

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144724.10 - phase: 0 /pseudo
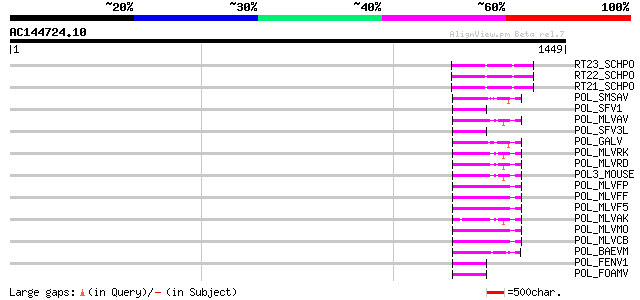
(1449 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
RT23_SCHPO (Q9UR07) Retrotransposable element Tf2 155 kDa protei... 122 7e-27
RT22_SCHPO (Q9C0R2) Retrotransposable element Tf2 155 kDa protei... 122 7e-27
RT21_SCHPO (Q05654) Retrotransposable element Tf2 155 kDa protei... 122 7e-27
POL_SMSAV (P03359) Pol polyprotein [Contains: Reverse transcript... 58 2e-07
POL_SFV1 (P23074) Pol polyprotein [Contains: Protease (EC 3.4.23... 55 1e-06
POL_MLVAV (P03356) Pol polyprotein [Contains: Protease (EC 3.4.2... 55 1e-06
POL_SFV3L (P27401) Pol polyprotein [Contains: Protease (EC 3.4.2... 55 2e-06
POL_GALV (P21414) Pol polyprotein [Contains: Protease (EC 3.4.23... 54 2e-06
POL_MLVRK (P31795) Pol polyprotein [Contains: Protease (EC 3.4.2... 52 9e-06
POL_MLVRD (P11227) Pol polyprotein [Contains: Protease (EC 3.4.2... 52 9e-06
POL3_MOUSE (P11367) Retrovirus-related Pol polyprotein (Endonucl... 52 9e-06
POL_MLVFP (P26808) Pol polyprotein [Contains: Protease (EC 3.4.2... 52 2e-05
POL_MLVFF (P26809) Pol polyprotein [Contains: Protease (EC 3.4.2... 52 2e-05
POL_MLVF5 (P26810) Pol polyprotein [Contains: Protease (EC 3.4.2... 52 2e-05
POL_MLVAK (P03357) Pol polyprotein [Contains: Reverse transcript... 52 2e-05
POL_MLVMO (P03355) Pol polyprotein [Contains: Protease (EC 3.4.2... 51 3e-05
POL_MLVCB (P08361) Pol polyprotein [Contains: Reverse transcript... 51 3e-05
POL_BAEVM (P10272) Pol polyprotein [Contains: Protease (EC 3.4.2... 50 4e-05
POL_FENV1 (P31792) Pol polyprotein [Contains: Reverse transcript... 50 6e-05
POL_FOAMV (P14350) Pol polyprotein [Contains: Reverse transcript... 49 1e-04
>RT23_SCHPO (Q9UR07) Retrotransposable element Tf2 155 kDa protein
type 3
Length = 1333
Score = 122 bits (306), Expect = 7e-27
Identities = 74/217 (34%), Positives = 112/217 (51%), Gaps = 7/217 (3%)
Query: 1153 EHLSDRDPRFTSRFWKSLQEALGSKLRLSSAYHPQTDGQSERTIQSLEDLLRVCVLEQGG 1212
E ++D D FTS+ WK ++ S Y PQTDGQ+ERT Q++E LLR
Sbjct: 1046 EIIADNDHIFTSQTWKDFAHKYNFVMKFSLPYRPQTDGQTERTNQTVEKLLRCVCSTHPN 1105
Query: 1213 TWDSHLPLIEFTYNNSYHSSIGMAPFEALYGRRCRTQLCWFESGGRVVLGPEIVQQTTEK 1272
TW H+ L++ +YNN+ HS+ M PFE ++ R L E E Q+T +
Sbjct: 1106 TWVDHISLVQQSYNNAIHSATQMTPFEIVH--RYSPALSPLELPSFSDKTDENSQETIQV 1163
Query: 1273 VKMIQEKMKASQSRQKSYHDKRRKDL-EFQEGDHVFLRVTPVTGVGRALKSKKLTPRFIG 1331
+ ++E + + + K Y D + +++ EFQ GD V ++ T G KS KL P F G
