

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144721.10 + phase: 0 /pseudo
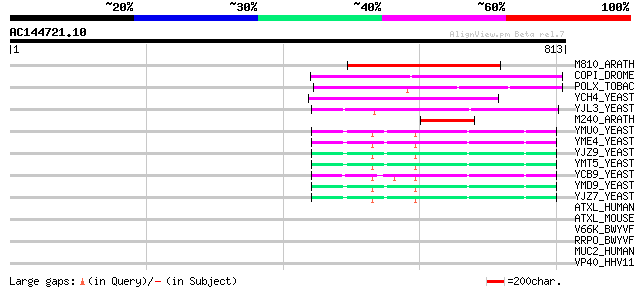
(813 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
M810_ARATH (P92519) Hypothetical mitochondrial protein AtMg00810... 246 2e-64
COPI_DROME (P04146) Copia protein (Gag-int-pol protein) [Contain... 244 7e-64
POLX_TOBAC (P10978) Retrovirus-related Pol polyprotein from tran... 219 2e-56
YCH4_YEAST (P25600) Transposon Ty5-1 34.5 kDa hypothetical protein 149 4e-35
YJL3_YEAST (P47024) Transposon Ty4 207.7 kDa hypothetical protein 98 8e-20
M240_ARATH (P93290) Hypothetical mitochondrial protein AtMg00240... 78 8e-14
YMU0_YEAST (Q04670) Transposon Ty1 protein B 73 3e-12
YME4_YEAST (Q04711) Transposon Ty1 protein B 72 8e-12
YJZ9_YEAST (P47100) Transposon Ty1 protein B 70 2e-11
YMT5_YEAST (Q04214) Transposon Ty1 protein B 70 3e-11
YCB9_YEAST (P25384) Transposon Ty2 protein B (Ty1-17 protein B) 67 2e-10
YMD9_YEAST (Q03434) Transposon Ty1 protein B 65 9e-10
YJZ7_YEAST (P47098) Transposon Ty1 protein B 62 6e-09
ATXL_HUMAN (Q8WWM7) Ataxin-2-like protein (Ataxin-2 domain prote... 35 0.79
ATXL_MOUSE (Q7TQH0) Ataxin-2-like protein 33 2.3
V66K_BWYVF (P09506) 66.2 kDa protein (ORF 2) 33 3.0
RRPO_BWYVF (P09507) Putative RNA-directed RNA polymerase (EC 2.7... 33 3.0
MUC2_HUMAN (Q02817) Mucin 2 precursor (Intestinal mucin 2) 32 5.2
VP40_HHV11 (P10210) Capsid protein P40 (Virion structural protei... 32 8.8
>M810_ARATH (P92519) Hypothetical mitochondrial protein AtMg00810
(ORF240b)
Length = 240
Score = 246 bits (627), Expect = 2e-64
Identities = 119/225 (52%), Positives = 160/225 (70%), Gaps = 1/225 (0%)
Query: 495 YILLYVDDIILTTSSHSLRDTIIAALRSEFSLTDLGPLNYFLGVSATRHSGGLFLHQSKY 554
Y+LLYVDDI+LT SS++L + +I L S FS+ DLGP++YFLG+ H GLFL Q+KY
Sbjct: 2 YLLLYVDDILLTGSSNTLLNMLIFQLSSTFSMKDLGPVHYFLGIQIKTHPSGLFLSQTKY 61
Query: 555 AEDIISRAKMVNCKPALTPVDTKSKLSATSGEKFDDSTLYRSLAGALQYLTFTRPDISYA 614
AE I++ A M++CKP TP+ K S+ S K+ D + +RS+ GALQYLT TRPDISYA
Sbjct: 62 AEQILNNAGMLDCKPMSTPLPLKLN-SSVSTAKYPDPSDFRSIVGALQYLTLTRPDISYA 120
Query: 615 VQQICLYMHDPRTSHFAALKRIIRYIQGTKEFGLHLSPSTVTNLSAYSDADWGGCPDTRR 674
V +C MH+P + F LKR++RY++GT GL++ ++ N+ A+ D+DW GC TRR
Sbjct: 121 VNIVCQRMHEPTLADFDLLKRVLRYVKGTIFHGLYIHKNSKLNVQAFCDSDWAGCTSTRR 180
Query: 675 STSGYCVYLGDNLISWSAKRQATLSRSSAEAEYRGVANVVAESCW 719
ST+G+C +LG N+ISWSAKRQ T+SRSS E EYR +A AE W
Sbjct: 181 STTGFCTFLGCNIISWSAKRQPTVSRSSTETEYRALALTAAELTW 225
>COPI_DROME (P04146) Copia protein (Gag-int-pol protein) [Contains:
Copia VLP protein; Copia protease (EC 3.4.23.-)]
Length = 1409
Score = 244 bits (623), Expect = 7e-64
Identities = 134/375 (35%), Positives = 205/375 (53%), Gaps = 7/375 (1%)
Query: 441 NPDHVCLLKKSLYGLKQAPRAWYQRFASFLSTIGFVNSKTDHSLFISTHGD--DTTYILL 498
N D+VC L K++YGLKQA R W++ F L FVNS D ++I G+ + Y+LL
Sbjct: 1027 NSDNVCKLNKAIYGLKQAARCWFEVFEQALKECEFVNSSVDRCIYILDKGNINENIYVLL 1086
