

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144721.6 + phase: 0 /pseudo
(748 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
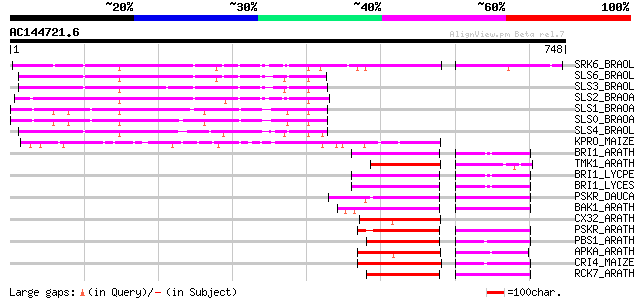
Score E
Sequences producing significant alignments: (bits) Value
SRK6_BRAOL (Q09092) Putative serine/threonine-protein kinase rec... 329 2e-89
SLS6_BRAOL (P07761) S-locus-specific glycoprotein S6 precursor (... 217 1e-55
SLS3_BRAOL (P17840) S-locus-specific glycoprotein S13 precursor ... 213 1e-54
SLS2_BRAOA (P22553) S-locus-specific glycoprotein BS29-2 precursor 210 1e-53
SLS1_BRAOA (P22552) S-locus-specific glycoprotein BS29-1 precursor 198 5e-50
SLS0_BRAOA (P22551) S-locus-specific glycoprotein precursor 194 7e-49
SLS4_BRAOL (P17841) S-locus-specific glycoprotein S14 precursor ... 180 1e-44
KPRO_MAIZE (P17801) Putative receptor protein kinase ZmPK1 precu... 172 4e-42
BRI1_ARATH (O22476) BRASSINOSTEROID INSENSITIVE 1 precursor (EC ... 105 4e-22
TMK1_ARATH (P43298) Putative receptor protein kinase TMK1 precur... 102 4e-21
BRI1_LYCPE (Q8L899) Systemin receptor SR160 precursor (EC 2.7.1.... 102 5e-21
BRI1_LYCES (Q8GUQ5) Brassinosteroid LRR receptor kinase precurso... 102 5e-21
PSKR_DAUCA (Q8LPB4) Phytosulfokine receptor precursor (EC 2.7.1.... 101 6e-21
BAK1_ARATH (Q94F62) BRASSINOSTEROID INSENSITIVE 1-associated rec... 99 5e-20
CX32_ARATH (P27450) Probable serine/threonine-protein kinase Cx3... 98 9e-20
PSKR_ARATH (Q9ZVR7) Putative phytosulfokine receptor precursor (... 91 1e-17
PBS1_ARATH (Q9FE20) Serine/threonine-protein kinase PBS1 (EC 2.7... 91 1e-17
APKA_ARATH (Q06548) Protein kinase APK1A, chloroplast precursor ... 90 2e-17
CRI4_MAIZE (O24585) Putative receptor protein kinase CRINKLY4 pr... 89 3e-17
RCK7_ARATH (Q9LQQ8) Putative serine/threonine-protein kinase RLC... 89 4e-17
>SRK6_BRAOL (Q09092) Putative serine/threonine-protein kinase
receptor precursor (EC 2.7.1.37) (S-receptor kinase)
(SRK)
Length = 849
Score = 329 bits (843), Expect = 2e-89
Identities = 203/626 (32%), Positives = 332/626 (52%), Gaps = 65/626 (10%)
Query: 4 LIHNTNHFFIITFIIFSTFYSCYSSTNDTITSSKSL--KDNETITSNNTNFKLGFFSPLN 61
+ H++ F++ F++ + S +T++S++SL N+T+ S + F++GFF
Sbjct: 7 IYHHSYMSFLLVFVVMILIHPALSIYINTLSSTESLTISSNKTLVSPGSIFEVGFFR--T 64
Query: 62 STNRYLGIWY--INETNNIWIANRDQPLKDSNGIVTIHKNGNLVILNKENGSIIWSTSIS 119
++ YLG+WY +++ +W+ANRD PL ++ G + I N NLV+L+ N + W+
Sbjct: 65 NSRWYLGMWYKKVSDRTYVWVANRDNPLSNAIGTLKISGN-NLVLLDHSNKPVWWTNLTR 123
Query: 120 SPNSINSTAQLVDVGNLILSDINSRST---IWDSFTHPADAAVPTMRIASNKATGKNISF 176
A+L+ GN ++ D ++ +W SF +P D +P M++ N TG N
Sbjct: 124 GNERSPVVAELLANGNFVMRDSSNNDASEYLWQSFDYPTDTLLPEMKLGYNLKTGLNRFL 183
Query: 177 VSRKSENDPSSGHYIGSLERLDAPEVFIWYDKRIHWRTGPWNGTVFLGSPRMLTEYLAGW 236
S +S +DPSSG++ LE PE ++ + R+GPWNG F G P +
Sbjct: 184 TSWRSSDDPSSGNFSYKLETQSLPEFYLSRENFPMHRSGPWNGIRFSGIPEDQKLSYMVY 243
Query: 237 RFDQDKDGTTYLTYDFAVKAMFGILSLTPNGTLKLVEFLNN----KEFLSLTVSQNECDF 292
F ++ + Y T+ + + L+L G + + + + F S V +CD
Sbjct: 244 NFIENNEEVAY-TFRMTNNSFYSRLTLISEGYFQRLTWYPSIRIWNRFWSSPVDP-QCDT 301
Query: 293 YGKCGPFGNCDISSVPNICSCFKGFEPKNLVEWSSRNWTNGCVRKEGMNLKCEMVKNGSS 352
Y CGP+ CD+++ P +C+C +GF P+N+ +W R W GC+R+ L C +G
Sbjct: 302 YIMCGPYAYCDVNTSP-VCNCIQGFNPRNIQQWDQRVWAGGCIRR--TQLSC----SGDG 354
Query: 353 VVKQDKFLVHPNTKPPDFAERSDVSRDKCRTDCLANCSCLAYAYDPFIR-----CMYWSS 407
+ K + T +RS + +C+ C+++C+C A+A + IR C+ W+
Sbjct: 355 FTRMKKMKLPETTMAT--VDRS-IGVKECKKRCISDCNCTAFA-NADIRNGGSGCVIWTE 410
Query: 408 ELIDLQKFPT---SGVDLFIRVPAELEKEKGNKSFLIIAIAGGLGAFILVICAYLLWRKW 464
L D++ + T G DL++R+ A +K N S II++ G+ +L+I + LW++
Sbjct: 411 RLEDIRNYATDAIDGQDLYVRLAAADIAKKRNASGKIISLTVGVSVLLLLI-MFCLWKRK 469
Query: 465 SAR---------------HTEQKEMKLD--------------ELPLYDFVKLENATNSFH 495
R + EM L ELPL + + AT +F
Sbjct: 470 QKRAKASAISIANTQRNQNLPMNEMVLSSKREFSGEYKFEELELPLIEMETVVKATENFS 529
Query: 496 NSNMLGKGGFGPVYKGILEDGQEVAVKRLSKSSGQGIEEFMNEVAVISKLQHRNLVRLLG 555
+ N LG+GGFG VYKG L DG+E+AVKRLSK+S QG +EFMNEV +I++LQH NLV++LG
Sbjct: 530 SCNKLGQGGFGIVYKGRLLDGKEIAVKRLSKTSVQGTDEFMNEVTLIARLQHINLVQVLG 589
