

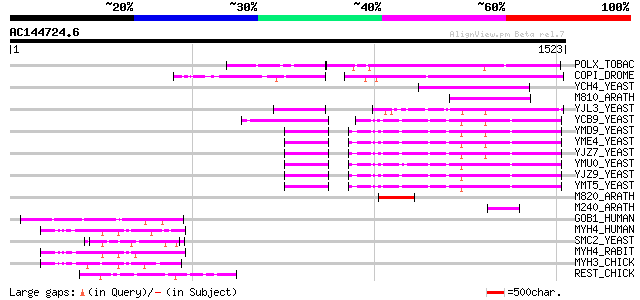
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144724.6 + phase: 0
(1523 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
POLX_TOBAC (P10978) Retrovirus-related Pol polyprotein from tran... 360 1e-98
COPI_DROME (P04146) Copia protein (Gag-int-pol protein) [Contain... 348 5e-95
YCH4_YEAST (P25600) Transposon Ty5-1 34.5 kDa hypothetical protein 181 2e-44
M810_ARATH (P92519) Hypothetical mitochondrial protein AtMg00810... 153 3e-36
YJL3_YEAST (P47024) Transposon Ty4 207.7 kDa hypothetical protein 138 1e-31
YCB9_YEAST (P25384) Transposon Ty2 protein B (Ty1-17 protein B) 127 2e-28
YMD9_YEAST (Q03434) Transposon Ty1 protein B 126 5e-28
YME4_YEAST (Q04711) Transposon Ty1 protein B 123 4e-27
YJZ7_YEAST (P47098) Transposon Ty1 protein B 122 8e-27
YMU0_YEAST (Q04670) Transposon Ty1 protein B 121 1e-26
YJZ9_YEAST (P47100) Transposon Ty1 protein B 118 1e-25
YMT5_YEAST (Q04214) Transposon Ty1 protein B 117 2e-25
M820_ARATH (P92520) Hypothetical mitochondrial protein AtMg00820... 99 9e-20
M240_ARATH (P93290) Hypothetical mitochondrial protein AtMg00240... 63 5e-09
GOB1_HUMAN (Q14789) Golgi autoantigen, golgin subfamily B member... 57 4e-07
MYH4_HUMAN (Q9Y623) Myosin heavy chain, skeletal muscle, fetal (... 54 4e-06
SMC2_YEAST (P38989) Structural maintenance of chromosome 2 (DA-b... 53 7e-06
MYH4_RABIT (Q28641) Myosin heavy chain, skeletal muscle, juvenile 51 2e-05
MYH3_CHICK (P02565) Myosin heavy chain, fast skeletal muscle, em... 51 3e-05
REST_CHICK (O42184) Restin (Cytoplasmic linker protein-170) (CLI... 50 5e-05
>POLX_TOBAC (P10978) Retrovirus-related Pol polyprotein from
transposon TNT 1-94 [Contains: Protease (EC 3.4.23.-);
Reverse transcriptase (EC 2.7.7.49); Endonuclease]
Length = 1328
Score = 360 bits (925), Expect = 1e-98
Identities = 224/676 (33%), Positives = 366/676 (54%), Gaps = 37/676 (5%)
Query: 870 VFGCYCYILNNKEKLGKFDPKSDKAIFLGYSTTSKGYRVYNLKTQTVEISMHIIFDEYDE 929
VFGC + KE+ K D KS IF+GY GYR+++ + V S ++F E +
Sbjct: 650 VFGCRAFAHVPKEQRTKLDDKSIPCIFIGYGDEEFGYRLWDPVKKKVIRSRDVVFRESEV 709
Query: 930 HSKPKENEDTE---------VPTLQNVPVQNTENTVEKEDDQNAQ-----DQSFQSPPRS 975
+ +E + +P+ N P + E+T ++ +Q Q +Q Q
Sbjct: 710 RTAADMSEKVKNGIIPNFVTIPSTSNNPT-SAESTTDEVSEQGEQPGEVIEQGEQLDEGV 768
Query: 976 WRMVGDHPTD------QIIGSTTDGVRTRLSFQDNNMAMISQMEPKSINEAIID---DSW 1026
+ +HPT + S V +R + + EP+S+ E + +
Sbjct: 769 EEV--EHPTQGEEQHQPLRRSERPRVESRRYPSTEYVLISDDREPESLKEVLSHPEKNQL 826
Query: 1027 IKAMKEELSQFERNKVWNLVPNNQDKTIIGTRWVFRNKLDEEGKVVRNKARLVAQGYNQQ 1086
+KAM+EE+ ++N + LV + K + +WVF+ K D + K+VR KARLV +G+ Q+
Sbjct: 827 MKAMQEEMESLQKNGTYKLVELPKGKRPLKCKWVFKLKKDGDCKLVRYKARLVVKGFEQK 886
Query: 1087 EGIDYDETFAPVARLEAIRILLAYAAHKSIKLFQMDVKSAFLNGFLNEEVYVSQPPGFIN 1146
+GID+DE F+PV ++ +IR +L+ AA +++ Q+DVK+AFL+G L EE+Y+ QP GF
Sbjct: 887 KGIDFDEIFSPVVKMTSIRTILSLAASLDLEVEQLDVKTAFLHGDLEEEIYMEQPEGFEV 946
Query: 1147 KEKPNHVFKLTKALYGLKQAPRAWYDRLSTFLIENGFSRGKIDTTL-FRKTHNTDLLIVQ 1205
K + V KL K+LYGLKQAPR WY + +F+ + + D + F++ + +I+
Sbjct: 947 AGKKHMVCKLNKSLYGLKQAPRQWYMKFDSFMKSQTYLKTYSDPCVYFKRFSENNFIILL 1006
Query: 1206 VYVDDIIFGATKIKMCEEFSNLMQSEFEMSMMGELGFFLGLQI--KQHSNGIFISQEKYV 1263
+YVDD++ + + + F+M +G LG++I ++ S +++SQEKY+
Sbjct: 1007 LYVDDMLIVGKDKGLIAKLKGDLSKSFDMKDLGPAQQILGMKIVRERTSRKLWLSQEKYI 1066
Query: 1264 KDILKKYKMNEAKIMSTPMHPSSSLDKDESGKSISEK------EYRGMIGSLLY-LTASR 1316
+ +L+++ M AK +STP+ L K ++ EK Y +GSL+Y + +R
Sbjct: 1067 ERVLERFNMKNAKPVSTPLAGHLKLSKKMCPTTVEEKGNMAKVPYSSAVGSLMYAMVCTR 1126
Query: 1317 PDIVFAVGLCARFQTCAKESHLTAVKRIFRYLVGTTDLGLWYRKGSSFDLVAYCDADYAG 1376
PDI AVG+ +RF + H AVK I RYL GTT L + GS L Y DAD AG
Sbjct: 1127 PDIAHAVGVVSRFLENPGKEHWEAVKWILRYLRGTTGDCLCF-GGSDPILKGYTDADMAG 1185
Query: 1377 DKVERKSTSGSCQFLGQALIGWSCRKQNTIALSTTEAEYVSAASCCSQILWVRNQLEDYS 1436
D RKS++G I W + Q +ALSTTEAEY++A +++W++ L++
Sbjct: 1186 DIDNRKSSTGYLFTFSGGAISWQSKLQKCVALSTTEAEYIAATETGKEMIWLKRFLQELG 1245
Query: 1437 LRYTSVPIYCDNTSAINLSKNPIQHSRSKHIEIKHHFIRDHVQKKNIALSFVDTENQLAD 1496
L +YCD+ SAI+LSKN + H+R+KHI++++H+IR+ V +++ + + T AD
Sbjct: 1246 LHQKEYVVYCDSQSAIDLSKNSMYHARTKHIDVRYHWIREMVDDESLKVLKISTNENPAD 1305
Query: 1497 IFTKPLVEDRFNFLKE 1512
+ TK + ++F KE
Sbjct: 1306 MLTKVVPRNKFELCKE 1321
Score = 123 bits (308), Expect = 4e-27
Identities = 90/280 (32%), Positives = 137/280 (48%), Gaps = 22/280 (7%)
Query: 596 LDSGCSRHMTGDRDCFLTFEKKDGGLVTFGNNDKGKIRGKGTI---GNLNSARI-ENVQY 651
+D+ S H T RD F + D G V GN KI G G I N+ + ++V++
Sbjct: 296 VDTAASHHATPVRDLFCRYVAGDFGTVKMGNTSYSKIAGIGDICIKTNVGCTLVLKDVRH 355
Query: 652 VEGLKHNLLSISQLCDSGFEVIFKPNICEVRQTSSNKLFFSGSRRKNLYVLELNDMPAES 711
V L+ NL+S L G+E F + R T + + G R LY E
Sbjct: 356 VPDLRMNLISGIALDRDGYESYFANQ--KWRLTKGSLVIAKGVARGTLYRTNAEICQGEL 413
Query: 712 CFMSLEKDKWIWHKRAGHISMKTIAKLSQLDLVRGLPKISFEKD---RICEACVKGKQVK 768
E +WHKR GH+S K + L++ L IS+ K + C+ C+ GKQ +
Sbjct: 414 NAAQDEISVDLWHKRMGHMSEKGLQILAKKSL------ISYAKGTTVKPCDYCLFGKQHR 467
Query: 769 SSFKTIEFISTQKPLELLHI---DLFGPVQTASLTGKRYGFVIVDDFSRFTWVLFLKHKD 825
SF+T S+++ L +L + D+ GP++ S+ G +Y +DD SR WV LK KD
Sbjct: 468 VSFQT----SSERKLNILDLVYSDVCGPMEIESMGGNKYFVTFIDDASRKLWVYILKTKD 523
Query: 826 ESFEAFQNFCKRVQNEKGYNIITVRSDHGGEFENASFKTF 865
+ F+ FQ F V+ E G + +RSD+GGE+ + F+ +
Sbjct: 524 QVFQVFQKFHALVERETGRKLKRLRSDNGGEYTSREFEEY 563
Score = 43.9 bits (102), Expect = 0.003
Identities = 57/266 (21%), Positives = 105/266 (39%), Gaps = 56/266 (21%)
Query: 52 VPTNKEGAVKAKSAWSTDEKAQVLLNSKARLFLSCALTMEESERVDECTNAKGVWDTLKI 111
V + K +KA+ DE+A S RL LS + + +DE T A+G+W L+
Sbjct: 39 VDSKKPDTMKAEDWADLDERAA----SAIRLHLSDDVV---NNIIDEDT-ARGIWTRLES 90
Query: 112 HHEGTSHVKETRIDIGVRKFEVFEMSENETIDEMYARFTTIVNEMRSLGKAYSTHDRIRK 171
+ + + + ++ MSE F ++ ++ +LG D+
Sbjct: 91 LYMSKTLTNKLYLK---KQLYALHMSEGTNFLSHLNVFNGLITQLANLGVKIEEEDKAIL 147
Query: 172 ILRCLPSMWRPMVTAITQAKDLKSMNLEDLIGSLRAHEVVLQGDKPVKKVRTLALKASQQ 231
+L LPS + + T I K ++ L+D+ +L +
Sbjct: 148 LLNSLPSSYDNLATTILHGK--TTIELKDVTSALLLN----------------------- 182
Query: 232 TSSVDDEDVQEPQELEEVHEEEAEDELALISKRIQRMMLRRNQIRKKFPKTNISIKTE-A 290
E++ ++ ALI++ R R + + ++ K++
Sbjct: 183 ---------------EKMRKKPENQGQALITEGRGRSYQRSS---NNYGRSGARGKSKNR 224
Query: 291 DKSQV-TCYGCNKTGHFKNECPDIKK 315
KS+V CY CN+ GHFK +CP+ +K
Sbjct: 225 SKSRVRNCYNCNQPGHFKRDCPNPRK 250
>COPI_DROME (P04146) Copia protein (Gag-int-pol protein) [Contains:
Copia VLP protein; Copia protease (EC 3.4.23.-)]
Length = 1409
Score = 348 bits (894), Expect = 5e-95
Identities = 214/635 (33%), Positives = 351/635 (54%), Gaps = 37/635 (5%)
Query: 919 SMHIIFDEYDEHSKPKENEDTEVPTLQNVPVQNTENTVEKEDDQNAQDQSFQSPPRS--- 975
S II E+ SK +N + ++ E+ K DD + + +P S
Sbjct: 772 SRKIIQTEFPNESKECDNIQFLKDSKESNKYFLNESKKRKRDDHLNESKGSGNPNESRES 831
Query: 976 -----WRMVG-DHPTD----QIIGSTTDGVRTR--LSF--QDN-------NMAMISQMEP 1014
+ +G D+PT +II ++ ++T+ +S+ +DN N I P
Sbjct: 832 ETAEHLKEIGIDNPTKNDGIEIINRRSERLKTKPQISYNEEDNSLNKVVLNAHTIFNDVP 891
Query: 1015 KSINEAIIDD---SWIKAMKEELSQFERNKVWNLVPNNQDKTIIGTRWVFRNKLDEEGKV 1071
S +E D SW +A+ EL+ + N W + ++K I+ +RWVF K +E G
Sbjct: 892 NSFDEIQYRDDKSSWEEAINTELNAHKINNTWTITKRPENKNIVDSRWVFSVKYNELGNP 951
Query: 1072 VRNKARLVAQGYNQQEGIDYDETFAPVARLEAIRILLAYAAHKSIKLFQMDVKSAFLNGF 1131
+R KARLVA+G+ Q+ IDY+ETFAPVAR+ + R +L+ ++K+ QMDVK+AFLNG
Sbjct: 952 IRYKARLVARGFTQKYQIDYEETFAPVARISSFRFILSLVIQYNLKVHQMDVKTAFLNGT 1011
Query: 1132 LNEEVYVSQPPGFINKEKPNHVFKLTKALYGLKQAPRAWYDRLSTFLIENGFSRGKIDTT 1191
L EE+Y+ P G ++V KL KA+YGLKQA R W++ L E F +D
Sbjct: 1012 LKEEIYMRLPQGI--SCNSDNVCKLNKAIYGLKQAARCWFEVFEQALKECEFVNSSVDRC 1069
Query: 1192 LF--RKTHNTDLLIVQVYVDDIIFGATKIKMCEEFSNLMQSEFEMSMMGELGFFLGLQIK 1249
++ K + + + V +YVDD++ + F + +F M+ + E+ F+G++I+
Sbjct: 1070 IYILDKGNINENIYVLLYVDDVVIATGDMTRMNNFKRYLMEKFRMTDLNEIKHFIGIRIE 1129
Query: 1250 QHSNGIFISQEKYVKDILKKYKMNEAKIMSTPMHPSSSLDKDESGKSISEKEYRGMIGSL 1309
+ I++SQ YVK IL K+ M +STP+ + + S + + R +IG L
Sbjct: 1130 MQEDKIYLSQSAYVKKILSKFNMENCNAVSTPLPSKINYELLNSDEDCN-TPCRSLIGCL 1188
Query: 1310 LY-LTASRPDIVFAVGLCARFQTCAKESHLTAVKRIFRYLVGTTDLGLWYRKGSSFD--L 1366
+Y + +RPD+ AV + +R+ + +KR+ RYL GT D+ L ++K +F+ +
Sbjct: 1189 MYIMLCTRPDLTTAVNILSRYSSKNNSELWQNLKRVLRYLKGTIDMKLIFKKNLAFENKI 1248
Query: 1367 VAYCDADYAGDKVERKSTSGSC-QFLGQALIGWSCRKQNTIALSTTEAEYVSAASCCSQI 1425
+ Y D+D+AG +++RKST+G + LI W+ ++QN++A S+TEAEY++ +
Sbjct: 1249 IGYVDSDWAGSEIDRKSTTGYLFKMFDFNLICWNTKRQNSVAASSTEAEYMALFEAVREA 1308
Query: 1426 LWVRNQLEDYSLRYTS-VPIYCDNTSAINLSKNPIQHSRSKHIEIKHHFIRDHVQKKNIA 1484
