTLGHYANDCKKDISCHKCGKAGHKAAECKDVARDVTCYNC 279
++ GG +++ CY CG+ GH A DC + C+ CGK GH++ EC+ + CY C
Sbjct: 103 QQSGGRFGGHRSNMNCYACGSYGHQARDCTMGVKCYSCGKIGHRSFECQQASDGQLCYKC 162
Query: 280 GEKGHISTKCTKP 292
+ GHI+ CT P
Sbjct: 163 NQPGHIAVNCTSP 175
Score = 68.9 bits (167), Expect = 9e-11
Identities = 25/56 (44%), Positives = 34/56 (60%)
Query: 234 KCYRCGTLGHYANDCKKDISCHKCGKAGHKAAECKDVARDVTCYNCGEKGHISTKC 289
+CY CG GH A +C K C+ C + GHKA+EC + ++ TCY CG GH+ C
Sbjct: 18 RCYNCGENGHQARECTKGSICYNCNQTGHKASECTEPQQEKTCYACGTAGHLVRDC 73
Score = 57.0 bits (136), Expect = 4e-07
Identities = 28/84 (33%), Positives = 40/84 (47%), Gaps = 19/84 (22%)
Query: 235 CYRCGTLGHYANDC------KKDISCHKCGKAGHKAAECKDVAR-----------DVTCY 277
CY CGT GH DC ++ C+KCG+ GH A +C+ + ++ CY
Sbjct: 60 CYACGTAGHLVRDCPSSPNPRQGAECYKCGRVGHIARDCRTNGQQSGGRFGGHRSNMNCY 119
Query: 278 NCGEKGHISTKCTKPKK--AAGKV 299
CG GH + CT K + GK+
Sbjct: 120 ACGSYGHQARDCTMGVKCYSCGKI 143
Score = 47.4 bits (111), Expect = 3e-04
Identities = 17/41 (41%), Positives = 28/41 (67%), Gaps = 3/41 (7%)
Query: 254 CHKCGKAGHKAAECKDVARDVTCYNCGEKGHISTKCTKPKK 294
C+ CG+ GH+A EC + CYNC + GH +++CT+P++
Sbjct: 19 CYNCGENGHQAREC---TKGSICYNCNQTGHKASECTEPQQ 56
>POL_CERV (P05400) Enzymatic polyprotein [Contains: Aspartic
protease (EC 3.4.23.-); Endonuclease; Reverse
transcriptase (EC 2.7.7.49)]
Length = 659
Score = 73.9 bits (180), Expect = 3e-12
Identities = 43/143 (30%), Positives = 76/143 (53%), Gaps = 4/143 (2%)
Query: 490 IDLVPGMSPIWITPYPMSASELGELKKQLEELLEKQFIRPSVSPWGAPVLLVK----KKD 545
I+L+ + + + P S S+ E +Q++ELLE + I+PS S +P LV+ ++
Sbjct: 220 IELIDPKTVVKVKPMSYSPSDREEFDRQIKELLELKVIKPSKSTHMSPAFLVENEAERRR 279
Query: 546 GSVRLCVDYRQLNKVTIKNRYPLLRIDDLMDQLVGAEVFSKIDFRSGYHHNRVKAEDISK 605
G R+ V+Y+ +NK T + + L D+L+ + G +++S D +SG + E
Sbjct: 280 GKKRMVVNYKAMNKATKGDAHNLPNKDELLTLVRGKKIYSSFDCKSGLWQVLLDKESQLL 339
Query: 606 TVFRTRYGHYEYFVMQFGVYNVP 628
T F GHY++ V+ FG+ P
Sbjct: 340 TAFTCPQGHYQWNVVPFGLKQAP 362
>CNBP_RAT (P62634) Cellular nucleic acid binding protein (CNBP)
(Zinc finger protein 9)
Length = 177
Score = 71.6 bits (174), Expect = 1e-11
Identities = 35/97 (36%), Positives = 51/97 (52%), Gaps = 11/97 (11%)
Query: 203 GKGHGVGKPYNKDKRKKREV--GGGSKPSLA-------DIKCYRCGTLGHYANDCKKDIS 253
G+G + K + KR++ + G LA + KCY CG GH DC K +
Sbjct: 78 GRGGHIAKDCKEPKREREQCCYNCGKPGHLARDCDHADEQKCYSCGEFGHIQKDCTK-VK 136
Query: 254 CHKCGKAGHKAAECKDVARDVTCYNCGEKGHISTKCT 290
C++CG+ GH A C + +V CY CGE GH++ +CT
Sbjct: 137 CYRCGETGHVAINCSKTS-EVNCYRCGESGHLARECT 172
Score = 69.3 bits (168), Expect = 7e-11
Identities = 33/69 (47%), Positives = 40/69 (57%), Gaps = 6/69 (8%)
Query: 230 LADIKCYRCGTLGHYANDCK-----KDISCHKCGKAGHKAAECKDVARDVTCYNCGEKGH 284
L + CY CG GH A DCK ++ C+ CGK GH A +C D A + CY+CGE GH
Sbjct: 69 LQEDACYNCGRGGHIAKDCKEPKREREQCCYNCGKPGHLARDC-DHADEQKCYSCGEFGH 127
Query: 285 ISTKCTKPK 293
I CTK K
Sbjct: 128 IQKDCTKVK 136
Score = 68.9 bits (167), Expect = 9e-11
Identities = 31/64 (48%), Positives = 42/64 (65%), Gaps = 4/64 (6%)
Query: 229 SLADIKCYRCGTLGHYANDCK-KDISCHKCGKAGHKAAECKDV--ARDVTCYNCGEKGHI 285
SL DI CYRCG GH A DC ++ +C+ CG+ GH A +CK+ R+ CYNCG+ GH+