Query: 556 CCVERGEQMLVYEFMPNKSLDAFLFG 581
CC+E E+ML+YE++ N SLD++LFG
Sbjct: 590 CCIEGDEKMLIYEYLENLSLDSYLFG 615
Score = 112 bits (281), Expect = 3e-24
Identities = 61/151 (40%), Positives = 90/151 (59%), Gaps = 8/151 (5%)
Query: 602 PEYAMEGLFSEKSDVYSFGVLLLEIVSGRRNSSFYHNEDSLSLVGFAWKLWLEENIISLI 661
PEYAM G+FSEKSDV+SFGV++LEIVSG++N FY+ + L+ + W W E + ++
Sbjct: 699 PEYAMYGIFSEKSDVFSFGVIVLEIVSGKKNRGFYNLDYENDLLSYVWSRWKEGRALEIV 758
Query: 662 DREVWDASFE-------SSMLRCIHIGLLCVQELPRDRPNISTVVLMLISEITHLPPPGK 714
D + D+ +L+CI IGLLCVQEL RP +S+VV M SE T +P P
Sbjct: 759 DPVIVDSLSSQPSIFQPQEVLKCIQIGLLCVQELAEHRPAMSSVVWMFGSEATEIPQPKP 818
Query: 715 VAFVHKKNSKSGESSQKSQQSNSNNSVTLSE 745
+ +++ + S S Q + N S T+++
Sbjct: 819 PGYCVRRSPYELDPS-SSWQCDENESWTVNQ 848
>SLS6_BRAOL (P07761) S-locus-specific glycoprotein S6 precursor
(SLSG-6)
Length = 436
Score = 217 bits (552), Expect = 1e-55
Identities = 133/434 (30%), Positives = 219/434 (49%), Gaps = 38/434 (8%)
Query: 12 FIITFIIFSTFYSCYSSTNDTITSSKSLKDNETITSNNTNFKLGFFSPLNSTNRYLGIWY 71
F++ F + F +S + T S + N T+ S NF+LGFF +S+ YLGIWY
Sbjct: 16 FLLVFFVLILFCPAFSINTLSSTESLRISSNRTLVSPGNNFELGFFRTNSSSRWYLGIWY 75
Query: 72 --INETNNIWIANRDQPLKDSNGIVTIHKNGNLVILNKENGSIIWSTSISSPNS-INSTA 128
+ + +W+ANRD PL ++ G + I N NLV+L N S+ WST+++ N + A
Sbjct: 76 KKLLDRTYVWVANRDNPLSNAIGTLKISGN-NLVLLGHTNKSV-WSTNLTRGNERLPVVA 133
Query: 129 QLVDVGNLILSDINSRST---IWDSFTHPADAAVPTMRIASNKATGKNISFVSRKSENDP 185
+L+ GN ++ D ++ +W SF +P D +P M++ + TG N S +S +DP
Sbjct: 134 ELLSNGNFVMRDSSNNDASEYLWQSFDYPTDTLLPEMKLGYDLKTGLNRFLTSWRSSDDP 193
Query: 186 SSGHYIGSLERLDAPEVFIWYDKRIHWRTGPWNGTVFLGSPRMLTEYLAGWRFDQDKDGT 245
SSG + LE PE ++W+ R+GPWNG F G P + F ++ +
Sbjct: 194 SSGDFSYKLETRSLPEFYLWHGIFPMHRSGPWNGVRFSGIPEDQKLSYMVYNFTENSEEV 253
Query: 246 TYLTYDFAVKAMFGILSLTPNGTLKLVEFLNN----KEFLSLTVSQNECDFYGKCGPFGN 301
Y T+ +++ L+L+ G + + + + F S V +CD Y CGP+
Sbjct: 254 AY-TFRMTNNSIYSRLTLSSEGYFQRLTWNPSIGIWNRFWSSPVDP-QCDTYIMCGPYAY 311
Query: 302 CDISSVPNICSCFKGFEPKNLVEWSSRNWTNGCVRKEGMNLKCEMVKNGSSVVKQDKFLV 361
C +++ P +C+C +GF P+N+ +W R W GC+R+ L C D F
Sbjct: 312 CGVNTSP-VCNCIQGFNPRNIQQWDQRVWAGGCIRR--TRLSC----------SGDGFTR 358
Query: 362 HPNTKPPD----FAERSDVSRDKCRTDCLANCSCLAYAYDPFIR-----CMYWSSELIDL 412
N K P+ +RS + +C CL++C+C A+A + IR C+ W+ L D+
Sbjct: 359 MKNMKLPETTMAIVDRS-IGVKECEKRCLSDCNCTAFA-NADIRNGGTGCVIWTGRLDDM 416
Query: 413 QKFPTSGVDLFIRV 426
+ + G DL++R+
Sbjct: 417 RNYVAHGQDLYVRL 430
>SLS3_BRAOL (P17840) S-locus-specific glycoprotein S13 precursor
(SLSG-13) (Fragment)
Length = 434
Score = 213 bits (542), Expect = 1e-54
Identities = 137/435 (31%), Positives = 221/435 (50%), Gaps = 37/435 (8%)
Query: 12 FIITFIIFSTFYSCYSSTNDTITSSKSLKDNETITSNNTNFKLGFFSPLNSTNRYLGIWY 71
F++ F + F +S + T S ++ N T+ S F+LGFF +S+ YLGIWY
Sbjct: 15 FLLVFFVLILFRPAFSINTLSSTESLTISSNRTLVSPGNVFELGFFKTTSSSRWYLGIWY 74
Query: 72 --INETNNIWIANRDQPLKDSNGIVTIHKNGNLVILNKENGSIIWSTSISSPNSINST-A 128
+W+ANRD PL + G + I N NLV+L+ N S+ WST+++ N + A
Sbjct: 75 KKFPYRTYVWVANRDNPLSNDIGTLKISGN-NLVLLDHSNKSV-WSTNVTRGNERSPVVA 132
Query: 129 QLVDVGNLILSDINSRST---IWDSFTHPADAAVPTMRIASNKATGKNISFVSRKSENDP 185
+L+D GN ++ D NS + +W SF +P D +P M++ + TG N S +S +DP
Sbjct: 133 ELLDNGNFVMRDSNSNNASQFLWQSFDYPTDTLLPEMKLGYDLKTGLNRFLTSWRSSDDP 192
Query: 186 SSGHYIGSLERLDAPEVFIWYDK-RIHWRTGPWNGTVFLGSPRMLTEYLAGWRFDQDKDG 244
SSG Y LE PE ++ R+H R+GPWNG G P + F ++ +
Sbjct: 193 SSGDYSYKLELRRLPEFYLSSGSFRLH-RSGPWNGFRISGIPEDQKLSYMVYNFTENSEE 251
Query: 245 TTYLTYDFAVKAMFGILSLTPNGTLKLVEFLNNKEFLSLTVS--QNECDFYGKCGPFGNC 302
Y T+ + + L+++ G + + + + ++ S ++CD Y CGP+ C
Sbjct: 252 AAY-TFLMTNNSFYSRLTISSTGYFERLTWAPSSVVWNVFWSSPNHQCDMYRMCGPYSYC 310
Query: 303 DISSVPNICSCFKGFEPKNLVEWSSRNWTNGCVRKEGMNLKCEMVKNGSSVVKQDKFLVH 362
D+++ P +C+C +GF PKN +W R T+GC+R+ L C D F
Sbjct: 311 DVNTSP-VCNCIQGFRPKNRQQWDLRIPTSGCIRR--TRLSC----------SGDGFTRM 357
Query: 363 PNTKPPD----FAERSDVSRDKCRTDCLANCSCLAYAYDPFIR-----CMYWSSELIDLQ 413
N K P+ RS + +C CL++C+C A+A + IR C+ W+ EL D++