LW++ L +++ + + IY DN I+++ NP H R+KHI+IK+HF R+ VQ I
Sbjct: 1309 LWLKFLLTSINIKLENPIKIYEDNQGCISIANNPSCHKRAKHIDIKYHFAREQVQNNVIC 1368
Query: 1485 LSFVDTENQLADIFTKPLVEDRFNFLKEKLLIMKN 1519
L ++ TENQLADIFTKPL RF L++KL ++++
Sbjct: 1369 LEYIPTENQLADIFTKPLPAARFVELRDKLGLLQD 1403
Score = 97.8 bits (242), Expect = 2e-19
Identities = 103/436 (23%), Positives = 181/436 (40%), Gaps = 65/436 (14%)
Query: 450 ENILLKKEVETLKKDLTGFIKSTETFQNIVGSQNKSTKKSGLGFKD----PSKIIGSFVP 505
+N LL +E++ +K D K N + N +T K+ L FK+ P KI F
Sbjct: 173 KNRLLDQEIK-IKNDHNDTSKKV---MNAIVHNNNNTYKNNL-FKNRVTKPKKI---FKG 224
Query: 506 KAKIRVKCCFCDKYGHNESVCHVKKKFIKQNNLNLSSERSHLNRSESSQKAEKAKKTCFY 565
+K +VKC C + GH + C K+ + N E+ ++ + A
Sbjct: 225 NSKYKVKCHHCGREGHIKKDCFHYKRILNNKN------------KENEKQVQTATS---- 268
Query: 566 CNKSDHKRQNVTFRKDLLEELTFKDPTLHGLDSGCSRHMTGDRDCFLTFEKKDGGL---- 621
+ F + + D LDSG S H+ D + + L
Sbjct: 269 --------HGIAFMVKEVNNTSVMDNCGFVLDSGASDHLINDESLYTDSVEVVPPLKIAV 320
Query: 622 VTFGNNDKGKIRGKGTIGNLNSARIENVQYVEGLKHNLLSISQLCDSGFEVIFKPNICEV 681
G RG + N + +E+V + + NL+S+ +L ++G + F + +
Sbjct: 321 AKQGEFIYATKRGIVRLRNDHEITLEDVLFCKEAAGNLMSVKRLQEAGMSIEFDKSGVTI 380
Query: 682 RQTSSNKLFFSGSRRKNLYVLELNDMP-----AESCFMSLEKDKWIWHKRAGHIS----- 731
+ + SG LN++P A S + + +WH+R GHIS
Sbjct: 381 SKNGLMVVKNSGM---------LNNVPVINFQAYSINAKHKNNFRLWHERFGHISDGKLL 431
Query: 732 -MKTIAKLSQLDLVRGLPKISFEKDRICEACVKGKQVKSSFKTI-EFISTQKPLELLHID 789
+K S L+ L ++S E ICE C+ GKQ + FK + + ++PL ++H D
Sbjct: 432 EIKRKNMFSDQSLLNNL-ELSCE---ICEPCLNGKQARLPFKQLKDKTHIKRPLFVVHSD 487
Query: 790 LFGPVQTASLTGKRYGFVIVDDFSRFTWVLFLKHKDESFEAFQNFCKRVQNEKGYNIITV 849
+ GP+ +L K Y + VD F+ + +K+K + F FQ+F + + ++ +
Sbjct: 488 VCGPITPVTLDDKNYFVIFVDQFTHYCVTYLIKYKSDVFSMFQDFVAKSEAHFNLKVVYL 547
Query: 850 RSDHGGEFENASFKTF 865
D+G E+ + + F
Sbjct: 548 YIDNGREYLSNEMRQF 563
Score = 38.5 bits (88), Expect = 0.14
Identities = 20/57 (35%), Positives = 34/57 (59%), Gaps = 3/57 (5%)
Query: 870 VFGCYCYILNNKEKLGKFDPKSDKAIFLGYSTTSKGYRVYNLKTQTVEISMHIIFDE 926
VFG Y+ + K K GKFD KS K+IF+GY G+++++ + ++ ++ DE
Sbjct: 652 VFGATVYV-HIKNKQGKFDDKSFKSIFVGYE--PNGFKLWDAVNEKFIVARDVVVDE 705
>YCH4_YEAST (P25600) Transposon Ty5-1 34.5 kDa hypothetical protein
Length = 308
Score = 181 bits (458), Expect = 2e-44
Identities = 104/308 (33%), Positives = 167/308 (53%), Gaps = 3/308 (0%)
Query: 1121 MDVKSAFLNGFLNEEVYVSQPPGFINKEKPNHVFKLTKALYGLKQAPRAWYDRLSTFLIE 1180
MDV +AFLN ++E +YV QPPGF+N+ P++V++L +YGLKQAP W + ++ L +
Sbjct: 1 MDVDTAFLNSTMDEPIYVKQPPGFVNERNPDYVWELYGGMYGLKQAPLLWNEHINNTLKK 60
Query: 1181 NGFSRGKIDTTLFRKTHNTDLLIVQVYVDDIIFGATKIKMCEEFSNLMQSEFEMSMMGEL 1240
GF R + + L+ ++ + + + VYVDD++ A K+ + + + M +G++
Sbjct: 61 IGFCRHEGEHGLYFRSTSDGPIYIGVYVDDLLVAAPSPKIYDRVKQELTKLYSMKDLGKV 120
Query: 1241 GFFLGLQIKQHSNG-IFISQEKYVKDILKKYKMNEAKIMSTPMHPSSSLDKDESGKSISE 1299
FLGL I Q +NG I +S + Y+ + ++N K+ TP+ S L + S
Sbjct: 121 DKFLGLNIHQSTNGDITLSLQDYIAKAASESEINTFKLTQTPLCNSKPLFETTSPHLKDI 180
Query: 1300 KEYRGMIGSLLY-LTASRPDIVFAVGLCARFQTCAKESHLTAVKRIFRYLVGTTDLGLWY 1358
Y+ ++G LL+ RPDI + V L +RF + HL + +R+ RYL T + L Y
Sbjct: 181 TPYQSIVGQLLFCANTGRPDISYPVSLLSRFLREPRAIHLESARRVLRYLYTTRSMCLKY 240
Query: 1359 RKGSSFDLVAYCDADYAGDKVERKSTSGSCQFLGQALIGWSCRK-QNTIALSTTEAEYVS 1417
R GS L YCDA + ST G L A + WS +K + I + +TEAEY++
Sbjct: 241 RSGSQVALTVYCDASHGAIHDLPHSTGGYVTLLAGAPVTWSSKKLKGVIPVPSTEAEYIT 300
Query: 1418 AASCCSQI 1425
A+ +I
Sbjct: 301 ASETVMEI 308
>M810_ARATH (P92519) Hypothetical mitochondrial protein AtMg00810
(ORF240b)
Length = 240
Score = 153 bits (387), Expect = 3e-36
Identities = 80/222 (36%), Positives = 127/222 (57%), Gaps = 1/222 (0%)
Query: 1206 VYVDDIIFGATKIKMCEEFSNLMQSEFEMSMMGELGFFLGLQIKQHSNGIFISQEKYVKD 1265
+YVDDI+ + + + S F M +G + +FLG+QIK H +G+F+SQ KY +
Sbjct: 5 LYVDDILLTGSSNTLLNMLIFQLSSTFSMKDLGPVHYFLGIQIKTHPSGLFLSQTKYAEQ 64
Query: 1266 ILKKYKMNEAKIMSTPMHPSSSLDKDESGKSISEKEYRGMIGSLLYLTASRPDIVFAVGL 1325
IL M + K MSTP+ P + K ++R ++G+L YLT +RPDI +AV +
Sbjct: 65 ILNNAGMLDCKPMSTPL-PLKLNSSVSTAKYPDPSDFRSIVGALQYLTLTRPDISYAVNI 123
Query: 1326 CARFQTCAKESHLTAVKRIFRYLVGTTDLGLWYRKGSSFDLVAYCDADYAGDKVERKSTS 1385
+ + +KR+ RY+ GT GL+ K S ++ A+CD+D+AG R+ST+
Sbjct: 124 VCQRMHEPTLADFDLLKRVLRYVKGTIFHGLYIHKNSKLNVQAFCDSDWAGCTSTRRSTT 183
Query: 1386 GSCQFLGQALIGWSCRKQNTIALSTTEAEYVSAASCCSQILW 1427
G C FLG +I WS ++Q T++ S+TE EY + A +++ W
Sbjct: 184 GFCTFLGCNIISWSAKRQPTVSRSSTETEYRALALTAAELTW 225
>YJL3_YEAST (P47024) Transposon Ty4 207.7 kDa hypothetical protein
Length = 1803
Score = 138 bits (348), Expect = 1e-31
Identities = 147/565 (26%), Positives = 251/565 (44%), Gaps = 65/565 (11%)
Query: 996 RTRLSFQDNNMAMISQMEPKSI--NEAIIDDSWIK-------AMKEELSQFERNKVWNL- 1045
+ R+ +NM +S + ++I NEAI + +K A +EL + KV+++
Sbjct: 1250 KNRVKLIPDNMETVSAPKIRAIYYNEAISKNPDLKEKHEYKQAYHKELQNLKDMKVFDVD 1309
Query: 1046 -------VPNNQDKTIIGTRWVFRNKLDEEGKVVRN---KARLVAQGYNQQEGIDYDETF 1095
+P+N I+ T +F K RN KAR+V +G Q Y
Sbjct: 1310 VKYSRSEIPDN---LIVPTNTIFTKK--------RNGIYKARIVCRGDTQSPDT-YSVIT 1357
Query: 1096 APVARLEAIRILLAYAAHKSIKLFQMDVKSAFLNGFLNEEVYVSQPPGFINKEKPNHVFK 1155
I+I L A ++++ + +D+ AFL L EE+Y+ P V K
Sbjct: 1358 TESLNHNHIKIFLMIANNRNMFMKTLDINHAFLYAKLEEEIYIPHP------HDRRCVVK 1411
Query: 1156 LTKALYGLKQAPRAWYDRLSTFLIENGFSRGKIDTTLFRKTHNTDLLIVQVYVDDIIFGA 1215
L KALYGLKQ+P+ W D L +L G L+ +T + +L+I VYVDD + A
Sbjct: 1412 LNKALYGLKQSPKEWNDHLRQYLNGIGLKDNSYTPGLY-QTEDKNLMIA-VYVDDCVIAA 1469
Query: 1216 TKIKMCEEFSNLMQSEFEMSMMGEL------GFFLGLQI--KQHSNGIFISQEKYVKDIL 1267
+ + +EF N ++S FE+ + G L LG+ + + I ++ + ++ +
Sbjct: 1470 SNEQRLDEFINKLKSNFELKITGTLIDDVLDTDILGMDLVYNKRLGTIDLTLKSFINRMD 1529
Query: 1268 KKYKMNEAKI--MSTPMHPSSSLDKDESGKSISEKEYR-------GMIGSLLYLT-ASRP 1317
KKY KI S P + +D + +SE+E+R ++G L Y+ R
Sbjct: 1530 KKYNEELKKIRKSSIPHMSTYKIDPKKDVLQMSEEEFRQGVLKLQQLLGELNYVRHKCRY 1589
Query: 1318 DIVFAVGLCARFQTCAKESHLTAVKRIFRYLVGTTDLGLWYRKGSSFD--LVAYCDADYA 1375
DI FAV AR E + +I +YLV D+G+ Y + + D ++A DA
Sbjct: 1590 DIEFAVKKVARLVNYPHERVFYMIYKIIQYLVRYKDIGIHYDRDCNKDKKVIAITDAS-V 1648
Query: 1376 GDKVERKSTSGSCQFLGQALIGWSCRKQNTIALSTTEAEYVSAASCCSQILWVRNQLEDY 1435
G + + +S G + G + K +S+TEAE + + ++ L++
Sbjct: 1649 GSEYDAQSRIGVILWYGMNIFNVYSNKSTNRCVSSTEAELHAIYEGYADSETLKVTLKEL 1708
Query: 1436 SL-RYTSVPIYCDNTSAINLSKNPIQHSRSKHIEIKHHFIRDHVQKKNIALSFVDTENQL 1494
+ + D+ AI Q + K IK I++ +++K+I L + + +
Sbjct: 1709 GEGDNNDIVMITDSKPAIQGLNRSYQQPKEKFTWIKTEIIKEKIKEKSIKLLKITGKGNI 1768
Query: 1495 ADIFTKPLVEDRFNFLKEKLLIMKN 1519
AD+ TKP+ F K + ++KN
Sbjct: 1769 ADLLTKPVSASDF---KRFIQVLKN 1790
Score = 47.0 bits (110), Expect = 4e-04
Identities = 36/149 (24%), Positives = 64/149 (42%), Gaps = 6/149 (4%)
Query: 724 HKRAGHISMKTIAK-LSQLDLVRGLPKISFEKDRICEACVKGKQVKSSFKTIEF---IST 779
HKR GH ++ I + L I + C+ C K K + T +
Sbjct: 562 HKRMGHTGIQQIENSIKHNHYEESLDLIKEPNEFWCQTCKISKATKRNHYTGSMNNHSTD 621
Query: 780 QKPLELLHIDLFGPVQTASLTGKRYGFVIVDDFSRF--TWVLFLKHKDESFEAFQNFCKR 837
+P +D+FGPV +++ KRY ++VD+ +R+ T F K+ + + +
Sbjct: 622 HEPGSSWCMDIFGPVSSSNADTKRYMLIMVDNNTRYCMTSTHFNKNAETILAQVRKNIQY 681
Query: 838 VQNEKGYNIITVRSDHGGEFENASFKTFF 866
V+ + + + SD G EF N + +F
Sbjct: 682 VETQFDRKVREINSDRGTEFTNDQIEEYF 710
>YCB9_YEAST (P25384) Transposon Ty2 protein B (Ty1-17 protein B)
Length = 1770
Score = 127 bits (320), Expect = 2e-28
Identities = 140/596 (23%), Positives = 264/596 (43%), Gaps = 56/596 (9%)
Query: 949 VQNTENTVE-KEDDQNAQDQSFQSPPRSWRMVGDHPTDQIIGSTTDGVRTRLSFQDNNMA 1007
+++ E +E D N ++ PPRS + + + + S VRT L + + A
Sbjct: 1197 LEDNETEIEVSRDTWNNKNMRSLEPPRSKKRINLIAAIKGVKSIKP-VRTTLRYDE---A 1252
Query: 1008 MISQMEPKSINEAIIDDSWIKAMKEELSQFERNKVW--NLVPNNQD---KTIIGTRWVFR 1062
+ + K D +++A +E+SQ + W N + D K +I + ++F
Sbjct: 1253 ITYNKDNKE------KDRYVEAYHKEISQLLKMNTWDTNKYYDRNDIDPKKVINSMFIFN 1306
Query: 1063 NKLDEEGKVVRNKARLVAQGYNQQEGIDYDETFAPVARLEAIRILLAYAAHKSIKLFQMD 1122
K D +KAR VA+G Q + + A+ L+ A + Q+D
Sbjct: 1307 KKRDGT-----HKARFVARGDIQHPDTYDSDMQSNTVHHYALMTSLSIALDNDYYITQLD 1361
Query: 1123 VKSAFLNGFLNEEVYVSQPPGFINKEKPNHVFKLTKALYGLKQAPRAWYDRLSTFLIEN- 1181
+ SA+L + EE+Y+ PP +K + +L K+LYGLKQ+ WY+ + ++LI
Sbjct: 1362 ISSAYLYADIKEELYIRPPPHLGLNDK---LLRLRKSLYGLKQSGANWYETIKSYLINCC 1418
Query: 1182 GFSRGKIDTTLFRKTHNTDLLIVQVYVDDIIFGATKIKMCEEFSNLMQSEFEMSMMG--- 1238
+ + +F+ + T + ++VDD+I + + ++ ++ +++ ++
Sbjct: 1419 DMQEVRGWSCVFKNSQVT----ICLFVDDMILFSKDLNANKKIITTLKKQYDTKIINLGE 1474
Query: 1239 ---ELGF-FLGLQIK-QHSNGIFISQEKYVKDILKKYKMN---EAKIMSTPMHPSSSLDK 1290
E+ + LGL+IK Q S + + EK + + L K + + K + P P +D+