Query: 499 YVDDIILTTSSHSLRDTIIAALRSEFSLTDLGPLNYFLGVSATRHSGGLFLHQSKYAEDI 558
YVDD+++ T + + L +F +TDL + +F+G+ ++L QS Y + I
Sbjct: 1087 YVDDVVIATGDMTRMNNFKRYLMEKFRMTDLNEIKHFIGIRIEMQEDKIYLSQSAYVKKI 1146
Query: 559 ISRAKMVNCKPALTPVDTKSKLSATSGEKFDDSTLYRSLAGALQYLTF-TRPDISYAVQQ 617
+S+ M NC TP+ +K + ++ D +T RSL G L Y+ TRPD++ AV
Sbjct: 1147 LSKFNMENCNAVSTPLPSKINYELLNSDE-DCNTPCRSLIGCLMYIMLCTRPDLTTAVNI 1205
Query: 618 ICLYMHDPRTSHFAALKRIIRYIQGTKEFGLHLSPSTV--TNLSAYSDADWGGCPDTRRS 675
+ Y + + LKR++RY++GT + L + + Y D+DW G R+S
Sbjct: 1206 LSRYSSKNNSELWQNLKRVLRYLKGTIDMKLIFKKNLAFENKIIGYVDSDWAGSEIDRKS 1265
Query: 676 TSGYCVYLGD-NLISWSAKRQATLSRSSAEAEYRGVANVVAESCWLRNLLLELHCPISKA 734
T+GY + D NLI W+ KRQ +++ SS EAEY + V E+ WL+ LL ++ +
Sbjct: 1266 TTGYLFKMFDFNLICWNTKRQNSVAASSTEAEYMALFEAVREALWLKFLLTSINIKLENP 1325
Query: 735 TVVYCDNVSAIYLSGNPVNHQRTKHIEMDIHFVREKVALGQVHVLHVPTRYQYADIFTKG 794
+Y DN I ++ NP H+R KHI++ HF RE+V + + ++PT Q ADIFTK
Sbjct: 1326 IKIYEDNQGCISIANNPSCHKRAKHIDIKYHFAREQVQNNVICLEYIPTENQLADIFTKP 1385
Query: 795 LPKPLFDEFRSSLSI 809
LP F E R L +
Sbjct: 1386 LPAARFVELRDKLGL 1400
>POLX_TOBAC (P10978) Retrovirus-related Pol polyprotein from
transposon TNT 1-94 [Contains: Protease (EC 3.4.23.-);
Reverse transcriptase (EC 2.7.7.49); Endonuclease]
Length = 1328
Score = 219 bits (559), Expect = 2e-56
Identities = 139/376 (36%), Positives = 200/376 (52%), Gaps = 12/376 (3%)
Query: 445 VCLLKKSLYGLKQAPRAWYQRFASFLSTIGFVNSKTDHSLFISTHGDDTTYILL-YVDDI 503
VC L KSLYGLKQAPR WY +F SF+ + ++ + +D ++ ++ ILL YVDD+
Sbjct: 953 VCKLNKSLYGLKQAPRQWYMKFDSFMKSQTYLKTYSDPCVYFKRFSENNFIILLLYVDDM 1012
Query: 504 ILTTSSHSLRDTIIAALRSEFSLTDLGPLNYFLGVSATRH--SGGLFLHQSKYAEDIISR 561
++ L + L F + DLGP LG+ R S L+L Q KY E ++ R
Sbjct: 1013 LIVGKDKGLIAKLKGDLSKSFDMKDLGPAQQILGMKIVRERTSRKLWLSQEKYIERVLER 1072
Query: 562 AKMVNCKPALTPVDTKSKLS-----ATSGEKFDDSTL-YRSLAGALQY-LTFTRPDISYA 614
M N KP TP+ KLS T EK + + + Y S G+L Y + TRPDI++A
Sbjct: 1073 FNMKNAKPVSTPLAGHLKLSKKMCPTTVEEKGNMAKVPYSSAVGSLMYAMVCTRPDIAHA 1132
Query: 615 VQQICLYMHDPRTSHFAALKRIIRYIQGTKEFGLHLSPSTVTNLSAYSDADWGGCPDTRR 674
V + ++ +P H+ A+K I+RY++GT L S L Y+DAD G D R+
Sbjct: 1133 VGVVSRFLENPGKEHWEAVKWILRYLRGTTGDCLCFGGSDPI-LKGYTDADMAGDIDNRK 1191
Query: 675 STSGYCVYLGDNLISWSAKRQATLSRSSAEAEYRGVANVVAESCWLRNLLLELHCPISKA 734
S++GY ISW +K Q ++ S+ EAEY E WL+ L EL K
Sbjct: 1192 SSTGYLFTFSGGAISWQSKLQKCVALSTTEAEYIAATETGKEMIWLKRFLQELGLH-QKE 1250
Query: 735 TVVYCDNVSAIYLSGNPVNHQRTKHIEMDIHFVREKVALGQVHVLHVPTRYQYADIFTKG 794
VVYCD+ SAI LS N + H RTKHI++ H++RE V + VL + T AD+ TK
Sbjct: 1251 YVVYCDSQSAIDLSKNSMYHARTKHIDVRYHWIREMVDDESLKVLKISTNENPADMLTKV 1310
Query: 795 LPKPLFDEFRSSLSIH 810
+P+ F+ + + +H
Sbjct: 1311 VPRNKFELCKELVGMH 1326
>YCH4_YEAST (P25600) Transposon Ty5-1 34.5 kDa hypothetical protein
Length = 308
Score = 149 bits (375), Expect = 4e-35
Identities = 88/282 (31%), Positives = 139/282 (49%), Gaps = 3/282 (1%)