Sbjct: 1164 FQTVKEHLNTNNIKMKKYFDMKIQEIEEFQPGDLVMVKRTK---TGFLHKSNKLAPSFAG 1220
Query: 1332 PYQISERVGTVAYRVGLPPHLSNL-HDVFHVSQLRKY 1367
P+ + ++ G Y + LP + ++ FHVS L KY
Sbjct: 1221 PFYVLQKSGPNNYELDLPDSIKHMFSSTFHVSHLEKY 1257
>RT22_SCHPO (Q9C0R2) Retrotransposable element Tf2 155 kDa protein
type 2
Length = 1333
Score = 122 bits (306), Expect = 7e-27
Identities = 74/217 (34%), Positives = 112/217 (51%), Gaps = 7/217 (3%)
Query: 1153 EHLSDRDPRFTSRFWKSLQEALGSKLRLSSAYHPQTDGQSERTIQSLEDLLRVCVLEQGG 1212
E ++D D FTS+ WK ++ S Y PQTDGQ+ERT Q++E LLR
Sbjct: 1046 EIIADNDHIFTSQTWKDFAHKYNFVMKFSLPYRPQTDGQTERTNQTVEKLLRCVCSTHPN 1105
Query: 1213 TWDSHLPLIEFTYNNSYHSSIGMAPFEALYGRRCRTQLCWFESGGRVVLGPEIVQQTTEK 1272
TW H+ L++ +YNN+ HS+ M PFE ++ R L E E Q+T +
Sbjct: 1106 TWVDHISLVQQSYNNAIHSATQMTPFEIVH--RYSPALSPLELPSFSDKTDENSQETIQV 1163
Query: 1273 VKMIQEKMKASQSRQKSYHDKRRKDL-EFQEGDHVFLRVTPVTGVGRALKSKKLTPRFIG 1331
+ ++E + + + K Y D + +++ EFQ GD V ++ T G KS KL P F G
Sbjct: 1164 FQTVKEHLNTNNIKMKKYFDMKIQEIEEFQPGDLVMVKRTK---TGFLHKSNKLAPSFAG 1220
Query: 1332 PYQISERVGTVAYRVGLPPHLSNL-HDVFHVSQLRKY 1367
P+ + ++ G Y + LP + ++ FHVS L KY
Sbjct: 1221 PFYVLQKSGPNNYELDLPDSIKHMFSSTFHVSHLEKY 1257
>RT21_SCHPO (Q05654) Retrotransposable element Tf2 155 kDa protein
type 1
Length = 1333
Score = 122 bits (306), Expect = 7e-27
Identities = 74/217 (34%), Positives = 112/217 (51%), Gaps = 7/217 (3%)
Query: 1153 EHLSDRDPRFTSRFWKSLQEALGSKLRLSSAYHPQTDGQSERTIQSLEDLLRVCVLEQGG 1212
E ++D D FTS+ WK ++ S Y PQTDGQ+ERT Q++E LLR
Sbjct: 1046 EIIADNDHIFTSQTWKDFAHKYNFVMKFSLPYRPQTDGQTERTNQTVEKLLRCVCSTHPN 1105
Query: 1213 TWDSHLPLIEFTYNNSYHSSIGMAPFEALYGRRCRTQLCWFESGGRVVLGPEIVQQTTEK 1272
TW H+ L++ +YNN+ HS+ M PFE ++ R L E E Q+T +
Sbjct: 1106 TWVDHISLVQQSYNNAIHSATQMTPFEIVH--RYSPALSPLELPSFSDKTDENSQETIQV 1163
Query: 1273 VKMIQEKMKASQSRQKSYHDKRRKDL-EFQEGDHVFLRVTPVTGVGRALKSKKLTPRFIG 1331
+ ++E + + + K Y D + +++ EFQ GD V ++ T G KS KL P F G
Sbjct: 1164 FQTVKEHLNTNNIKMKKYFDMKIQEIEEFQPGDLVMVKRTK---TGFLHKSNKLAPSFAG 1220
Query: 1332 PYQISERVGTVAYRVGLPPHLSNL-HDVFHVSQLRKY 1367
P+ + ++ G Y + LP + ++ FHVS L KY
Sbjct: 1221 PFYVLQKSGPNNYELDLPDSIKHMFSSTFHVSHLEKY 1257
>POL_SMSAV (P03359) Pol polyprotein [Contains: Reverse
transcriptase/ribonuclease H (EC 2.7.7.49) (EC 3.1.26.4)
(RT); Integrase (IN)] (Fragment)
Length = 294
Score = 58.2 bits (139), Expect = 2e-07
Identities = 55/190 (28%), Positives = 84/190 (43%), Gaps = 31/190 (16%)
Query: 1156 SDRDPRFTSRFWKSLQEALGSKLRLSSAYHPQTDGQSERTIQSLEDLLRVCVLEQGG-TW 1214