Sbjct: 1475 SDNEIQYDILGLEIKYQRSKYMKLGMEKSLTEKLPKLNVPLNPKGKKLRAPGQPGHYIDQ 1534
Query: 1291 DESGKSISEKEYRG-------MIGSLLYLTAS-RPDIVFAVGLCARFQTCAKESHLTAVK 1342
DE I E EY+ +IG Y+ R D+++ + A+ L
Sbjct: 1535 DEL--EIDEDEYKEKVHEMQKLIGLASYVGYKFRFDLLYYINTLAQHILFPSRQVLDMTY 1592
Query: 1343 RIFRYLVGTTDLGLWYRKGSSFD----LVAYCDADYAGDKVERKSTSGSCQFLGQALIGW 1398
+ +++ T D L + K LVA DA Y G++ KS G+ L +IG
Sbjct: 1593 ELIQFMWDTRDKQLIWHKNKPTKPDNKLVAISDASY-GNQPYYKSQIGNIFLLNGKVIGG 1651
Query: 1399 SCRKQNTIALSTTEAEYVSAASCCSQILWVRNQLEDYSLRYTSVPIYCDNTSAINLSKNP 1458
K + STTEAE + + + + + +++ + + + D+ S I++ K+
Sbjct: 1652 KSTKASLTCTSTTEAEIHAVSEAIPLLNNLSHLVQELNKKPIIKGLLTDSRSTISIIKST 1711
Query: 1459 IQHS-RSKHIEIKHHFIRDHVQKKNIALSFVDTENQLADIFTKPLVEDRFNFLKEK 1513
+ R++ K +RD V N+ + +++T+ +AD+ TKPL F L K
Sbjct: 1712 NEEKFRNRFFGTKAMRLRDEVSGNNLYVYYIETKKNIADVMTKPLPIKTFKLLTNK 1767
Score = 66.2 bits (160), Expect = 6e-10
Identities = 56/257 (21%), Positives = 105/257 (40%), Gaps = 24/257 (9%)
Query: 636 GTIGNLNSARIENVQYVEGLKHNLLSISQLCDSGFEVIFKPNICE-----VRQTSSNKLF 690
GT ++ + N+ Y +LLS+S+L + F N E V
Sbjct: 505 GTKTSIKALHTPNIAY------DLLSLSELANQNITACFTRNTLERSDGTVLAPIVKHGD 558
Query: 691 FSGSRRKNLYVLELNDMPAESCFMSLEKDKWIW---HKRAGHISMKTIAKLSQLDLVRGL 747
F +K L ++ + + S +K+ + H+ GH + ++I K + + V L
Sbjct: 559 FYWLSKKYLIPSHISKLTINNVNKSKSVNKYPYPLIHRMLGHANFRSIQKSLKKNAVTYL 618
Query: 748 PKISFEKDRI----CEACVKGKQVKSSF---KTIEFISTQKPLELLHIDLFGPVQTASLT 800
+ E C C+ GK K +++ + +P + LH D+FGPV +
Sbjct: 619 KESDIEWSNASTYQCPDCLIGKSTKHRHVKGSRLKYQESYEPFQYLHTDIFGPVHHLPKS 678
Query: 801 GKRYGFVIVDDFSRFTWV--LFLKHKDESFEAFQNFCKRVQNEKGYNIITVRSDHGGEFE 858
Y D+ +RF WV L + ++ F + ++N+ ++ ++ D G E+
Sbjct: 679 APSYFISFTDEKTRFQWVYPLHDRREESILNVFTSILAFIKNQFNARVLVIQMDRGSEYT 738
Query: 859 NASFKTFF-DENVFGCY 874
N + FF + + CY
Sbjct: 739 NKTLHKFFTNRGITACY 755
>YMD9_YEAST (Q03434) Transposon Ty1 protein B
Length = 1328
Score = 126 bits (316), Expect = 5e-28
Identities = 142/614 (23%), Positives = 269/614 (43%), Gaps = 67/614 (10%)
Query: 930 HSKPKENEDTEVPTLQNVPVQNTENTVEKEDDQNAQDQSFQSPPRSWRMVGDHPTDQIIG 989
+SK + ED E TE V + D N ++ PPRS + + + +
Sbjct: 749 NSKKRSLEDNE-----------TEIKVSR-DTWNTKNMRSLEPPRSKKRIHLIAAVKAVK 796
Query: 990 STTDGVRTRLSFQDNNMAMISQMEPKSINEAIIDDSWIKAMKEELSQFERNKVWNLVP-- 1047
S +RT L + + + K I E + +I+A +E++Q + K W+
Sbjct: 797 SIKP-IRTTLRYDE------AITYNKDIKEK---EKYIEAYHKEVNQLLKMKTWDTDEYY 846
Query: 1048 ---NNQDKTIIGTRWVFRNKLDEEGKVVRNKARLVAQGYNQQEGIDYDETFAPVARLEAI 1104
K +I + ++F K D +KAR VA+G Q + A+
Sbjct: 847 DRKEIDPKRVINSMFIFNKKRDGT-----HKARFVARGDIQHPDTYDSGMQSNTVHHYAL 901
Query: 1105 RILLAYAAHKSIKLFQMDVKSAFLNGFLNEEVYVSQPPGFINKEKPNHVFKLTKALYGLK 1164
L+ A + + Q+D+ SA+L + EE+Y+ PP +K + +L K+LYGLK
Sbjct: 902 MTSLSLALDNNYYITQLDISSAYLYADIKEELYIRPPPHLGMNDK---LIRLKKSLYGLK 958
Query: 1165 QAPRAWYDRLSTFLIEN-GFSRGKIDTTLFRKTHNTDLLIVQVYVDDIIFGATKIKMCEE 1223
Q+ WY+ + ++LI+ G + + +F+ + T + ++VDD+I + + ++
Sbjct: 959 QSGANWYETIKSYLIKQCGMEEVRGWSCVFKNSQVT----ICLFVDDMILFSKDLNANKK 1014
Query: 1224 FSNLMQSEFEMSMMG------ELGF-FLGLQIK-QHSNGIFISQEKYVKDILKKYKMN-- 1273
++ +++ ++ E+ + LGL+IK Q + + E + + + K +
Sbjct: 1015 IITTLKKQYDTKIINLGESDNEIQYDILGLEIKYQRGKYMKLGMENSLTEKIPKLNVPLN 1074
Query: 1274 -EAKIMSTPMHPSSSLDKDESGKSISEKEYRG-------MIGSLLYLTAS-RPDIVFAVG 1324
+ + +S P P +D+DE I E EY+ +IG Y+ R D+++ +
Sbjct: 1075 PKGRKLSAPGQPGLYIDQDEL--EIDEDEYKEKVHEMQKLIGLASYVGYKFRFDLLYYIN 1132
Query: 1325 LCARFQTCAKESHLTAVKRIFRYLVGTTDLGLWYRKGSSFD----LVAYCDADYAGDKVE 1380
A+ L + +++ T D L + K + LVA DA Y G++
Sbjct: 1133 TLAQHILFPSRQVLDMTYELIQFMWDTRDKQLIWHKNKPTEPDNKLVAISDASY-GNQPY 1191
Query: 1381 RKSTSGSCQFLGQALIGWSCRKQNTIALSTTEAEYVSAASCCSQILWVRNQLEDYSLRYT 1440
KS G+ L +IG K + STTEAE + + + + +++ + +
Sbjct: 1192 YKSQIGNIYLLNGKVIGGKSTKASLTCTSTTEAEIHAISESVPLLNNLSYLIQELNKKPI 1251
Query: 1441 SVPIYCDNTSAINLSKNPIQHS-RSKHIEIKHHFIRDHVQKKNIALSFVDTENQLADIFT 1499
+ D+ S I++ K+ + R++ K +RD V N+ + +++T+ +AD+ T
Sbjct: 1252 IKGLLTDSRSTISIIKSTNEEKFRNRFFGTKAMRLRDEVSGNNLYVYYIETKKNIADVMT 1311
Query: 1500 KPLVEDRFNFLKEK 1513
KPL F L K
Sbjct: 1312 KPLPIKTFKLLTNK 1325
Score = 59.7 bits (143), Expect = 6e-08
Identities = 31/126 (24%), Positives = 60/126 (47%), Gaps = 6/126 (4%)
Query: 755 DRICEACVKGKQVKSSF---KTIEFISTQKPLELLHIDLFGPVQTASLTGKRYGFVIVDD 811
D C C+ GK K +++ ++ +P + LH D+FGPV + Y D+
Sbjct: 207 DYQCPDCLIGKSTKHRHIKGSRLKYQNSYEPFQYLHTDIFGPVHNLPKSAPSYFISFTDE 266
Query: 812 FSRFTWV--LFLKHKDESFEAFQNFCKRVQNEKGYNIITVRSDHGGEFENASFKTFFDEN 869
++F WV L + +D + F ++N+ +++ ++ D G E+ N + F ++N
Sbjct: 267 TTKFRWVYPLHDRREDSILDVFTTILAFIKNQFQASVLVIQMDRGSEYTNRTLHKFLEKN 326
Query: 870 -VFGCY 874
+ CY
Sbjct: 327 GITPCY 332
>YME4_YEAST (Q04711) Transposon Ty1 protein B
Length = 1328
Score = 123 bits (308), Expect = 4e-27
Identities = 140/614 (22%), Positives = 269/614 (43%), Gaps = 67/614 (10%)
Query: 930 HSKPKENEDTEVPTLQNVPVQNTENTVEKEDDQNAQDQSFQSPPRSWRMVGDHPTDQIIG 989
+SK + ED E TE V + D N ++ PPRS + + + +
Sbjct: 749 NSKKRSLEDNE-----------TEIKVSR-DTWNTKNMRSLEPPRSKKRIHLIAAVKAVK 796
Query: 990 STTDGVRTRLSFQDNNMAMISQMEPKSINEAIIDDSWIKAMKEELSQFERNKVWNLVP-- 1047
S +RT L + + + K I E + +I+A +E++Q + W+
Sbjct: 797 SIKP-IRTTLRYDE------AITYNKDIKEK---EKYIEAYHKEVNQLLKMNTWDTDKYY 846
Query: 1048 ---NNQDKTIIGTRWVFRNKLDEEGKVVRNKARLVAQGYNQQEGIDYDETFAPVARLEAI 1104
K +I + ++F K D +KAR VA+G Q + A+
Sbjct: 847 DRKEIDPKRVINSMFIFNRKRDGT-----HKARFVARGDIQHPDTYDSGMQSNTVHHYAL 901
Query: 1105 RILLAYAAHKSIKLFQMDVKSAFLNGFLNEEVYVSQPPGFINKEKPNHVFKLTKALYGLK 1164
L+ A + + Q+D+ SA+L + EE+Y+ PP +K + +L K+LYGLK
Sbjct: 902 MTSLSLALDNNYYITQLDISSAYLYADIKEELYIRPPPHLGMNDK---LIRLKKSLYGLK 958
Query: 1165 QAPRAWYDRLSTFLIEN-GFSRGKIDTTLFRKTHNTDLLIVQVYVDDIIFGATKIKMCEE 1223
Q+ WY+ + ++LI+ G + + +F+ + T + ++VDD+I + + ++
Sbjct: 959 QSGANWYETIKSYLIKQCGMEEVRGWSCVFKNSQVT----ICLFVDDMILFSKDLNANKK 1014
Query: 1224 FSNLMQSEFEMSMMG------ELGF-FLGLQIK-QHSNGIFISQEKYVKDILKKYKMN-- 1273
++ +++ ++ E+ + LGL+IK Q + + E + + + K +
Sbjct: 1015 IITTLKKQYDTKIINLGESDNEIQYDILGLEIKYQRGKYMKLGMENSLTEKIPKLNVPLN 1074
Query: 1274 -EAKIMSTPMHPSSSLDKDESGKSISEKEYRG-------MIGSLLYLTAS-RPDIVFAVG 1324
+ + +S P P +D+DE I E EY+ +IG Y+ R D+++ +
Sbjct: 1075 PKGRKLSAPGQPGLYIDQDEL--EIDEDEYKEKVHEMQKLIGLASYVGYKFRFDLLYYIN 1132
Query: 1325 LCARFQTCAKESHLTAVKRIFRYLVGTTDLGLWYRKGSSFD----LVAYCDADYAGDKVE 1380
A+ L + +++ T D L + K + LVA DA Y G++
Sbjct: 1133 TLAQHILFPSRQVLDMTYELIQFMWDTRDKQLIWHKNKPTEPDNKLVAISDASY-GNQPY 1191
Query: 1381 RKSTSGSCQFLGQALIGWSCRKQNTIALSTTEAEYVSAASCCSQILWVRNQLEDYSLRYT 1440
KS G+ L +IG K + STTEAE + + + + + +++ + +
Sbjct: 1192 YKSQIGNIYLLNGKVIGGKSTKASLTCTSTTEAEIHAISESVPLLNNLSHLVQELNKKPI 1251
Query: 1441 SVPIYCDNTSAINLS-KNPIQHSRSKHIEIKHHFIRDHVQKKNIALSFVDTENQLADIFT 1499
+ + D+ S I++ N + R++ K +RD V ++ + +++T+ +AD+ T
Sbjct: 1252 TKGLLTDSKSTISIIISNNEEKFRNRFFGTKAMRLRDEVSGNHLHVCYIETKKNIADVMT 1311
Query: 1500 KPLVEDRFNFLKEK 1513
KPL F L K
Sbjct: 1312 KPLPIKTFKLLTNK 1325
Score = 59.7 bits (143), Expect = 6e-08
Identities = 31/126 (24%), Positives = 60/126 (47%), Gaps = 6/126 (4%)
Query: 755 DRICEACVKGKQVKSSF---KTIEFISTQKPLELLHIDLFGPVQTASLTGKRYGFVIVDD 811
D C C+ GK K +++ ++ +P + LH D+FGPV + Y D+
Sbjct: 207 DYQCPDCLIGKSTKHRHIKGSRLKYQNSYEPFQYLHTDIFGPVHNLPKSAPSYFISFTDE 266
Query: 812 FSRFTWV--LFLKHKDESFEAFQNFCKRVQNEKGYNIITVRSDHGGEFENASFKTFFDEN 869
++F WV L + +D + F ++N+ +++ ++ D G E+ N + F ++N
Sbjct: 267 TTKFRWVYPLHDRREDSILDVFTTILAFIKNQFQASVLVIQMDRGSEYTNRTLHKFLEKN 326
Query: 870 -VFGCY 874
+ CY
Sbjct: 327 GITPCY 332
>YJZ7_YEAST (P47098) Transposon Ty1 protein B
Length = 1755
Score = 122 bits (306), Expect = 8e-27
Identities = 141/614 (22%), Positives = 268/614 (42%), Gaps = 67/614 (10%)
Query: 930 HSKPKENEDTEVPTLQNVPVQNTENTVEKEDDQNAQDQSFQSPPRSWRMVGDHPTDQIIG 989
+SK + ED E TE V + D N ++ PPRS + + + +
Sbjct: 1176 NSKKRSLEDNE-----------TEIKVSR-DTWNTKNMRSLEPPRSKKRIHLIAAVKAVK 1223
Query: 990 STTDGVRTRLSFQDNNMAMISQMEPKSINEAIIDDSWIKAMKEELSQFERNKVWNLVP-- 1047
S +RT L + + + K I E + +I+A +E++Q + K W+
Sbjct: 1224 SIKP-IRTTLRYDE------AITYNKDIKEK---EKYIEAYHKEVNQLLKMKTWDTDEYY 1273
Query: 1048 ---NNQDKTIIGTRWVFRNKLDEEGKVVRNKARLVAQGYNQQEGIDYDETFAPVARLEAI 1104
K +I + ++F K D +KAR VA+G Q + A+
Sbjct: 1274 DRKEIDPKRVINSMFIFNKKRDGT-----HKARFVARGDIQHPDTYDTGMQSNTVHHYAL 1328
Query: 1105 RILLAYAAHKSIKLFQMDVKSAFLNGFLNEEVYVSQPPGFINKEKPNHVFKLTKALYGLK 1164
L+ A + + Q+D+ SA+L + EE+Y+ PP +K + +L K+ YGLK
Sbjct: 1329 MTSLSLALDNNYYITQLDISSAYLYADIKEELYIRPPPHLGMNDK---LIRLKKSHYGLK 1385