Sbjct: 358 KNMKLPETTMAIVHRS-IGLKECEKRCLSDCNCTAFA-NADIRNRGTGCVIWTGELEDIR 415
Query: 414 KFPTSGVDLFIRVPA 428
+ G DL++R+ A
Sbjct: 416 TYFADGQDLYVRLAA 430
>SLS2_BRAOA (P22553) S-locus-specific glycoprotein BS29-2 precursor
Length = 435
Score = 210 bits (534), Expect = 1e-53
Identities = 136/441 (30%), Positives = 233/441 (51%), Gaps = 33/441 (7%)
Query: 7 NTNHFFIITFIIFSTFYSCYSSTNDTITSSKSLK--DNETITSNNTNFKLGFFSPLNSTN 64
N++ F++ F + + F +S +T++S +SLK ++ T+ S +LGFF +S+
Sbjct: 10 NSHTSFLLVFFVLTLFSPAFSI--NTLSSIESLKISNSRTLVSPGNVLELGFFRTPSSSR 67
Query: 65 RYLGIWY--INETNNIWIANRDQPLKDSNGIVTIHKNGNLVILNKENGSIIWSTSISSPN 122
YLG+WY ++E +W+ANRD PL S G + I N NLV+L+ N S+ WST+ + N
Sbjct: 68 WYLGMWYKKLSERTYVWVANRDNPLSCSIGTLKI-SNMNLVLLDHSNKSL-WSTNHTRGN 125
Query: 123 SINST-AQLVDVGNLILSDINSRST---IWDSFTHPADAAVPTMRIASNKATGKNISFVS 178
+ A+L+ GN +L D N +W SF +P D +P M++ + TG N S
Sbjct: 126 ERSPVVAELLANGNFVLRDSNKNDRSGFLWQSFDYPTDTLLPEMKLGYDLRTGLNRFLTS 185
Query: 179 RKSENDPSSGHYIGSLERLDAPEVFIWYDKRIHWRTGPWNGTVFLGSPRMLTEYLAGWRF 238
+S +DPSSG + L+ PE +++ D + R+GPWNG F G P + F
Sbjct: 186 WRSSDDPSSGDFSYKLQTRRLPEFYLFKDDFLVHRSGPWNGVGFSGMPEDQKLSYMVYNF 245
Query: 239 DQDKDGTTYLTYDFAVKAMFGILSLTPNGTLKLVEFLNNKEFLSLTVSQNE---CDFYGK 295
Q+ + Y T+ +++ L+++ +G + + + + ++ S E CD Y
Sbjct: 246 TQNSEEVAY-TFLMTNNSIYSRLTISSSGYFERLTWTPSSGMWNVFWSSPEDFQCDVYKI 304
Query: 296 CGPFGNCDISSVPNICSCFKGFEPKNLVEWSSRNWTNGCVRKEGMNLKCEMVKNGSSVVK 355
CG + CD+++ P +C+C + F+P N+ EW R W+ GC R+ L C +G +
Sbjct: 305 CGAYSYCDVNTSP-VCNCIQRFDPSNVQEWGLRAWSGGCRRR--TRLSC----SGDGFTR 357
Query: 356 QDKFLVHPNTKPPDFAERSDVSRDKCRTDCLANCSCLAYAYDPFIR-----CMYWSSELI 410
K + T +RS + +C CL++C+C A+A + IR C+ W+ +L
Sbjct: 358 MKKMKLPETTMA--IVDRS-IGLKECEKRCLSDCNCTAFA-NADIRNGGTGCVIWTGQLE 413
Query: 411 DLQKFPTSGVDLFIRV-PAEL 430
D++ + +G DL++R+ PA+L
Sbjct: 414 DIRTYFANGQDLYVRLAPADL 434
>SLS1_BRAOA (P22552) S-locus-specific glycoprotein BS29-1 precursor
Length = 444
Score = 198 bits (503), Expect = 5e-50
Identities = 138/457 (30%), Positives = 222/457 (48%), Gaps = 48/457 (10%)
Query: 1 MAFLIHNTNHFFIITFIIFSTFYSCYSSTNDTITSSKSL--KDNETITSNNTNFKLGFF- 57
M +I N +H + + F + + STN T++ +++L N+T+ S F+LGFF
Sbjct: 1 MRGVIPNYHHSYTLLFFVILVLFPHVFSTN-TLSPNEALTISSNKTLVSPGDVFELGFFK 59
Query: 58 -----SPLNSTNRYLGIWYINETNN---IWIANRDQPLKDSNGIVTIHKNGNLVILNKEN 109
SP + YLGIWY + + +W+ANRD L +S G + I + +LV+L+ N
Sbjct: 60 TTTRNSPDGTDRWYLGIWYKTTSGHRTYVWVANRDNALHNSMGTLKI-SHASLVLLDHSN 118
Query: 110 GSIIWSTSISSPNSINSTAQLVDVGNLILSDINSRST---IWDSFTHPADAAVPTMRIAS 166
+ +WST+ + + TA+L+ GN +L D + + +W SF +P D +P M++
Sbjct: 119 -TPVWSTNFTGVAHLPVTAELLANGNFVLRDSKTTALDRFMWQSFDYPVDTLLPEMKLGR 177
Query: 167 NK-ATGKNISFVSRKSENDPSSGHYIGSLERLDAPEVFIWYDKRIH-WRTGPWNGTVFLG 224
N+ +G S KS DPSSG Y LE F + +RTGPWNG F G
Sbjct: 178 NRNGSGNEKILTSWKSPTDPSSGDYSFILETEGFLHEFYLLNNEFKVYRTGPWNGVRFNG 237
Query: 225 SPRMLT-EYLAGWRFDQDKDGTTYLTYDFAVKAMFGI---LSLTPNGTLKLVEFLNN--K 278
P+M Y+ D +K+ + Y F V I ++ G L+++ + +
Sbjct: 238 IPKMQNWSYIDNSFIDNNKE----VAYSFQVNNNHNIHTRFRMSSTGYLQVITWTKTVPQ 293
Query: 279 EFLSLTVSQNECDFYGKCGPFGNCDISSVPNICSCFKGFEPKNLVEWSSRNWTNGCVRKE 338
+ + ++ CD Y CGP+ CD+ + P C+C KGF PKN W R+ + GCVR
Sbjct: 294 RNMFWSFPEDTCDLYKVCGPYAYCDMHTSPT-CNCIKGFVPKNAGRWDLRDMSGGCVRSS 352
Query: 339 GMNLKCEMVKNGSSVVKQDKFLVHPNTKPPDFAER---SDVSRDKCRTDCLANCSCLAYA 395
+ S + D FL K P+ +E + +CR C+ +C+C YA
Sbjct: 353 KL-----------SCGEGDGFLRMSQMKLPETSEAVVDKRIGLKECREKCVRDCNCTGYA 401
Query: 396 YDPFIR----CMYWSSELIDLQKFPTSGVDLFIRVPA 428
+ C+ W+ EL D++K+ G DL+++V A
Sbjct: 402 NMDIMNGGSGCVMWTGELDDMRKYNAGGQDLYLKVAA 438
>SLS0_BRAOA (P22551) S-locus-specific glycoprotein precursor
Length = 444
Score = 194 bits (493), Expect = 7e-49
Identities = 136/458 (29%), Positives = 222/458 (47%), Gaps = 50/458 (10%)
Query: 1 MAFLIHNTNHFFIITFIIFSTFYSCYSSTNDTITSSKSL--KDNETITSNNTNFKLGFF- 57
M +I N +H + + F + + STN T++ +++L N+T+ S F+LGFF
Sbjct: 1 MRGVIPNYHHSYTLFFFVILVLFPHVFSTN-TLSPNEALTISSNKTLVSPGDVFELGFFK 59
Query: 58 -----SPLNSTNRYLGIWYINETNN---IWIANRDQPLKDSNGIVTIHKNGNLVILNKEN 109
SP + YLGIWY + + +W+ANRD L +S G + I + +LV+L+ N