SD P F ++ + L LG +L AY PQ+ GQ ER +++++ L LE GG W
Sbjct: 70 SDNGPAFVAQVSQGLATQLGINWKLHCAYRPQSSGQVERMNRTIKETLTKLALETGGKDW 129
Query: 1215 DSHLPLIEFTYNNSYHSSIGMAPFEALYGRRCRTQLCWFESGGRVVLGPEIVQQTTEKVK 1274
+ LPL N+ S G+ P+E LYG ESGG LGP+ +
Sbjct: 130 VALLPLALLRAKNT-PSRFGLTPYEILYGGPPPI----LESGG--TLGPD-----DNFLP 177
Query: 1275 MIQEKMKASQSRQKSYHDKRRKDLE---------FQEGDHVFLRVTPVTGVGRALKSKKL 1325
++ +KA + + D+ ++ + FQ GD V + R + L
Sbjct: 178 VLFTHLKALEVVRTQIWDQIKEVYKPGTVAIPHPFQVGDQVLV---------RRHRPGSL 228
Query: 1326 TPRFIGPYQI 1335
PR+ GPY +
Sbjct: 229 EPRWKGPYLV 238
>POL_SFV1 (P23074) Pol polyprotein [Contains: Protease (EC 3.4.23.-);
Reverse transcriptase/ribonuclease H (EC 2.7.7.49) (EC
3.1.26.4) (RT); Integrase (IN)]
Length = 1161
Score = 55.5 bits (132), Expect = 1e-06
Identities = 28/88 (31%), Positives = 45/88 (50%)
Query: 1156 SDRDPRFTSRFWKSLQEALGSKLRLSSAYHPQTDGQSERTIQSLEDLLRVCVLEQGGTWD 1215
SD+ FTS + + G +L S+ YHPQ+ G+ ER ++ LL ++ + W
Sbjct: 946 SDQGAAFTSSTFADWAKEKGIQLEFSTPYHPQSSGKVERKNSDIKRLLTKLLIGRPAKWY 1005
Query: 1216 SHLPLIEFTYNNSYHSSIGMAPFEALYG 1243
LP+++ NNSY S P + L+G
Sbjct: 1006 DLLPVVQLALNNSYSPSSKYTPHQLLFG 1033
>POL_MLVAV (P03356) Pol polyprotein [Contains: Protease (EC 3.4.23.-);
Reverse transcriptase/ribonuclease H (EC 2.7.7.49) (EC
3.1.26.4) (RT); Integrase (IN)]
Length = 1196
Score = 55.5 bits (132), Expect = 1e-06
Identities = 54/192 (28%), Positives = 88/192 (45%), Gaps = 33/192 (17%)
Query: 1156 SDRDPRFTSRFWKSLQEALGSKLRLSSAYHPQTDGQSERTIQSLEDLLRVCVLEQG-GTW 1214
SD P FTS+ +S+ + LG +L AY PQ+ GQ ER +++++ L L G W
Sbjct: 974 SDNGPAFTSQVSQSVADLLGIDWKLHCAYRPQSSGQVERMNRTIKETLTKLTLAAGTRDW 1033
Query: 1215 DSHLPLIEFTYNNSYHSSIGMAPFEALYGRRCRTQLCWFESGGRVVLGPEIVQQTTEKVK 1274
LPL + N+ G+ P+E LYG L F P++ + T
Sbjct: 1034 VLLLPLALYRARNT-PGPHGLTPYEILYG--APPPLVNFHD-------PDMSELTNS--P 1081
Query: 1275 MIQEKMKASQSRQK--------SYHDKRRKDL---EFQEGDHVFLRVTPVTGVGRALKSK 1323
+Q ++A Q+ Q+ +Y D+ + + F+ GD V++ R ++K
Sbjct: 1082 SLQAHLQALQTVQREIWKPLAEAYRDQLDQPVIPHPFRIGDSVWV---------RRHQTK 1132
Query: 1324 KLTPRFIGPYQI 1335
L PR+ GPY +
Sbjct: 1133 NLEPRWKGPYTV 1144
>POL_SFV3L (P27401) Pol polyprotein [Contains: Protease (EC 3.4.23.-);
Reverse transcriptase/ribonuclease H (EC 2.7.7.49) (EC
3.1.26.4) (RT); Integrase (IN)]
Length = 1157
Score = 54.7 bits (130), Expect = 2e-06
Identities = 28/88 (31%), Positives = 45/88 (50%)
Query: 1156 SDRDPRFTSRFWKSLQEALGSKLRLSSAYHPQTDGQSERTIQSLEDLLRVCVLEQGGTWD 1215