Query: 1165 QAPRAWYDRLSTFLIEN-GFSRGKIDTTLFRKTHNTDLLIVQVYVDDIIFGATKIKMCEE 1223
Q+ WY+ + ++LI+ G + + +F+ + T + ++VDD+I + + ++
Sbjct: 1386 QSGANWYETIKSYLIKQCGMEEVRGWSCVFKNSQVT----ICLFVDDMILFSKDLNANKK 1441
Query: 1224 FSNLMQSEFEMSMMG------ELGF-FLGLQIK-QHSNGIFISQEKYVKDILKKYKMN-- 1273
++ +++ ++ E+ + LGL+IK Q + + E + + + K +
Sbjct: 1442 IITTLKKQYDTKIINLGESDNEIQYDILGLEIKYQRGKYMKLGMENSLTEKIPKLNVPLN 1501
Query: 1274 -EAKIMSTPMHPSSSLDKDESGKSISEKEYRG-------MIGSLLYLTAS-RPDIVFAVG 1324
+ + +S P P +D+DE I E EY+ +IG Y+ R D+++ +
Sbjct: 1502 PKGRKLSAPGQPGLYIDQDEL--EIDEDEYKEKVHEMQKLIGLASYVGYKFRFDLLYYIN 1559
Query: 1325 LCARFQTCAKESHLTAVKRIFRYLVGTTDLGLWYRKGSSFD----LVAYCDADYAGDKVE 1380
A+ L + +++ T D L + K + LVA DA Y G++
Sbjct: 1560 TLAQHILFPSRQVLDMTYELIQFMWDTRDKQLIWHKNKPTEPDNKLVAISDASY-GNQPY 1618
Query: 1381 RKSTSGSCQFLGQALIGWSCRKQNTIALSTTEAEYVSAASCCSQILWVRNQLEDYSLRYT 1440
KS G+ L +IG K + STTEAE + + + + +++ + +
Sbjct: 1619 YKSQIGNIFLLNGKVIGGKSTKASLTCTSTTEAEIHAISESVPLLNNLSYLIQELNKKPI 1678
Query: 1441 SVPIYCDNTSAINLSKNPIQHS-RSKHIEIKHHFIRDHVQKKNIALSFVDTENQLADIFT 1499
+ D+ S I++ K+ + R++ K +RD V N+ + +++T+ +AD+ T
Sbjct: 1679 IKGLLTDSRSTISIIKSTNEEKFRNRFFGTKAMRLRDEVSGNNLYVYYIETKKNIADVMT 1738
Query: 1500 KPLVEDRFNFLKEK 1513
KPL F L K
Sbjct: 1739 KPLPIKTFKLLTNK 1752
Score = 59.7 bits (143), Expect = 6e-08
Identities = 31/126 (24%), Positives = 60/126 (47%), Gaps = 6/126 (4%)
Query: 755 DRICEACVKGKQVKSSF---KTIEFISTQKPLELLHIDLFGPVQTASLTGKRYGFVIVDD 811
D C C+ GK K +++ ++ +P + LH D+FGPV + Y D+
Sbjct: 634 DYQCPDCLIGKSTKHRHIKGSRLKYQNSYEPFQYLHTDIFGPVHNLPKSAPSYFISFTDE 693
Query: 812 FSRFTWV--LFLKHKDESFEAFQNFCKRVQNEKGYNIITVRSDHGGEFENASFKTFFDEN 869
++F WV L + +D + F ++N+ +++ ++ D G E+ N + F ++N
Sbjct: 694 TTKFRWVYPLHDRREDSILDVFTTILAFIKNQFQASVLVIQMDRGSEYTNRTLHKFLEKN 753
Query: 870 -VFGCY 874
+ CY
Sbjct: 754 GITPCY 759
>YMU0_YEAST (Q04670) Transposon Ty1 protein B
Length = 1328
Score = 121 bits (304), Expect = 1e-26
Identities = 138/617 (22%), Positives = 272/617 (43%), Gaps = 73/617 (11%)
Query: 930 HSKPKENEDTEVPTLQNVPVQNTENTVEKEDDQNAQDQSFQSPPRSWRMVGDHPTDQIIG 989
+SK + ED E TE V + D N ++ PPRS + + + +
Sbjct: 749 NSKKRSLEDNE-----------TEIKVSR-DTWNTKNMRSLEPPRSKKRIHLIAAVKAVK 796
Query: 990 STTDGVRTRLSFQDNNMAMISQMEPKSINEAIIDDSWIKAMKEELSQFERNKVWNLV--- 1046
S +RT L + + + K I E + +I+A +E++Q + K W+
Sbjct: 797 SIKP-IRTTLRYDE------AITYNKDIKEK---EKYIQAYHKEVNQLLKMKTWDTDRYY 846
Query: 1047 --PNNQDKTIIGTRWVFRNKLDEEGKVVRNKARLVAQGYNQQEGIDYDETFAPVARLE-- 1102
K +I + ++F K D +KAR VA+G I + +T+ P +
Sbjct: 847 DRKEIDPKRVINSMFIFNRKRDGT-----HKARFVARG-----DIQHPDTYDPGMQSNTV 896
Query: 1103 ---AIRILLAYAAHKSIKLFQMDVKSAFLNGFLNEEVYVSQPPGFINKEKPNHVFKLTKA 1159
A+ L+ A + + Q+D+ SA+L + EE+Y+ PP +K + +L K+
Sbjct: 897 HHYALMTSLSLALDNNYYITQLDISSAYLYADIKEELYIRPPPHLGMNDK---LIRLKKS 953
Query: 1160 LYGLKQAPRAWYDRLSTFLIEN-GFSRGKIDTTLFRKTHNTDLLIVQVYVDDIIFGATKI 1218
LYGLKQ+ WY+ + ++LI+ G + + +F+ + T + ++VDD+I + +
Sbjct: 954 LYGLKQSGANWYETIKSYLIKQCGMEEVRGWSCVFKNSQVT----ICLFVDDMILFSKDL 1009
Query: 1219 KMCEEFSNLMQSEFEMSMMG------ELGF-FLGLQIK-QHSNGIFISQEKYVKDILKKY 1270
++ ++ +++ ++ E+ + LGL+IK Q + + E + + + K
Sbjct: 1010 NANKKIITTLKKQYDTKIINLGESDNEIQYDILGLEIKYQRGKYMKLGMENSLTEKIPKL 1069
Query: 1271 KMN---EAKIMSTPMHPSSSLDK-----DESGKSISEKEYRGMIGSLLYLTAS-RPDIVF 1321
+ + + +S P P +D+ +E + E + +IG Y+ R D+++
Sbjct: 1070 NVPLNPKGRKLSAPGQPGLYIDQQELELEEDDYKMKVHEMQKLIGLASYVGYKFRFDLLY 1129
Query: 1322 AVGLCARFQTCAKESHLTAVKRIFRYLVGTTDLGLWYRKGSSFD----LVAYCDADYAGD 1377
+ A+ + L + +++ T D L + K LV DA Y G+
Sbjct: 1130 YINTLAQHILFPSKQVLDMTYELIQFIWNTRDKQLIWHKSKPVKPTNKLVVISDASY-GN 1188
Query: 1378 KVERKSTSGSCQFLGQALIGWSCRKQNTIALSTTEAEYVSAASCCSQILWVRNQLEDYSL 1437
+ KS G+ L +IG K + STTEAE + + + + + +++ +
Sbjct: 1189 QPYYKSQIGNIYLLNGKVIGGKSTKASLTCTSTTEAEIHAISESVPLLNNLSHLVQELNK 1248
Query: 1438 RYTSVPIYCDNTSAINLS-KNPIQHSRSKHIEIKHHFIRDHVQKKNIALSFVDTENQLAD 1496
+ + + D+ S I++ N + R++ K +RD V ++ + +++T+ +AD
Sbjct: 1249 KPITKGLLTDSKSTISIIISNNEEKFRNRFFGTKAMRLRDEVSGNHLHVCYIETKKNIAD 1308
Query: 1497 IFTKPLVEDRFNFLKEK 1513
+ TKPL F L K
Sbjct: 1309 VMTKPLPIKTFKLLTNK 1325
Score = 59.7 bits (143), Expect = 6e-08
Identities = 31/126 (24%), Positives = 60/126 (47%), Gaps = 6/126 (4%)
Query: 755 DRICEACVKGKQVKSSF---KTIEFISTQKPLELLHIDLFGPVQTASLTGKRYGFVIVDD 811
D C C+ GK K +++ ++ +P + LH D+FGPV + Y D+
Sbjct: 207 DYQCPDCLIGKSTKHRHIKGSRLKYQNSYEPFQYLHTDIFGPVHNLPKSAPSYFISFTDE 266
Query: 812 FSRFTWV--LFLKHKDESFEAFQNFCKRVQNEKGYNIITVRSDHGGEFENASFKTFFDEN 869
++F WV L + +D + F ++N+ +++ ++ D G E+ N + F ++N
Sbjct: 267 TTKFRWVYPLHDRREDSILDVFTTILAFIKNQFQASVLVIQMDRGSEYTNRTLHKFLEKN 326
Query: 870 -VFGCY 874
+ CY
Sbjct: 327 GITPCY 332
>YJZ9_YEAST (P47100) Transposon Ty1 protein B
Length = 1755
Score = 118 bits (295), Expect = 1e-25
Identities = 135/612 (22%), Positives = 264/612 (43%), Gaps = 63/612 (10%)
Query: 930 HSKPKENEDTEVPTLQNVPVQNTENTVEKEDDQNAQDQSFQSPPRSWRMVGDHPTDQIIG 989
+SK + ED E TE V + D N ++ PPRS + + + +
Sbjct: 1176 NSKKRSLEDNE-----------TEIKVSR-DTWNTKNMRSLEPPRSKKRIHLIAAVKAVK 1223
Query: 990 STTDGVRTRLSFQDNNMAMISQMEPKSINEAIIDDSWIKAMKEELSQFERNKVWNLVP-- 1047
S +RT L + + + K I E + +I+A +E++Q + K W+
Sbjct: 1224 SIKP-IRTTLRYDE------AITYNKDIKEK---EKYIEAYHKEVNQLLKMKTWDTDEYY 1273
Query: 1048 ---NNQDKTIIGTRWVFRNKLDEEGKVVRNKARLVAQGYNQQEGIDYDETFAPVARLEAI 1104
K +I + ++F K D +KAR VA+G Q + A+
Sbjct: 1274 DRKEIDPKRVINSMFIFNKKRDGT-----HKARFVARGDIQHPDTYDSGMQSNTVHHYAL 1328
Query: 1105 RILLAYAAHKSIKLFQMDVKSAFLNGFLNEEVYVSQPPGFINKEKPNHVFKLTKALYGLK 1164
L+ A + + Q+D+ SA+L + EE+Y+ PP +K + +L K+LYGLK
Sbjct: 1329 MTSLSLALDNNYYITQLDISSAYLYADIKEELYIRPPPHLGMNDK---LIRLKKSLYGLK 1385
Query: 1165 QAPRAWYDRLSTFLIEN-GFSRGKIDTTLFRKTHNTDLLIVQVYVDDIIFGATKIKMCEE 1223
Q+ WY+ + ++LI+ G + + +F+ + T + ++VDD++ + + +
Sbjct: 1386 QSGANWYETIKSYLIQQCGMEEVRGWSCVFKNSQVT----ICLFVDDMVLFSKNLNSNKR 1441
Query: 1224 FSNLMQSEFEMSMMG------ELGF-FLGLQIK-QHSNGIFISQEKYVKDILKKYKMN-- 1273
++ +++ ++ E+ + LGL+IK Q + + E + + + K +
Sbjct: 1442 IIEKLKMQYDTKIINLGESDEEIQYDILGLEIKYQRGKYMKLGMENSLTEKIPKLNVPLN 1501
Query: 1274 -EAKIMSTPMHPSSSLDK-----DESGKSISEKEYRGMIGSLLYLTAS-RPDIVFAVGLC 1326
+ + +S P P +D+ +E + E + +IG Y+ R D+++ +
Sbjct: 1502 PKGRKLSAPGQPGLYIDQQELELEEDDYKMKVHEMQKLIGLASYVGYKFRFDLLYYINTL 1561
Query: 1327 ARFQTCAKESHLTAVKRIFRYLVGTTDLGLWYRKGSSFD----LVAYCDADYAGDKVERK 1382
A+ + L + +++ T D L + K LV DA Y G++ K
Sbjct: 1562 AQHILFPSKQVLDMTYELIQFIWNTRDKQLIWHKSKPVKPTNKLVVISDASY-GNQPYYK 1620
Query: 1383 STSGSCQFLGQALIGWSCRKQNTIALSTTEAEYVSAASCCSQILWVRNQLEDYSLRYTSV 1442
S G+ L +IG K + STTEAE + + + + +++ + +
Sbjct: 1621 SQIGNIYLLNGKVIGGKSTKASLTCTSTTEAEIHAISESVPLLNNLSYLIQELDKKPITK 1680
Query: 1443 PIYCDNTSAINLS-KNPIQHSRSKHIEIKHHFIRDHVQKKNIALSFVDTENQLADIFTKP 1501
+ D+ S I++ N + R++ K +RD V ++ + +++T+ +AD+ TKP
Sbjct: 1681 GLLTDSKSTISIIISNNEEKFRNRFFGTKAMRLRDEVSGNHLHVCYIETKKNIADVMTKP 1740
Query: 1502 LVEDRFNFLKEK 1513
L F L K
Sbjct: 1741 LPIKTFKLLTNK 1752
Score = 59.7 bits (143), Expect = 6e-08
Identities = 31/126 (24%), Positives = 60/126 (47%), Gaps = 6/126 (4%)
Query: 755 DRICEACVKGKQVKSSF---KTIEFISTQKPLELLHIDLFGPVQTASLTGKRYGFVIVDD 811
D C C+ GK K +++ ++ +P + LH D+FGPV + Y D+
Sbjct: 634 DYQCPDCLIGKSTKHRHIKGSRLKYQNSYEPFQYLHTDIFGPVHNLPKSAPSYFISFTDE 693
Query: 812 FSRFTWV--LFLKHKDESFEAFQNFCKRVQNEKGYNIITVRSDHGGEFENASFKTFFDEN 869
++F WV L + +D + F ++N+ +++ ++ D G E+ N + F ++N
Sbjct: 694 TTKFRWVYPLHDRREDSILDVFTTILAFIKNQFQASVLVIQMDRGSEYTNRTLHKFLEKN 753
Query: 870 -VFGCY 874
+ CY
Sbjct: 754 GITPCY 759
>YMT5_YEAST (Q04214) Transposon Ty1 protein B
Length = 1328
Score = 117 bits (293), Expect = 2e-25
Identities = 135/612 (22%), Positives = 264/612 (43%), Gaps = 63/612 (10%)
Query: 930 HSKPKENEDTEVPTLQNVPVQNTENTVEKEDDQNAQDQSFQSPPRSWRMVGDHPTDQIIG 989
+SK + ED E TE V + D N ++ PPRS + + + +
Sbjct: 749 NSKKRSLEDNE-----------TEIKVSR-DTWNTKNMRSLEPPRSKKRIHLIAAVKAVK 796
Query: 990 STTDGVRTRLSFQDNNMAMISQMEPKSINEAIIDDSWIKAMKEELSQFERNKVWNLVP-- 1047
S +RT L + + + K I E + +I+A +E++Q + K W+
Sbjct: 797 SIKP-IRTTLRYDE------AITYNKDIKEK---EKYIEAYHKEVNQLLKMKTWDTDKYY 846
Query: 1048 ---NNQDKTIIGTRWVFRNKLDEEGKVVRNKARLVAQGYNQQEGIDYDETFAPVARLEAI 1104
K +I + ++F K D +KAR VA+G Q + A+
Sbjct: 847 DRKEIDPKRVINSMFIFNRKRDGT-----HKARFVARGDIQHPDTYDSGMQSNTVHHYAL 901
Query: 1105 RILLAYAAHKSIKLFQMDVKSAFLNGFLNEEVYVSQPPGFINKEKPNHVFKLTKALYGLK 1164
L+ A + + Q+D+ SA+L + EE+Y+ PP +K + +L K+LYGLK
Sbjct: 902 MTSLSLALDNNYYITQLDISSAYLYADIKEELYIRPPPHLGMNDK---LIRLKKSLYGLK 958