Sbjct: 60 TTTRNSPDGTDRWYLGIWYKTTSGHRTYVWVANRDNALHNSMGTLKI-SHASLVLLDHSN 118
Query: 110 GSIIWSTSISSPNSINSTAQLVDVGNLILSDINSRST---IWDSFTHPADAAVPTMRIAS 166
+ +WST+ + + TA+L+ GN +L D + +W SF +P D +P M++
Sbjct: 119 -TPVWSTNFTGVAHLPVTAELLANGNFVLRDSKTNDLDRFMWQSFDYPVDTLLPEMKLGR 177
Query: 167 NKATGKNISFV-SRKSENDPSSGHYIGSLERLD-APEVFIWYDKRIHWRTGPWNGTVFLG 224
N +N + S KS DPSSG + LE E ++ ++ +RTGPWNG F G
Sbjct: 178 NLIGSENEKILTSWKSPTDPSSGDFSFILETEGFLHEFYLLKNEFKVYRTGPWNGVRFNG 237
Query: 225 SPRMLTEYLAGWRFDQDK--DGTTYLTYDFAVKAMFGI---LSLTPNGTLKLVEFLNN-- 277
P+M W + + D + Y F V I ++ G L+++ +
Sbjct: 238 IPKM-----QNWSYIDNSFIDNNEEVAYSFQVNNNHNIHTRFRMSSTGYLQVITWTKTVP 292
Query: 278 KEFLSLTVSQNECDFYGKCGPFGNCDISSVPNICSCFKGFEPKNLVEWSSRNWTNGCVRK 337
+ + + ++ CD Y CGP+ CD+ + P C+C KGF PKN W R+ + GCVR
Sbjct: 293 QRNMFWSFPEDTCDLYKVCGPYAYCDMHTSPT-CNCIKGFVPKNAGRWDLRDMSGGCVRS 351
Query: 338 EGMNLKCEMVKNGSSVVKQDKFLVHPNTKPPDFAER---SDVSRDKCRTDCLANCSCLAY 394
+ S + D FL K P+ +E + +CR C+ +C+C Y
Sbjct: 352 SKL-----------SCGEGDGFLRMSQMKLPETSEAVVDKRIGLKECREKCVRDCNCTGY 400
Query: 395 AYDPFIR----CMYWSSELIDLQKFPTSGVDLFIRVPA 428
A + C+ W+ EL D++K+ G DL+++V A
Sbjct: 401 ANMDIMNGGSGCVMWTGELDDMRKYNAGGQDLYVKVAA 438
>SLS4_BRAOL (P17841) S-locus-specific glycoprotein S14 precursor
(SLSG-14) (Fragment)
Length = 434
Score = 180 bits (457), Expect = 1e-44
Identities = 131/447 (29%), Positives = 209/447 (46%), Gaps = 59/447 (13%)
Query: 12 FIITFIIFSTFYSCYSSTNDTITSSKSLKDNETITSNNTNFKLGFFSPLNSTNRYLGIWY 71
F++ F + F ++ + S ++ N T+ S F+LGFF +S+ YLGIWY
Sbjct: 13 FLLVFFVLILFPPAFTINTLSSIESLTISSNRTLVSPGNVFELGFFRTNSSSRWYLGIWY 72
Query: 72 --INETNNIWIANRDQPLKDSNGIVTIHKNGNLVILNKENGSIIWSTSISSPNSINST-A 128
+++ +W+ANRD PL S G + I N N L+ N S+ WST+++ N + A
Sbjct: 73 KKVSDRTYVWVANRDNPLSSSIGTLKISGN-NPCHLDHSNKSV-WSTNLTRGNERSPVVA 130
Query: 129 QLVDVGNLILSDINSRST---IWDSFTHPADAAVPTMRIASNKATGKNISFVSRKSENDP 185
++ GN ++ D N+ +W SF P D +P M+++ + TG N SR+S +DP
Sbjct: 131 DVLANGNFVMRDSNNNDASGFLWQSFDFPTDTLLPEMKLSYDLKTGLNRFLTSRRSSDDP 190
Query: 186 SSGHYIGSLERLDAPEVFIWYDKRIHWRTGPWNGTVFLGSPRMLTEYLAGWRFDQDKDGT 245
SSG + LE PE ++ + +R+GPWNG F G P DQ
Sbjct: 191 SSGDFSYKLEPRRLPEFYLSSGVFLLYRSGPWNGIRFSGLPD-----------DQKLSYL 239
Query: 246 TYLTYDFAVKAMFGILSLTPNGTLKLVEFLNNKEFLSLTVS------------QNECDFY 293
Y++ D V F + + + L V F E + S ++C Y
Sbjct: 240 VYISQDMRVAYKFRMTNNSFYSRL-FVSFSGYIEQQTWNPSSQMWNSFWAFPLDSQCYTY 298
Query: 294 GKCGPFGNCDISSVPNICSCFKGFEPKNLVEWSSRNWTNGCVRKEGMNLKCEMVKNGSSV 353
CGP+ C + + IC+C +GF P N+ +W R W GC+R+ + +GS
Sbjct: 299 RACGPYSYC-VVNTSAICNCIQGFNPSNVQQWDQRVWAGGCIRRTRL--------SGSG- 348
Query: 354 VKQDKFLVHPNTKPPD----FAERSDVSRDKCRTDCLANCSCLAYAYDPFIR-----CMY 404
D F N K P+ +RS + +C CL +C+C A+A + IR C+
Sbjct: 349 ---DGFTRMKNMKLPETTMAIVDRS-IGVKECEKRCLNDCNCTAFA-NADIRNGGTGCVI 403
Query: 405 WSSELIDLQKFPTSGV---DLFIRVPA 428
+ EL D++ + T DL++R+ A
Sbjct: 404 NTGELEDMRSYATGATDSQDLYVRLAA 430
>KPRO_MAIZE (P17801) Putative receptor protein kinase ZmPK1
precursor (EC 2.7.1.37)
Length = 817
Score = 172 bits (435), Expect = 4e-42
Identities = 176/641 (27%), Positives = 278/641 (42%), Gaps = 107/641 (16%)
Query: 15 TFIIFSTFYSCY---SSTNDTITSSKSLK----DNETITSNNTNFKLGFFSPLNSTNRYL 67
T I S F + + +S+ D + SL ++ T+ S++ F GF+ +
Sbjct: 11 TACILSFFIALFPRAASSRDILPLGSSLVVESYESSTLQSSDGTFSSGFYEVYTHAFTF- 69
Query: 68 GIWY-------INETNNIWIANRDQPLKDSNGIVTIHKNGNLVILNKENGSIIWSTSISS 120
+WY N +W AN D+P+ +T+ K+GN+V+ + + G+ +W ++
Sbjct: 70 SVWYSKTEAAAANNKTIVWSANPDRPVHARRSALTLQKDGNMVLTDYD-GAAVWRADGNN 128
Query: 121 PNSINSTAQLVDVGNLILSDINSRSTIWDSFTHPADAAVPTMRIASNKATGKNISFVSRK 180
+ A+L+D GNL++ D + +T+W SF P D +PT I + AT + SR
Sbjct: 129 FTGVQR-ARLLDTGNLVIED-SGGNTVWQSFDSPTDTFLPTQLITA--ATRLVPTTQSR- 183
Query: 181 SENDPSSGHYIGSLERLDAPEVFIWYDK--RIHWRTGPWN----GTVFLGSPR--MLTEY 232
S G+YI L + + I+W N G S R MLT+
Sbjct: 184 -----SPGNYIFRFSDLSVLSLIYHVPQVSDIYWPDPDQNLYQDGRNQYNSTRLGMLTD- 237
Query: 233 LAGWRFDQD-KDGTTYLTYDFAVKAMFGILSLTPNGTLKLVEFLNNKE---FLSLTVSQN 288
+G D DG + D + L+L P+G L+L +N+ + +S+