SD+ FTS + + G +L S+ YHPQ+ G+ ER ++ LL ++ + W
Sbjct: 948 SDQGAAFTSATFADWAKNKGIQLEFSTPYHPQSSGKVERKNSDIKRLLTKLLVGRPAKWY 1007
Query: 1216 SHLPLIEFTYNNSYHSSIGMAPFEALYG 1243
LP+++ NNSY S P + L+G
Sbjct: 1008 DLLPVVQLALNNSYSPSSKYTPHQLLFG 1035
>POL_GALV (P21414) Pol polyprotein [Contains: Protease (EC 3.4.23.-);
Reverse transcriptase/ribonuclease H (EC 2.7.7.49) (EC
3.1.26.4) (RT); Integrase (IN)]
Length = 1165
Score = 54.3 bits (129), Expect = 2e-06
Identities = 53/190 (27%), Positives = 82/190 (42%), Gaps = 31/190 (16%)
Query: 1156 SDRDPRFTSRFWKSLQEALGSKLRLSSAYHPQTDGQSERTIQSLEDLLRVCVLEQGG-TW 1214
SD P F ++ + L LG +L AY PQ+ GQ ER +++++ L LE GG W
Sbjct: 941 SDNGPAFVAQVSQGLATQLGINWKLHCAYRPQSSGQVERMNRTIKETLTKLALETGGKDW 1000
Query: 1215 DSHLPLIEFTYNNSYHSSIGMAPFEALYGRRCRTQLCWFESGGRVVLGPEIVQQTTEKVK 1274
+ LPL N+ G+ P+E LYG ESG LGP+ +
Sbjct: 1001 VTLLPLALLRARNT-PGRFGLTPYEILYGGPPPI----LESG--ETLGPD-----DRFLP 1048
Query: 1275 MIQEKMKASQSRQKSYHDKRRKDLE---------FQEGDHVFLRVTPVTGVGRALKSKKL 1325
++ +KA + + D+ ++ + FQ GD V + R + L
Sbjct: 1049 VLFTHLKALEIVRTQIWDQIKEVYKPGTVTIPHPFQVGDQVLV---------RRHRPSSL 1099
Query: 1326 TPRFIGPYQI 1335
PR+ GPY +
Sbjct: 1100 EPRWKGPYLV 1109
>POL_MLVRK (P31795) Pol polyprotein [Contains: Protease (EC 3.4.23.-);
Reverse transcriptase/ribonuclease H (EC 2.7.7.49) (EC
3.1.26.4) (RT); Integrase (IN)] (Fragment)
Length = 581
Score = 52.4 bits (124), Expect = 9e-06
Identities = 53/192 (27%), Positives = 87/192 (44%), Gaps = 33/192 (17%)
Query: 1156 SDRDPRFTSRFWKSLQEALGSKLRLSSAYHPQTDGQSERTIQSLEDLLRVCVLEQG-GTW 1214
+D P F S+ +S+ + LG +L AY PQ+ GQ ER +++++ L L G W
Sbjct: 359 TDNGPAFVSQVSQSVAKLLGIDWKLHCAYRPQSSGQVERMNRTIKETLTKLTLATGTRDW 418
Query: 1215 DSHLPLIEFTYNNSYHSSIGMAPFEALYGRRCRTQLCWFESGGRVVLGPEIVQQTTEKVK 1274
LPL + N+ G+ P+E LYG L F PE+ + T
Sbjct: 419 VLLLPLALYRARNT-PGPHGLTPYEILYG--APPPLVNFHD-------PEMSKFTNS--P 466
Query: 1275 MIQEKMKASQSRQK--------SYHDKRRKDL---EFQEGDHVFLRVTPVTGVGRALKSK 1323
+Q ++A Q+ Q+ +Y D+ + + F+ GD V++ R ++K
Sbjct: 467 SLQAHLQALQAVQREVWKPLAAAYQDQLDQPVIPHPFRVGDTVWV---------RRHQTK 517
Query: 1324 KLTPRFIGPYQI 1335
L PR+ GPY +
Sbjct: 518 NLEPRWKGPYTV 529
>POL_MLVRD (P11227) Pol polyprotein [Contains: Protease (EC 3.4.23.-);
Reverse transcriptase/ribonuclease H (EC 2.7.7.49) (EC
3.1.26.4) (RT); Integrase (IN)]
Length = 1196
Score = 52.4 bits (124), Expect = 9e-06
Identities = 53/192 (27%), Positives = 87/192 (44%), Gaps = 33/192 (17%)
Query: 1156 SDRDPRFTSRFWKSLQEALGSKLRLSSAYHPQTDGQSERTIQSLEDLLRVCVLEQG-GTW 1214