Query: 1165 QAPRAWYDRLSTFLIEN-GFSRGKIDTTLFRKTHNTDLLIVQVYVDDIIFGATKIKMCEE 1223
Q+ WY+ + ++LI+ G + + +F + T + ++VDD++ + + +
Sbjct: 959 QSGANWYETIKSYLIKQCGMEEVRGWSCVFENSQVT----ICLFVDDMVLFSKNLNSNKR 1014
Query: 1224 FSNLMQSEFEMSMMG------ELGF-FLGLQIK-QHSNGIFISQEKYVKDILKKYKMN-- 1273
+ ++ +++ ++ E+ + LGL+IK Q + + E + + + K +
Sbjct: 1015 IIDKLKMQYDTKIINLGESDEEIQYDILGLEIKYQRGKYMKLGMENSLTEKIPKLNVPLN 1074
Query: 1274 -EAKIMSTPMHPSSSLDK-----DESGKSISEKEYRGMIGSLLYLTAS-RPDIVFAVGLC 1326
+ + +S P P +D+ +E + E + +IG Y+ R D+++ +
Sbjct: 1075 PKGRKLSAPGQPGLYIDQQELELEEDDYKMKVHEMQKLIGLASYVGYKFRFDLLYYINTL 1134
Query: 1327 ARFQTCAKESHLTAVKRIFRYLVGTTDLGLWYRKGSSFD----LVAYCDADYAGDKVERK 1382
A+ + L + +++ T D L + K LV DA Y G++ K
Sbjct: 1135 AQHILFPSKQVLDMTYELIQFIWNTRDKQLIWHKSKPVKPTNKLVVISDASY-GNQPYYK 1193
Query: 1383 STSGSCQFLGQALIGWSCRKQNTIALSTTEAEYVSAASCCSQILWVRNQLEDYSLRYTSV 1442
S G+ L +IG K + STTEAE + + + + +++ + +
Sbjct: 1194 SQIGNIYLLNGKVIGGKSTKASLTCTSTTEAEIHAISESVPLLNNLSYLIQELDKKPITK 1253
Query: 1443 PIYCDNTSAINLS-KNPIQHSRSKHIEIKHHFIRDHVQKKNIALSFVDTENQLADIFTKP 1501
+ D+ S I++ N + R++ K +RD V ++ + +++T+ +AD+ TKP
Sbjct: 1254 GLLTDSKSTISIIISNNEEKFRNRFFGTKAMRLRDEVSGNHLHVCYIETKKNIADVMTKP 1313
Query: 1502 LVEDRFNFLKEK 1513
L F L K
Sbjct: 1314 LPIKTFKLLTNK 1325
Score = 59.7 bits (143), Expect = 6e-08
Identities = 31/126 (24%), Positives = 60/126 (47%), Gaps = 6/126 (4%)
Query: 755 DRICEACVKGKQVKSSF---KTIEFISTQKPLELLHIDLFGPVQTASLTGKRYGFVIVDD 811
D C C+ GK K +++ ++ +P + LH D+FGPV + Y D+
Sbjct: 207 DYQCPDCLIGKSTKHRHIKGSRLKYQNSYEPFQYLHTDIFGPVHNLPKSAPSYFISFTDE 266
Query: 812 FSRFTWV--LFLKHKDESFEAFQNFCKRVQNEKGYNIITVRSDHGGEFENASFKTFFDEN 869
++F WV L + +D + F ++N+ +++ ++ D G E+ N + F ++N
Sbjct: 267 TTKFRWVYPLHDRREDSILDVFTTILAFIKNQFQASVLVIQMDRGSEYTNRTLHKFLEKN 326
Query: 870 -VFGCY 874
+ CY
Sbjct: 327 GITPCY 332
>M820_ARATH (P92520) Hypothetical mitochondrial protein AtMg00820
(ORF170)
Length = 170
Score = 99.0 bits (245), Expect = 9e-20
Identities = 48/99 (48%), Positives = 67/99 (67%)
Query: 1013 EPKSINEAIIDDSWIKAMKEELSQFERNKVWNLVPNNQDKTIIGTRWVFRNKLDEEGKVV 1072
EPKS+ A+ D W +AM+EEL RNK W LVP ++ I+G +WVF+ KL +G +
Sbjct: 27 EPKSVIFALKDPGWCQAMQEELDALSRNKTWILVPPPVNQNILGCKWVFKTKLHSDGTLD 86
Query: 1073 RNKARLVAQGYNQQEGIDYDETFAPVARLEAIRILLAYA 1111
R KARLVA+G++Q+EGI + ET++PV R IR +L A
Sbjct: 87 RLKARLVAKGFHQEEGIYFVETYSPVVRTATIRTILNVA 125
>M240_ARATH (P93290) Hypothetical mitochondrial protein AtMg00240
(ORF111a)
Length = 111
Score = 63.2 bits (152), Expect = 5e-09
Identities = 29/88 (32%), Positives = 50/88 (55%)
Query: 1310 LYLTASRPDIVFAVGLCARFQTCAKESHLTAVKRIFRYLVGTTDLGLWYRKGSSFDLVAY 1369
+YLT +RPD+ FAV ++F + ++ + + AV ++ Y+ GT GL+Y S L A+
Sbjct: 1 MYLTITRPDLTFAVNRLSQFSSASRTAQMQAVYKVLHYVKGTVGQGLFYSATSDLQLKAF 60
Query: 1370 CDADYAGDKVERKSTSGSCQFLGQALIG 1397
D+D+A R+S +G C + +G
Sbjct: 61 ADSDWASCPDTRRSVTGFCSLVPLWFLG 88
>GOB1_HUMAN (Q14789) Golgi autoantigen, golgin subfamily B member 1
(Giantin) (Macrogolgin) (Golgi complex-associated
protein, 372-kDa) (GCP372)
Length = 3259
Score = 57.0 bits (136), Expect = 4e-07
Identities = 93/479 (19%), Positives = 203/479 (41%), Gaps = 47/479 (9%)
Query: 29 GKMELFLRSQDNDMWAVITNGDFVPTNKEGAVKAKSAWSTDEKAQVLLNSKA--RLFLSC 86
G++++ L +D ++ + N D T K+ S+ D++ +V+ +K R F
Sbjct: 2160 GEIQVTLNKKDKEVQQLQENLDSTVTQLAAFTKSMSSLQ-DDRDRVIDEAKKWERKFSDA 2218
Query: 87 ALTMEESERV--DECTNAKGVWDTLKIHHEGTSHVKETRID----IGVRKFEVFEMSENE 140
+ EE R+ D C+ K + IH E + +R++ I K + + +
Sbjct: 2219 IQSKEEEIRLKEDNCSVLKDQLRQMSIHMEELK-INISRLEHDKQIWESKAQTEVQLQQK 2277
Query: 141 TIDEMYARFTTIVNEMRSLGKAY-STHDRIRKI---LRCLPSMWRPMVTAITQAKDLKSM 196
D + +++++ Y S+ + + K+ L+ L + ++ + K+ K
Sbjct: 2278 VCDTLQGENKELLSQLEETRHLYHSSQNELAKLESELKSLKDQLTDLSNSLEKCKEQKG- 2336
Query: 197 NLEDLIGSLRAHEVVLQGDKPVKKVRTLALKASQQTSSVDDEDVQEPQE----LEEVHEE 252
NLE G +R E +Q K + L+AS++ +S E++ ++ L EE
Sbjct: 2337 NLE---GIIRQQEADIQNSKFSYEQLETDLQASRELTSRLHEEINMKEQKIISLLSGKEE 2393
Query: 253 EAEDELALISKRIQRMMLRRNQIRKKFPKTNISIKTEADKSQVTCYGCNKTGHFKNECPD 312
+ +A + ++ + + + + + NI ++ E K+ +KT N+ +
Sbjct: 2394 AIQVAIAELRQQHDKEIKELENLLSQEEEENIVLEEENKKA------VDKT----NQLME 2443
Query: 313 IKKVQRKPPFKKKAMITWDDMEESDSQEDADTDMGLMAQSDDEEEVIIYKTDSLYKD--- 369
K +K ++KA + S Q D D +G Q ++ II + D L ++
Sbjct: 2444 TLKTIKKENIQQKAQLDSFVKSMSSLQNDRDRIVGDYQQLEERHLSIILEKDQLIQEAAA 2503
Query: 370 ----LENKIDSLLYDSNFLTNRCHSLIKELSEIKEE-KEILQNKYDESRKTIKI------ 418
L+ +I L + L + L EL + +E+ +++ K + ++ +++
Sbjct: 2504 ENNKLKEEIRGLRSHMDDLNSENAKLDAELIQYREDLNQVITIKDSQQKQLLEVQLQQNK 2563
Query: 419 -LQDSHFDMSEKQREINRKQKGIMSVPSEVQKENILLKKEVETLKKDLTGFIKSTETFQ 476
L++ + + EK +E + + + +Q+E L KE+E+LK ++ + Q
Sbjct: 2564 ELENKYAKLEEKLKESEEANEDLRRSFNALQEEKQDLSKEIESLKVSISQLTRQVTALQ 2622
Score = 44.7 bits (104), Expect = 0.002
Identities = 58/257 (22%), Positives = 111/257 (42%), Gaps = 42/257 (16%)
Query: 221 VRTLALKASQQTSSVDDEDVQEPQELEEVHEEEAE-DELALISKRIQRMMLRRNQIRKKF 279
++ L + Q D+ Q EL E+ ++ E E + ++IQR + RK+
Sbjct: 1436 IKALHTQLEMQAKEHDERIKQLQVELCEMKQKPEEIGEESRAKQQIQRKLQAALISRKEA 1495
Query: 280 PKTNISIKTEADKSQVTCYGCNKTGHFKNECPDIKKVQRKPPFKKKAMITWDDMEESDSQ 339
K N S++ E ++ T K+ D++ + S
Sbjct: 1496 LKENKSLQEELSLARGTIERLTKS------LADVES------------------QVSAQN 1531
Query: 340 EDADTDMGLMAQSDDEEEVIIYKTDSLYKDLENKIDSLLYDSNFLTNRCHSLIKELSEIK 399
++ DT +G +A +E + +I ++D L ++ L++ C SL L +
Sbjct: 1532 KEKDTVLGRLALLQEERDKLI-----------TEMDRSLLENQSLSSSCESLKLALEGLT 1580
Query: 400 EEKEILQNKYDESRKTIKILQDSHFDMSEKQREINRKQKGIMSVPSEVQKENILLKKEVE 459
E+KE L K ES K+ KI + + + EK +E+ ++ + ++ V E ++ VE
Sbjct: 1581 EDKEKLV-KEIESLKSSKIAEST--EWQEKHKELQKEYEILLQSYENVSNEAERIQHVVE 1637
Query: 460 TL---KKDLTGFIKSTE 473
+ K++L G ++STE
Sbjct: 1638 AVRQEKQELYGKLRSTE 1654
Score = 40.4 bits (93), Expect = 0.038
Identities = 35/140 (25%), Positives = 64/140 (45%), Gaps = 17/140 (12%)
Query: 362 KTDSLYKDLENKIDSLLYDSNFLTNRCHSLIKELSEIKEEKEI-------LQNKYDESRK 414
K D L K + K + + Y S L+ + +L K +EI E++++ L+ + E +
Sbjct: 1393 KLDELQKLISKKEEDVSYLSGQLSEKEAALTKIQTEIIEQEDLIKALHTQLEMQAKEHDE 1452
Query: 415 TIKILQDSHFDMSEKQREINRKQKGIMSVPSEVQ----------KENILLKKEVETLKKD 464
IK LQ +M +K EI + + + ++Q KEN L++E+ +
Sbjct: 1453 RIKQLQVELCEMKQKPEEIGEESRAKQQIQRKLQAALISRKEALKENKSLQEELSLARGT 1512
Query: 465 LTGFIKSTETFQNIVGSQNK 484
+ KS ++ V +QNK
Sbjct: 1513 IERLTKSLADVESQVSAQNK 1532
Score = 36.6 bits (83), Expect = 0.54
Identities = 74/332 (22%), Positives = 132/332 (39%), Gaps = 59/332 (17%)
Query: 194 KSMNLEDLIGSLRAHEVVLQGDKPVKK--------VRTLALKASQQTSSVDDED--VQEP 243
+ ++LE+ IG ++ + + DK V++ V LA +S DD D + E
Sbjct: 2151 EKVHLEETIGEIQV--TLNKKDKEVQQLQENLDSTVTQLAAFTKSMSSLQDDRDRVIDEA 2208
Query: 244 QELEE-----VHEEEAEDELALISKRIQRMMLRRNQIRKKFPKTNIS------------I 286
++ E + +E E L + + + LR+ I + K NIS
Sbjct: 2209 KKWERKFSDAIQSKEEEIRLKEDNCSVLKDQLRQMSIHMEELKINISRLEHDKQIWESKA 2268
Query: 287 KTEADKSQVTC---YGCNK--------TGH-FKNECPDIKKVQRKPPFKKKAMITWDDME 334
+TE Q C G NK T H + + ++ K++ + K + +
Sbjct: 2269 QTEVQLQQKVCDTLQGENKELLSQLEETRHLYHSSQNELAKLESELKSLKDQLT-----D 2323
Query: 335 ESDSQEDADTDMGLMAQSDDEEEVIIYKTDSLYKDLENKIDSLLYDSNFLTNRCHSLI-- 392
S+S E G + ++E I + Y+ LE + + S LT+R H I
Sbjct: 2324 LSNSLEKCKEQKGNLEGIIRQQEADIQNSKFSYEQLETDLQA----SRELTSRLHEEINM 2379
Query: 393 ---KELSEIKEEKEILQNKYDESR----KTIKILQDSHFDMSEKQREINRKQKGIMSVPS 445
K +S + ++E +Q E R K IK L++ E+ + + K + +
Sbjct: 2380 KEQKIISLLSGKEEAIQVAIAELRQQHDKEIKELENLLSQEEEENIVLEEENKKAVDKTN 2439
Query: 446 EVQKENILLKKEVETLKKDLTGFIKSTETFQN 477
++ + +KKE K L F+KS + QN
Sbjct: 2440 QLMETLKTIKKENIQQKAQLDSFVKSMSSLQN 2471
Score = 36.6 bits (83), Expect = 0.54
Identities = 66/340 (19%), Positives = 144/340 (41%), Gaps = 47/340 (13%)
Query: 248 EVHEEEAEDELALISKRIQRMMLRRNQIRKKFPKTNISIKTE-ADKSQVTCYGCNKTGHF 306
++ E E E+ + K + + + Q+ K+ K I+ E +K Q H
Sbjct: 1948 QIKNELLESEMKNLKKCVSELEEEKQQLVKEKTKVESEIRKEYLEKIQGAQKEPGNKSHA 2007
Query: 307 KNECPDIKKVQRKPPFKKKAMITWDD-------------MEESDSQEDADTDMGLMAQSD 353
K +K+ Q++ +K I + + +++SQ+D + +AQ+
Sbjct: 2008 KELQELLKEKQQEVKQLQKDCIRYQEKISALERTVKALEFVQTESQKDLEITKENLAQAV 2067
Query: 354 D-----EEEVIIYKTDSLYKDLENKIDSLLYDSNFLTNRCHSLIKEL-SEIKEEKEILQN 407
+ + E+ +K L D +++ +L D+ L S + + S++K++ E L+
Sbjct: 2068 EHRKKAQAELASFKV--LLDDTQSEAARVLADNLKLKKELQSNKESVKSQMKQKDEDLER 2125
Query: 408 KYDESRKTIKILQDSHFDMSEKQREINRKQKGIMSVPSEVQKENILLKKEVETLKKD--- 464
+ +++ + K L++ +M EK + R++ + E+Q KEV+ L+++
