

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144720.4 + phase: 0
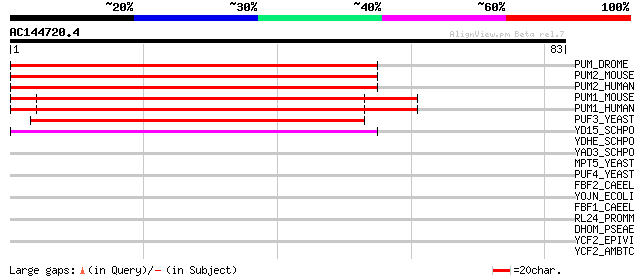
(83 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
PUM_DROME (P25822) Maternal pumilio protein 61 4e-10
PUM2_MOUSE (Q80U58) Pumilio homolog 2 61 6e-10
PUM2_HUMAN (Q8TB72) Pumilio homolog 2 (Pumilio2) 61 6e-10
PUM1_MOUSE (Q80U78) Pumilio homolog 1 60 8e-10
PUM1_HUMAN (Q14671) Pumilio homolog 1 (Pumilio1) (HsPUM) 60 8e-10
PUF3_YEAST (Q07807) PUF3 protein 43 2e-04
YD15_SCHPO (Q10238) Putative regulatory protein C4G9.05 42 2e-04
YDHE_SCHPO (Q92359) Hypothetical protein C6G9.14 in chromosome I 36 0.020
YAD3_SCHPO (Q09829) Hypothetical protein C4G8.03c in chromosome I 34 0.076
MPT5_YEAST (P39016) Suppressor protein MPT5 (HTR1 protein) 33 0.13
PUF4_YEAST (P25339) PUF4 protein 29 1.9
FBF2_CAEEL (Q09312) Fem-3 mRNA binding factor 2 28 3.2
YOJN_ECOLI (P39838) Putative sensor-like histidine kinase yojN (... 28 4.2
FBF1_CAEEL (Q9N5M6) Fem-3 mRNA binding factor 1 28 4.2
RL24_PROMM (Q7V534) 50S ribosomal protein L24 27 7.1
DHOM_PSEAE (P29365) Homoserine dehydrogenase (EC 1.1.1.3) (HDH) 27 7.1
YCF2_EPIVI (P30072) Protein ycf2 27 9.3
YCF2_AMBTC (P61241) Protein ycf2 27 9.3
>PUM_DROME (P25822) Maternal pumilio protein
Length = 1533
Score = 61.2 bits (147), Expect = 4e-10
Identities = 24/55 (43%), Positives = 42/55 (75%)
Query: 1 MQAMMKDPFGNYVVQKVLETCDDQSLELILSRIKVHLNALKRYTYGKHIVSRVEK 55
+ MMKD + NYVVQK+++ + L+ ++++I+ H+ AL++YTYGKHI +++EK
Sbjct: 1372 LHVMMKDQYANYVVQKMIDVSEPTQLKKLMTKIRPHMAALRKYTYGKHINAKLEK 1426
Score = 39.3 bits (90), Expect = 0.002
Identities = 17/49 (34%), Positives = 30/49 (60%)
Query: 5 MKDPFGNYVVQKVLETCDDQSLELILSRIKVHLNALKRYTYGKHIVSRV 53
+KD GN+VVQK +E D +L+ I++ K + +L + YG ++ R+
Sbjct: 1228 VKDQNGNHVVQKCIECVDPVALQFIINAFKGQVYSLSTHPYGCRVIQRI 1276
Score = 34.7 bits (78), Expect = 0.045
Identities = 18/73 (24%), Positives = 38/73 (51%), Gaps = 1/73 (1%)
Query: 3 AMMKDPFGNYVVQKVLETCDDQSLELILSRIKVHLNALKRYTYGKHIVSRVEKLITTGGN 62
++M D FGNYV+QK E + + ++K H+ L YG ++ + + I+