+D P F S+ +S+ + LG +L AY PQ+ GQ ER +++++ L L G W
Sbjct: 974 TDNGPAFVSQVSQSVAKLLGIDWKLHCAYRPQSSGQVERMNRTIKETLTKLTLATGTRDW 1033
Query: 1215 DSHLPLIEFTYNNSYHSSIGMAPFEALYGRRCRTQLCWFESGGRVVLGPEIVQQTTEKVK 1274
LPL + N+ G+ P+E LYG L F PE+ + T
Sbjct: 1034 VLLLPLALYRARNT-PGPHGLTPYEILYG--APPPLVNFHD-------PEMSKFTNS--P 1081
Query: 1275 MIQEKMKASQSRQK--------SYHDKRRKDL---EFQEGDHVFLRVTPVTGVGRALKSK 1323
+Q ++A Q+ Q+ +Y D+ + + F+ GD V++ R ++K
Sbjct: 1082 SLQAHLQALQAVQREVWKPLAAAYQDQLDQPVIPHPFRVGDTVWV---------RRHQTK 1132
Query: 1324 KLTPRFIGPYQI 1335
L PR+ GPY +
Sbjct: 1133 NLEPRWKGPYTV 1144
>POL3_MOUSE (P11367) Retrovirus-related Pol polyprotein (Endonuclease)
(Fragment)
Length = 390
Score = 52.4 bits (124), Expect = 9e-06
Identities = 53/192 (27%), Positives = 87/192 (44%), Gaps = 33/192 (17%)
Query: 1156 SDRDPRFTSRFWKSLQEALGSKLRLSSAYHPQTDGQSERTIQSLEDLLRVCVLEQG-GTW 1214
+D P F S+ +S+ + LG +L AY PQ+ GQ ER +++++ L L G W
Sbjct: 183 TDNGPAFVSQVSQSVAKLLGIDWKLHCAYRPQSSGQVERMNRTIKETLTKLTLATGTRDW 242
Query: 1215 DSHLPLIEFTYNNSYHSSIGMAPFEALYGRRCRTQLCWFESGGRVVLGPEIVQQTTEKVK 1274
LPL + N+ G+ P+E LYG L F PE+ + T
Sbjct: 243 VLLLPLALYRARNT-PGPHGLTPYEILYG--APPPLVNFHD-------PEMSKFTNS--P 290
Query: 1275 MIQEKMKASQSRQK--------SYHDKRRKDL---EFQEGDHVFLRVTPVTGVGRALKSK 1323
+Q ++A Q+ Q+ +Y D+ + + F+ GD V++ R ++K
Sbjct: 291 SLQAHLQALQAVQREVWKPLAAAYQDQLDQPVIPHPFRVGDTVWV---------RRHQTK 341
Query: 1324 KLTPRFIGPYQI 1335
L PR+ GPY +
Sbjct: 342 NLEPRWKGPYTV 353
>POL_MLVFP (P26808) Pol polyprotein [Contains: Protease (EC 3.4.23.-);
Reverse transcriptase/ribonuclease H (EC 2.7.7.49) (EC
3.1.26.4) (RT); Integrase (IN)]
Length = 1204
Score = 51.6 bits (122), Expect = 2e-05
Identities = 45/181 (24%), Positives = 76/181 (41%), Gaps = 11/181 (6%)
Query: 1156 SDRDPRFTSRFWKSLQEALGSKLRLSSAYHPQTDGQSERTIQSLEDLLRVCVLEQGG-TW 1214
+D P F S+ +++ + LG +L AY PQ+ GQ ER +++++ L L G W
Sbjct: 979 TDNGPAFVSKVSQTVADLLGVDWKLHCAYRPQSSGQVERMNRTIKETLTKLTLATGSRDW 1038
Query: 1215 DSHLPLIEFTYNNSYHSSIGMAPFEALYGRRCRTQLCWFESGGRVVLGPEIVQQTTEKVK 1274
LPL + N+ G+ P+E LYG +V P +
Sbjct: 1039 VLLLPLALYRARNT-PGPHGLTPYEILYGAPPPLVNFPDPDMAKVTHNPSLQAHLQALYL 1097
Query: 1275 MIQEKMKASQSRQKSYHDKRRKDLEFQEGDHVFLRVTPVTGVGRALKSKKLTPRFIGPYQ 1334
+ E + + + D+ F+ GD V++ R ++K L PR+ GPY
Sbjct: 1098 VQHEVWRPLAAAYQEQLDRPVVPHPFRVGDTVWV---------RRHQTKNLEPRWKGPYT 1148
Query: 1335 I 1335
+
Sbjct: 1149 V 1149
>POL_MLVFF (P26809) Pol polyprotein [Contains: Protease (EC 3.4.23.-);