Sbjct: 2126 RLEQAEE--KHLKEKK-NMQEKLDALRREKVHLEETIGEIQVTLNKKDKEVQQLQENLDS 2182
Query: 465 ----LTGFIKSTETFQNIVGSQNKSTKKSGLGFKDPSKIIGSFVPKAKIRVKCCFCDKYG 520
L F KS + Q+ KK F D + + +IR+K
Sbjct: 2183 TVTQLAAFTKSMSSLQDDRDRVIDEAKKWERKFSDAIQ-----SKEEEIRLK-------- 2229
Query: 521 HNESVCHVKKKFIKQNNLNLSSERSHLNRSESSQKAEKAK 560
E C V K ++Q ++++ + +++R E ++ ++K
Sbjct: 2230 --EDNCSVLKDQLRQMSIHMEELKINISRLEHDKQIWESK 2267
Score = 35.0 bits (79), Expect = 1.6
Identities = 22/96 (22%), Positives = 49/96 (50%), Gaps = 4/96 (4%)
Query: 367 YKDLENKIDSLLYDSNFLTNRCHSLIKELSEIKEEKEILQNKYDES----RKTIKILQDS 422
+K+L+ + + LL ++N + + +++EK+ L K + ++T K LQ++
Sbjct: 1608 HKELQKEYEILLQSYENVSNEAERIQHVVEAVRQEKQELYGKLRSTEANKKETEKQLQEA 1667
Query: 423 HFDMSEKQREINRKQKGIMSVPSEVQKENILLKKEV 458
+M E + ++ + K E+++EN L+ EV
Sbjct: 1668 EQEMEEMKEKMRKFAKSKQQKILELEEENDRLRAEV 1703
Score = 34.7 bits (78), Expect = 2.1
Identities = 29/126 (23%), Positives = 58/126 (46%), Gaps = 9/126 (7%)
Query: 382 NFLTNRCHSLIKELSEIKEEKEILQNKYDESR----KTIKILQDSHFDMSEKQREINRKQ 437
N LT + HSL E + E+LQN+ D+ + + +++ + K+ E+
Sbjct: 792 NLLTEQIHSLSIEAKSKDVKIEVLQNELDDVQLQFSEQSTLIRSLQSQLQNKESEVLEGA 851
Query: 438 KGIMSVPSEVQK-ENILLKKEVETLKKD--LTGFIKSTETFQNIVGSQNKSTKKSGLGFK 494
+ + + S+V++ L +KE+E K D L + ET Q + + K + + + F
Sbjct: 852 ERVRHISSKVEELSQALSQKELEITKMDQLLLEKKRDVETLQQTI--EEKDQQVTEISFS 909
Query: 495 DPSKII 500
K++
Sbjct: 910 MTEKMV 915
Score = 33.9 bits (76), Expect = 3.5
Identities = 34/146 (23%), Positives = 64/146 (43%), Gaps = 16/146 (10%)
Query: 349 MAQSDDEEEVIIYKTDSLY----KDLENKIDSLLYDSNFLTNRCHSLIKELSEIKEEKEI 404
++Q+ ++E+ I K D L +D+E ++ +T S+ +++ ++ EEK
Sbjct: 864 LSQALSQKELEITKMDQLLLEKKRDVETLQQTIEEKDQQVTEISFSMTEKMVQLNEEKFS 923
Query: 405 LQNKYDESRKTIKILQDSHFDMSEKQREINRKQKGI------MSVPSEVQKENI-----L 453
L + ++ + +L + E+ E N G+ MS ++ KE + L
Sbjct: 924 LGVEIKTLKEQLNLLSRAEEAKKEQVEEDNEVSSGLKQNYDEMSPAGQISKEELQHEFDL 983
Query: 454 LKKEVETLKKDL-TGFIKSTETFQNI 478
LKKE E K+ L I E Q +
Sbjct: 984 LKKENEQRKRKLQAALINRKELLQRV 1009
>MYH4_HUMAN (Q9Y623) Myosin heavy chain, skeletal muscle, fetal
(Myosin heavy chain IIb) (MyHC-IIb)
Length = 1939
Score = 53.5 bits (127), Expect = 4e-06
Identities = 99/437 (22%), Positives = 184/437 (41%), Gaps = 72/437 (16%)
Query: 84 LSCALTMEESERVDECTNAKGVWDTLKI---HHEGTSHVKETRIDIGVRKFEVFEMSENE 140
++ LT ++ + DEC+ K D L++ E H E ++ K EM+
Sbjct: 936 INAELTAKKRKLEDECSELKKDIDDLELTLAKVEKEKHATENKV-----KNLTEEMAG-- 988
Query: 141 TIDEMYARFTTIVNEMRSLGKAYS-THDRIRKILRCLPSMWRPMVTAITQAKDLKSMNLE 199
+DE A+ T E ++L +A+ T D ++ M V +T+AK ++
Sbjct: 989 -LDETIAKLT---KEKKALQEAHQQTLDDLQ--------MEEDKVNTLTKAKTKLEQQVD 1036
Query: 200 DLIGSLRAHEVVLQGDKPVKKVRTLALKASQQTSSVDDEDVQEPQELEEVHEEE------ 253
DL GSL + + + K+ LK +Q+++ + D Q+ E + E E
Sbjct: 1037 DLEGSLEQEKKLCMDLERAKRKLEGDLKLAQESTMDTENDKQQLNEKLKKKEFEMSNLQG 1096
Query: 254 -AEDELAL---ISKRIQRMMLRRNQIRKKFPKTNIS-IKTEADKSQVT------CYGCNK 302
EDE AL + K+I+ + R ++ ++ S K E +S ++ +
Sbjct: 1097 KIEDEQALAMQLQKKIKELQARIEELEEEIEAERASRAKAEKQRSDLSRELEEISERLEE 1156
Query: 303 TGHFKNECPDIKKVQRKPPFKKKAMITWDDMEESDSQEDADTDMGLMAQSDDEEEVIIYK 362
G + ++ K +R+ F+K D+EES Q +A T L + D + +
Sbjct: 1157 AGGATSAQIELNK-KREAEFQKMRR----DLEESTLQHEA-TAAALRKKHADSVAELGKQ 1210
Query: 363 TDSLYK---DLENKIDSLLYDSNFLTNR--------------CHSLIKELSEIKEEKEIL 405
DSL + LE + L + N L + C +L +LSEIK ++E
Sbjct: 1211 IDSLQRVKQKLEKEKSELKMEINDLASNMETVSKAKANFEKMCRTLEDQLSEIKTKEEEQ 1270
Query: 406 QNKYDE-SRKTIKILQDSHFDMSEKQREINRKQKGIMSVPSEVQKENILLKKEVETLKKD 464
Q +E S + ++ H + E R+++ K ++ S++ + +++E LK+
Sbjct: 1271 QRLINELSAQKARL----HTESGEFSRQLDEKD----AMVSQLSRGKQAFTQQIEELKRQ 1322
Query: 465 LTGFIKSTETFQNIVGS 481
L K+ T + + S
Sbjct: 1323 LEEETKAKSTLAHALQS 1339
Score = 37.4 bits (85), Expect = 0.32
Identities = 44/232 (18%), Positives = 96/232 (40%), Gaps = 21/232 (9%)
Query: 341 DADTDMGLMAQSDDEEEVIIYKTDSLYKDLENKIDSLLYDSNFLTNRCHSLIKELSEIKE 400
+ + +M M + ++ + + KT++ K+LE K+ +L+ + N L + + L++ +E
Sbjct: 848 ETEKEMANMKEEFEKTKEELAKTEAKRKELEEKMVTLMQEKNDLQLQVQAEADALADAEE 907
Query: 401 EKEILQNKYDESRKTIKILQDSHFDMSEKQREINRKQKGIMSVPSEVQKENILLKKEVET 460
+ L + IK + + D E E+ K++ + SE LKK+++
Sbjct: 908 RCDQLIKTKIQLEAKIKEVTERAEDEEEINAELTAKKRKLEDECSE-------LKKDIDD 960
Query: 461 LKKDLTGFIKSTETFQNIVGSQNKSTKKSGLGFKDPSKIIGSFVPKAKIRVKCCFCDKYG 520
L+ L K +N V +N + + +GL + I + K +
Sbjct: 961 LELTLAKVEKEKHATENKV--KNLTEEMAGL-----DETIAKLTKEKKAL-------QEA 1006
Query: 521 HNESVCHVKKKFIKQNNLNLSSERSHLNRSESSQKAEKAKKTCFYCNKSDHK 572
H +++ ++ + K N L + + + E+ KK C ++ K
Sbjct: 1007 HQQTLDDLQMEEDKVNTLTKAKTKLEQQVDDLEGSLEQEKKLCMDLERAKRK 1058
Score = 34.7 bits (78), Expect = 2.1
Identities = 60/340 (17%), Positives = 140/340 (40%), Gaps = 28/340 (8%)
Query: 150 TTIVNEMRSLGKAYSTHDRIRKILRCLPSMWRPMVTAITQAKDLKSMNLEDLIGSLRAHE 209
+T+ +E + D + ++ + + S+ R + +LK M + DL ++ E
Sbjct: 1185 STLQHEATAAALRKKHADSVAELGKQIDSLQRVKQKLEKEKSELK-MEINDLASNM---E 1240
Query: 210 VVLQGDKPVKKV-RTLA-------LKASQQTSSVDDEDVQEPQELEEVHE-----EEAED 256
V + +K+ RTL K +Q +++ Q+ + E E +E +
Sbjct: 1241 TVSKAKANFEKMCRTLEDQLSEIKTKEEEQQRLINELSAQKARLHTESGEFSRQLDEKDA 1300
Query: 257 ELALISKRIQRMMLRRNQIRKKFPKTNISIKTEADKSQVTCYGCNKTGHFKNECPDIK-K 315
++ +S+ Q + +++++ + + T A Q + C+ E + K +
Sbjct: 1301 MVSQLSRGKQAFTQQIEELKRQLEEETKAKSTLAHALQSARHDCDLLREQYEEEQEAKAE 1360
Query: 316 VQRKPPFKKKAMITW------DDMEESDSQEDADTDMGLMAQSDDEE-EVIIYKTDSLYK 368
+QR + W D ++ ++ E+A + Q +E E + K SL K
Sbjct: 1361 LQRGMSKANSEVAQWRTKYETDAIQRTEELEEAKKKLAQRLQDAEEHVEAVNSKCASLEK 1420
Query: 369 D---LENKIDSLLYDSNFLTNRCHSLIKELSEIKEEKEILQNKYDESRKTIKILQDSHFD 425
L+N+++ L+ D C +L K+ + + KY+E++ ++ Q
Sbjct: 1421 TKQRLQNEVEDLMIDVERSNAACIALDKKQRNFDKVLAEWKQKYEETQAELEASQKESRS 1480
Query: 426 MSEKQREINRKQKGIMSVPSEVQKENILLKKEVETLKKDL 465
+S + ++ + + +++EN L++E+ L + +
Sbjct: 1481 LSTELFKVKNAYEESLDHLETLKRENKNLQQEISDLTEQI 1520
Score = 34.7 bits (78), Expect = 2.1
Identities = 43/270 (15%), Positives = 110/270 (39%), Gaps = 40/270 (14%)
Query: 220 KVRTLALKASQQTSSVDDEDVQEPQELEEVHEEEAEDELALISKRIQRMMLRRNQIRKKF 279
K T A++ +++ + Q Q+ EE H E + A + K QR+ +N++
Sbjct: 1378 KYETDAIQRTEELEEAKKKLAQRLQDAEE-HVEAVNSKCASLEKTKQRL---QNEVE--- 1430
Query: 280 PKTNISIKTEADKSQVTCYGCNKTGHFKNECPDIKKVQRKPPFKKKAMITWDDMEESDSQ 339
+ + ++S C +K K K + W
Sbjct: 1431 -----DLMIDVERSNAACIALDK----------------KQRNFDKVLAEW-----KQKY 1464
Query: 340 EDADTDMGLMAQSDDEEEVIIYKTDSLYKDLENKIDSLLYDSNFLTNRCHSLIKELSE-- 397
E+ ++ + ++K + Y++ + +++L ++ L L ++++E
Sbjct: 1465 EETQAELEASQKESRSLSTELFKVKNAYEESLDHLETLKRENKNLQQEISDLTEQIAEGG 1524
Query: 398 --IKEEKEILQNKYDESRKTIKILQDSHFDMSEKQREINRKQKGIMSVPSEVQKENILLK 455
I E +++ + E + L+++ + ++ +I R Q + V SE+ ++
Sbjct: 1525 KHIHELEKVKKQLDHEKSELQTSLEEAEASLEHEEGKILRIQLELNQVKSEIDRKIAEKD 1584
Query: 456 KEVETLKKDLTGFIKSTETFQNIVGSQNKS 485
+E++ LK++ ++ E+ Q+ + ++ +S
Sbjct: 1585 EELDQLKRN---HLRVVESMQSTLDAEIRS 1611
>SMC2_YEAST (P38989) Structural maintenance of chromosome 2 (DA-box
protein SMC2)
Length = 1170
Score = 52.8 bits (125), Expect = 7e-06
Identities = 58/264 (21%), Positives = 123/264 (45%), Gaps = 34/264 (12%)
Query: 220 KVRTLALKASQQTSSVDDEDVQEPQELEEVHEEEAEDELALISKRIQRMMLRRNQIRKKF 279
K + ++LK Q+ S ++D++E + E + EL L++K ++ + ++ +K+
Sbjct: 762 KTKQMSLKKCQEEVSTIEKDMKEYDSDKGSKLNELKKELKLLAKELEE---QESESERKY 818
Query: 280 PK-TNISIKTEADKSQVTCYGCNKTGHFKNECPDIKKVQ-RKPPFKKKAMITWDDM---- 333
N+ ++TE S++ NKT N I+ ++ + K DD+
Sbjct: 819 DLFQNLELETEQLSSELDS---NKT-LLHNHLKSIESLKLENSDLEGKIRGVEDDLVTVQ 874
Query: 334 ----EESDSQEDADTDMGLMAQSDDEEEVIIYKTDSLYKDLENKIDSLLYDSNFLTNRCH 389
EE D D ++ +E E +I K K E ++ L++D N + +
Sbjct: 875 TELNEEKKRLMDIDDEL-------NELETLIKKKQDEKKSSELELQKLVHDLNKYKSNTN 927
Query: 390 SLIKELSEIKEEKEILQNKYDESRKTIKILQDSHFD--------MSEKQREINRK-QKGI 440
++ K + +++++ E L++ +D R +K + D ++EK +E+ +K I
Sbjct: 928 NMEKIIEDLRQKHEFLED-FDLVRNIVKQNEGIDLDTYRERSKQLNEKFQELRKKVNPNI 986
Query: 441 MSVPSEVQKENILLKKEVETLKKD 464
M++ V+K+ LK ++T++KD
Sbjct: 987 MNMIENVEKKEAALKTMIKTIEKD 1010
Score = 48.5 bits (114), Expect = 1e-04
Identities = 63/300 (21%), Positives = 127/300 (42%), Gaps = 38/300 (12%)
Query: 205 LRAHEVVLQGDKPVKKVRTLALKASQQTSSVDDEDVQEPQELEEVHE----------EEA 254
+RA + LQGD V S+ TS D+Q+ ++++ E EE
Sbjct: 647 IRARSITLQGD--VYDPEGTLSGGSRNTSESLLVDIQKYNQIQKQIETIQADLNHVTEEL 704
Query: 255 EDELALI--SKRIQ---RMMLRRNQIRKKFPKTNISIKTEADKSQVTCYGCNKTGHFKNE 309
+ + A +K IQ + L + + K+ N S + A ++ K +
Sbjct: 705 QTQYATSQKTKTIQSDLNLSLHKLDLAKRNLDANPSSQIIARNEEILRDIGECENEIKTK 764