Sbjct: 238 -SGVLASSDFADGQALVASDVG-PGVKRRLTLDPDGNLRLYS-MNDSDGSWSVSMVAMTQ 294
Query: 289 ECDFYGKCGPFGNCDISSVPNICSCFKGFEPKNLVEWSSRNWTNGCVRKEGMNLKCEMVK 348
C+ +G CGP G C S P CSC G+ +N NWT GC+ +N C+
Sbjct: 295 PCNIHGLCGPNGICHYSPTPT-CSCPPGYATRN-----PGNWTEGCMAI--VNTTCDRYD 346
Query: 349 NGSSVVKQDKFLVHPNTK--PPDFAERSDVSRDKCRTDCLANCSCLAYAYDPFIRCMYWS 406
S +F+ PNT D VS CR C+++C+C + Y Y
Sbjct: 347 KRSM-----RFVRLPNTDFWGSDQQHLLSVSLRTCRDICISDCTCKGFQYQEGTGSCYPK 401
Query: 407 SELIDLQKFPTSGV---------------------DLFIRVPAELEKEKGNKS----FLI 441
+ L + +PTS V D+F VP L+ ++ NKS F
Sbjct: 402 AYLFSGRTYPTSDVRTIYLKLPTGVSVSNALIPRSDVFDSVPRRLDCDRMNKSIREPFPD 461
Query: 442 IAIAGG-----------LGAFILVICAYLLWRKWSARHTEQKEMKL-----------DEL 479
+ GG + AF +V +++ + + E + +L
Sbjct: 462 VHKTGGGESKWFYFYGFIAAFFVVEVSFISFAWFFVLKRELRPSELWASEKGYKAMTSNF 521
Query: 480 PLYDFVKLENATNSFHNSNMLGKGGFGPVYKGILEDGQEVAVKRLSKSSGQGIEEFMNEV 539
Y + +L AT F LG+G G VYKG+LED + VAVK+L ++ QG E F E+
Sbjct: 522 RRYSYRELVKATRKFKVE--LGRGESGTVYKGVLEDDRHVAVKKL-ENVRQGKEVFQAEL 578
Query: 540 AVISKLQHRNLVRLLGCCVERGEQMLVYEFMPNKSLDAFLF 580
+VI ++ H NLVR+ G C E ++LV E++ N SL LF
Sbjct: 579 SVIGRINHMNLVRIWGFCSEGSHRLLVSEYVENGSLANILF 619
Score = 34.7 bits (78), Expect = 0.95
Identities = 31/116 (26%), Positives = 51/116 (43%), Gaps = 18/116 (15%)
Query: 602 PEYAMEGLFSEKSDVYSFGVLLLEIVSGRRNSSFYHNED-------------SLSLVGFA 648
PE+ + K DVYS+GV+LLE+++G R S D S L G
Sbjct: 704 PEWVSSLPITAKVDVYSYGVVLLELLTGTRVSELVGGTDEVHSMLRKLVRMLSAKLEG-E 762
Query: 649 WKLWLEENIISLIDREVWDASFESSMLRCIHIGLLCVQELPRDRPNISTVVLMLIS 704
+ W++ + S ++R V + I + + C++E RP + V L+S
Sbjct: 763 EQSWIDGYLDSKLNRPVNYVQART----LIKLAVSCLEEDRSKRPTMEHAVQTLLS 814
>BRI1_ARATH (O22476) BRASSINOSTEROID INSENSITIVE 1 precursor (EC
2.7.1.37) (AtBRI1) (Brassinosteroid LRR receptor kinase)
Length = 1196
Score = 105 bits (262), Expect = 4e-22
Identities = 57/121 (47%), Positives = 72/121 (59%), Gaps = 2/121 (1%)
Query: 461 WRKWSARHTEQKEMKLDELPL--YDFVKLENATNSFHNSNMLGKGGFGPVYKGILEDGQE 518
W+ + + E PL F L ATN FHN +++G GGFG VYK IL+DG
Sbjct: 848 WKLTGVKEALSINLAAFEKPLRKLTFADLLQATNGFHNDSLIGSGGFGDVYKAILKDGSA 907
Query: 519 VAVKRLSKSSGQGIEEFMNEVAVISKLQHRNLVRLLGCCVERGEQMLVYEFMPNKSLDAF 578
VA+K+L SGQG EFM E+ I K++HRNLV LLG C E++LVYEFM SL+
Sbjct: 908 VAIKKLIHVSGQGDREFMAEMETIGKIKHRNLVPLLGYCKVGDERLLVYEFMKYGSLEDV 967
Query: 579 L 579
L
Sbjct: 968 L 968
Score = 45.8 bits (107), Expect = 4e-04
Identities = 32/102 (31%), Positives = 55/102 (53%), Gaps = 4/102 (3%)
Query: 602 PEYAMEGLFSEKSDVYSFGVLLLEIVSGRRNSSFYHNEDSLSLVGFAWKLWLEENIISLI 661
PEY S K DVYS+GV+LLE+++G+R + D+ +LVG+ K + I +
Sbjct: 1055 PEYYQSFRCSTKGDVYSYGVVLLELLTGKRPTDSPDFGDN-NLVGWV-KQHAKLRISDVF 1112
Query: 662 DREVW--DASFESSMLRCIHIGLLCVQELPRDRPNISTVVLM 701
D E+ D + E +L+ + + + C+ + RP + V+ M
Sbjct: 1113 DPELMKEDPALEIELLQHLKVAVACLDDRAWRRPTMVQVMAM 1154
>TMK1_ARATH (P43298) Putative receptor protein kinase TMK1 precursor
(EC 2.7.1.-)
Length = 942
Score = 102 bits (254), Expect = 4e-21
Identities = 48/96 (50%), Positives = 70/96 (72%), Gaps = 2/96 (2%)
Query: 487 LENATNSFHNSNMLGKGGFGPVYKGILEDGQEVAVKRLSKS--SGQGIEEFMNEVAVISK 544
L + TN+F + N+LG GGFG VYKG L DG ++AVKR+ +G+G EF +E+AV++K
Sbjct: 581 LRSVTNNFSSDNILGSGGFGVVYKGELHDGTKIAVKRMENGVIAGKGFAEFKSEIAVLTK 640
Query: 545 LQHRNLVRLLGCCVERGEQMLVYEFMPNKSLDAFLF 580
++HR+LV LLG C++ E++LVYE+MP +L LF
Sbjct: 641 VRHRHLVTLLGYCLDGNEKLLVYEYMPQGTLSRHLF 676
Score = 53.1 bits (126), Expect = 3e-06
Identities = 35/110 (31%), Positives = 59/110 (52%), Gaps = 11/110 (10%)
Query: 602 PEYAMEGLFSEKSDVYSFGVLLLEIVSGRRNSSFYHNEDSLSLVGFAWKLWL--EENIIS 659
PEYA+ G + K DVYSFGV+L+E+++GR++ E+S+ LV + ++++ E +
Sbjct: 762 PEYAVTGRVTTKVDVYSFGVILMELITGRKSLDESQPEESIHLVSWFKRMYINKEASFKK 821
Query: 660 LIDREVWDASFESSMLRCIH-----IGLLCVQELPRDRPNISTVVLMLIS 704
ID + + L +H G C +E P RP++ V +L S
Sbjct: 822 AIDTTI---DLDEETLASVHTVAELAGHCCARE-PYQRPDMGHAVNILSS 867
>BRI1_LYCPE (Q8L899) Systemin receptor SR160 precursor (EC 2.7.1.37)
(Brassinosteroid LRR receptor kinase)
Length = 1207