Query: 438 DRNNPDHVCLLKKSLYGLKQAPRAWYQRFASFLSTIGFVNSKTDHSLFISTHGDDTTYIL 497
+ NPD+V L +YGLKQAP W + + L IGF + +H L+ + D YI
Sbjct: 26 NERNPDYVWELYGGMYGLKQAPLLWNEHINNTLKKIGFCRHEGEHGLYFRSTSDGPIYIG 85
Query: 498 LYVDDIILTTSSHSLRDTIIAALRSEFSLTDLGPLNYFLGVSATRHSGG-LFLHQSKYAE 556
+YVDD+++ S + D + L +S+ DLG ++ FLG++ + + G + L Y
Sbjct: 86 VYVDDLLVAAPSPKIYDRVKQELTKLYSMKDLGKVDKFLGLNIHQSTNGDITLSLQDYIA 145
Query: 557 DIISRAKMVNCKPALTPVDTKSKLSATSGEKFDDSTLYRSLAGALQYLTFT-RPDISYAV 615
S +++ K TP+ L T+ D T Y+S+ G L + T RPDISY V
Sbjct: 146 KAASESEINTFKLTQTPLCNSKPLFETTSPHLKDITPYQSIVGQLLFCANTGRPDISYPV 205
Query: 616 QQICLYMHDPRTSHFAALKRIIRYIQGTKEFGLHLSPSTVTNLSAYSDADWGGCPDTRRS 675
+ ++ +PR H + +R++RY+ T+ L + L+ Y DA G D S
Sbjct: 206 SLLSRFLREPRAIHLESARRVLRYLYTTRSMCLKYRSGSQVALTVYCDASHGAIHDLPHS 265
Query: 676 TSGYCVYLGDNLISWSAKR-QATLSRSSAEAEYRGVANVVAE 716
T GY L ++WS+K+ + + S EAEY + V E
Sbjct: 266 TGGYVTLLAGAPVTWSSKKLKGVIPVPSTEAEYITASETVME 307
>YJL3_YEAST (P47024) Transposon Ty4 207.7 kDa hypothetical protein
Length = 1803
Score = 98.2 bits (243), Expect = 8e-20
Identities = 95/383 (24%), Positives = 159/383 (40%), Gaps = 25/383 (6%)
Query: 443 DHVCLLK--KSLYGLKQAPRAWYQRFASFLSTIGFVNSKTDHSLFISTHGDDTTYILLYV 500
D C++K K+LYGLKQ+P+ W +L+ IG ++ L+ + D I +YV
Sbjct: 1405 DRRCVVKLNKALYGLKQSPKEWNDHLRQYLNGIGLKDNSYTPGLYQTE--DKNLMIAVYV 1462
Query: 501 DDIILTTSSHSLRDTIIAALRSEFSLTDLGPL----------------NYFLGVSATRHS 544
DD ++ S+ D I L+S F L G L N LG
Sbjct: 1463 DDCVIAASNEQRLDEFINKLKSNFELKITGTLIDDVLDTDILGMDLVYNKRLGTIDLTLK 1522
Query: 545 GGLFLHQSKYAEDIISRAKMVNCKPALTPVDTKSKLSATSGEKFDDSTL-YRSLAGALQY 603
+ KY E++ K + +D K + S E+F L + L G L Y
Sbjct: 1523 SFINRMDKKYNEELKKIRKSSIPHMSTYKIDPKKDVLQMSEEEFRQGVLKLQQLLGELNY 1582
Query: 604 LTFT-RPDISYAVQQICLYMHDPRTSHFAALKRIIRYIQGTKEFGLHLSPSTVTN--LSA 660
+ R DI +AV+++ ++ P F + +II+Y+ K+ G+H + + A
Sbjct: 1583 VRHKCRYDIEFAVKKVARLVNYPHERVFYMIYKIIQYLVRYKDIGIHYDRDCNKDKKVIA 1642
Query: 661 YSDADWGGCPDTRRSTSGYCVYLGDNLISWSAKRQATLSRSSAEAEYRGVANVVAESCWL 720
+DA G D + S G ++ G N+ + + + SS EAE + A+S L
Sbjct: 1643 ITDASVGSEYDAQ-SRIGVILWYGMNIFNVYSNKSTNRCVSSTEAELHAIYEGYADSETL 1701
Query: 721 RNLLLELHCPISKATVVYCDNVSAIYLSGNPVNHQRTKHIEMDIHFVREKVALGQVHVLH 780
+ L EL + V+ D+ AI + K + ++EK+ + +L
Sbjct: 1702 KVTLKELGEGDNNDIVMITDSKPAIQGLNRSYQQPKEKFTWIKTEIIKEKIKEKSIKLLK 1761
Query: 781 VPTRYQYADIFTKGLPKPLFDEF 803
+ + AD+ TK + F F
Sbjct: 1762 ITGKGNIADLLTKPVSASDFKRF 1784
>M240_ARATH (P93290) Hypothetical mitochondrial protein AtMg00240
(ORF111a)
Length = 111
Score = 78.2 bits (191), Expect = 8e-14
Identities = 32/78 (41%), Positives = 52/78 (66%)
Query: 603 YLTFTRPDISYAVQQICLYMHDPRTSHFAALKRIIRYIQGTKEFGLHLSPSTVTNLSAYS 662
YLT TRPD+++AV ++ + RT+ A+ +++ Y++GT GL S ++ L A++
Sbjct: 2 YLTITRPDLTFAVNRLSQFSSASRTAQMQAVYKVLHYVKGTVGQGLFYSATSDLQLKAFA 61
Query: 663 DADWGGCPDTRRSTSGYC 680
D+DW CPDTRRS +G+C
Sbjct: 62 DSDWASCPDTRRSVTGFC 79
>YMU0_YEAST (Q04670) Transposon Ty1 protein B