Sbjct: 1154 SLMTDVFGNYVIQKFFEFGTPEQKNTLGMQVKGHVLQLALQMYGCRVIQKALESISPEQQ 1213
Query: 63 ECKLLRYMNSHLL 75
+ +++ ++ H+L
Sbjct: 1214 Q-EIVHELDGHVL 1225
Score = 33.9 bits (76), Expect = 0.076
Identities = 13/59 (22%), Positives = 33/59 (55%)
Query: 2 QAMMKDPFGNYVVQKVLETCDDQSLELILSRIKVHLNALKRYTYGKHIVSRVEKLITTG 60
+ +++D +GNYV+Q VLE + ++++ ++ + L ++ + ++V + T G
Sbjct: 1297 EQLIQDQYGNYVIQHVLEHGKQEDKSILINSVRGKVLVLSQHKFASNVVEKCVTHATRG 1355
Score = 33.1 bits (74), Expect = 0.13
Identities = 14/46 (30%), Positives = 24/46 (51%)
Query: 8 PFGNYVVQKVLETCDDQSLELILSRIKVHLNALKRYTYGKHIVSRV 53
P+G V+Q++LE C + IL + H L + YG +++ V
Sbjct: 1267 PYGCRVIQRILEHCTAEQTTPILDELHEHTEQLIQDQYGNYVIQHV 1312
>PUM2_MOUSE (Q80U58) Pumilio homolog 2
Length = 1066
Score = 60.8 bits (146), Expect = 6e-10
Identities = 23/55 (41%), Positives = 41/55 (73%)
Query: 1 MQAMMKDPFGNYVVQKVLETCDDQSLELILSRIKVHLNALKRYTYGKHIVSRVEK 55
+ MMKD + NYVVQK+++ + ++I+ +I+ H+ L++YTYGKHI++++EK
Sbjct: 992 LYTMMKDQYANYVVQKMIDMAEPAQRKIIMHKIRPHITTLRKYTYGKHILAKLEK 1046
Score = 39.7 bits (91), Expect = 0.001
Identities = 18/49 (36%), Positives = 28/49 (56%)
Query: 5 MKDPFGNYVVQKVLETCDDQSLELILSRIKVHLNALKRYTYGKHIVSRV 53
+KD GN+VVQK +E QSL+ I+ K + L + YG ++ R+
Sbjct: 845 VKDQNGNHVVQKCIECVQPQSLQFIIDAFKGQVFVLSTHPYGCRVIQRI 893
Score = 35.0 bits (79), Expect = 0.034
Identities = 18/57 (31%), Positives = 34/57 (59%), Gaps = 3/57 (5%)
Query: 2 QAMMKDPFGNYVVQKVLETCDDQSLELILSRIKVHLNALKRYTYGKHIVSRVEKLIT 58
+ +++D +GNYV+Q VLE + I+S I+ + AL ++ + ++ VEK +T
Sbjct: 914 EQLVQDQYGNYVIQHVLEHGRPEDKSKIVSEIRGKVLALSQHKFASNV---VEKCVT 967
Score = 35.0 bits (79), Expect = 0.034
Identities = 23/74 (31%), Positives = 41/74 (55%), Gaps = 3/74 (4%)
Query: 4 MMKDPFGNYVVQKVLETCD-DQSLELILSRIKVHLNALKRYTYGKHIVSRVEKLITTGGN 62
+M D FGNYV+QK E DQ L L +RI+ H+ L YG ++ + + I++
Sbjct: 770 LMTDVFGNYVIQKFFEFGSLDQKLAL-ATRIRGHVLPLALQMYGCRVIQKALESISSDQQ 828
Query: 63 E-CKLLRYMNSHLL 75
++++ ++ H+L
Sbjct: 829 VISEMVKELDGHVL 842
Score = 32.7 bits (73), Expect = 0.17
Identities = 14/46 (30%), Positives = 24/46 (51%)
