

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144721.7 + phase: 0 /pseudo
(817 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
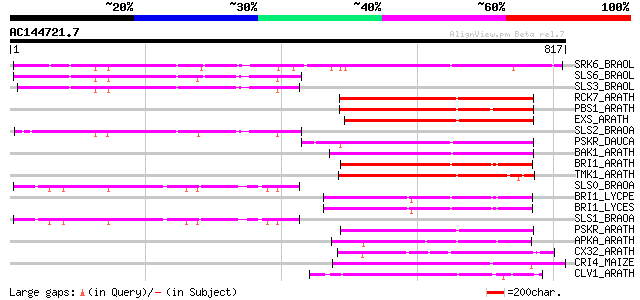
Score E
Sequences producing significant alignments: (bits) Value
SRK6_BRAOL (Q09092) Putative serine/threonine-protein kinase rec... 544 e-154
SLS6_BRAOL (P07761) S-locus-specific glycoprotein S6 precursor (... 223 2e-57
SLS3_BRAOL (P17840) S-locus-specific glycoprotein S13 precursor ... 222 3e-57
RCK7_ARATH (Q9LQQ8) Putative serine/threonine-protein kinase RLC... 221 5e-57
PBS1_ARATH (Q9FE20) Serine/threonine-protein kinase PBS1 (EC 2.7... 218 7e-56
EXS_ARATH (Q9LYN8) Leucine-rich repeat receptor protein kinase E... 216 2e-55
SLS2_BRAOA (P22553) S-locus-specific glycoprotein BS29-2 precursor 214 7e-55
PSKR_DAUCA (Q8LPB4) Phytosulfokine receptor precursor (EC 2.7.1.... 211 6e-54
BAK1_ARATH (Q94F62) BRASSINOSTEROID INSENSITIVE 1-associated rec... 209 2e-53
BRI1_ARATH (O22476) BRASSINOSTEROID INSENSITIVE 1 precursor (EC ... 208 4e-53
TMK1_ARATH (P43298) Putative receptor protein kinase TMK1 precur... 206 2e-52
SLS0_BRAOA (P22551) S-locus-specific glycoprotein precursor 199 2e-50
BRI1_LYCPE (Q8L899) Systemin receptor SR160 precursor (EC 2.7.1.... 199 3e-50
BRI1_LYCES (Q8GUQ5) Brassinosteroid LRR receptor kinase precurso... 199 3e-50
SLS1_BRAOA (P22552) S-locus-specific glycoprotein BS29-1 precursor 198 4e-50
PSKR_ARATH (Q9ZVR7) Putative phytosulfokine receptor precursor (... 196 2e-49
APKA_ARATH (Q06548) Protein kinase APK1A, chloroplast precursor ... 191 9e-48
CX32_ARATH (P27450) Probable serine/threonine-protein kinase Cx3... 190 1e-47
CRI4_MAIZE (O24585) Putative receptor protein kinase CRINKLY4 pr... 190 1e-47
CLV1_ARATH (Q9SYQ8) Receptor protein kinase CLAVATA1 precursor (... 185 4e-46
>SRK6_BRAOL (Q09092) Putative serine/threonine-protein kinase
receptor precursor (EC 2.7.1.37) (S-receptor kinase)
(SRK)
Length = 849
Score = 544 bits (1402), Expect = e-154
Identities = 324/866 (37%), Positives = 489/866 (56%), Gaps = 83/866 (9%)
Query: 6 HNSNYFFIITFLIFCTIYSCYSAINDTITSSKSL--KDNETITSNNTNFKLGFFSPLNST 63
H+S F++ F++ I+ S +T++S++SL N+T+ S + F++GFF ++
Sbjct: 9 HHSYMSFLLVFVVMILIHPALSIYINTLSSTESLTISSNKTLVSPGSIFEVGFFR--TNS 66
Query: 64 NRYLGIWY--INKTNNIWIANRDQPLKDSNGIVTIHKDGNFIILNKPNGVIIWSTNISSS 121
YLG+WY ++ +W+ANRD PL SN I T+ GN ++L + +W TN++
Sbjct: 67 RWYLGMWYKKVSDRTYVWVANRDNPL--SNAIGTLKISGNNLVLLDHSNKPVWWTNLTRG 124
Query: 122 TNST---AQLADSGNLILRDISSGAT---IWDSFTHPADAAVPTMRIAANQVTGKKISFV 175
+ A+L +GN ++RD S+ +W SF +P D +P M++ N TG
Sbjct: 125 NERSPVVAELLANGNFVMRDSSNNDASEYLWQSFDYPTDTLLPEMKLGYNLKTGLNRFLT 184
Query: 176 SRKSDNDPSSGHYSASLERLDAPEVFIWKDKNIHWRTGPWNGRVFLGSPRMLTEYLAGWR 235
S +S +DPSSG++S LE PE ++ ++ R+GPWNG F G P +
Sbjct: 185 SWRSSDDPSSGNFSYKLETQSLPEFYLSRENFPMHRSGPWNGIRFSGIPEDQKLSYMVYN 244
Query: 236 FDQDTDGTTYITYNFADKTMFGILSLTPHGTLKLIEYMNKKELFRL----EVDQNECDFY 291
F ++ + Y T+ + + + L+L G + + + ++ VD +CD Y
Sbjct: 245 FIENNEEVAY-TFRMTNNSFYSRLTLISEGYFQRLTWYPSIRIWNRFWSSPVDP-QCDTY 302
Query: 292 GKCGPFGNCDNSTVPICSCFDGFEPKNSVEWSLGNWTNGCVRKEGMNLKCEMVKNGSSIV 351
CGP+ CD +T P+C+C GF P+N +W W GC+R+ L C
Sbjct: 303 IMCGPYAYCDVNTSPVCNCIQGFNPRNIQQWDQRVWAGGCIRRT--QLSCS--------- 351
Query: 352 KQDGFKVYHNMKPPDFNVRT--NNADQDKCGADCLANCSCLAYAY----DPSIFCMYWTG 405
DGF MK P+ + T + +C C+++C+C A+A + C+ WT
Sbjct: 352 -GDGFTRMKKMKLPETTMATVDRSIGVKECKKRCISDCNCTAFANADIRNGGSGCVIWTE 410
Query: 406 ELIDLQKFPNG---GVDLFVRVPAELVAVKKEKGHNKSFLIIVIAGVIGALILVICAYLL 462
L D++ + G DL+VR+ A +A K+ N S II + + L+L+I + L
Sbjct: 411 RLEDIRNYATDAIDGQDLYVRLAAADIAKKR----NASGKIISLTVGVSVLLLLIM-FCL 465
Query: 463 WRKCSARHKA--------REHQQMKLDELPL---------YDFEKLET----------AT 495
W++ R KA + +Q + ++E+ L Y FE+LE AT
Sbjct: 466 WKRKQKRAKASAISIANTQRNQNLPMNEMVLSSKREFSGEYKFEELELPLIEMETVVKAT 525
Query: 496 NCFHFNNMLGKGGFGPVYKGVMEDGQEIAVKRLSKASGQGIEEFMNEVVVISKLQHRNLV 555
F N LG+GGFG VYKG + DG+EIAVKRLSK S QG +EFMNEV +I++LQH NLV
Sbjct: 526 ENFSSCNKLGQGGFGIVYKGRLLDGKEIAVKRLSKTSVQGTDEFMNEVTLIARLQHINLV 585