Length = 1328
Score = 73.2 bits (178), Expect = 3e-12
Identities = 93/384 (24%), Positives = 158/384 (40%), Gaps = 35/384 (9%)
Query: 443 DHVCLLKKSLYGLKQAPRAWYQRFASFL-STIGFVNSKTDHSLFISTHGDDTTYILLYVD 501
D + LKKSLYGLKQ+ WY+ S+L G + +F ++ I L+VD
Sbjct: 945 DKLIRLKKSLYGLKQSGANWYETIKSYLIKQCGMEEVRGWSCVFKNSQ----VTICLFVD 1000
Query: 502 DIILTTSSHSLRDTIIAALRSEF--SLTDLG----PLNYFLGVSATRHSGGLFLHQSKYA 555
D+IL + + II L+ ++ + +LG + Y + ++ G ++ K
Sbjct: 1001 DMILFSKDLNANKKIITTLKKQYDTKIINLGESDNEIQYDILGLEIKYQRGKYM---KLG 1057
Query: 556 EDIISRAKMVNCKPALTPVDTKSKLSATSGEKFDDSTL-------------YRSLAGALQ 602
+ K+ L P K G D L + L G
Sbjct: 1058 MENSLTEKIPKLNVPLNPKGRKLSAPGQPGLYIDQQELELEEDDYKMKVHEMQKLIGLAS 1117
Query: 603 YLTFT-RPDISYAVQQICLYMHDPRTSHFAALKRIIRYIQGTKEFGL--HLS-PSTVTN- 657
Y+ + R D+ Y + + ++ P +I++I T++ L H S P TN
Sbjct: 1118 YVGYKFRFDLLYYINTLAQHILFPSKQVLDMTYELIQFIWNTRDKQLIWHKSKPVKPTNK 1177
Query: 658 LSAYSDADWGGCPDTRRSTSGYCVYLGDNLISWSAKRQATLSRSSAEAEYRGVANVVAES 717
L SDA +G P +S G L +I + + + S+ EAE ++ V
Sbjct: 1178 LVVISDASYGNQP-YYKSQIGNIYLLNGKVIGGKSTKASLTCTSTTEAEIHAISESVPLL 1236
Query: 718 CWLRNLLLELH-CPISKATVVYCDNVSAIYLSGNPVNHQRTKHIEMDIHFVREKVALGQV 776
L +L+ EL+ PI+K + + +I +S N R + +R++V+ +
Sbjct: 1237 NNLSHLVQELNKKPITKGLLTDSKSTISIIISNNEEKF-RNRFFGTKAMRLRDEVSGNHL 1295
Query: 777 HVLHVPTRYQYADIFTKGLPKPLF 800
HV ++ T+ AD+ TK LP F
Sbjct: 1296 HVCYIETKKNIADVMTKPLPIKTF 1319
>YME4_YEAST (Q04711) Transposon Ty1 protein B
Length = 1328
Score = 71.6 bits (174), Expect = 8e-12
Identities = 90/384 (23%), Positives = 157/384 (40%), Gaps = 35/384 (9%)
Query: 443 DHVCLLKKSLYGLKQAPRAWYQRFASFL-STIGFVNSKTDHSLFISTHGDDTTYILLYVD 501
D + LKKSLYGLKQ+ WY+ S+L G + +F ++ I L+VD
Sbjct: 945 DKLIRLKKSLYGLKQSGANWYETIKSYLIKQCGMEEVRGWSCVFKNSQ----VTICLFVD 1000
Query: 502 DIILTTSSHSLRDTIIAALRSEF--SLTDLG----PLNYFLGVSATRHSGGLFLHQSKYA 555
D+IL + + II L+ ++ + +LG + Y + ++ G ++ K
Sbjct: 1001 DMILFSKDLNANKKIITTLKKQYDTKIINLGESDNEIQYDILGLEIKYQRGKYM---KLG 1057
Query: 556 EDIISRAKMVNCKPALTPVDTKSKLSATSGEKFDDSTL-------------YRSLAGALQ 602
+ K+ L P K G D L + L G
Sbjct: 1058 MENSLTEKIPKLNVPLNPKGRKLSAPGQPGLYIDQDELEIDEDEYKEKVHEMQKLIGLAS 1117
Query: 603 YLTFT-RPDISYAVQQICLYMHDPRTSHFAALKRIIRYIQGTKEFGL--HLSPSTV--TN 657
Y+ + R D+ Y + + ++ P +I+++ T++ L H + T
Sbjct: 1118 YVGYKFRFDLLYYINTLAQHILFPSRQVLDMTYELIQFMWDTRDKQLIWHKNKPTEPDNK 1177
Query: 658 LSAYSDADWGGCPDTRRSTSGYCVYLGDNLISWSAKRQATLSRSSAEAEYRGVANVVAES 717
L A SDA +G P +S G L +I + + + S+ EAE ++ V
Sbjct: 1178 LVAISDASYGNQP-YYKSQIGNIYLLNGKVIGGKSTKASLTCTSTTEAEIHAISESVPLL 1236
Query: 718 CWLRNLLLELH-CPISKATVVYCDNVSAIYLSGNPVNHQRTKHIEMDIHFVREKVALGQV 776
L +L+ EL+ PI+K + + +I +S N R + +R++V+ +
Sbjct: 1237 NNLSHLVQELNKKPITKGLLTDSKSTISIIISNNEEKF-RNRFFGTKAMRLRDEVSGNHL 1295
Query: 777 HVLHVPTRYQYADIFTKGLPKPLF 800
HV ++ T+ AD+ TK LP F
Sbjct: 1296 HVCYIETKKNIADVMTKPLPIKTF 1319
>YJZ9_YEAST (P47100) Transposon Ty1 protein B
Length = 1755