Reverse transcriptase/ribonuclease H (EC 2.7.7.49) (EC
3.1.26.4) (RT); Integrase (IN)]
Length = 1204
Score = 51.6 bits (122), Expect = 2e-05
Identities = 45/181 (24%), Positives = 76/181 (41%), Gaps = 11/181 (6%)
Query: 1156 SDRDPRFTSRFWKSLQEALGSKLRLSSAYHPQTDGQSERTIQSLEDLLRVCVLEQGG-TW 1214
+D P F S+ +++ + LG +L AY PQ+ GQ ER +++++ L L G W
Sbjct: 979 TDNGPAFVSKVSQTVADLLGVDWKLHCAYRPQSSGQVERMNRTIKETLTKLTLATGSRDW 1038
Query: 1215 DSHLPLIEFTYNNSYHSSIGMAPFEALYGRRCRTQLCWFESGGRVVLGPEIVQQTTEKVK 1274
LPL + N+ G+ P+E LYG +V P +
Sbjct: 1039 VLLLPLALYRARNT-PGPHGLTPYEILYGAPPPLVNFPDPDMAKVTHNPSLQAHLQALYL 1097
Query: 1275 MIQEKMKASQSRQKSYHDKRRKDLEFQEGDHVFLRVTPVTGVGRALKSKKLTPRFIGPYQ 1334
+ E + + + D+ F+ GD V++ R ++K L PR+ GPY
Sbjct: 1098 VQHEVWRPLAAAYQEQLDRPVVPHPFRVGDTVWV---------RRHQTKNLEPRWKGPYT 1148
Query: 1335 I 1335
+
Sbjct: 1149 V 1149
>POL_MLVF5 (P26810) Pol polyprotein [Contains: Protease (EC 3.4.23.-);
Reverse transcriptase/ribonuclease H (EC 2.7.7.49) (EC
3.1.26.4) (RT); Integrase (IN)]
Length = 1204
Score = 51.6 bits (122), Expect = 2e-05
Identities = 45/181 (24%), Positives = 76/181 (41%), Gaps = 11/181 (6%)
Query: 1156 SDRDPRFTSRFWKSLQEALGSKLRLSSAYHPQTDGQSERTIQSLEDLLRVCVLEQGG-TW 1214
+D P F S+ +++ + LG +L AY PQ+ GQ ER +++++ L L G W
Sbjct: 979 TDNGPAFVSKVSQTVADLLGVDWKLHCAYRPQSSGQVERMNRTIKETLTKLTLATGSRDW 1038
Query: 1215 DSHLPLIEFTYNNSYHSSIGMAPFEALYGRRCRTQLCWFESGGRVVLGPEIVQQTTEKVK 1274
LPL + N+ G+ P+E LYG +V P +
Sbjct: 1039 VLLLPLALYRARNT-PGPHGLTPYEILYGAPPPLVNFPDPDMAKVTHNPSLQAHLQALYL 1097
Query: 1275 MIQEKMKASQSRQKSYHDKRRKDLEFQEGDHVFLRVTPVTGVGRALKSKKLTPRFIGPYQ 1334
+ E + + + D+ F+ GD V++ R ++K L PR+ GPY
Sbjct: 1098 VQHEVWRPLAAAYQEQLDRPVVPHPFRVGDTVWV---------RRHQTKNLEPRWKGPYT 1148
Query: 1335 I 1335
+
Sbjct: 1149 V 1149
>POL_MLVAK (P03357) Pol polyprotein [Contains: Reverse
transcriptase/ribonuclease H (EC 2.7.7.49) (EC 3.1.26.4)
(RT); Integrase (IN)] (Fragment)
Length = 843
Score = 51.6 bits (122), Expect = 2e-05
Identities = 54/192 (28%), Positives = 88/192 (45%), Gaps = 34/192 (17%)
Query: 1156 SDRDPRFTSRFWKSLQEALGSKLRLSSAYHPQTDGQSERTIQSLEDLLRVCVLEQG-GTW 1214
SD P FTS+ +S+ + LG +L AY PQ+ GQ ER +++++ L L G W
Sbjct: 622 SDNGPAFTSQVSQSVADLLGID-KLHCAYRPQSSGQVERMNRTIKETLTKLTLAAGTRDW 680
Query: 1215 DSHLPLIEFTYNNSYHSSIGMAPFEALYGRRCRTQLCWFESGGRVVLGPEIVQQTTEKVK 1274
LPL + N+ G+ P+E LYG L F P++ + T
Sbjct: 681 VLLLPLALYRARNT-PGPHGLTPYEILYG--APPPLVNFHD-------PDMSELTNS--P 728
Query: 1275 MIQEKMKASQSRQK--------SYHDKRRKDL---EFQEGDHVFLRVTPVTGVGRALKSK 1323