Query: 556 RLLGCCVERGEQILVYEFMPNKSLDAFLFDPLQKKNLDWRKRSNIIEGIARGIMYLHRDS 615
++LGCC+E E++L+YE++ N SLD++LF ++ L+W +R +I G+ARG++YLH+DS
Sbjct: 586 QVLGCCIEGDEKMLIYEYLENLSLDSYLFGKTRRSKLNWNERFDITNGVARGLLYLHQDS 645
Query: 616 RLRIIHRDLKASNILLDSDMIPKISDFGLARIVKFGEDDEANTKRVVGTYGYMPPEYAME 675
R RIIHRDLK SNILLD +MIPKISDFG+ARI + ++ EANT +VVGTYGYM PEYAM
Sbjct: 646 RFRIIHRDLKVSNILLDKNMIPKISDFGMARIFE-RDETEANTMKVVGTYGYMSPEYAMY 704
Query: 676 GLFSEKSDVYSFGVLLLEIVSGRRNSSFSHHEDTLSLVGFAWKLWLEENIISLIDPEVWD 735
G+FSEKSDV+SFGV++LEIVSG++N F + + L+ + W W E + ++DP + D
Sbjct: 705 GIFSEKSDVFSFGVIVLEIVSGKKNRGFYNLDYENDLLSYVWSRWKEGRALEIVDPVIVD 764
Query: 736 ACFE-------SSMLRCIHIGLLCVQELPRDRPNISTVVLMLVSEITHLPPPGRVAFVHK 788
+ +L+CI IGLLCVQEL RP +S+VV M SE T +P P + +
Sbjct: 765 SLSSQPSIFQPQEVLKCIQIGLLCVQELAEHRPAMSSVVWMFGSEATEIPQPKPPGYCVR 824
Query: 789 QSSKSTTESSQKSHQSNSNNNVTLSE 814
+S SS S Q + N + T+++
Sbjct: 825 RSPYELDPSS--SWQCDENESWTVNQ 848
>SLS6_BRAOL (P07761) S-locus-specific glycoprotein S6 precursor
(SLSG-6)
Length = 436
Score = 223 bits (567), Expect = 2e-57
Identities = 138/445 (31%), Positives = 221/445 (49%), Gaps = 37/445 (8%)
Query: 6 HNSNYFFIITFLIFCTIYSCYSAINDTITSSKSLK--DNETITSNNTNFKLGFFSPLNST 63
++++Y + F I C + +T++S++SL+ N T+ S NF+LGFF +S+
Sbjct: 8 YDNSYTLSFLLVFFVLILFCPAFSINTLSSTESLRISSNRTLVSPGNNFELGFFRTNSSS 67
Query: 64 NRYLGIWYINKTNN--IWIANRDQPLKDSNGIVTIHKDGNFIILNKPNGVIIWSTNISSS 121
YLGIWY + +W+ANRD PL SN I T+ GN ++L +WSTN++
Sbjct: 68 RWYLGIWYKKLLDRTYVWVANRDNPL--SNAIGTLKISGNNLVLLGHTNKSVWSTNLTRG 125
Query: 122 TNS---TAQLADSGNLILRDISSGAT---IWDSFTHPADAAVPTMRIAANQVTGKKISFV 175
A+L +GN ++RD S+ +W SF +P D +P M++ + TG
Sbjct: 126 NERLPVVAELLSNGNFVMRDSSNNDASEYLWQSFDYPTDTLLPEMKLGYDLKTGLNRFLT 185
Query: 176 SRKSDNDPSSGHYSASLERLDAPEVFIWKDKNIHWRTGPWNGRVFLGSPRMLTEYLAGWR 235
S +S +DPSSG +S LE PE ++W R+GPWNG F G P +
Sbjct: 186 SWRSSDDPSSGDFSYKLETRSLPEFYLWHGIFPMHRSGPWNGVRFSGIPEDQKLSYMVYN 245
Query: 236 FDQDTDGTTYITYNFADKTMFGILSLTPHGTLKLIEYMNK----KELFRLEVDQNECDFY 291
F ++++ Y T+ + +++ L+L+ G + + + + VD +CD Y
Sbjct: 246 FTENSEEVAY-TFRMTNNSIYSRLTLSSEGYFQRLTWNPSIGIWNRFWSSPVDP-QCDTY 303
Query: 292 GKCGPFGNCDNSTVPICSCFDGFEPKNSVEWSLGNWTNGCVRKEGMNLKCEMVKNGSSIV 351
CGP+ C +T P+C+C GF P+N +W W GC+R+ L C
Sbjct: 304 IMCGPYAYCGVNTSPVCNCIQGFNPRNIQQWDQRVWAGGCIRR--TRLSC---------- 351
Query: 352 KQDGFKVYHNMKPPD--FNVRTNNADQDKCGADCLANCSCLAYA----YDPSIFCMYWTG 405
DGF NMK P+ + + +C CL++C+C A+A + C+ WTG
Sbjct: 352 SGDGFTRMKNMKLPETTMAIVDRSIGVKECEKRCLSDCNCTAFANADIRNGGTGCVIWTG 411
Query: 406 ELIDLQKFPNGGVDLFVRVP-AELV 429
L D++ + G DL+VR+ A+LV
Sbjct: 412 RLDDMRNYVAHGQDLYVRLAVADLV 436
>SLS3_BRAOL (P17840) S-locus-specific glycoprotein S13 precursor
(SLSG-13) (Fragment)
Length = 434
Score = 222 bits (566), Expect = 3e-57
Identities = 133/431 (30%), Positives = 213/431 (48%), Gaps = 31/431 (7%)
Query: 12 FIITFLIFCTIYSCYSAINDTITSSKSLKDNETITSNNTNFKLGFFSPLNSTNRYLGIWY 71
F++ F + +S + T S ++ N T+ S F+LGFF +S+ YLGIWY
Sbjct: 15 FLLVFFVLILFRPAFSINTLSSTESLTISSNRTLVSPGNVFELGFFKTTSSSRWYLGIWY 74
Query: 72 --INKTNNIWIANRDQPLKDSNGIVTIHKDGNFIILNKPNGVIIWSTNISSSTNST---A 126
+W+ANRD PL SN I T+ GN ++L + +WSTN++ + A
Sbjct: 75 KKFPYRTYVWVANRDNPL--SNDIGTLKISGNNLVLLDHSNKSVWSTNVTRGNERSPVVA 132
Query: 127 QLADSGNLILRDISSGAT---IWDSFTHPADAAVPTMRIAANQVTGKKISFVSRKSDNDP 183
+L D+GN ++RD +S +W SF +P D +P M++ + TG S +S +DP
Sbjct: 133 ELLDNGNFVMRDSNSNNASQFLWQSFDYPTDTLLPEMKLGYDLKTGLNRFLTSWRSSDDP 192
Query: 184 SSGHYSASLERLDAPEVFIWKDKNIHWRTGPWNGRVFLGSPRMLTEYLAGWRFDQDTDGT 243
SSG YS LE PE ++ R+GPWNG G P + F ++++
Sbjct: 193 SSGDYSYKLELRRLPEFYLSSGSFRLHRSGPWNGFRISGIPEDQKLSYMVYNFTENSEEA 252
Query: 244 TYITYNFADKTMFGILSLTPHGTLKLIEYMNKKELFRL--EVDQNECDFYGKCGPFGNCD 301
Y T+ + + + L+++ G + + + ++ + ++CD Y CGP+ CD
Sbjct: 253 AY-TFLMTNNSFYSRLTISSTGYFERLTWAPSSVVWNVFWSSPNHQCDMYRMCGPYSYCD 311
Query: 302 NSTVPICSCFDGFEPKNSVEWSLGNWTNGCVRKEGMNLKCEMVKNGSSIVKQDGFKVYHN 361
+T P+C+C GF PKN +W L T+GC+R+ L C DGF N
Sbjct: 312 VNTSPVCNCIQGFRPKNRQQWDLRIPTSGCIRR--TRLSC----------SGDGFTRMKN 359
Query: 362 MKPPDFNVRT--NNADQDKCGADCLANCSCLAYA----YDPSIFCMYWTGELIDLQKFPN 415