Query: 8 PFGNYVVQKVLETCDDQSLELILSRIKVHLNALKRYTYGKHIVSRV 53
P+G V+Q++LE C + IL + H L + YG +++ V
Sbjct: 884 PYGCRVIQRILEHCTAEQTLPILEELHQHTEQLVQDQYGNYVIQHV 929
>PUM2_HUMAN (Q8TB72) Pumilio homolog 2 (Pumilio2)
Length = 1066
Score = 60.8 bits (146), Expect = 6e-10
Identities = 23/55 (41%), Positives = 41/55 (73%)
Query: 1 MQAMMKDPFGNYVVQKVLETCDDQSLELILSRIKVHLNALKRYTYGKHIVSRVEK 55
+ MMKD + NYVVQK+++ + ++I+ +I+ H+ L++YTYGKHI++++EK
Sbjct: 992 LYTMMKDQYANYVVQKMIDMAEPAQRKIIMHKIRPHITTLRKYTYGKHILAKLEK 1046
Score = 39.7 bits (91), Expect = 0.001
Identities = 18/49 (36%), Positives = 28/49 (56%)
Query: 5 MKDPFGNYVVQKVLETCDDQSLELILSRIKVHLNALKRYTYGKHIVSRV 53
+KD GN+VVQK +E QSL+ I+ K + L + YG ++ R+
Sbjct: 845 VKDQNGNHVVQKCIECVQPQSLQFIIDAFKGQVFVLSTHPYGCRVIQRI 893
Score = 35.0 bits (79), Expect = 0.034
Identities = 18/57 (31%), Positives = 34/57 (59%), Gaps = 3/57 (5%)
Query: 2 QAMMKDPFGNYVVQKVLETCDDQSLELILSRIKVHLNALKRYTYGKHIVSRVEKLIT 58
+ +++D +GNYV+Q VLE + I+S I+ + AL ++ + ++ VEK +T
Sbjct: 914 EQLVQDQYGNYVIQHVLEHGRPEDKSKIVSEIRGKVLALSQHKFASNV---VEKCVT 967
Score = 35.0 bits (79), Expect = 0.034
Identities = 23/74 (31%), Positives = 41/74 (55%), Gaps = 3/74 (4%)
Query: 4 MMKDPFGNYVVQKVLETCD-DQSLELILSRIKVHLNALKRYTYGKHIVSRVEKLITTGGN 62
+M D FGNYV+QK E DQ L L +RI+ H+ L YG ++ + + I++
Sbjct: 770 LMTDVFGNYVIQKFFEFGSLDQKLAL-ATRIRGHVLPLALQMYGCRVIQKALESISSDQQ 828
Query: 63 E-CKLLRYMNSHLL 75
++++ ++ H+L
Sbjct: 829 VISEMVKELDGHVL 842
Score = 32.7 bits (73), Expect = 0.17
Identities = 14/46 (30%), Positives = 24/46 (51%)
Query: 8 PFGNYVVQKVLETCDDQSLELILSRIKVHLNALKRYTYGKHIVSRV 53
P+G V+Q++LE C + IL + H L + YG +++ V
Sbjct: 884 PYGCRVIQRILEHCTAEQTLPILEELHQHTEQLVQDQYGNYVIQHV 929
>PUM1_MOUSE (Q80U78) Pumilio homolog 1
Length = 1189
Score = 60.5 bits (145), Expect = 8e-10
Identities = 23/61 (37%), Positives = 42/61 (68%)
Query: 1 MQAMMKDPFGNYVVQKVLETCDDQSLELILSRIKVHLNALKRYTYGKHIVSRVEKLITTG 60
+ MMKD + NYVVQK+++ + ++++ +I+ H+ L++YTYGKHI++++EK
Sbjct: 1115 LYTMMKDQYANYVVQKMIDVAEPGQRKIVMHKIRPHIATLRKYTYGKHILAKLEKYYMKN 1174
Query: 61 G 61
G
Sbjct: 1175 G 1175