Score = 102 bits (253), Expect = 5e-21
Identities = 57/121 (47%), Positives = 72/121 (59%), Gaps = 2/121 (1%)
Query: 461 WRKWSARHTEQKEMKLDELPL--YDFVKLENATNSFHNSNMLGKGGFGPVYKGILEDGQE 518
W+ SAR + E PL F L ATN FHN +++G GGFG VYK L+DG
Sbjct: 853 WKFTSAREALSINLAAFEKPLRKLTFADLLEATNGFHNDSLVGSGGFGDVYKAQLKDGSV 912
Query: 519 VAVKRLSKSSGQGIEEFMNEVAVISKLQHRNLVRLLGCCVERGEQMLVYEFMPNKSLDAF 578
VA+K+L SGQG EF E+ I K++HRNLV LLG C E++LVYE+M SL+
Sbjct: 913 VAIKKLIHVSGQGDREFTAEMETIGKIKHRNLVPLLGYCKVGEERLLVYEYMKYGSLEDV 972
Query: 579 L 579
L
Sbjct: 973 L 973
Score = 52.4 bits (124), Expect = 4e-06
Identities = 35/102 (34%), Positives = 57/102 (55%), Gaps = 4/102 (3%)
Query: 602 PEYAMEGLFSEKSDVYSFGVLLLEIVSGRRNSSFYHNEDSLSLVGFAWKLWLEENIISLI 661
PEY S K DVYS+GV+LLE+++G++ + D+ +LVG+ KL + I +
Sbjct: 1060 PEYYQSFRCSTKGDVYSYGVVLLELLTGKQPTDSADFGDN-NLVGWV-KLHAKGKITDVF 1117
Query: 662 DREVW--DASFESSMLRCIHIGLLCVQELPRDRPNISTVVLM 701
DRE+ DAS E +L+ + + C+ + RP + V+ M
Sbjct: 1118 DRELLKEDASIEIELLQHLKVACACLDDRHWKRPTMIQVMAM 1159
>BRI1_LYCES (Q8GUQ5) Brassinosteroid LRR receptor kinase precursor
(EC 2.7.1.37) (tBRI1) (Altered brassinolide sensitivity
1) (Systemin receptor SR160)
Length = 1207
Score = 102 bits (253), Expect = 5e-21
Identities = 57/121 (47%), Positives = 72/121 (59%), Gaps = 2/121 (1%)
Query: 461 WRKWSARHTEQKEMKLDELPL--YDFVKLENATNSFHNSNMLGKGGFGPVYKGILEDGQE 518
W+ SAR + E PL F L ATN FHN +++G GGFG VYK L+DG
Sbjct: 853 WKFTSAREALSINLAAFEKPLRKLTFADLLEATNGFHNDSLVGSGGFGDVYKAQLKDGSV 912
Query: 519 VAVKRLSKSSGQGIEEFMNEVAVISKLQHRNLVRLLGCCVERGEQMLVYEFMPNKSLDAF 578
VA+K+L SGQG EF E+ I K++HRNLV LLG C E++LVYE+M SL+
Sbjct: 913 VAIKKLIHVSGQGDREFTAEMETIGKIKHRNLVPLLGYCKVGEERLLVYEYMKYGSLEDV 972
Query: 579 L 579
L
Sbjct: 973 L 973
Score = 52.4 bits (124), Expect = 4e-06
Identities = 35/102 (34%), Positives = 57/102 (55%), Gaps = 4/102 (3%)
Query: 602 PEYAMEGLFSEKSDVYSFGVLLLEIVSGRRNSSFYHNEDSLSLVGFAWKLWLEENIISLI 661
PEY S K DVYS+GV+LLE+++G++ + D+ +LVG+ KL + I +
Sbjct: 1060 PEYYQSFRCSTKGDVYSYGVVLLELLTGKQPTDSADFGDN-NLVGWV-KLHAKGKITDVF 1117
Query: 662 DREVW--DASFESSMLRCIHIGLLCVQELPRDRPNISTVVLM 701
DRE+ DAS E +L+ + + C+ + RP + V+ M
Sbjct: 1118 DRELLKEDASIEIELLQHLKVACACLDDRHWKRPTMIQVMAM 1159
>PSKR_DAUCA (Q8LPB4) Phytosulfokine receptor precursor (EC 2.7.1.37)
(Phytosulfokine LRR receptor kinase)
Length = 1021
Score = 101 bits (252), Expect = 6e-21
Identities = 61/171 (35%), Positives = 92/171 (53%), Gaps = 24/171 (14%)
Query: 430 LEKEKGNKSFLIIAIAGGLGAFILVICAYLLWRKWSARHTEQKEMKLD------------ 477
++ +K + + +A+ GLG L+ L+ + ++R E K D
Sbjct: 661 VKSKKNIRKIVAVAVGTGLGTVFLLTVTLLIILRTTSRGEVDPEKKADADEIELGSRSVV 720
Query: 478 ---------ELPLYDFVKLENATNSFHNSNMLGKGGFGPVYKGILEDGQEVAVKRLSKSS 528
EL L D +K +T+SF+ +N++G GGFG VYK L DG +VA+KRLS +
Sbjct: 721 LFHNKDSNNELSLDDILK---STSSFNQANIIGCGGFGLVYKATLPDGTKVAIKRLSGDT 777
Query: 529 GQGIEEFMNEVAVISKLQHRNLVRLLGCCVERGEQMLVYEFMPNKSLDAFL 579
GQ EF EV +S+ QH NLV LLG C + +++L+Y +M N SLD +L
Sbjct: 778 GQMDREFQAEVETLSRAQHPNLVHLLGYCNYKNDKLLIYSYMDNGSLDYWL 828
Score = 60.1 bits (144), Expect = 2e-08
Identities = 32/101 (31%), Positives = 50/101 (48%)
Query: 602 PEYAMEGLFSEKSDVYSFGVLLLEIVSGRRNSSFYHNEDSLSLVGFAWKLWLEENIISLI 661
PEY + + K DVYSFGV+LLE+++GRR S L+ + ++ E+ +
Sbjct: 914 PEYGQASVATYKGDVYSFGVVLLELLTGRRPMDVCKPRGSRDLISWVLQMKTEKRESEIF 973
Query: 662 DREVWDASFESSMLRCIHIGLLCVQELPRDRPNISTVVLML 702
D ++D ML + I C+ E P+ RP +V L
Sbjct: 974 DPFIYDKDHAEEMLLVLEIACRCLGENPKTRPTTQQLVSWL 1014
>BAK1_ARATH (Q94F62) BRASSINOSTEROID INSENSITIVE 1-associated
receptor kinase 1 precursor (EC 2.7.1.37)
(BRI1-associated receptor kinase 1) (Somatic
embryogenesis receptor-like kinase 3)
Length = 615
Score = 98.6 bits (244), Expect = 5e-20
Identities = 62/150 (41%), Positives = 86/150 (57%), Gaps = 13/150 (8%)
Query: 443 AIAGGLGA-----FILVICAYLLWRK-------WSARHTEQKEMKLDELPLYDFVKLENA 490
AIAGG+ A F + A WR+ + E E+ L +L + +L+ A
Sbjct: 226 AIAGGVAAGAALLFAVPAIALAWWRRKKPQDHFFDVPAEEDPEVHLGQLKRFSLRELQVA 285
Query: 491 TNSFHNSNMLGKGGFGPVYKGILEDGQEVAVKRLSKSSGQGIE-EFMNEVAVISKLQHRN 549