MK P+ + + +C CL++C+C A+A + C+ WTGEL D++ +
Sbjct: 360 MKLPETTMAIVHRSIGLKECEKRCLSDCNCTAFANADIRNRGTGCVIWTGELEDIRTYFA 419
Query: 416 GGVDLFVRVPA 426
G DL+VR+ A
Sbjct: 420 DGQDLYVRLAA 430
>RCK7_ARATH (Q9LQQ8) Putative serine/threonine-protein kinase
RLCKVII (EC 2.7.1.37)
Length = 423
Score = 221 bits (564), Expect = 5e-57
Identities = 126/288 (43%), Positives = 177/288 (60%), Gaps = 4/288 (1%)
Query: 486 YDFEKLETATNCFHFNNMLGKGGFGPVYKGVMED-GQEIAVKRLSKASGQGIEEFMNEVV 544
+ F++L AT F + LG+GGFG V+KG +E Q +A+K+L + QGI EF+ EV+
Sbjct: 91 FTFQELAEATGNFRSDCFLGEGGFGKVFKGTIEKLDQVVAIKQLDRNGVQGIREFVVEVL 150
Query: 545 VISKLQHRNLVRLLGCCVERGEQILVYEFMPNKSLDAFLFD-PLQKKNLDWRKRSNIIEG 603
+S H NLV+L+G C E +++LVYE+MP SL+ L P KK LDW R I G
Sbjct: 151 TLSLADHPNLVKLIGFCAEGDQRLLVYEYMPQGSLEDHLHVLPSGKKPLDWNTRMKIAAG 210
Query: 604 IARGIMYLHRDSRLRIIHRDLKASNILLDSDMIPKISDFGLARIVKFGEDDEANTKRVVG 663
ARG+ YLH +I+RDLK SNILL D PK+SDFGLA++ G+ +T RV+G
Sbjct: 211 AARGLEYLHDRMTPPVIYRDLKCSNILLGEDYQPKLSDFGLAKVGPSGDKTHVST-RVMG 269
Query: 664 TYGYMPPEYAMEGLFSEKSDVYSFGVLLLEIVSGRRNSSFSHHEDTLSLVGFAWKLWLE- 722
TYGY P+YAM G + KSD+YSFGV+LLE+++GR+ + +LVG+A L+ +
Sbjct: 270 TYGYCAPDYAMTGQLTFKSDIYSFGVVLLELITGRKAIDNTKTRKDQNLVGWARPLFKDR 329
Query: 723 ENIISLIDPEVWDACFESSMLRCIHIGLLCVQELPRDRPNISTVVLML 770
N ++DP + + + + I +CVQE P RP +S VVL L
Sbjct: 330 RNFPKMVDPLLQGQYPVRGLYQALAISAMCVQEQPTMRPVVSDVVLAL 377
>PBS1_ARATH (Q9FE20) Serine/threonine-protein kinase PBS1 (EC
2.7.1.37) (AvrPphB susceptible protein 1)
Length = 456
Score = 218 bits (554), Expect = 7e-56
Identities = 126/290 (43%), Positives = 179/290 (61%), Gaps = 8/290 (2%)
Query: 486 YDFEKLETATNCFHFNNMLGKGGFGPVYKGVMED-GQEIAVKRLSKASGQGIEEFMNEVV 544
+ F +L AT FH + LG+GGFG VYKG ++ GQ +AVK+L + QG EF+ EV+
Sbjct: 74 FAFRELAAATMNFHPDTFLGEGGFGRVYKGRLDSTGQVVAVKQLDRNGLQGNREFLVEVL 133
Query: 545 VISKLQHRNLVRLLGCCVERGEQILVYEFMPNKSLDAFLFD-PLQKKNLDWRKRSNIIEG 603
++S L H NLV L+G C + +++LVYEFMP SL+ L D P K+ LDW R I G
Sbjct: 134 MLSLLHHPNLVNLIGYCADGDQRLLVYEFMPLGSLEDHLHDLPPDKEALDWNMRMKIAAG 193
Query: 604 IARGIMYLHRDSRLRIIHRDLKASNILLDSDMIPKISDFGLARIVKFGEDDEANTKRVVG 663
A+G+ +LH + +I+RD K+SNILLD PK+SDFGLA++ G+ +T RV+G
Sbjct: 194 AAKGLEFLHDKANPPVIYRDFKSSNILLDEGFHPKLSDFGLAKLGPTGDKSHVST-RVMG 252
Query: 664 TYGYMPPEYAMEGLFSEKSDVYSFGVLLLEIVSGRR--NSSFSHHEDTLSLVGFAWKLWL 721
TYGY PEYAM G + KSDVYSFGV+ LE+++GR+ +S H E +LV +A L+
Sbjct: 253 TYGYCAPEYAMTGQLTVKSDVYSFGVVFLELITGRKAIDSEMPHGEQ--NLVAWARPLFN 310
Query: 722 E-ENIISLIDPEVWDACFESSMLRCIHIGLLCVQELPRDRPNISTVVLML 770
+ I L DP + ++ + + + +C+QE RP I+ VV L
Sbjct: 311 DRRKFIKLADPRLKGRFPTRALYQALAVASMCIQEQAATRPLIADVVTAL 360
>EXS_ARATH (Q9LYN8) Leucine-rich repeat receptor protein kinase EXS
precursor (EC 2.7.1.37) (Extra sporogenous cells protein)
(EXCESS MICROSPOROCYTES1 protein)
Length = 1192
Score = 216 bits (550), Expect = 2e-55
Identities = 121/279 (43%), Positives = 171/279 (60%), Gaps = 4/279 (1%)
Query: 494 ATNCFHFNNMLGKGGFGPVYKGVMEDGQEIAVKRLSKASGQGIEEFMNEVVVISKLQHRN 553
AT+ F N++G GGFG VYK + + +AVK+LS+A QG EFM E+ + K++H N
Sbjct: 913 ATDHFSKKNIIGDGGFGTVYKACLPGEKTVAVKKLSEAKTQGNREFMAEMETLGKVKHPN 972
Query: 554 LVRLLGCCVERGEQILVYEFMPNKSLDAFLFDPL-QKKNLDWRKRSNIIEGIARGIMYLH 612
LV LLG C E++LVYE+M N SLD +L + + LDW KR I G ARG+ +LH
Sbjct: 973 LVSLLGYCSFSEEKLLVYEYMVNGSLDHWLRNQTGMLEVLDWSKRLKIAVGAARGLAFLH 1032
Query: 613 RDSRLRIIHRDLKASNILLDSDMIPKISDFGLARIVKFGEDDEANTKRVVGTYGYMPPEY 672
IIHRD+KASNILLD D PK++DFGLAR++ E + + GT+GY+PPEY
Sbjct: 1033 HGFIPHIIHRDIKASNILLDGDFEPKVADFGLARLISACESHVSTV--IAGTFGYIPPEY 1090
Query: 673 AMEGLFSEKSDVYSFGVLLLEIVSGRRNSSFSHHE-DTLSLVGFAWKLWLEENIISLIDP 731
+ K DVYSFGV+LLE+V+G+ + E + +LVG+A + + + +IDP
Sbjct: 1091 GQSARATTKGDVYSFGVILLELVTGKEPTGPDFKESEGGNLVGWAIQKINQGKAVDVIDP 1150
Query: 732 EVWDACFESSMLRCIHIGLLCVQELPRDRPNISTVVLML 770
+ ++S LR + I +LC+ E P RPN+ V+ L
Sbjct: 1151 LLVSVALKNSQLRLLQIAMLCLAETPAKRPNMLDVLKAL 1189
>SLS2_BRAOA (P22553) S-locus-specific glycoprotein BS29-2 precursor
Length = 435
Score = 214 bits (545), Expect = 7e-55
Identities = 138/443 (31%), Positives = 229/443 (51%), Gaps = 37/443 (8%)