Score = 70.1 bits (170), Expect = 2e-11
Identities = 92/384 (23%), Positives = 157/384 (39%), Gaps = 35/384 (9%)
Query: 443 DHVCLLKKSLYGLKQAPRAWYQRFASFL-STIGFVNSKTDHSLFISTHGDDTTYILLYVD 501
D + LKKSLYGLKQ+ WY+ S+L G + +F ++ I L+VD
Sbjct: 1372 DKLIRLKKSLYGLKQSGANWYETIKSYLIQQCGMEEVRGWSCVFKNSQ----VTICLFVD 1427
Query: 502 DIILTTSSHSLRDTIIAALRSEF--SLTDLG----PLNYFLGVSATRHSGGLFLHQSKYA 555
D++L + + + II L+ ++ + +LG + Y + ++ G ++ K
Sbjct: 1428 DMVLFSKNLNSNKRIIEKLKMQYDTKIINLGESDEEIQYDILGLEIKYQRGKYM---KLG 1484
Query: 556 EDIISRAKMVNCKPALTPVDTKSKLSATSGEKFDDSTL-------------YRSLAGALQ 602
+ K+ L P K G D L + L G
Sbjct: 1485 MENSLTEKIPKLNVPLNPKGRKLSAPGQPGLYIDQQELELEEDDYKMKVHEMQKLIGLAS 1544
Query: 603 YLTFT-RPDISYAVQQICLYMHDPRTSHFAALKRIIRYIQGTKEFGL--HLS-PSTVTN- 657
Y+ + R D+ Y + + ++ P +I++I T++ L H S P TN
Sbjct: 1545 YVGYKFRFDLLYYINTLAQHILFPSKQVLDMTYELIQFIWNTRDKQLIWHKSKPVKPTNK 1604
Query: 658 LSAYSDADWGGCPDTRRSTSGYCVYLGDNLISWSAKRQATLSRSSAEAEYRGVANVVAES 717
L SDA +G P +S G L +I + + + S+ EAE ++ V
Sbjct: 1605 LVVISDASYGNQP-YYKSQIGNIYLLNGKVIGGKSTKASLTCTSTTEAEIHAISESVPLL 1663
Query: 718 CWLRNLLLEL-HCPISKATVVYCDNVSAIYLSGNPVNHQRTKHIEMDIHFVREKVALGQV 776
L L+ EL PI+K + + +I +S N R + +R++V+ +
Sbjct: 1664 NNLSYLIQELDKKPITKGLLTDSKSTISIIISNNEEKF-RNRFFGTKAMRLRDEVSGNHL 1722
Query: 777 HVLHVPTRYQYADIFTKGLPKPLF 800
HV ++ T+ AD+ TK LP F
Sbjct: 1723 HVCYIETKKNIADVMTKPLPIKTF 1746
>YMT5_YEAST (Q04214) Transposon Ty1 protein B
Length = 1328
Score = 69.7 bits (169), Expect = 3e-11
Identities = 92/384 (23%), Positives = 157/384 (39%), Gaps = 35/384 (9%)
Query: 443 DHVCLLKKSLYGLKQAPRAWYQRFASFL-STIGFVNSKTDHSLFISTHGDDTTYILLYVD 501
D + LKKSLYGLKQ+ WY+ S+L G + +F ++ I L+VD
Sbjct: 945 DKLIRLKKSLYGLKQSGANWYETIKSYLIKQCGMEEVRGWSCVFENSQ----VTICLFVD 1000
Query: 502 DIILTTSSHSLRDTIIAALRSEF--SLTDLG----PLNYFLGVSATRHSGGLFLHQSKYA 555
D++L + + + II L+ ++ + +LG + Y + ++ G ++ K
Sbjct: 1001 DMVLFSKNLNSNKRIIDKLKMQYDTKIINLGESDEEIQYDILGLEIKYQRGKYM---KLG 1057
Query: 556 EDIISRAKMVNCKPALTPVDTKSKLSATSGEKFDDSTL-------------YRSLAGALQ 602
+ K+ L P K G D L + L G
Sbjct: 1058 MENSLTEKIPKLNVPLNPKGRKLSAPGQPGLYIDQQELELEEDDYKMKVHEMQKLIGLAS 1117
Query: 603 YLTFT-RPDISYAVQQICLYMHDPRTSHFAALKRIIRYIQGTKEFGL--HLS-PSTVTN- 657
Y+ + R D+ Y + + ++ P +I++I T++ L H S P TN
Sbjct: 1118 YVGYKFRFDLLYYINTLAQHILFPSKQVLDMTYELIQFIWNTRDKQLIWHKSKPVKPTNK 1177
Query: 658 LSAYSDADWGGCPDTRRSTSGYCVYLGDNLISWSAKRQATLSRSSAEAEYRGVANVVAES 717
L SDA +G P +S G L +I + + + S+ EAE ++ V
Sbjct: 1178 LVVISDASYGNQP-YYKSQIGNIYLLNGKVIGGKSTKASLTCTSTTEAEIHAISESVPLL 1236
Query: 718 CWLRNLLLEL-HCPISKATVVYCDNVSAIYLSGNPVNHQRTKHIEMDIHFVREKVALGQV 776
L L+ EL PI+K + + +I +S N R + +R++V+ +
Sbjct: 1237 NNLSYLIQELDKKPITKGLLTDSKSTISIIISNNEEKF-RNRFFGTKAMRLRDEVSGNHL 1295
Query: 777 HVLHVPTRYQYADIFTKGLPKPLF 800
HV ++ T+ AD+ TK LP F
Sbjct: 1296 HVCYIETKKNIADVMTKPLPIKTF 1319
>YCB9_YEAST (P25384) Transposon Ty2 protein B (Ty1-17 protein B)
Length = 1770