+++F N N+LG+GGFG VYKG L DG VAVKRL + QG E +F EV +IS HRN
Sbjct: 286 SDNFSNKNILGRGGFGKVYKGRLADGTLVAVKRLKEERTQGGELQFQTEVEMISMAVHRN 345
Query: 550 LVRLLGCCVERGEQMLVYEFMPNKSLDAFL 579
L+RL G C+ E++LVY +M N S+ + L
Sbjct: 346 LLRLRGFCMTPTERLLVYPYMANGSVASCL 375
Score = 67.0 bits (162), Expect = 2e-10
Identities = 36/103 (34%), Positives = 62/103 (59%), Gaps = 2/103 (1%)
Query: 602 PEYAMEGLFSEKSDVYSFGVLLLEIVSGRR--NSSFYHNEDSLSLVGFAWKLWLEENIIS 659
PEY G SEK+DV+ +GV+LLE+++G+R + + N+D + L+ + L E+ + +
Sbjct: 461 PEYLSTGKSSEKTDVFGYGVMLLELITGQRAFDLARLANDDDVMLLDWVKGLLKEKKLEA 520
Query: 660 LIDREVWDASFESSMLRCIHIGLLCVQELPRDRPNISTVVLML 702
L+D ++ + + + I + LLC Q P +RP +S VV ML
Sbjct: 521 LVDVDLQGNYKDEEVEQLIQVALLCTQSSPMERPKMSEVVRML 563
>CX32_ARATH (P27450) Probable serine/threonine-protein kinase Cx32,
chloroplast precursor (EC 2.7.1.37)
Length = 419
Score = 97.8 bits (242), Expect = 9e-20
Identities = 53/119 (44%), Positives = 74/119 (61%), Gaps = 10/119 (8%)
Query: 472 KEMKLDELPLYDFVKLENATNSFHNSNMLGKGGFGPVYKGILE----------DGQEVAV 521
K ++ L +Y+F+ L+ AT +F +MLG+GGFG VY+G ++ G VA+
Sbjct: 64 KLLESPNLKVYNFLDLKTATKNFKPDSMLGQGGFGKVYRGWVDATTLAPSRVGSGMIVAI 123
Query: 522 KRLSKSSGQGIEEFMNEVAVISKLQHRNLVRLLGCCVERGEQMLVYEFMPNKSLDAFLF 580
KRL+ S QG E+ +EV + L HRNLV+LLG C E E +LVYEFMP SL++ LF
Sbjct: 124 KRLNSESVQGFAEWRSEVNFLGMLSHRNLVKLLGYCREDKELLLVYEFMPKGSLESHLF 182
Score = 44.3 bits (103), Expect = 0.001
Identities = 30/102 (29%), Positives = 51/102 (49%), Gaps = 1/102 (0%)
Query: 602 PEYAMEGLFSEKSDVYSFGVLLLEIVSGRRNSSFYHNEDSLSLVGFAW-KLWLEENIISL 660
PEY G KSDV++FGV+LLEI++G + SLV + +L + + +
Sbjct: 264 PEYMATGHLYVKSDVFAFGVVLLEIMTGLTAHNTKRPRGQESLVDWLRPELSNKHRVKQI 323
Query: 661 IDREVWDASFESSMLRCIHIGLLCVQELPRDRPNISTVVLML 702
+D+ + I L C++ P++RP++ VV +L
Sbjct: 324 MDKGIKGQYTTKVATEMARITLSCIEPDPKNRPHMKEVVEVL 365
>PSKR_ARATH (Q9ZVR7) Putative phytosulfokine receptor precursor (EC
2.7.1.37) (Phytosulfokine LRR receptor kinase)
Length = 1008
Score = 90.9 bits (224), Expect = 1e-17
Identities = 48/111 (43%), Positives = 69/111 (61%), Gaps = 8/111 (7%)
Query: 469 TEQKEMKLDELPLYDFVKLENATNSFHNSNMLGKGGFGPVYKGILEDGQEVAVKRLSKSS 528
+ KE+ D+L ++TNSF +N++G GGFG VYK L DG++VA+K+LS
Sbjct: 717 SNDKELSYDDLL--------DSTNSFDQANIIGCGGFGMVYKATLPDGKKVAIKKLSGDC 768
Query: 529 GQGIEEFMNEVAVISKLQHRNLVRLLGCCVERGEQMLVYEFMPNKSLDAFL 579
GQ EF EV +S+ QH NLV L G C + +++L+Y +M N SLD +L
Sbjct: 769 GQIEREFEAEVETLSRAQHPNLVLLRGFCFYKNDRLLIYSYMENGSLDYWL 819
Score = 57.8 bits (138), Expect = 1e-07
Identities = 30/101 (29%), Positives = 49/101 (47%)
Query: 602 PEYAMEGLFSEKSDVYSFGVLLLEIVSGRRNSSFYHNEDSLSLVGFAWKLWLEENIISLI 661
PEY + + K DVYSFGV+LLE+++ +R + L+ + K+ E +
Sbjct: 905 PEYGQASVATYKGDVYSFGVVLLELLTDKRPVDMCKPKGCRDLISWVVKMKHESRASEVF 964
Query: 662 DREVWDASFESSMLRCIHIGLLCVQELPRDRPNISTVVLML 702
D ++ + M R + I LC+ E P+ RP +V L
Sbjct: 965 DPLIYSKENDKEMFRVLEIACLCLSENPKQRPTTQQLVSWL 1005
>PBS1_ARATH (Q9FE20) Serine/threonine-protein kinase PBS1 (EC
2.7.1.37) (AvrPphB susceptible protein 1)
Length = 456
Score = 90.9 bits (224), Expect = 1e-17
Identities = 49/99 (49%), Positives = 66/99 (66%), Gaps = 1/99 (1%)
Query: 482 YDFVKLENATNSFHNSNMLGKGGFGPVYKGILED-GQEVAVKRLSKSSGQGIEEFMNEVA 540
+ F +L AT +FH LG+GGFG VYKG L+ GQ VAVK+L ++ QG EF+ EV
Sbjct: 74 FAFRELAAATMNFHPDTFLGEGGFGRVYKGRLDSTGQVVAVKQLDRNGLQGNREFLVEVL 133
Query: 541 VISKLQHRNLVRLLGCCVERGEQMLVYEFMPNKSLDAFL 579
++S L H NLV L+G C + +++LVYEFMP SL+ L
Sbjct: 134 MLSLLHHPNLVNLIGYCADGDQRLLVYEFMPLGSLEDHL 172
Score = 58.9 bits (141), Expect = 5e-08
Identities = 38/104 (36%), Positives = 58/104 (55%), Gaps = 5/104 (4%)
Query: 602 PEYAMEGLFSEKSDVYSFGVLLLEIVSGRR--NSSFYHNEDSLSLVGFAWKLWLE-ENII 658
PEYAM G + KSDVYSFGV+ LE+++GR+ +S H E +LV +A L+ + I
Sbjct: 259 PEYAMTGQLTVKSDVYSFGVVFLELITGRKAIDSEMPHGEQ--NLVAWARPLFNDRRKFI 316
Query: 659 SLIDREVWDASFESSMLRCIHIGLLCVQELPRDRPNISTVVLML 702
L D + ++ + + + +C+QE RP I+ VV L
Sbjct: 317 KLADPRLKGRFPTRALYQALAVASMCIQEQAATRPLIADVVTAL 360
>APKA_ARATH (Q06548) Protein kinase APK1A, chloroplast precursor (EC