Query: 7 NSNYFFIITFLIFCTIYSCYSAINDTITSSKSLK--DNETITSNNTNFKLGFFSPLNSTN 64
NS+ F++ F + T++S +IN T++S +SLK ++ T+ S +LGFF +S+
Sbjct: 10 NSHTSFLLVFFVL-TLFSPAFSIN-TLSSIESLKISNSRTLVSPGNVLELGFFRTPSSSR 67
Query: 65 RYLGIWY--INKTNNIWIANRDQPLKDSNGIVTIHKDGNFIILNKPNGVIIWSTNISSST 122
YLG+WY +++ +W+ANRD PL S G + I + N ++L+ N + WSTN +
Sbjct: 68 WYLGMWYKKLSERTYVWVANRDNPLSCSIGTLKI-SNMNLVLLDHSNKSL-WSTNHTRGN 125
Query: 123 NST---AQLADSGNLILRDISSG---ATIWDSFTHPADAAVPTMRIAANQVTGKKISFVS 176
+ A+L +GN +LRD + +W SF +P D +P M++ + TG S
Sbjct: 126 ERSPVVAELLANGNFVLRDSNKNDRSGFLWQSFDYPTDTLLPEMKLGYDLRTGLNRFLTS 185
Query: 177 RKSDNDPSSGHYSASLERLDAPEVFIWKDKNIHWRTGPWNGRVFLGSPRMLTEYLAGWRF 236
+S +DPSSG +S L+ PE +++KD + R+GPWNG F G P + F
Sbjct: 186 WRSSDDPSSGDFSYKLQTRRLPEFYLFKDDFLVHRSGPWNGVGFSGMPEDQKLSYMVYNF 245
Query: 237 DQDTDGTTYITYNFADKTMFGILSLTPHGTLKLIEYMNKK---ELFRLEVDQNECDFYGK 293
Q+++ Y T+ + +++ L+++ G + + + +F + +CD Y
Sbjct: 246 TQNSEEVAY-TFLMTNNSIYSRLTISSSGYFERLTWTPSSGMWNVFWSSPEDFQCDVYKI 304
Query: 294 CGPFGNCDNSTVPICSCFDGFEPKNSVEWSLGNWTNGCVRKEGMNLKCEMVKNGSSIVKQ 353
CG + CD +T P+C+C F+P N EW L W+ GC R+ L C
Sbjct: 305 CGAYSYCDVNTSPVCNCIQRFDPSNVQEWGLRAWSGGCRRR--TRLSC----------SG 352
Query: 354 DGFKVYHNMKPPD--FNVRTNNADQDKCGADCLANCSCLAYA----YDPSIFCMYWTGEL 407
DGF MK P+ + + +C CL++C+C A+A + C+ WTG+L
Sbjct: 353 DGFTRMKKMKLPETTMAIVDRSIGLKECEKRCLSDCNCTAFANADIRNGGTGCVIWTGQL 412
Query: 408 IDLQKFPNGGVDLFVRV-PAELV 429
D++ + G DL+VR+ PA+LV
Sbjct: 413 EDIRTYFANGQDLYVRLAPADLV 435
>PSKR_DAUCA (Q8LPB4) Phytosulfokine receptor precursor (EC 2.7.1.37)
(Phytosulfokine LRR receptor kinase)
Length = 1021
Score = 211 bits (537), Expect = 6e-54
Identities = 123/358 (34%), Positives = 192/358 (53%), Gaps = 20/358 (5%)
Query: 430 AVKKEKGHNKSFLIIVIAGVIGALILVICAYLLWRKCSARHKAREHQQMKLDELPL---- 485
AVK +K K + V G +G + L+ L+ + ++R + ++ DE+ L
Sbjct: 660 AVKSKKNIRKIVAVAVGTG-LGTVFLLTVTLLIILRTTSRGEVDPEKKADADEIELGSRS 718
Query: 486 ------------YDFEKLETATNCFHFNNMLGKGGFGPVYKGVMEDGQEIAVKRLSKASG 533
+ + +T+ F+ N++G GGFG VYK + DG ++A+KRLS +G
Sbjct: 719 VVLFHNKDSNNELSLDDILKSTSSFNQANIIGCGGFGLVYKATLPDGTKVAIKRLSGDTG 778
Query: 534 QGIEEFMNEVVVISKLQHRNLVRLLGCCVERGEQILVYEFMPNKSLDAFLFDPLQ-KKNL 592
Q EF EV +S+ QH NLV LLG C + +++L+Y +M N SLD +L + + +L
Sbjct: 779 QMDREFQAEVETLSRAQHPNLVHLLGYCNYKNDKLLIYSYMDNGSLDYWLHEKVDGPPSL 838
Query: 593 DWRKRSNIIEGIARGIMYLHRDSRLRIIHRDLKASNILLDSDMIPKISDFGLARIVKFGE 652
DW+ R I G A G+ YLH+ I+HRD+K+SNILL + ++DFGLAR++
Sbjct: 839 DWKTRLRIARGAAEGLAYLHQSCEPHILHRDIKSSNILLSDTFVAHLADFGLARLIL--P 896
Query: 653 DDEANTKRVVGTYGYMPPEYAMEGLFSEKSDVYSFGVLLLEIVSGRRNSSFSHHEDTLSL 712
D T +VGT GY+PPEY + + K DVYSFGV+LLE+++GRR + L
Sbjct: 897 YDTHVTTDLVGTLGYIPPEYGQASVATYKGDVYSFGVVLLELLTGRRPMDVCKPRGSRDL 956
Query: 713 VGFAWKLWLEENIISLIDPEVWDACFESSMLRCIHIGLLCVQELPRDRPNISTVVLML 770
+ + ++ E+ + DP ++D ML + I C+ E P+ RP +V L
Sbjct: 957 ISWVLQMKTEKRESEIFDPFIYDKDHAEEMLLVLEIACRCLGENPKTRPTTQQLVSWL 1014
>BAK1_ARATH (Q94F62) BRASSINOSTEROID INSENSITIVE 1-associated
receptor kinase 1 precursor (EC 2.7.1.37)
(BRI1-associated receptor kinase 1) (Somatic
embryogenesis receptor-like kinase 3)
Length = 615
Score = 209 bits (533), Expect = 2e-53
Identities = 121/303 (39%), Positives = 183/303 (59%), Gaps = 6/303 (1%)
Query: 472 AREHQQMKLDELPLYDFEKLETATNCFHFNNMLGKGGFGPVYKGVMEDGQEIAVKRLSKA 531
A E ++ L +L + +L+ A++ F N+LG+GGFG VYKG + DG +AVKRL +
Sbjct: 263 AEEDPEVHLGQLKRFSLRELQVASDNFSNKNILGRGGFGKVYKGRLADGTLVAVKRLKEE 322
Query: 532 SGQGIE-EFMNEVVVISKLQHRNLVRLLGCCVERGEQILVYEFMPNKSLDAFLFD-PLQK 589
QG E +F EV +IS HRNL+RL G C+ E++LVY +M N S+ + L + P +
Sbjct: 323 RTQGGELQFQTEVEMISMAVHRNLLRLRGFCMTPTERLLVYPYMANGSVASCLRERPESQ 382
Query: 590 KNLDWRKRSNIIEGIARGIMYLHRDSRLRIIHRDLKASNILLDSDMIPKISDFGLARIVK 649
LDW KR I G ARG+ YLH +IIHRD+KA+NILLD + + DFGLA+++
Sbjct: 383 PPLDWPKRQRIALGSARGLAYLHDHCDPKIIHRDVKAANILLDEEFEAVVGDFGLAKLMD 442
Query: 650 FGEDDEANTKRVVGTYGYMPPEYAMEGLFSEKSDVYSFGVLLLEIVSGRRNSSFSH--HE 707
+ D T V GT G++ PEY G SEK+DV+ +GV+LLE+++G+R + ++
Sbjct: 443 Y--KDTHVTTAVRGTIGHIAPEYLSTGKSSEKTDVFGYGVMLLELITGQRAFDLARLAND 500