Query: 310 CPDIKKVQRKPPFKKKAMITWDDMEESDSQEDADTDMGLMAQSDDEEEVIIYKTDSLYKD 369
+KK Q + +K M +D ++ + ++ L+A+ +E+E + L+++
Sbjct: 765 QMSLKKCQEEVSTIEKDMKEYDS-DKGSKLNELKKELKLLAKELEEQESESERKYDLFQN 823
Query: 370 LENKIDSLLYDSNFLTNRCHSLIKELSEIKEEKEILQNKYDESRKTIKILQDSHFDMSEK 429
LE + + L + + H+ +K + +K E L+ K I+ ++D D+
Sbjct: 824 LELETEQLSSELDSNKTLLHNHLKSIESLKLENSDLEGK-------IRGVED---DLVTV 873
Query: 430 QREINRKQKGIMSVPSEVQKENILLKK----------EVETLKKDLTGFIKSTETFQNIV 479
Q E+N ++K +M + E+ + L+KK E++ L DL + +T + I+
Sbjct: 874 QTELNEEKKRLMDIDDELNELETLIKKKQDEKKSSELELQKLVHDLNKYKSNTNNMEKII 933
Score = 36.2 bits (82), Expect = 0.71
Identities = 55/251 (21%), Positives = 102/251 (39%), Gaps = 48/251 (19%)
Query: 352 SDDEEEVIIYKTDSLYKD-----LENKIDSLLYDSNFLTNRCHSLIKELSEIKEEKEILQ 406
++D EE+ + K L+K+ LENK + LL + + L ++ L++ E+ + L+
Sbjct: 278 NEDVEEIKLQKEKELHKEGTISKLENKENGLLNEISRLKTSLSIKVENLNDTTEKSKALE 337
Query: 407 NKYDESRKTIKILQDSHFDMSEKQREINRKQKGIMSVPSEVQKENILLKKEVETLKKDLT 466
++ + S + EK+ +K V ++ K+ L K++ E L LT
Sbjct: 338 SE----------IASSSAKLIEKKSAYANTEKDYKMVQEQLSKQRDLYKRK-EELVSTLT 386
Query: 467 GFIKSTETFQNIVGSQ-----------NKSTKKSGLGFKDPSKIIGSFVPKAKIRVKCCF 515
I ST +Q + + KKS + + K + + PK K K
Sbjct: 387 TGISSTGAADGGYNAQLAKAKTELNEVSLAIKKSSMKMELLKKELLTIEPKLKEATK--- 443
Query: 516 CDKYGHNESVCHVKKKFIKQNNLNLSSERSHL-------NRSESSQKAEKAKKTCFY--C 566
NE + K +KQ R+ L +R + ++ E K+ +Y C
Sbjct: 444 -----DNE----LNVKHVKQCQETCDKLRARLVEYGFDPSRIKDLKQREDKLKSHYYQTC 494
Query: 567 NKSDHKRQNVT 577
S++ ++ VT
Sbjct: 495 KNSEYLKRRVT 505
>MYH4_RABIT (Q28641) Myosin heavy chain, skeletal muscle, juvenile
Length = 1938
Score = 51.2 bits (121), Expect = 2e-05
Identities = 87/428 (20%), Positives = 178/428 (41%), Gaps = 54/428 (12%)
Query: 84 LSCALTMEESERVDECTNAKGVWDTLKI---HHEGTSHVKETRIDIGVRKFEVFEMSENE 140
++ LT ++ + DEC+ K D L++ E H E ++ K EM+
Sbjct: 935 INAELTAKKRKLEDECSELKKDIDDLELTLAKVEKEKHATENKV-----KNLTEEMAG-- 987
Query: 141 TIDEMYARFTTIVNEMRSLGKAYS-THDRIRKILRCLPSMWRPMVTAITQAKDLKSMNLE 199
+DE A+ T E ++L +A+ T D ++ V +T+AK ++
Sbjct: 988 -LDETIAKLT---KEKKALQEAHQQTLDDLQAE--------EDKVNTLTKAKTKLEQQVD 1035
Query: 200 DLIGSLRAHEVVLQGDKPVKKVRTLALKASQQTSSVDDEDVQEPQELEEVHEEE------ 253
DL GSL + + + K+ LK +Q+++ + D Q+ E + E E
Sbjct: 1036 DLEGSLEQEKKIRMDLERAKRKLEGDLKLAQESTMDIENDKQQLDEKLKKKEFEMSNLQS 1095
Query: 254 -AEDELAL---ISKRIQRMMLRRNQIRKKFPKTNIS-IKTEADKSQVT------CYGCNK 302
EDE AL + K+I+ + R ++ ++ S K E +S ++ +
Sbjct: 1096 KIEDEQALAMQLQKKIKELQARIEELEEEIEAERASRAKAEKQRSDLSRELEEISERLEE 1155
Query: 303 TGHFKNECPDIKKVQRKPPFKKKAMITWDDMEESDSQEDADT---------DMGLMAQSD 353
G + ++ K +R+ F+K D+EE+ Q +A + + +
Sbjct: 1156 AGGATSAQIEMNK-KREAEFQKMRR----DLEEATLQHEATAATLRKKHADSVAELGEQI 1210
Query: 354 DEEEVIIYKTDSLYKDLENKIDSLLYDSNFLTNRCHSLIKELSEIKEEKEILQNKYDESR 413
D + + K + +L+ +ID L + ++ +L K ++++ L+ K +E +
Sbjct: 1211 DNLQRVKQKLEKEKSELKMEIDDLASNMETVSKAKGNLEKMCRTLEDQVSELKTKEEEHQ 1270
Query: 414 KTIKILQDSHFDMSEKQREINRKQKGIMSVPSEVQKENILLKKEVETLKKDLTGFIKSTE 473
+ I L + + E +R+ S+ S++ + +++E LK+ L IK+
Sbjct: 1271 RLINDLSAQRARLQTESGEFSRQLDEKDSLVSQLSRGKQAFTQQIEELKRQLEEEIKAKS 1330
Query: 474 TFQNIVGS 481
+ + S
Sbjct: 1331 ALAHALQS 1338
Score = 36.6 bits (83), Expect = 0.54
Identities = 44/263 (16%), Positives = 110/263 (41%), Gaps = 26/263 (9%)
Query: 227 KASQQTSSVDDEDVQEPQELEEVHEEEAEDELALISKRIQRMMLRRNQIRKKFPKTNISI 286
+ +Q + + + +Q +ELEE A+ +LA + + + N KT +
Sbjct: 1370 EVAQWRTKYETDAIQRTEELEE-----AKKKLAQRLQDAEEHVEAVNAKCASLEKTKQRL 1424
Query: 287 KTEADKSQVTCYGCNKTGHFKNECPDIKKVQRKPPFKKKAMITWDDMEESDSQEDADTDM 346
+ E + + N C + K QR K + W E+ ++
Sbjct: 1425 QNEVEDLMIDVERTNAA------CAALDKKQRN---FDKILAEW-----KHKYEETHAEL 1470
Query: 347 GLMAQSDDEEEVIIYKTDSLYKDLENKIDSLLYDSNFLTNRCHSLIKELSE----IKEEK 402
+ ++K + Y++ +++++L ++ L L ++++E I E +
Sbjct: 1471 EASQKESRSLSTEVFKVKNAYEESLDQLETLKRENKNLQQEISDLTEQIAEGGKRIHELE 1530
Query: 403 EILQNKYDESRKTIKILQDSHFDMSEKQREINRKQKGIMSVPSEVQKENILLKKEVETLK 462
++ + E + L+++ + ++ +I R Q + V SE+ ++ +E++ LK
Sbjct: 1531 KVKKQVEQEKSELQAALEEAEASLEHEEGKILRIQLELNQVKSEIDRKIAEKDEEIDQLK 1590
Query: 463 KDLTGFIKSTETFQNIVGSQNKS 485
++ I+ E+ Q+ + ++ +S
Sbjct: 1591 RN---HIRVVESMQSTLDAEIRS 1610
Score = 35.0 bits (79), Expect = 1.6
Identities = 85/404 (21%), Positives = 150/404 (37%), Gaps = 56/404 (13%)
Query: 72 AQVLLNSKARL-FLSCALTMEESERVDECTNAKGVWDTLKIHHEGTSHVKETRIDIGVRK 130
AQ+ +N K F +EE+ E T A TL+ H + +ID R
Sbjct: 1162 AQIEMNKKREAEFQKMRRDLEEATLQHEATAA-----TLRKKHADSVAELGEQIDNLQRV 1216
Query: 131 FEVFEMSENETIDEMYARFTTIVNEMRSLGKAYSTHDRIRKILRCLPSMWRPMVTAITQA 190
+ E ++E E+ + + M ++ KA L M R + +++
Sbjct: 1217 KQKLEKEKSELKMEI----DDLASNMETVSKAKGN----------LEKMCRTLEDQVSEL 1262
Query: 191 KDLKSMNLEDLIGSLRAHEVVLQGDKPVKKVRTLALKAS--QQTSSVDDEDVQEPQELEE 248
K K + LI L A LQ + + R L K S Q S Q+ +EL+
Sbjct: 1263 KT-KEEEHQRLINDLSAQRARLQTESG-EFSRQLDEKDSLVSQLSRGKQAFTQQIEELKR 1320
Query: 249 VHEEEAEDELALI----SKRIQRMMLRRNQIRKKFPKTNISIKTEADKSQVTCYGCNKTG 304
EEE + + AL S R +LR ++ K + S+V +
Sbjct: 1321 QLEEEIKAKSALAHALQSARHDCDLLREQYEEEQEAKAELQRAMSKANSEVAQW------ 1374
Query: 305 HFKNECPDIKKVQRKPPFKKKAMITWDDMEESDSQEDADTDMGLMAQSDDEEEVIIYKTD 364
K E I++ + KKK D EE E + K
Sbjct: 1375 RTKYETDAIQRTEELEEAKKKLAQRLQDAEE-------------------HVEAVNAKCA 1415
Query: 365 SLYKD---LENKIDSLLYDSNFLTNRCHSLIKELSEIKEEKEILQNKYDESRKTIKILQD 421
SL K L+N+++ L+ D C +L K+ + ++KY+E+ ++ Q
Sbjct: 1416 SLEKTKQRLQNEVEDLMIDVERTNAACAALDKKQRNFDKILAEWKHKYEETHAELEASQK 1475
Query: 422 SHFDMSEKQREINRKQKGIMSVPSEVQKENILLKKEVETLKKDL 465
+S + ++ + + +++EN L++E+ L + +
Sbjct: 1476 ESRSLSTEVFKVKNAYEESLDQLETLKRENKNLQQEISDLTEQI 1519
>MYH3_CHICK (P02565) Myosin heavy chain, fast skeletal muscle,
embryonic
Length = 1940
Score = 50.8 bits (120), Expect = 3e-05
Identities = 92/429 (21%), Positives = 183/429 (42%), Gaps = 76/429 (17%)
Query: 84 LSCALTMEESERVDECTNAKGVWDTLKI---HHEGTSHVKETRIDIGVRKFEVFEMSENE 140
++ LT ++ + DEC+ K D L++ E H E ++ K EM+
Sbjct: 937 MNAELTAKKRKLEDECSELKKDIDDLELTLAKVEKEKHATENKV-----KNLTEEMA--- 988
Query: 141 TIDEMYARFTTIVNEMRSLGKAYS-THDRIRKILRCLPSMWRPMVTAITQAKDLKSMNLE 199
+DE A+ T E ++L +A+ T D ++ V +T+AK ++
Sbjct: 989 ALDETIAKLT---KEKKALQEAHQQTLDDLQAE--------EDKVNTLTKAKTKLEQQVD 1037
Query: 200 DLIGSLRAHEVV----------LQGDKPVKKVRTLALKASQQTSSVDDEDVQEPQELEEV 249
DL GSL + + L+GD + + T+ L+ +Q +D++ ++ E+ ++
Sbjct: 1038 DLEGSLEQEKKLRMDLERAKRKLEGDLKMTQESTMDLENDKQ--QLDEKLKKKDFEISQI 1095
Query: 250 HEEEAEDELAL---ISKRIQRMMLRRNQIRKKFPKTNIS-IKTEADKSQVT------CYG 299
+ + EDE AL + K+I+ + R ++ ++ S K E ++ ++
Sbjct: 1096 -QSKIEDEQALGMQLQKKIKELQARIEELEEEIEAERTSRAKAEKHRADLSRELEEISER 1154
Query: 300 CNKTGHFKNECPDIKKVQRKPPFKKKAMITWDDMEESDSQEDADTDMGLMAQSDDEEEVI 359
+ G D+ K +R+ F+K D+EE+ Q +A T L + D +
Sbjct: 1155 LEEAGGATAAQIDMNK-KREAEFQKMRR----DLEEATLQHEA-TAAALRKKHADSTADV 1208
Query: 360 IYKTDSLYK-----------------DLENKIDSLLYDSNFLTNRCHSLIKELSEIKEEK 402
+ D+L + DL + ++S+ L C SL +LSEIK ++
Sbjct: 1209 GEQIDNLQRVKQKLEKEKSELKMEIDDLASNMESVSKAKANLEKMCRSLEDQLSEIKTKE 1268
Query: 403 EILQNKYDESRKTIKILQDSHFDMSEKQREINRKQKGIMSVPSEVQKENILLKKEVETLK 462
E E ++TI + + + E +R+ + ++ S++ + +++E LK
Sbjct: 1269 E-------EQQRTINDISAQKARLQTESGEYSRQVEEKDALISQLSRGKQAFTQQIEELK 1321
Query: 463 KDLTGFIKS 471
+ L IK+
Sbjct: 1322 RHLEEEIKA 1330
Score = 37.4 bits (85), Expect = 0.32
Identities = 32/139 (23%), Positives = 64/139 (46%), Gaps = 7/139 (5%)
Query: 341 DADTDMGLMAQSDDEEEVIIYKTDSLYKDLENKIDSLLYDSNFLTNRCHSLIKELSEIKE 400
+++ +M M + ++ + + K+++ K+LE K+ SLL + N L + + L++ +E
Sbjct: 849 ESEKEMANMKEEFEKTKEELAKSEAKRKELEEKMVSLLQEKNDLQLQVQAEADGLADAEE 908
Query: 401 EKEILQNKYDESRKTIKILQDSHFDMSEKQREINRKQKGIMSVPSEVQKENILLKKEVET 460
+ L + IK L + D E E+ K++ + SE LKK+++
Sbjct: 909 RCDQLIKTKIQLEAKIKELTERAEDEEEMNAELTAKKRKLEDECSE-------LKKDIDD 961
Query: 461 LKKDLTGFIKSTETFQNIV 479
L+ L K +N V
Sbjct: 962 LELTLAKVEKEKHATENKV 980
Score = 36.6 bits (83), Expect = 0.54
Identities = 46/255 (18%), Positives = 106/255 (41%), Gaps = 16/255 (6%)
Query: 227 KASQQTSSVDDEDVQEPQELEEVHE-----EEAEDELALISKRIQRMMLRRNQIRKKFPK 281
K +Q +++D Q+ + E E EE + ++ +S+ Q + ++++ +
Sbjct: 1267 KEEEQQRTINDISAQKARLQTESGEYSRQVEEKDALISQLSRGKQAFTQQIEELKRHLEE 1326
Query: 282 TNISIKTEADKSQVTCYGCNKTGHFKNECPDIK-KVQRKPPFKKKAMITW------DDME 334
+ K A Q + C+ E + K ++QR + W D ++
Sbjct: 1327 EIKAKKCPAHALQSARHDCDLLREQYEEEQEAKGELQRALSKANSEVAQWRTKYETDAIQ 1386
Query: 335 ESDSQEDADTDMGLMAQSDDEE-EVIIYKTDSLYKD---LENKIDSLLYDSNFLTNRCHS 390