Sbjct: 49 SLPDI-CYRCGESGHLAKDCDLQEDACYNCGRGGHIAKDCKEPKREREQCCYNCGKPGHL 107
Query: 286 STKC 289
+ C
Sbjct: 108 ARDC 111
Score = 55.5 bits (132), Expect = 1e-06
Identities = 29/91 (31%), Positives = 43/91 (46%), Gaps = 33/91 (36%)
Query: 234 KCYRCGTLGHYANDCKK------------------------------DISCHKCGKAGHK 263
+C++CG GH+A +C DI C++CG++GH
Sbjct: 5 ECFKCGRSGHWARECPTGGGRGRGMRSRGRGGFTSDRGFQFVSSSLPDI-CYRCGESGHL 63
Query: 264 AAECKDVARDVTCYNCGEKGHISTKCTKPKK 294
A +C D+ D CYNCG GHI+ C +PK+
Sbjct: 64 AKDC-DLQEDA-CYNCGRGGHIAKDCKEPKR 92
>CNBP_MOUSE (P53996) Cellular nucleic acid binding protein (CNBP)
(Zinc finger protein 9)
Length = 178
Score = 71.6 bits (174), Expect = 1e-11
Identities = 35/97 (36%), Positives = 51/97 (52%), Gaps = 11/97 (11%)
Query: 203 GKGHGVGKPYNKDKRKKREV--GGGSKPSLA-------DIKCYRCGTLGHYANDCKKDIS 253
G+G + K + KR++ + G LA + KCY CG GH DC K +
Sbjct: 79 GRGGHIAKDCKEPKREREQCCYNCGKPGHLARDCDHADEQKCYSCGEFGHIQKDCTK-VK 137
Query: 254 CHKCGKAGHKAAECKDVARDVTCYNCGEKGHISTKCT 290
C++CG+ GH A C + +V CY CGE GH++ +CT
Sbjct: 138 CYRCGETGHVAINCSKTS-EVNCYRCGESGHLARECT 173
Score = 70.9 bits (172), Expect = 2e-11
Identities = 32/65 (49%), Positives = 43/65 (65%), Gaps = 5/65 (7%)
Query: 229 SLADIKCYRCGTLGHYANDC--KKDISCHKCGKAGHKAAECKDV--ARDVTCYNCGEKGH 284
SL DI CYRCG GH A DC ++D +C+ CG+ GH A +CK+ R+ CYNCG+ GH
Sbjct: 49 SLPDI-CYRCGESGHLAKDCDLQEDEACYNCGRGGHIAKDCKEPKREREQCCYNCGKPGH 107
Query: 285 ISTKC 289
++ C
Sbjct: 108 LARDC 112
Score = 69.7 bits (169), Expect = 5e-11
Identities = 33/67 (49%), Positives = 39/67 (57%), Gaps = 6/67 (8%)
Query: 232 DIKCYRCGTLGHYANDCK-----KDISCHKCGKAGHKAAECKDVARDVTCYNCGEKGHIS 286
D CY CG GH A DCK ++ C+ CGK GH A +C D A + CY+CGE GHI
Sbjct: 72 DEACYNCGRGGHIAKDCKEPKREREQCCYNCGKPGHLARDC-DHADEQKCYSCGEFGHIQ 130
Query: 287 TKCTKPK 293
CTK K
Sbjct: 131 KDCTKVK 137
Score = 59.3 bits (142), Expect = 7e-08
Identities = 29/91 (31%), Positives = 43/91 (46%), Gaps = 32/91 (35%)
Query: 234 KCYRCGTLGHYANDCKK------------------------------DISCHKCGKAGHK 263
+C++CG GH+A +C DI C++CG++GH
Sbjct: 5 ECFKCGRSGHWARECPTGGGRGRGMRSRGRGGFTSDRGFQFVSSSLPDI-CYRCGESGHL 63
Query: 264 AAECKDVARDVTCYNCGEKGHISTKCTKPKK 294
A +C D+ D CYNCG GHI+ C +PK+
Sbjct: 64 AKDC-DLQEDEACYNCGRGGHIAKDCKEPKR 93
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.350 0.157 0.564
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 148,016,833
Number of Sequences: 164201
Number of extensions: 5867463
Number of successful extensions: 23151
Number of sequences better than 10.0: 229
Number of HSP's better than 10.0 without gapping: 130
Number of HSP's successfully gapped in prelim test: 100
Number of HSP's that attempted gapping in prelim test: 22337
Number of HSP's gapped (non-prelim): 542
length of query: 1419
length of database: 59,974,054
effective HSP length: 123
effective length of query: 1296
effective length of database: 39,777,331
effective search space: 51551420976
effective search space used: 51551420976
T: 11
A: 40
X1: 14 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 38 (21.9 bits)
S2: 72 (32.3 bits)
Medicago: description of AC144724.7