Query: 708 DTLSLVGFAWKLWLEENIISLIDPEVWDACFESSMLRCIHIGLLCVQELPRDRPNISTVV 767
D + L+ + L E+ + +L+D ++ + + + I + LLC Q P +RP +S VV
Sbjct: 501 DDVMLLDWVKGLLKEKKLEALVDVDLQGNYKDEEVEQLIQVALLCTQSSPMERPKMSEVV 560
Query: 768 LML 770
ML
Sbjct: 561 RML 563
>BRI1_ARATH (O22476) BRASSINOSTEROID INSENSITIVE 1 precursor (EC
2.7.1.37) (AtBRI1) (Brassinosteroid LRR receptor kinase)
Length = 1196
Score = 208 bits (530), Expect = 4e-53
Identities = 117/285 (41%), Positives = 172/285 (60%), Gaps = 6/285 (2%)
Query: 488 FEKLETATNCFHFNNMLGKGGFGPVYKGVMEDGQEIAVKRLSKASGQGIEEFMNEVVVIS 547
F L ATN FH ++++G GGFG VYK +++DG +A+K+L SGQG EFM E+ I
Sbjct: 873 FADLLQATNGFHNDSLIGSGGFGDVYKAILKDGSAVAIKKLIHVSGQGDREFMAEMETIG 932
Query: 548 KLQHRNLVRLLGCCVERGEQILVYEFMPNKSLDAFLFDPLQK-KNLDWRKRSNIIEGIAR 606
K++HRNLV LLG C E++LVYEFM SL+ L DP + L+W R I G AR
Sbjct: 933 KIKHRNLVPLLGYCKVGDERLLVYEFMKYGSLEDVLHDPKKAGVKLNWSTRRKIAIGSAR 992
Query: 607 GIMYLHRDSRLRIIHRDLKASNILLDSDMIPKISDFGLARIVKFGEDDEANTKRVVGTYG 666
G+ +LH + IIHRD+K+SN+LLD ++ ++SDFG+AR++ D + + GT G
Sbjct: 993 GLAFLHHNCSPHIIHRDMKSSNVLLDENLEARVSDFGMARLMS-AMDTHLSVSTLAGTPG 1051
Query: 667 YMPPEYAMEGLFSEKSDVYSFGVLLLEIVSGRRNSSFSHHEDTLSLVGFAWKLWLEENII 726
Y+PPEY S K DVYS+GV+LLE+++G+R + D +LVG+ K + I
Sbjct: 1052 YVPPEYYQSFRCSTKGDVYSYGVVLLELLTGKRPTDSPDFGDN-NLVGWV-KQHAKLRIS 1109
Query: 727 SLIDPEVW--DACFESSMLRCIHIGLLCVQELPRDRPNISTVVLM 769
+ DPE+ D E +L+ + + + C+ + RP + V+ M
Sbjct: 1110 DVFDPELMKEDPALEIELLQHLKVAVACLDDRAWRRPTMVQVMAM 1154
>TMK1_ARATH (P43298) Putative receptor protein kinase TMK1 precursor
(EC 2.7.1.-)
Length = 942
Score = 206 bits (524), Expect = 2e-52
Identities = 118/299 (39%), Positives = 181/299 (60%), Gaps = 17/299 (5%)
Query: 485 LYDFEKLETATNCFHFNNMLGKGGFGPVYKGVMEDGQEIAVKRLSKA--SGQGIEEFMNE 542
L + L + TN F +N+LG GGFG VYKG + DG +IAVKR+ +G+G EF +E
Sbjct: 575 LISIQVLRSVTNNFSSDNILGSGGFGVVYKGELHDGTKIAVKRMENGVIAGKGFAEFKSE 634
Query: 543 VVVISKLQHRNLVRLLGCCVERGEQILVYEFMPNKSLDAFLFDPLQK--KNLDWRKRSNI 600
+ V++K++HR+LV LLG C++ E++LVYE+MP +L LF+ ++ K L W++R +
Sbjct: 635 IAVLTKVRHRHLVTLLGYCLDGNEKLLVYEYMPQGTLSRHLFEWSEEGLKPLLWKQRLTL 694
Query: 601 IEGIARGIMYLHRDSRLRIIHRDLKASNILLDSDMIPKISDFGLARIVKFGEDDEANTKR 660
+ARG+ YLH + IHRDLK SNILL DM K++DFGL R+ G+ R
Sbjct: 695 ALDVARGVEYLHGLAHQSFIHRDLKPSNILLGDDMRAKVADFGLVRLAPEGKGSIET--R 752
Query: 661 VVGTYGYMPPEYAMEGLFSEKSDVYSFGVLLLEIVSGRRNSSFSHHEDTLSLVGFAWKLW 720
+ GT+GY+ PEYA+ G + K DVYSFGV+L+E+++GR++ S E+++ LV + +++
Sbjct: 753 IAGTFGYLAPEYAVTGRVTTKVDVYSFGVILMELITGRKSLDESQPEESIHLVSWFKRMY 812
Query: 721 L--EENIISLIDPEVWDACFESSMLRCIH-----IGLLCVQELPRDRPNISTVVLMLVS 772
+ E + ID + + L +H G C +E P RP++ V +L S
Sbjct: 813 INKEASFKKAIDTTI---DLDEETLASVHTVAELAGHCCARE-PYQRPDMGHAVNILSS 867
>SLS0_BRAOA (P22551) S-locus-specific glycoprotein precursor
Length = 444
Score = 199 bits (507), Expect = 2e-50
Identities = 143/456 (31%), Positives = 223/456 (48%), Gaps = 62/456 (13%)
Query: 6 HNSNYFFIITFLIFCTIYSCYS-AINDTITSSKSLKDNETITSNNTNFKLGFF------S 58
H+ FF + ++F ++S + + N+ +T S N+T+ S F+LGFF S
Sbjct: 10 HSYTLFFFVILVLFPHVFSTNTLSPNEALTISS----NKTLVSPGDVFELGFFKTTTRNS 65
Query: 59 PLNSTNRYLGIWYINKTNN---IWIANRDQPLKDSNGIVTIHKDGNFIILNKPNGVIIWS 115
P + YLGIWY + + +W+ANRD L +S G + I + ++L+ N +WS
Sbjct: 66 PDGTDRWYLGIWYKTTSGHRTYVWVANRDNALHNSMGTLKI-SHASLVLLDHSN-TPVWS 123
Query: 116 TNISSSTNS--TAQLADSGNLILRDISSGAT---IWDSFTHPADAAVPTMRIAANQVTGK 170
TN + + TA+L +GN +LRD + +W SF +P D +P M++ N + +
Sbjct: 124 TNFTGVAHLPVTAELLANGNFVLRDSKTNDLDRFMWQSFDYPVDTLLPEMKLGRNLIGSE 183
Query: 171 KISFV-SRKSDNDPSSGHYSASLERLD-APEVFIWKDKNIHWRTGPWNGRVFLGSPRMLT 228
+ S KS DPSSG +S LE E ++ K++ +RTGPWNG F G P+M
Sbjct: 184 NEKILTSWKSPTDPSSGDFSFILETEGFLHEFYLLKNEFKVYRTGPWNGVRFNGIPKM-- 241
Query: 229 EYLAGWRFDQDT--DGTTYITYNFADKTMFGI---LSLTPHGTLKLIEYMN---KKELFR 280
W + ++ D + Y+F I ++ G L++I + ++ +F
Sbjct: 242 ---QNWSYIDNSFIDNNEEVAYSFQVNNNHNIHTRFRMSSTGYLQVITWTKTVPQRNMF- 297
Query: 281 LEVDQNECDFYGKCGPFGNCDNSTVPICSCFDGFEPKNSVEWSLGNWTNGCVRKEGMNLK 340
++ CD Y CGP+ CD T P C+C GF PKN+ W L + + GCVR +
Sbjct: 298 WSFPEDTCDLYKVCGPYAYCDMHTSPTCNCIKGFVPKNAGRWDLRDMSGGCVRSSKL--- 354