+Q ++A Q+ Q+ +Y D+ + + F+ GD V++ R ++K
Sbjct: 729 SLQAHLQALQTVQREIWKPLAEAYRDQLDQPVIPHPFRIGDSVWV---------RRHQTK 779
Query: 1324 KLTPRFIGPYQI 1335
L PR+ GPY +
Sbjct: 780 NLEPRWKGPYTV 791
>POL_MLVMO (P03355) Pol polyprotein [Contains: Protease (EC 3.4.23.-);
Reverse transcriptase/ribonuclease H (EC 2.7.7.49) (EC
3.1.26.4) (RT); Integrase (IN)]
Length = 1199
Score = 50.8 bits (120), Expect = 3e-05
Identities = 45/181 (24%), Positives = 76/181 (41%), Gaps = 11/181 (6%)
Query: 1156 SDRDPRFTSRFWKSLQEALGSKLRLSSAYHPQTDGQSERTIQSLEDLLRVCVLEQGG-TW 1214
+D P F S+ +++ + LG +L AY PQ+ GQ ER +++++ L L G W
Sbjct: 974 TDNGPAFVSKVSQTVADLLGIDWKLHCAYRPQSSGQVERMNRTIKETLTKLTLATGSRDW 1033
Query: 1215 DSHLPLIEFTYNNSYHSSIGMAPFEALYGRRCRTQLCWFESGGRVVLGPEIVQQTTEKVK 1274
LPL + N+ G+ P+E LYG RV P +
Sbjct: 1034 VLLLPLALYRARNT-PGPHGLTPYEILYGAPPPLVNFPDPDMTRVTNSPSLQAHLQALYL 1092
Query: 1275 MIQEKMKASQSRQKSYHDKRRKDLEFQEGDHVFLRVTPVTGVGRALKSKKLTPRFIGPYQ 1334
+ E + + + D+ ++ GD V++ R ++K L PR+ GPY
Sbjct: 1093 VQHEVWRPLAAAYQEQLDRPVVPHPYRVGDTVWV---------RRHQTKNLEPRWKGPYT 1143
Query: 1335 I 1335
+
Sbjct: 1144 V 1144
>POL_MLVCB (P08361) Pol polyprotein [Contains: Reverse
transcriptase/ribonuclease H (EC 2.7.7.49) (EC 3.1.26.4)
(RT); Integrase (IN)] (Fragment)
Length = 282
Score = 50.8 bits (120), Expect = 3e-05
Identities = 45/181 (24%), Positives = 76/181 (41%), Gaps = 11/181 (6%)
Query: 1156 SDRDPRFTSRFWKSLQEALGSKLRLSSAYHPQTDGQSERTIQSLEDLLRVCVLEQGG-TW 1214
+D P F S+ +++ + LG +L AY PQ+ GQ ER +++++ L L G W
Sbjct: 60 TDNGPAFVSKVSQTVADLLGIDWKLHCAYRPQSSGQVERMNRTIKETLTKLTLATGSRDW 119
Query: 1215 DSHLPLIEFTYNNSYHSSIGMAPFEALYGRRCRTQLCWFESGGRVVLGPEIVQQTTEKVK 1274
LPL + N+ G+ P+E LYG RV P +
Sbjct: 120 VLLLPLALYRARNT-PGPHGLTPYEILYGAPPPLVNFPDPDMTRVTNSPSLQAHLQALYL 178
Query: 1275 MIQEKMKASQSRQKSYHDKRRKDLEFQEGDHVFLRVTPVTGVGRALKSKKLTPRFIGPYQ 1334
+ E + + + D+ ++ GD V++ R ++K L PR+ GPY
Sbjct: 179 VQHEVWRPLAAAYQEQLDRPVVPHPYRVGDTVWV---------RRHQTKNLEPRWKGPYT 229
Query: 1335 I 1335
+
Sbjct: 230 V 230
>POL_BAEVM (P10272) Pol polyprotein [Contains: Protease (EC 3.4.23.-);
Reverse transcriptase/ribonuclease H (EC 2.7.7.49) (EC
3.1.26.4) (RT); Integrase (IN)]
Length = 1189
Score = 50.1 bits (118), Expect = 4e-05
Identities = 51/182 (28%), Positives = 79/182 (43%), Gaps = 20/182 (10%)
Query: 1156 SDRDPRFTSRFWKSLQEALGSKLRLSSAYHPQTDGQSERTIQSLEDLLRVCVLEQG-GTW 1214
SD P F S+ + L LG +L AY PQ+ GQ ER +++++ L LE G W
Sbjct: 969 SDNGPAFVSQVSQGLARILGINWKLHCAYRPQSSGQVERMNRTIKETLTKLTLETGLKDW 1028