2.7.1.-)
Length = 410
Score = 89.7 bits (221), Expect = 2e-17
Identities = 46/122 (37%), Positives = 77/122 (62%), Gaps = 10/122 (8%)
Query: 469 TEQKEMKLDELPLYDFVKLENATNSFHNSNMLGKGGFGPVYKGILED----------GQE 518
TE + ++ L + F +L++AT +F ++LG+GGFG V+KG +++ G
Sbjct: 43 TEGEILQSPNLKSFSFAELKSATRNFRPDSVLGEGGFGCVFKGWIDEKSLTASRPGTGLV 102
Query: 519 VAVKRLSKSSGQGIEEFMNEVAVISKLQHRNLVRLLGCCVERGEQMLVYEFMPNKSLDAF 578
+AVK+L++ QG +E++ EV + + HR+LV+L+G C+E ++LVYEFMP SL+
Sbjct: 103 IAVKKLNQDGWQGHQEWLAEVNYLGQFSHRHLVKLIGYCLEDEHRLLVYEFMPRGSLENH 162
Query: 579 LF 580
LF
Sbjct: 163 LF 164
Score = 53.1 bits (126), Expect = 3e-06
Identities = 37/100 (37%), Positives = 52/100 (52%), Gaps = 3/100 (3%)
Query: 602 PEYAMEGLFSEKSDVYSFGVLLLEIVSGRRNSSFYHNEDSLSLVGFAWKLWL--EENIIS 659
PEY G + KSDVYSFGV+LLE++SGRR +LV +A K +L + I
Sbjct: 249 PEYLATGHLTTKSDVYSFGVVLLELLSGRRAVDKNRPSGERNLVEWA-KPYLVNKRKIFR 307
Query: 660 LIDREVWDASFESSMLRCIHIGLLCVQELPRDRPNISTVV 699
+ID + D + + L C+ + RPN+S VV
Sbjct: 308 VIDNRLQDQYSMEEACKVATLSLRCLTTEIKLRPNMSEVV 347
>CRI4_MAIZE (O24585) Putative receptor protein kinase CRINKLY4
precursor (EC 2.7.1.-)
Length = 901
Score = 89.4 bits (220), Expect = 3e-17
Identities = 47/114 (41%), Positives = 71/114 (62%), Gaps = 2/114 (1%)
Query: 470 EQKEMKLDELPLYDFVKLENATNSFHNSNMLGKGGFGPVYKGILEDGQEVAVKRLSKSSG 529
+ +++K+ + + +LE AT F + +GKG F V+KGIL DG VAVKR K+S
Sbjct: 481 DMEDLKIRRAQEFSYEELEQATGGFSEDSQVGKGSFSCVFKGILRDGTVVAVKRAIKASD 540
Query: 530 --QGIEEFMNEVAVISKLQHRNLVRLLGCCVERGEQMLVYEFMPNKSLDAFLFG 581
+ +EF NE+ ++S+L H +L+ LLG C + E++LVYEFM + SL L G
Sbjct: 541 VKKSSKEFHNELDLLSRLNHAHLLNLLGYCEDGSERLLVYEFMAHGSLYQHLHG 594
Score = 50.8 bits (120), Expect = 1e-05
Identities = 33/101 (32%), Positives = 53/101 (51%), Gaps = 2/101 (1%)
Query: 602 PEYAMEGLFSEKSDVYSFGVLLLEIVSGRRNSSFYHNEDSLSLVGFAWKLWLEENIISLI 661
PEY + KSDVYSFGV+LLEI+SGR+ E ++V +A L +I +++
Sbjct: 680 PEYYRLHYLTTKSDVYSFGVVLLEILSGRKAIDMQFEEG--NIVEWAVPLIKAGDIFAIL 737
Query: 662 DREVWDASFESSMLRCIHIGLLCVQELPRDRPNISTVVLML 702
D + S ++ + + CV+ +DRP++ V L
Sbjct: 738 DPVLSPPSDLEALKKIASVACKCVRMRGKDRPSMDKVTTAL 778
>RCK7_ARATH (Q9LQQ8) Putative serine/threonine-protein kinase
RLCKVII (EC 2.7.1.37)
Length = 423
Score = 89.0 bits (219), Expect = 4e-17
Identities = 45/99 (45%), Positives = 65/99 (65%), Gaps = 1/99 (1%)
Query: 482 YDFVKLENATNSFHNSNMLGKGGFGPVYKGILED-GQEVAVKRLSKSSGQGIEEFMNEVA 540
+ F +L AT +F + LG+GGFG V+KG +E Q VA+K+L ++ QGI EF+ EV
Sbjct: 91 FTFQELAEATGNFRSDCFLGEGGFGKVFKGTIEKLDQVVAIKQLDRNGVQGIREFVVEVL 150
Query: 541 VISKLQHRNLVRLLGCCVERGEQMLVYEFMPNKSLDAFL 579
+S H NLV+L+G C E +++LVYE+MP SL+ L
Sbjct: 151 TLSLADHPNLVKLIGFCAEGDQRLLVYEYMPQGSLEDHL 189
Score = 65.5 bits (158), Expect = 5e-10
Identities = 38/102 (37%), Positives = 58/102 (56%), Gaps = 1/102 (0%)
Query: 602 PEYAMEGLFSEKSDVYSFGVLLLEIVSGRRNSSFYHNEDSLSLVGFAWKLWLE-ENIISL 660
P+YAM G + KSD+YSFGV+LLE+++GR+ +LVG+A L+ + N +
Sbjct: 276 PDYAMTGQLTFKSDIYSFGVVLLELITGRKAIDNTKTRKDQNLVGWARPLFKDRRNFPKM 335
Query: 661 IDREVWDASFESSMLRCIHIGLLCVQELPRDRPNISTVVLML 702
+D + + + + I +CVQE P RP +S VVL L
Sbjct: 336 VDPLLQGQYPVRGLYQALAISAMCVQEQPTMRPVVSDVVLAL 377
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.323 0.138 0.430
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 91,092,033
Number of Sequences: 164201
Number of extensions: 4021900
Number of successful extensions: 11439
Number of sequences better than 10.0: 779
Number of HSP's better than 10.0 without gapping: 406
Number of HSP's successfully gapped in prelim test: 373
Number of HSP's that attempted gapping in prelim test: 10120
Number of HSP's gapped (non-prelim): 1437
length of query: 748
length of database: 59,974,054
effective HSP length: 118
effective length of query: 630
effective length of database: 40,598,336
effective search space: 25576951680
effective search space used: 25576951680
T: 11
A: 40
X1: 16 ( 7.5 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (22.0 bits)
S2: 70 (31.6 bits)
Medicago: description of AC144721.6