Query: 341 CEMVKNGSSIVKQDGFKVYHNMKPPDFNVRTNNADQDK------CGADCLANCSCLAYA- 393
S + DGF MK P+ T+ A DK C C+ +C+C YA
Sbjct: 355 --------SCGEGDGFLRMSQMKLPE----TSEAVVDKRIGLKECREKCVRDCNCTGYAN 402
Query: 394 ---YDPSIFCMYWTGELIDLQKFPNGGVDLFVRVPA 426
+ C+ WTGEL D++K+ GG DL+V+V A
Sbjct: 403 MDIMNGGSGCVMWTGELDDMRKYNAGGQDLYVKVAA 438
>BRI1_LYCPE (Q8L899) Systemin receptor SR160 precursor (EC 2.7.1.37)
(Brassinosteroid LRR receptor kinase)
Length = 1207
Score = 199 bits (505), Expect = 3e-50
Identities = 121/312 (38%), Positives = 179/312 (56%), Gaps = 10/312 (3%)
Query: 463 WRKCSARHKAREHQQMKLDELPLYDFEKLETATNCFHFNNMLGKGGFGPVYKGVMEDGQE 522
W+ SAR + L F L ATN FH ++++G GGFG VYK ++DG
Sbjct: 853 WKFTSAREALSINLAAFEKPLRKLTFADLLEATNGFHNDSLVGSGGFGDVYKAQLKDGSV 912
Query: 523 IAVKRLSKASGQGIEEFMNEVVVISKLQHRNLVRLLGCCVERGEQILVYEFMPNKSLDAF 582
+A+K+L SGQG EF E+ I K++HRNLV LLG C E++LVYE+M SL+
Sbjct: 913 VAIKKLIHVSGQGDREFTAEMETIGKIKHRNLVPLLGYCKVGEERLLVYEYMKYGSLEDV 972
Query: 583 LFDPLQKK---NLDWRKRSNIIEGIARGIMYLHRDSRLRIIHRDLKASNILLDSDMIPKI 639
L D +KK L+W R I G ARG+ +LH + IIHRD+K+SN+LLD ++ ++
Sbjct: 973 LHD--RKKTGIKLNWPARRKIAIGAARGLAFLHHNCIPHIIHRDMKSSNVLLDENLEARV 1030
Query: 640 SDFGLARIVKFGEDDEANTKRVVGTYGYMPPEYAMEGLFSEKSDVYSFGVLLLEIVSGRR 699
SDFG+AR++ D + + GT GY+PPEY S K DVYS+GV+LLE+++G++
Sbjct: 1031 SDFGMARLMS-AMDTHLSVSTLAGTPGYVPPEYYQSFRCSTKGDVYSYGVVLLELLTGKQ 1089
Query: 700 NSSFSHHEDTLSLVGFAWKLWLEENIISLIDPEVW--DACFESSMLRCIHIGLLCVQELP 757
+ + D +LVG+ KL + I + D E+ DA E +L+ + + C+ +
Sbjct: 1090 PTDSADFGDN-NLVGWV-KLHAKGKITDVFDRELLKEDASIEIELLQHLKVACACLDDRH 1147
Query: 758 RDRPNISTVVLM 769
RP + V+ M
Sbjct: 1148 WKRPTMIQVMAM 1159
>BRI1_LYCES (Q8GUQ5) Brassinosteroid LRR receptor kinase precursor (EC
2.7.1.37) (tBRI1) (Altered brassinolide sensitivity 1)
(Systemin receptor SR160)
Length = 1207
Score = 199 bits (505), Expect = 3e-50
Identities = 121/312 (38%), Positives = 179/312 (56%), Gaps = 10/312 (3%)
Query: 463 WRKCSARHKAREHQQMKLDELPLYDFEKLETATNCFHFNNMLGKGGFGPVYKGVMEDGQE 522
W+ SAR + L F L ATN FH ++++G GGFG VYK ++DG
Sbjct: 853 WKFTSAREALSINLAAFEKPLRKLTFADLLEATNGFHNDSLVGSGGFGDVYKAQLKDGSV 912
Query: 523 IAVKRLSKASGQGIEEFMNEVVVISKLQHRNLVRLLGCCVERGEQILVYEFMPNKSLDAF 582
+A+K+L SGQG EF E+ I K++HRNLV LLG C E++LVYE+M SL+
Sbjct: 913 VAIKKLIHVSGQGDREFTAEMETIGKIKHRNLVPLLGYCKVGEERLLVYEYMKYGSLEDV 972
Query: 583 LFDPLQKK---NLDWRKRSNIIEGIARGIMYLHRDSRLRIIHRDLKASNILLDSDMIPKI 639
L D +KK L+W R I G ARG+ +LH + IIHRD+K+SN+LLD ++ ++
Sbjct: 973 LHD--RKKIGIKLNWPARRKIAIGAARGLAFLHHNCIPHIIHRDMKSSNVLLDENLEARV 1030
Query: 640 SDFGLARIVKFGEDDEANTKRVVGTYGYMPPEYAMEGLFSEKSDVYSFGVLLLEIVSGRR 699
SDFG+AR++ D + + GT GY+PPEY S K DVYS+GV+LLE+++G++
Sbjct: 1031 SDFGMARLMS-AMDTHLSVSTLAGTPGYVPPEYYQSFRCSTKGDVYSYGVVLLELLTGKQ 1089
Query: 700 NSSFSHHEDTLSLVGFAWKLWLEENIISLIDPEVW--DACFESSMLRCIHIGLLCVQELP 757
+ + D +LVG+ KL + I + D E+ DA E +L+ + + C+ +
Sbjct: 1090 PTDSADFGDN-NLVGWV-KLHAKGKITDVFDRELLKEDASIEIELLQHLKVACACLDDRH 1147
Query: 758 RDRPNISTVVLM 769
RP + V+ M
Sbjct: 1148 WKRPTMIQVMAM 1159
>SLS1_BRAOA (P22552) S-locus-specific glycoprotein BS29-1 precursor
Length = 444
Score = 198 bits (504), Expect = 4e-50
Identities = 143/456 (31%), Positives = 223/456 (48%), Gaps = 62/456 (13%)
Query: 6 HNSNYFFIITFLIFCTIYSCYS-AINDTITSSKSLKDNETITSNNTNFKLGFF------S 58
H+ F + ++F ++S + + N+ +T S N+T+ S F+LGFF S
Sbjct: 10 HSYTLLFFVILVLFPHVFSTNTLSPNEALTISS----NKTLVSPGDVFELGFFKTTTRNS 65
Query: 59 PLNSTNRYLGIWYINKTNN---IWIANRDQPLKDSNGIVTIHKDGNFIILNKPNGVIIWS 115
P + YLGIWY + + +W+ANRD L +S G + I + ++L+ N +WS
Sbjct: 66 PDGTDRWYLGIWYKTTSGHRTYVWVANRDNALHNSMGTLKI-SHASLVLLDHSN-TPVWS 123
Query: 116 TNISSSTNS--TAQLADSGNLILRDISSGAT---IWDSFTHPADAAVPTMRIAANQV-TG 169
TN + + TA+L +GN +LRD + A +W SF +P D +P M++ N+ +G
Sbjct: 124 TNFTGVAHLPVTAELLANGNFVLRDSKTTALDRFMWQSFDYPVDTLLPEMKLGRNRNGSG 183
Query: 170 KKISFVSRKSDNDPSSGHYSASLERLD-APEVFIWKDKNIHWRTGPWNGRVFLGSPRMLT 228
+ S KS DPSSG YS LE E ++ ++ +RTGPWNG F G P+M
Sbjct: 184 NEKILTSWKSPTDPSSGDYSFILETEGFLHEFYLLNNEFKVYRTGPWNGVRFNGIPKM-- 241