Score = 67.0 bits (162), Expect = 2e-10
Identities = 90/387 (23%), Positives = 157/387 (40%), Gaps = 41/387 (10%)
Query: 443 DHVCLLKKSLYGLKQAPRAWYQRFASFLSTIGFVNSKTDHSLFISTHGDDTTYILLYVDD 502
D + L+KSLYGLKQ+ WY+ S+L + S + I L+VDD
Sbjct: 1387 DKLLRLRKSLYGLKQSGANWYETIKSYLINCCDMQEVRGWSCVFK---NSQVTICLFVDD 1443
Query: 503 IILTTSSHSLRDTIIAALRSEF--SLTDLG----PLNY-FLGVSATRHSGGLFLHQSKYA 555
+IL + + II L+ ++ + +LG + Y LG+ + +SKY
Sbjct: 1444 MILFSKDLNANKKIITTLKKQYDTKIINLGESDNEIQYDILGLE-------IKYQRSKYM 1496
Query: 556 EDIISRA---KMVNCKPALTPVDTKSKLSATSGEKFDDSTL-------------YRSLAG 599
+ + ++ K+ L P K + G D L + L G
Sbjct: 1497 KLGMEKSLTEKLPKLNVPLNPKGKKLRAPGQPGHYIDQDELEIDEDEYKEKVHEMQKLIG 1556
Query: 600 ALQYLTFT-RPDISYAVQQICLYMHDPRTSHFAALKRIIRYIQGTKEFGL--HLSPSTV- 655
Y+ + R D+ Y + + ++ P +I+++ T++ L H + T
Sbjct: 1557 LASYVGYKFRFDLLYYINTLAQHILFPSRQVLDMTYELIQFMWDTRDKQLIWHKNKPTKP 1616
Query: 656 -TNLSAYSDADWGGCPDTRRSTSGYCVYLGDNLISWSAKRQATLSRSSAEAEYRGVANVV 714
L A SDA +G P +S G L +I + + + S+ EAE V+ +
Sbjct: 1617 DNKLVAISDASYGNQP-YYKSQIGNIFLLNGKVIGGKSTKASLTCTSTTEAEIHAVSEAI 1675
Query: 715 AESCWLRNLLLELH-CPISKATVVYCDNVSAIYLSGNPVNHQRTKHIEMDIHFVREKVAL 773
L +L+ EL+ PI K + + +I S N R + +R++V+
Sbjct: 1676 PLLNNLSHLVQELNKKPIIKGLLTDSRSTISIIKSTNEEKF-RNRFFGTKAMRLRDEVSG 1734
Query: 774 GQVHVLHVPTRYQYADIFTKGLPKPLF 800
++V ++ T+ AD+ TK LP F
Sbjct: 1735 NNLYVYYIETKKNIADVMTKPLPIKTF 1761
>YMD9_YEAST (Q03434) Transposon Ty1 protein B
Length = 1328
Score = 64.7 bits (156), Expect = 9e-10
Identities = 89/384 (23%), Positives = 154/384 (39%), Gaps = 35/384 (9%)
Query: 443 DHVCLLKKSLYGLKQAPRAWYQRFASFL-STIGFVNSKTDHSLFISTHGDDTTYILLYVD 501
D + LKKSLYGLKQ+ WY+ S+L G + +F ++ I L+VD
Sbjct: 945 DKLIRLKKSLYGLKQSGANWYETIKSYLIKQCGMEEVRGWSCVFKNSQ----VTICLFVD 1000
Query: 502 DIILTTSSHSLRDTIIAALRSEF--SLTDLG----PLNYFLGVSATRHSGGLFLHQSKYA 555
D+IL + + II L+ ++ + +LG + Y + ++ G ++ K
Sbjct: 1001 DMILFSKDLNANKKIITTLKKQYDTKIINLGESDNEIQYDILGLEIKYQRGKYM---KLG 1057
Query: 556 EDIISRAKMVNCKPALTPVDTKSKLSATSGEKFDDSTL-------------YRSLAGALQ 602
+ K+ L P K G D L + L G
Sbjct: 1058 MENSLTEKIPKLNVPLNPKGRKLSAPGQPGLYIDQDELEIDEDEYKEKVHEMQKLIGLAS 1117
Query: 603 YLTFT-RPDISYAVQQICLYMHDPRTSHFAALKRIIRYIQGTKEFGL--HLSPSTV--TN 657
Y+ + R D+ Y + + ++ P +I+++ T++ L H + T
Sbjct: 1118 YVGYKFRFDLLYYINTLAQHILFPSRQVLDMTYELIQFMWDTRDKQLIWHKNKPTEPDNK 1177
Query: 658 LSAYSDADWGGCPDTRRSTSGYCVYLGDNLISWSAKRQATLSRSSAEAEYRGVANVVAES 717
L A SDA +G P +S G L +I + + + S+ EAE ++ V
Sbjct: 1178 LVAISDASYGNQP-YYKSQIGNIYLLNGKVIGGKSTKASLTCTSTTEAEIHAISESVPLL 1236
Query: 718 CWLRNLLLELH-CPISKATVVYCDNVSAIYLSGNPVNHQRTKHIEMDIHFVREKVALGQV 776
L L+ EL+ PI K + + +I S N R + +R++V+ +
Sbjct: 1237 NNLSYLIQELNKKPIIKGLLTDSRSTISIIKSTNEEKF-RNRFFGTKAMRLRDEVSGNNL 1295
Query: 777 HVLHVPTRYQYADIFTKGLPKPLF 800
+V ++ T+ AD+ TK LP F
Sbjct: 1296 YVYYIETKKNIADVMTKPLPIKTF 1319
>YJZ7_YEAST (P47098) Transposon Ty1 protein B
Length = 1755