Query: 1215 DSHLPLIEFTYNNSYHSSIGMAPFEALYG--RRCRTQLCWFESGGRVVLGPEIVQQTTEK 1272
L L N+ + G+ P+E LYG T L F +Q +
Sbjct: 1029 RRLLSLALLRARNT-PNRFGLTPYEILYGGPPPLSTLLNSFSPSN----SKTDLQARLKG 1083
Query: 1273 VKMIQEKMKASQSR-QKSYHDKRRKDLEFQEGDHVFLRVTPVTGVGRALKSKKLTPRFIG 1331
++ +Q ++ A + + H + FQ GD V++ R +S+ L PR+ G
Sbjct: 1084 LQAVQAQIWAPLAELYRPGHSQTSH--PFQVGDSVYV---------RRHRSQGLEPRWKG 1132
Query: 1332 PY 1333
PY
Sbjct: 1133 PY 1134
>POL_FENV1 (P31792) Pol polyprotein [Contains: Reverse
transcriptase/ribonuclease H (EC 2.7.7.49) (EC 3.1.26.4)
(RT); Integrase (IN)] (Fragment)
Length = 1046
Score = 49.7 bits (117), Expect = 6e-05
Identities = 31/89 (34%), Positives = 44/89 (48%), Gaps = 2/89 (2%)
Query: 1156 SDRDPRFTSRFWKSLQEALGSKLRLSSAYHPQTDGQSERTIQSLEDLLRVCVLEQG-GTW 1214
SD P F S+ + L LG +L AY PQ+ GQ ER +++++ L LE G W
Sbjct: 826 SDNGPAFVSQVSQGLARTLGINWKLHCAYRPQSSGQVERMNRTIKETLTKLTLETGLKDW 885
Query: 1215 DSHLPLIEFTYNNSYHSSIGMAPFEALYG 1243
L L N+ + G+ P+E LYG
Sbjct: 886 RRLLSLALLRARNT-PNRFGLTPYEILYG 913
>POL_FOAMV (P14350) Pol polyprotein [Contains: Reverse
transcriptase/ribonuclease H (EC 2.7.7.49) (EC 3.1.26.4)
(RT); Integrase (IN)]
Length = 886
Score = 48.9 bits (115), Expect = 1e-04
Identities = 25/88 (28%), Positives = 43/88 (48%)
Query: 1156 SDRDPRFTSRFWKSLQEALGSKLRLSSAYHPQTDGQSERTIQSLEDLLRVCVLEQGGTWD 1215
SD+ FTS + + G L S+ YHPQ+ + ER ++ LL ++ + W
Sbjct: 738 SDQGAAFTSSTFAEWAKERGIHLEFSTPYHPQSGSKVERKNSDIKRLLTKLLVGRPTKWY 797
Query: 1216 SHLPLIEFTYNNSYHSSIGMAPFEALYG 1243
LP+++ NN+Y + P + L+G
Sbjct: 798 DLLPVVQLALNNTYSPVLKYTPHQLLFG 825
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.362 0.162 0.603
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 133,111,004
Number of Sequences: 164201
Number of extensions: 4697357
Number of successful extensions: 19049
Number of sequences better than 10.0: 31
Number of HSP's better than 10.0 without gapping: 25
Number of HSP's successfully gapped in prelim test: 6
Number of HSP's that attempted gapping in prelim test: 18967
Number of HSP's gapped (non-prelim): 52
length of query: 1449
length of database: 59,974,054
effective HSP length: 123
effective length of query: 1326
effective length of database: 39,777,331
effective search space: 52744740906
effective search space used: 52744740906
T: 11
A: 40
X1: 14 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 37 (22.0 bits)
S2: 72 (32.3 bits)
Medicago: description of AC144724.10