Query: 229 EYLAGWRFDQDT--DGTTYITYNFADKTMFGI---LSLTPHGTLKLIEYMN---KKELFR 280
W + ++ D + Y+F I ++ G L++I + ++ +F
Sbjct: 242 ---QNWSYIDNSFIDNNKEVAYSFQVNNNHNIHTRFRMSSTGYLQVITWTKTVPQRNMF- 297
Query: 281 LEVDQNECDFYGKCGPFGNCDNSTVPICSCFDGFEPKNSVEWSLGNWTNGCVRKEGMNLK 340
++ CD Y CGP+ CD T P C+C GF PKN+ W L + + GCVR +
Sbjct: 298 WSFPEDTCDLYKVCGPYAYCDMHTSPTCNCIKGFVPKNAGRWDLRDMSGGCVRSSKL--- 354
Query: 341 CEMVKNGSSIVKQDGFKVYHNMKPPDFNVRTNNADQDK------CGADCLANCSCLAYA- 393
S + DGF MK P+ T+ A DK C C+ +C+C YA
Sbjct: 355 --------SCGEGDGFLRMSQMKLPE----TSEAVVDKRIGLKECREKCVRDCNCTGYAN 402
Query: 394 ---YDPSIFCMYWTGELIDLQKFPNGGVDLFVRVPA 426
+ C+ WTGEL D++K+ GG DL+++V A
Sbjct: 403 MDIMNGGSGCVMWTGELDDMRKYNAGGQDLYLKVAA 438
>PSKR_ARATH (Q9ZVR7) Putative phytosulfokine receptor precursor (EC
2.7.1.37) (Phytosulfokine LRR receptor kinase)
Length = 1008
Score = 196 bits (499), Expect = 2e-49
Identities = 107/284 (37%), Positives = 163/284 (56%), Gaps = 3/284 (1%)
Query: 488 FEKLETATNCFHFNNMLGKGGFGPVYKGVMEDGQEIAVKRLSKASGQGIEEFMNEVVVIS 547
++ L +TN F N++G GGFG VYK + DG+++A+K+LS GQ EF EV +S
Sbjct: 724 YDDLLDSTNSFDQANIIGCGGFGMVYKATLPDGKKVAIKKLSGDCGQIEREFEAEVETLS 783
Query: 548 KLQHRNLVRLLGCCVERGEQILVYEFMPNKSLDAFLFDPLQ-KKNLDWRKRSNIIEGIAR 606
+ QH NLV L G C + +++L+Y +M N SLD +L + L W+ R I +G A+
Sbjct: 784 RAQHPNLVLLRGFCFYKNDRLLIYSYMENGSLDYWLHERNDGPALLKWKTRLRIAQGAAK 843
Query: 607 GIMYLHRDSRLRIIHRDLKASNILLDSDMIPKISDFGLARIVKFGEDDEANTKRVVGTYG 666
G++YLH I+HRD+K+SNILLD + ++DFGLAR++ E + +VGT G
Sbjct: 844 GLLYLHEGCDPHILHRDIKSSNILLDENFNSHLADFGLARLMSPYETHVSTD--LVGTLG 901
Query: 667 YMPPEYAMEGLFSEKSDVYSFGVLLLEIVSGRRNSSFSHHEDTLSLVGFAWKLWLEENII 726
Y+PPEY + + K DVYSFGV+LLE+++ +R + L+ + K+ E
Sbjct: 902 YIPPEYGQASVATYKGDVYSFGVVLLELLTDKRPVDMCKPKGCRDLISWVVKMKHESRAS 961
Query: 727 SLIDPEVWDACFESSMLRCIHIGLLCVQELPRDRPNISTVVLML 770
+ DP ++ + M R + I LC+ E P+ RP +V L
Sbjct: 962 EVFDPLIYSKENDKEMFRVLEIACLCLSENPKQRPTTQQLVSWL 1005
>APKA_ARATH (Q06548) Protein kinase APK1A, chloroplast precursor (EC
2.7.1.-)
Length = 410
Score = 191 bits (484), Expect = 9e-48
Identities = 120/307 (39%), Positives = 181/307 (58%), Gaps = 16/307 (5%)
Query: 474 EHQQMKLDELPLYDFEKLETATNCFHFNNMLGKGGFGPVYKGVMED----------GQEI 523
E + ++ L + F +L++AT F +++LG+GGFG V+KG +++ G I
Sbjct: 44 EGEILQSPNLKSFSFAELKSATRNFRPDSVLGEGGFGCVFKGWIDEKSLTASRPGTGLVI 103
Query: 524 AVKRLSKASGQGIEEFMNEVVVISKLQHRNLVRLLGCCVERGEQILVYEFMPNKSLDAFL 583
AVK+L++ QG +E++ EV + + HR+LV+L+G C+E ++LVYEFMP SL+ L
Sbjct: 104 AVKKLNQDGWQGHQEWLAEVNYLGQFSHRHLVKLIGYCLEDEHRLLVYEFMPRGSLENHL 163
Query: 584 F-DPLQKKNLDWRKRSNIIEGIARGIMYLHRDSRLRIIHRDLKASNILLDSDMIPKISDF 642
F L + L W+ R + G A+G+ +LH S R+I+RD K SNILLDS+ K+SDF
Sbjct: 164 FRRGLYFQPLSWKLRLKVALGAAKGLAFLH-SSETRVIYRDFKTSNILLDSEYNAKLSDF 222
Query: 643 GLARIVKFGEDDEANTKRVVGTYGYMPPEYAMEGLFSEKSDVYSFGVLLLEIVSGRRNSS 702
GLA+ G+ +T RV+GT+GY PEY G + KSDVYSFGV+LLE++SGRR
Sbjct: 223 GLAKDGPIGDKSHVST-RVMGTHGYAAPEYLATGHLTTKSDVYSFGVVLLELLSGRRAVD 281
Query: 703 FSHHEDTLSLVGFAWKLWL--EENIISLIDPEVWDACFESSMLRCIHIGLLCVQELPRDR 760
+ +LV +A K +L + I +ID + D + + L C+ + R
Sbjct: 282 KNRPSGERNLVEWA-KPYLVNKRKIFRVIDNRLQDQYSMEEACKVATLSLRCLTTEIKLR 340
Query: 761 PNISTVV 767
PN+S VV
Sbjct: 341 PNMSEVV 347
>CX32_ARATH (P27450) Probable serine/threonine-protein kinase Cx32,
chloroplast precursor (EC 2.7.1.37)
Length = 419
Score = 190 bits (483), Expect = 1e-47
Identities = 128/332 (38%), Positives = 184/332 (54%), Gaps = 25/332 (7%)
Query: 483 LPLYDFEKLETATNCFHFNNMLGKGGFGPVYKGVME----------DGQEIAVKRLSKAS 532
L +Y+F L+TAT F ++MLG+GGFG VY+G ++ G +A+KRL+ S
Sbjct: 71 LKVYNFLDLKTATKNFKPDSMLGQGGFGKVYRGWVDATTLAPSRVGSGMIVAIKRLNSES 130
Query: 533 GQGIEEFMNEVVVISKLQHRNLVRLLGCCVERGEQILVYEFMPNKSLDAFLFDPLQKKNL 592
QG E+ +EV + L HRNLV+LLG C E E +LVYEFMP SL++ LF +
Sbjct: 131 VQGFAEWRSEVNFLGMLSHRNLVKLLGYCREDKELLLVYEFMPKGSLESHLF--RRNDPF 188
Query: 593 DWRKRSNIIEGIARGIMYLHRDSRLRIIHRDLKASNILLDSDMIPKISDFGLARIVKFGE 652
W R I+ G ARG+ +LH R +I+RD KASNILLDS+ K+SDFGLA K G
Sbjct: 189 PWDLRIKIVIGAARGLAFLHSLQR-EVIYRDFKASNILLDSNYDAKLSDFGLA---KLGP 244
Query: 653 DDEAN--TKRVVGTYGYMPPEYAMEGLFSEKSDVYSFGVLLLEIVSGRRNSSFSHHEDTL 710