Score = 62.0 bits (149), Expect = 6e-09
Identities = 88/384 (22%), Positives = 153/384 (38%), Gaps = 35/384 (9%)
Query: 443 DHVCLLKKSLYGLKQAPRAWYQRFASFL-STIGFVNSKTDHSLFISTHGDDTTYILLYVD 501
D + LKKS YGLKQ+ WY+ S+L G + +F ++ I L+VD
Sbjct: 1372 DKLIRLKKSHYGLKQSGANWYETIKSYLIKQCGMEEVRGWSCVFKNSQ----VTICLFVD 1427
Query: 502 DIILTTSSHSLRDTIIAALRSEF--SLTDLG----PLNYFLGVSATRHSGGLFLHQSKYA 555
D+IL + + II L+ ++ + +LG + Y + ++ G ++ K
Sbjct: 1428 DMILFSKDLNANKKIITTLKKQYDTKIINLGESDNEIQYDILGLEIKYQRGKYM---KLG 1484
Query: 556 EDIISRAKMVNCKPALTPVDTKSKLSATSGEKFDDSTL-------------YRSLAGALQ 602
+ K+ L P K G D L + L G
Sbjct: 1485 MENSLTEKIPKLNVPLNPKGRKLSAPGQPGLYIDQDELEIDEDEYKEKVHEMQKLIGLAS 1544
Query: 603 YLTFT-RPDISYAVQQICLYMHDPRTSHFAALKRIIRYIQGTKEFGL--HLSPSTV--TN 657
Y+ + R D+ Y + + ++ P +I+++ T++ L H + T
Sbjct: 1545 YVGYKFRFDLLYYINTLAQHILFPSRQVLDMTYELIQFMWDTRDKQLIWHKNKPTEPDNK 1604
Query: 658 LSAYSDADWGGCPDTRRSTSGYCVYLGDNLISWSAKRQATLSRSSAEAEYRGVANVVAES 717
L A SDA +G P +S G L +I + + + S+ EAE ++ V
Sbjct: 1605 LVAISDASYGNQP-YYKSQIGNIFLLNGKVIGGKSTKASLTCTSTTEAEIHAISESVPLL 1663
Query: 718 CWLRNLLLELH-CPISKATVVYCDNVSAIYLSGNPVNHQRTKHIEMDIHFVREKVALGQV 776
L L+ EL+ PI K + + +I S N R + +R++V+ +
Sbjct: 1664 NNLSYLIQELNKKPIIKGLLTDSRSTISIIKSTNEEKF-RNRFFGTKAMRLRDEVSGNNL 1722
Query: 777 HVLHVPTRYQYADIFTKGLPKPLF 800
+V ++ T+ AD+ TK LP F
Sbjct: 1723 YVYYIETKKNIADVMTKPLPIKTF 1746
>ATXL_HUMAN (Q8WWM7) Ataxin-2-like protein (Ataxin-2 domain protein)
(Ataxin-2 related protein)
Length = 1075
Score = 35.0 bits (79), Expect = 0.79
Identities = 27/95 (28%), Positives = 41/95 (42%), Gaps = 11/95 (11%)
Query: 168 PLPVSSSAIRHIIVASSVSTYPHVRLSRP-VM*PSMSQSFPYHIT--HLHQPLLTLF*TA 224
P ++S + IV+SS YP P + ++ QS+P+H T H HQP T
Sbjct: 822 PRMLTSGSHPQAIVSSSTPQYPSAEQPTPQALYATVHQSYPHHATQLHAHQPQPATTPTG 881
Query: 225 THPRI*HTLSTLRLHLTSKQ*HL--------LHHP 251
+ P+ H + H + HL L+HP
Sbjct: 882 SQPQSQHAAPSPVQHQAGQAPHLGSGQPQQNLYHP 916
>ATXL_MOUSE (Q7TQH0) Ataxin-2-like protein
Length = 1049
Score = 33.5 bits (75), Expect = 2.3
Identities = 27/95 (28%), Positives = 41/95 (42%), Gaps = 11/95 (11%)
Query: 168 PLPVSSSAIRHIIVASSVSTYPHVRLSRP-VM*PSMSQSFPYHIT--HLHQPLLTLF*TA 224
P ++S + IV+SS YP P + ++ QS+P+H T H HQP T
Sbjct: 826 PRMLTSGSHPQAIVSSSTPQYPAAEQPTPQALYATVHQSYPHHATQLHGHQPQPATTPTG 885
Query: 225 THPRI*HTLSTLRLHLTSKQ*HL--------LHHP 251
+ P+ H + H + HL L+HP
Sbjct: 886 SQPQSQHAAPSPVQHQAGQAPHLGSGQPQQNLYHP 920
>V66K_BWYVF (P09506) 66.2 kDa protein (ORF 2)
Length = 607
Score = 33.1 bits (74), Expect = 3.0
Identities = 38/150 (25%), Positives = 59/150 (39%), Gaps = 15/150 (10%)
Query: 447 LLKKSLYGLKQAPRAWYQRFASFLSTIGFVNSKTDHSLFISTH--GDDTTY---ILLYVD 501
LLKKS L+ P+ Y++ + S + TH G+ Y I LY
Sbjct: 186 LLKKSFSALRSTPKCLYEKAIDGFKSFTIPQSPPKSCVIPITHASGNHAGYASCIKLYNG 245
Query: 502 DIILTTSSHSLRDTIIAALRSEFSLTDLGPLNYFLGVSATRHSGGLFLHQSK-------- 553
+ L T++H LRD A S L PL F + A G + L +