Score = 41.2 bits (95), Expect = 5e-04
Identities = 19/49 (38%), Positives = 29/49 (58%)
Query: 5 MKDPFGNYVVQKVLETCDDQSLELILSRIKVHLNALKRYTYGKHIVSRV 53
+KD GN+VVQK +E QSL+ I+ K + AL + YG ++ R+
Sbjct: 968 VKDQNGNHVVQKCIECVQPQSLQFIIDAFKGQVFALSTHPYGCRVIQRI 1016
Score = 37.4 bits (85), Expect = 0.007
Identities = 20/73 (27%), Positives = 38/73 (51%), Gaps = 1/73 (1%)
Query: 4 MMKDPFGNYVVQKVLETCDDQSLELILSRIKVHLNALKRYTYGKHIVSRVEKLITTGGNE 63
+M D FGNYV+QK E + + RI+ H+ +L YG ++ + + I +
Sbjct: 893 LMVDVFGNYVIQKFFEFGSHEQKLALAERIRGHVLSLALQMYGCRVIQKALEFIPSDQQV 952
Query: 64 C-KLLRYMNSHLL 75
+++R ++ H+L
Sbjct: 953 INEMVRELDGHVL 965
Score = 33.1 bits (74), Expect = 0.13
Identities = 18/52 (34%), Positives = 29/52 (55%), Gaps = 2/52 (3%)
Query: 3 AMMKDPFGNYVVQKVLETC-DDQSLELILSRIKVHLNALKRYTYGKHIVSRV 53
A+ P+G V+Q++LE C DQ+L IL + H L + YG +++ V
Sbjct: 1002 ALSTHPYGCRVIQRILEHCLPDQTLP-ILEELHQHTEQLVQDQYGNYVIQHV 1052
Score = 33.1 bits (74), Expect = 0.13
Identities = 16/57 (28%), Positives = 34/57 (59%), Gaps = 3/57 (5%)
Query: 2 QAMMKDPFGNYVVQKVLETCDDQSLELILSRIKVHLNALKRYTYGKHIVSRVEKLIT 58
+ +++D +GNYV+Q VLE + I++ I+ ++ L ++ + ++ VEK +T
Sbjct: 1037 EQLVQDQYGNYVIQHVLEHGRPEDKSKIVAEIRGNVLVLSQHKFASNV---VEKCVT 1090
>PUM1_HUMAN (Q14671) Pumilio homolog 1 (Pumilio1) (HsPUM)
Length = 1186
Score = 60.5 bits (145), Expect = 8e-10
Identities = 23/61 (37%), Positives = 42/61 (68%)
Query: 1 MQAMMKDPFGNYVVQKVLETCDDQSLELILSRIKVHLNALKRYTYGKHIVSRVEKLITTG 60
+ MMKD + NYVVQK+++ + ++++ +I+ H+ L++YTYGKHI++++EK
Sbjct: 1112 LYTMMKDQYANYVVQKMIDVAEPGQRKIVMHKIRPHIATLRKYTYGKHILAKLEKYYMKN 1171
Query: 61 G 61
G
Sbjct: 1172 G 1172
Score = 41.2 bits (95), Expect = 5e-04
Identities = 19/49 (38%), Positives = 29/49 (58%)
Query: 5 MKDPFGNYVVQKVLETCDDQSLELILSRIKVHLNALKRYTYGKHIVSRV 53
+KD GN+VVQK +E QSL+ I+ K + AL + YG ++ R+
Sbjct: 965 VKDQNGNHVVQKCIECVQPQSLQFIIDAFKGQVFALSTHPYGCRVIQRI 1013
Score = 37.7 bits (86), Expect = 0.005
Identities = 20/72 (27%), Positives = 39/72 (53%), Gaps = 1/72 (1%)
Query: 4 MMKDPFGNYVVQKVLETCDDQSLELILSRIKVHLNALKRYTYGKHIVSRVEKLITTGGNE 63