++ E+A + Q +E E + K SL K L+N+++ L+ D C +
Sbjct: 1387 RTEELEEAKKKLAQRLQDAEEHVEAVNSKCASLEKTKQRLQNEVEDLMIDVERSNAACAA 1446
Query: 391 LIKELSEIKEEKEILQNKYDESRKTIKILQDSHFDMSEKQREINRKQKGIMSVPSEVQKE 450
L K+ + + KY+E++ ++ Q +S + ++ + + +++E
Sbjct: 1447 LDKKQKNFDKILSEWKQKYEETQAELEASQKESRSLSTELFKMKNAYEESLDHLETLKRE 1506
Query: 451 NILLKKEVETLKKDL 465
N L++E+ L + +
Sbjct: 1507 NKNLQQEISDLTEQI 1521
Score = 35.0 bits (79), Expect = 1.6
Identities = 44/238 (18%), Positives = 93/238 (38%), Gaps = 21/238 (8%)
Query: 227 KASQQTSSVDDEDVQEPQELEEVHEEEAEDELALISKRIQRMMLRRNQIRKKFPKTNISI 286
KA + V + QE EL+ EE AE L +I R+ L NQ++ +
Sbjct: 1526 KAIHELEKVKKQIEQEKSELQTALEE-AEASLEHEEGKILRVQLELNQVKSDIDRKIAEK 1584
Query: 287 KTEADKSQVTCYGCNKTGHFK-----NECPDIKKVQRKPPFKKKAMITWDDMEESDSQED 341
E D+ K H + D + R + K + D+ E + Q
Sbjct: 1585 DEEIDQL--------KRNHLRVVDSMQSTLDAEIRSRNEALRLKKKME-GDLNEIEIQLS 1635
Query: 342 ADTDMGLMAQSDDEEEVIIYKTDSLYKDLENKIDSLLYDSNFLTNRCHSLIKELSEIKEE 401
AQ + + T + KD + +D L L + + + + ++ E
Sbjct: 1636 HANRQAAEAQKN------LRNTQGVLKDTQIHLDDALRSQEDLKEQVAMVERRANLLQAE 1689
Query: 402 KEILQNKYDESRKTIKILQDSHFDMSEKQREINRKQKGIMSVPSEVQKENILLKKEVE 459
E L+ +++ ++ K+ + D SE+ + ++ + +++ +++ + ++ E+E
Sbjct: 1690 IEELRAALEQTERSRKVAEQELLDASERVQLLHTQNTSLINTKKKLESDISQIQSEME 1747
>REST_CHICK (O42184) Restin (Cytoplasmic linker protein-170)
(CLIP-170)
Length = 1433
Score = 50.1 bits (118), Expect = 5e-05
Identities = 92/458 (20%), Positives = 188/458 (40%), Gaps = 58/458 (12%)
Query: 193 LKSMNLEDLIGSL-RAHEVVLQGDKPVKKVRTLALKASQQTSSVDDEDVQEPQEL----- 246
LK LE + L +A+E +Q K V++ A ++ Q+T E++++ Q+
Sbjct: 937 LKERQLEQIQLELTKANEKAVQLQKNVEQTAQKAEQSQQETLKTHQEELKKMQDQLTDMK 996
Query: 247 --EEVHEEEAEDELALISKRIQRMMLRRNQIRKKFPKTNISIKTEADKSQVTCYGCNKTG 304
E + + +D A K M+ + + K F K N+ EA K+ K
Sbjct: 997 KQMETSQNQYKDLQAKYEKETSEMITKHDADIKGF-KQNLLDAEEALKAAQ-----KKND 1050
Query: 305 HFKNECPDIKKVQRKPPFKKKAMITWDDMEESDSQEDADTDMGLMAQSDDEEEVIIYKTD 364
+ + ++KK + K+A ME+ ++DA + Q E + +
Sbjct: 1051 ELETQAEELKKQAEQAKADKRAEEVLQTMEKVTKEKDA------IHQEKIETLASLENSR 1104
Query: 365 SLYKDLENKIDSL----LYDSNFLTNRCHSLI---KELSEIKEEKEILQNKYDESRKTIK 417
+ L+N++D L L + LT L K++ E+K+E E L+ + + +
Sbjct: 1105 QTNEKLQNELDMLKQNNLKNEEELTKSKELLNLENKKVEELKKEFEALKLAAAQKSQQLA 1164
Query: 418 ILQDSHFDMSEKQREINRKQKGIMSVPSEVQKENIL------LKKEVETLKKDL----TG 467
LQ+ + ++E E+ R + + S ++ ++L +KK TLKK++
Sbjct: 1165 ALQEENVKLAE---ELGRSRDEVTSHQKLEEERSVLNNQLLEMKKRESTLKKEIDEERAS 1221
Query: 468 FIKSTETFQNIVGSQNKSTKKSGLGFKDPSKIIGSFVPKAKIR---VKCCFCDKYGHNES 524
KS ++ +++ +K ++ ++ AK VK DK E
Sbjct: 1222 LQKSISDTSALITQKDEELEK----LRNEITVLRGENASAKTLQSVVKTLESDKLKLEEK 1277
Query: 525 VCHVKKKFIKQNNLNLSSERSHLNRSESSQKAEKAKKTCFYCNKSDHKRQNVTFRKDLLE 584
V ++++K L SE+ S S A + ++ K+Q + F ++
Sbjct: 1278 VKNLEQK------LKAKSEQPLTVTSPSGDIAANLLQD----ESAEDKQQEIDFLNSVIV 1327
Query: 585 ELTFKDPTLH-GLDSGCSRHMTGDRDCFLTFEKKDGGL 621
+L ++ L+ + C + G+ + + ++ ++ GL
Sbjct: 1328 DLQRRNEELNLKIQRMCEAALNGNEEETINYDSEEEGL 1365
Score = 40.4 bits (93), Expect = 0.038
Identities = 82/392 (20%), Positives = 160/392 (39%), Gaps = 65/392 (16%)
Query: 225 ALKASQQ--TSSVDDEDVQEPQELEEV-HEEEAEDELALISKRIQRMMLRRNQIRKKFPK 281
ALK QQ + + D++ + + H E E ELAL+ R +L ++ K +
Sbjct: 357 ALKEKQQHIEQLLAERDLERAEVAKATSHVGEIEQELALVRDGHDRHVL---EMEAKMDQ 413
Query: 282 TNISIKTEADKSQVTCYGCNKTGHFKNECPDIKKVQRKPPFKKKAMITWDDMEESDSQED 341
++ AD+ +V N+ + K+ F+ + EES ++ D
Sbjct: 414 LRAMVEA-ADREKV---------ELLNQLEEEKRKVEDLQFRVE--------EESITKGD 455
Query: 342 ADTDMGLM-AQSDDEEEVIIY---KTDSLYKDLENKIDSLLYDSNFLTNRCHSLIKELSE 397
+T L A+ + E+ +++ K D L ++LE+ + + + + + L + E
Sbjct: 456 LETQTKLEHARIKELEQSLLFEKTKADKLQRELEDTRVATVSEKSRIMELERDLALRVKE 515
Query: 398 IKEEKEILQNK--YDESRKTIKILQDSHFDMSEKQREINRKQKGIMSVPSEVQKENILLK 455
+ E + L++ D+ ++ +LQ EI+ Q+ + + E Q+E LK
Sbjct: 516 VAELRGRLESSKHIDDVDTSLSLLQ-----------EISSLQEKMAAAGKEHQREMSSLK 564
Query: 456 KEVETLKKDLTGFIKSTETFQNIVGSQNKSTK-KSGLGFKDPSKIIGSFVPK-------- 506
++ E+ ++ L IK+ +G +N+S K K K+ S +I + K
Sbjct: 565 EKFESSEEALRKEIKTLSASNERMGKENESLKTKLDHANKENSDVIELWKSKLESAIASH 624
Query: 507 --AKIRVKCCFCDKYGHNESVCHVKKKFIKQNNLNLSSERSHLNRSESSQKAEKAKKTCF 564
A +K F G + K +++ L+ +E S+L + ++
Sbjct: 625 QQAMEELKVSFNKGVGAQTAEFAELKTQMEKVKLDYENEMSNLKLKQENE---------- 674
Query: 565 YCNKSDHKRQNVTFRKDLLEELTFKDPTLHGL 596
KS H ++ + LLE K+ TL L
Sbjct: 675 ---KSQHLKEIEALKAKLLEVTEEKEQTLENL 703
Score = 38.9 bits (89), Expect = 0.11
Identities = 85/492 (17%), Positives = 189/492 (38%), Gaps = 66/492 (13%)
Query: 59 AVKAKSAWSTDEKAQVLLNSKARLFL---SCALTMEES-ERVDECTNAKGVWDTLKIH-H 113
A+KAK T+EK Q L N KA+L + ME++ ++ E D L+ +
Sbjct: 684 ALKAKLLEVTEEKEQTLENLKAKLESVEDQHLVEMEDTLNKLQEAEIKVKELDVLQAKCN 743
Query: 114 EGTSHV----------KETRIDIGV-------RKFEVFEMSE-----------------N 139
E T + +E +D+ K E+ ++SE +
Sbjct: 744 EQTKLIGSLTQQIRASEEKLLDLAALQKANSEGKLEIQKLSEQLQAAEKQIQNLETEKVS 803
Query: 140 ETIDEMYARFTTIVNEMRSLGKAYSTHDRIRKILRCLPSMWRPMVTAITQAKDLKSMNLE 199
E+ + +++ ++L D + K L+ L + V A+ +
Sbjct: 804 NLTKELQGKEQKLLDLEKNLSAVNQVKDSLEKELQLLKEKFTSAVDGAENAQRAMQETIN 863
Query: 200 DL-------------IGSLRAHEVVLQGDKPVKKVRTLALKASQQTSSVDDEDVQEPQEL 246
L + L+++ V++ ++ R L ++ D ++ +
Sbjct: 864 KLNQKEEQFALMSSELEQLKSNLTVMETKLKEREEREQQLTEAKVKLENDIAEIMKSSGD 923
Query: 247 EEVHEEEAEDELALISKRIQRMMLRRNQIRKKFPKTNISIKTEADKSQVTCYGCNKTGH- 305
+ DEL L ++++++ L + +K + +++ A K++ + KT
Sbjct: 924 SSAQLMKMNDELRLKERQLEQIQLELTKANEKAVQLQKNVEQTAQKAEQSQQETLKTHQE 983
Query: 306 ----FKNECPDIKKVQRKPPFKKKAMITWDDMEESDSQEDADTDMGLMAQSDDEEEVIIY 361
+++ D+KK + K + + E S+ D D+ Q+ + E +
Sbjct: 984 ELKKMQDQLTDMKKQMETSQNQYKDLQAKYEKETSEMITKHDADIKGFKQNLLDAEEALK 1043
Query: 362 KTDSLYKDLENKIDSLLYDSN--FLTNRCHSLIKELSEIKEEKEILQNKYDESRKTIKIL 419
+LE + + L + R +++ + ++ +EK+ + + E +T+ L
Sbjct: 1044 AAQKKNDELETQAEELKKQAEQAKADKRAEEVLQTMEKVTKEKDAI---HQEKIETLASL 1100
Query: 420 QDSHFDMSEKQREINRKQKGIMSVPSEVQKENILL---KKEVETLKKDLTGF-IKSTETF 475
++S + Q E++ ++ + E+ K LL K+VE LKK+ + + +
Sbjct: 1101 ENSRQTNEKLQNELDMLKQNNLKNEEELTKSKELLNLENKKVEELKKEFEALKLAAAQKS 1160
Query: 476 QNIVGSQNKSTK 487
Q + Q ++ K
Sbjct: 1161 QQLAALQEENVK 1172
Score = 37.4 bits (85), Expect = 0.32
Identities = 82/417 (19%), Positives = 165/417 (38%), Gaps = 67/417 (16%)
Query: 129 RKFEVFEMSENETIDEMYARFTTIVNEMRSLGKAYSTHDRIRKILRCLPSMWRPMVTAIT 188
R EV + E T E A + + SL + T+++++ L L +T
Sbjct: 1071 RAEEVLQTMEKVT-KEKDAIHQEKIETLASLENSRQTNEKLQNELDMLKQNNLKNEEELT 1129
Query: 189 QAKDLKSMNLEDLIGSLRAHEVVLQGDKPVKKVRTLALKASQQTSSVDDEDVQEPQEL-- 246
++K+L +NLE+ + V + K + ++ A + SQQ +++ +E+V+ +EL
Sbjct: 1130 KSKEL--LNLEN--------KKVEELKKEFEALKLAAAQKSQQLAALQEENVKLAEELGR 1179
Query: 247 ---EEVHEEEAEDELALISKRIQRMMLRRNQIRKKFPKTNISI-KTEADKSQVTCYGCNK 302
E ++ E+E ++++ ++ M R + ++K+ + S+ K+ +D S + +
Sbjct: 1180 SRDEVTSHQKLEEERSVLNNQLLEMKKRESTLKKEIDEERASLQKSISDTSALITQKDEE 1239
Query: 303 TGHFKNECPDIKKVQRKPPFKKKAMITWDDMEESDSQEDADTDMGLMAQSDDEEEVIIYK 362
+NE V R K + + ESD K
Sbjct: 1240 LEKLRNEI----TVLRGENASAKTLQSVVKTLESDK----------------------LK 1273
Query: 363 TDSLYKDLENKIDSLLYDSNFLTNRCHSLIKELSEIKEEKEILQNKYDESRKTIKILQDS 422
+ K+LE K+ + +T+ + L + ++ ++ I L
Sbjct: 1274 LEEKVKNLEQKLKAKSEQPLTVTSPSGDIAANL--------LQDESAEDKQQEIDFLNSV 1325
Query: 423 HFDMSEKQREINRK-QKGIMSVPSEVQKENILLKKEVETLKKDLTGFIKSTETFQNIVG- 480
D+ + E+N K Q+ + + ++E I E E L K K+ F +I G
Sbjct: 1326 IVDLQRRNEELNLKIQRMCEAALNGNEEETINYDSEEEGLSK------KTPRLFCDICGC 1379
Query: 481 -----SQNKSTKKSGLGFKDPSKIIGSFVPKAKIRVKCCFCDKYGHNESVCHVKKKF 532
+++ T+ L S GS + + R C C+ +GH + C+ + F
Sbjct: 1380 FDLHDTEDCPTQAQMLEEPPHSTYHGS---RREERPYCDTCEMFGHWTADCNDDETF 1433
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.317 0.133 0.386
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 175,582,144
Number of Sequences: 164201
Number of extensions: 7617867
Number of successful extensions: 28859
Number of sequences better than 10.0: 400
Number of HSP's better than 10.0 without gapping: 66
Number of HSP's successfully gapped in prelim test: 351
Number of HSP's that attempted gapping in prelim test: 27401
Number of HSP's gapped (non-prelim): 1430
length of query: 1523
length of database: 59,974,054
effective HSP length: 123
effective length of query: 1400
effective length of database: 39,777,331
effective search space: 55688263400
effective search space used: 55688263400
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 73 (32.7 bits)
Medicago: description of AC144724.6