Sbjct: 246 ENALMTATHVLRDCPNAVAVSAKGLKTRIPLAEFKTI-AKSDKGDVTLLRGPPNWEGLLG 304
Query: 554 -YAEDIISRAKMVNCKPALTPVDTKSKLSA 582
A ++I+ A + CK ++ D +S+
Sbjct: 305 CKAANVITAANLAKCKASIYSFDRDGWVSS 334
>RRPO_BWYVF (P09507) Putative RNA-directed RNA polymerase (EC
2.7.7.48) (ORF 3)
Length = 1035
Score = 33.1 bits (74), Expect = 3.0
Identities = 38/150 (25%), Positives = 59/150 (39%), Gaps = 15/150 (10%)
Query: 447 LLKKSLYGLKQAPRAWYQRFASFLSTIGFVNSKTDHSLFISTH--GDDTTY---ILLYVD 501
LLKKS L+ P+ Y++ + S + TH G+ Y I LY
Sbjct: 186 LLKKSFSALRSTPKCLYEKAIDGFKSFTIPQSPPKSCVIPITHASGNHAGYASCIKLYNG 245
Query: 502 DIILTTSSHSLRDTIIAALRSEFSLTDLGPLNYFLGVSATRHSGGLFLHQSK-------- 553
+ L T++H LRD A S L PL F + A G + L +
Sbjct: 246 ENALMTATHVLRDCPNAVAVSAKGLKTRIPLAEFKTI-AKSDKGDVTLLRGPPNWEGLLG 304
Query: 554 -YAEDIISRAKMVNCKPALTPVDTKSKLSA 582
A ++I+ A + CK ++ D +S+
Sbjct: 305 CKAANVITAANLAKCKASIYSFDRDGWVSS 334
>MUC2_HUMAN (Q02817) Mucin 2 precursor (Intestinal mucin 2)
Length = 5179
Score = 32.3 bits (72), Expect = 5.2
Identities = 35/129 (27%), Positives = 48/129 (37%), Gaps = 9/129 (6%)
Query: 71 PVLTPPHKMEKLNVKLEHSTTR*EPFCFTPTYPLQNGTTPSLW----QPVYTISFPASQT 126
P T P +TT P TP+ P TTPS P+ T + P+S T
Sbjct: 1660 PTTTTPSPPITTTTTPPPTTTPSSPITTTPSPPTTTMTTPSPTTTPSSPITTTTTPSSTT 1719
Query: 127 NLSPPLLLSTSDTRITVH*KPLVVYAI--PTSRRRQLINSMHAPLPVSSSAIRHIIVASS 184
SPP T+ + T P PT+ L + PLP S + +++
Sbjct: 1720 TPSPPPTTMTTPSPTTTPSPPTTTMTTLPPTTTSSPLTTT---PLPPSITPPTFSPFSTT 1776
Query: 185 VSTYPHVRL 193
T P V L
Sbjct: 1777 TPTTPCVPL 1785
>VP40_HHV11 (P10210) Capsid protein P40 (Virion structural protein
UL26) [Contains: UL26.5 protein; Capsid protein VP24 (EC
3.4.21.97) (Assemblin) (Protease); Capsid protein VP22A]
Length = 635
Score = 31.6 bits (70), Expect = 8.8
Identities = 21/66 (31%), Positives = 28/66 (41%), Gaps = 8/66 (12%)
Query: 153 IPTSRRRQLINSMHAPLPVSSSAIRHIIVASSVSTYPHVRLSRPVM*PSMSQSFPYHITH 212
IP S QL+ AP P SA A SV+ PH +SQ +P H+ H
Sbjct: 329 IPASHYNQLVAGHAAPQPQPHSAFGFPAAAGSVAYGPHG--------AGLSQHYPPHVAH 380
Query: 213 LHQPLL 218
+ +L
Sbjct: 381 QYPGVL 386
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.333 0.143 0.459
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 90,959,097
Number of Sequences: 164201
Number of extensions: 3691562
Number of successful extensions: 11695
Number of sequences better than 10.0: 19
Number of HSP's better than 10.0 without gapping: 13
Number of HSP's successfully gapped in prelim test: 6
Number of HSP's that attempted gapping in prelim test: 11636
Number of HSP's gapped (non-prelim): 29
length of query: 813
length of database: 59,974,054
effective HSP length: 119
effective length of query: 694
effective length of database: 40,434,135
effective search space: 28061289690
effective search space used: 28061289690
T: 11
A: 40
X1: 15 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (21.5 bits)
S2: 70 (31.6 bits)
Medicago: description of AC144721.10