DE + T R++GTYGY PEY G KSDV++FGV+LLEI++G +
Sbjct: 245 ADEKSHVTTRIMGTYGYAAPEYMATGHLYVKSDVFAFGVVLLEIMTGLTAHNTKRPRGQE 304
Query: 711 SLVGFAW-KLWLEENIISLIDPEVWDACFESSMLRCIHIGLLCVQELPRDRPNISTVVLM 769
SLV + +L + + ++D + I L C++ P++RP++ VV +
Sbjct: 305 SLVDWLRPELSNKHRVKQIMDKGIKGQYTTKVATEMARITLSCIEPDPKNRPHMKEVVEV 364
Query: 770 LVSEITHLPPPGRVAFVHKQSSKSTTESSQKS 801
L H+ G ++ S+K +S +S
Sbjct: 365 L----EHI--QGLNVVPNRSSTKQAVANSSRS 390
>CRI4_MAIZE (O24585) Putative receptor protein kinase CRINKLY4
precursor (EC 2.7.1.-)
Length = 901
Score = 190 bits (483), Expect = 1e-47
Identities = 126/352 (35%), Positives = 192/352 (53%), Gaps = 13/352 (3%)
Query: 476 QQMKLDELPLYDFEKLETATNCFHFNNMLGKGGFGPVYKGVMEDGQEIAVKRLSKASG-- 533
+ +K+ + +E+LE AT F ++ +GKG F V+KG++ DG +AVKR KAS
Sbjct: 483 EDLKIRRAQEFSYEELEQATGGFSEDSQVGKGSFSCVFKGILRDGTVVAVKRAIKASDVK 542
Query: 534 QGIEEFMNEVVVISKLQHRNLVRLLGCCVERGEQILVYEFMPNKSLDAFLF--DPLQKKN 591
+ +EF NE+ ++S+L H +L+ LLG C + E++LVYEFM + SL L DP KK
Sbjct: 543 KSSKEFHNELDLLSRLNHAHLLNLLGYCEDGSERLLVYEFMAHGSLYQHLHGKDPNLKKR 602
Query: 592 LDWRKRSNIIEGIARGIMYLHRDSRLRIIHRDLKASNILLDSDMIPKISDFGLARIVKFG 651
L+W +R I ARGI YLH + +IHRD+K+SNIL+D D +++DFGL+ I+
Sbjct: 603 LNWARRVTIAVQAARGIEYLHGYACPPVIHRDIKSSNILIDEDHNARVADFGLS-ILGPA 661
Query: 652 EDDEANTKRVVGTYGYMPPEYAMEGLFSEKSDVYSFGVLLLEIVSGRRNSSFSHHEDTLS 711
+ ++ GT GY+ PEY + KSDVYSFGV+LLEI+SGR+ E +
Sbjct: 662 DSGTPLSELPAGTLGYLDPEYYRLHYLTTKSDVYSFGVVLLEILSGRKAIDMQFEEG--N 719
Query: 712 LVGFAWKLWLEENIISLIDPEVWDACFESSMLRCIHIGLLCVQELPRDRPNISTV----- 766
+V +A L +I +++DP + ++ + + CV+ +DRP++ V
Sbjct: 720 IVEWAVPLIKAGDIFAILDPVLSPPSDLEALKKIASVACKCVRMRGKDRPSMDKVTTALE 779
Query: 767 -VLMLVSEITHLPPPGRVAFVHKQSSKSTTESSQKSHQSNSNNNVTLSEVQG 817
L L+ + P V SS+ S S+ S S N + E QG
Sbjct: 780 HALALLMGSPCIEQPILPTEVVLGSSRMHKVSQMSSNHSCSENELADGEDQG 831
>CLV1_ARATH (Q9SYQ8) Receptor protein kinase CLAVATA1 precursor (EC
2.7.1.-)
Length = 980
Score = 185 bits (470), Expect = 4e-46
Identities = 120/354 (33%), Positives = 188/354 (52%), Gaps = 31/354 (8%)
Query: 442 LIIVIAGVIGALILVICAYLLWRKCSARHKAREHQQMKLDELPLYDFEKLETATNCFHFN 501
+I VIA + G +++ + + +K K ++ KL DF K E C
Sbjct: 642 VITVIAAITGLILISVAIRQMNKK-----KNQKSLAWKLTAFQKLDF-KSEDVLECLKEE 695
Query: 502 NMLGKGGFGPVYKGVMEDGQEIAVKRL-SKASGQGIEEFMNEVVVISKLQHRNLVRLLGC 560
N++GKGG G VY+G M + ++A+KRL + +G+ F E+ + +++HR++VRLLG
Sbjct: 696 NIIGKGGAGIVYRGSMPNNVDVAIKRLVGRGTGRSDHGFTAEIQTLGRIRHRHIVRLLGY 755
Query: 561 CVERGEQILVYEFMPNKSLDAFLFDPLQKKNLDWRKRSNIIEGIARGIMYLHRDSRLRII 620
+ +L+YE+MPN SL L + +L W R + A+G+ YLH D I+
Sbjct: 756 VANKDTNLLLYEYMPNGSLGELLHGS-KGGHLQWETRHRVAVEAAKGLCYLHHDCSPLIL 814
Query: 621 HRDLKASNILLDSDMIPKISDFGLARIVKFGEDDEANTKRVVGTYGYMPPEYAMEGLFSE 680
HRD+K++NILLDSD ++DFGLA+ + G E + + G+YGY+ PEYA E
Sbjct: 815 HRDVKSNNILLDSDFEAHVADFGLAKFLVDGAASECMSS-IAGSYGYIAPEYAYTLKVDE 873
Query: 681 KSDVYSFGVLLLEIVSGRRN-SSFSHHEDTLSLVGFAWKLWLEENI---------ISLID 730
KSDVYSFGV+LLE+++G++ F D + W EE I ++++D
Sbjct: 874 KSDVYSFGVVLLELIAGKKPVGEFGEGVDIV-----RWVRNTEEEITQPSDAAIVVAIVD 928
Query: 731 PEVWDACFESSMLRCIHIGLLCVQELPRDRPNISTVVLMLVSEITHLPPPGRVA 784
P + +S++ I ++CV+E RP + VV ML + PP VA
Sbjct: 929 PRLTGYPL-TSVIHVFKIAMMCVEEEAAARPTMREVVHMLTN------PPKSVA 975
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.320 0.137 0.422
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 100,781,544
Number of Sequences: 164201
Number of extensions: 4552939
Number of successful extensions: 14517
Number of sequences better than 10.0: 1640
Number of HSP's better than 10.0 without gapping: 1135
Number of HSP's successfully gapped in prelim test: 505
Number of HSP's that attempted gapping in prelim test: 10619
Number of HSP's gapped (non-prelim): 1972
length of query: 817
length of database: 59,974,054
effective HSP length: 119
effective length of query: 698
effective length of database: 40,434,135
effective search space: 28223026230
effective search space used: 28223026230
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 70 (31.6 bits)
Medicago: description of AC144721.7