+M D FGNYV+QK E + + RI+ H+ +L YG ++ + + I + +
Sbjct: 892 LMVDVFGNYVIQKFFEFGSLEQKLALAERIRGHVLSLALQMYGCRVIQKALEFIPS-DQQ 950
Query: 64 CKLLRYMNSHLL 75
+++R ++ H+L
Sbjct: 951 NEMVRELDGHVL 962
Score = 33.1 bits (74), Expect = 0.13
Identities = 18/52 (34%), Positives = 29/52 (55%), Gaps = 2/52 (3%)
Query: 3 AMMKDPFGNYVVQKVLETC-DDQSLELILSRIKVHLNALKRYTYGKHIVSRV 53
A+ P+G V+Q++LE C DQ+L IL + H L + YG +++ V
Sbjct: 999 ALSTHPYGCRVIQRILEHCLPDQTLP-ILEELHQHTEQLVQDQYGNYVIQHV 1049
Score = 33.1 bits (74), Expect = 0.13
Identities = 16/57 (28%), Positives = 34/57 (59%), Gaps = 3/57 (5%)
Query: 2 QAMMKDPFGNYVVQKVLETCDDQSLELILSRIKVHLNALKRYTYGKHIVSRVEKLIT 58
+ +++D +GNYV+Q VLE + I++ I+ ++ L ++ + ++ VEK +T
Sbjct: 1034 EQLVQDQYGNYVIQHVLEHGRPEDKSKIVAEIRGNVLVLSQHKFASNV---VEKCVT 1087
>PUF3_YEAST (Q07807) PUF3 protein
Length = 879
Score = 42.7 bits (99), Expect = 2e-04
Identities = 19/50 (38%), Positives = 31/50 (62%)
Query: 4 MMKDPFGNYVVQKVLETCDDQSLELILSRIKVHLNALKRYTYGKHIVSRV 53
M+KD GN+V+QK +ET + L ILS + H+ L ++YG ++ R+
Sbjct: 654 MIKDQNGNHVIQKAIETIPIEKLPFILSSLTGHIYHLSTHSYGCRVIQRL 703
Score = 39.3 bits (90), Expect = 0.002
Identities = 21/57 (36%), Positives = 35/57 (60%), Gaps = 1/57 (1%)
Query: 1 MQAMMKDPFGNYVVQKVLETCDDQSLELILSRIKVHLNAL-KRYTYGKHIVSRVEKL 56
M M+KD F NYV+QK++ + + +LI+ I+ +L+ L K + G ++ VEKL
Sbjct: 814 MILMIKDQFANYVIQKLVNVSEGEGKKLIVIAIRAYLDKLNKSNSLGNRHLASVEKL 870
Score = 28.9 bits (63), Expect = 2.5
Identities = 23/83 (27%), Positives = 40/83 (47%), Gaps = 9/83 (10%)
Query: 4 MMKDPFGNYVVQKVLETCDDQSLELILSR---IKVHLNALKRYTYGKHIVSRVEKLITTG 60
+++D +GNYV+Q VL+ + E++ + I+ N + Y+ K + VEK I G
Sbjct: 726 LIQDQYGNYVIQYVLQQDQFTNKEMVDIKQEIIETVANNVVEYSKHKFASNVVEKSILYG 785
Query: 61 GNE------CKLLRYMNSHLLSL 77
K+L +H L+L
Sbjct: 786 SKNQKDLIISKILPRDKNHALNL 808
Score = 27.7 bits (60), Expect = 5.5
Identities = 13/54 (24%), Positives = 24/54 (44%)
Query: 4 MMKDPFGNYVVQKVLETCDDQSLELILSRIKVHLNALKRYTYGKHIVSRVEKLI 57
+ D FGNYV+QK E ++ + K ++ L Y ++ + + I
Sbjct: 582 LSNDVFGNYVIQKFFEFGSKIQKNTLVDQFKGNMKQLSLQMYACRVIQKALEYI 635
>YD15_SCHPO (Q10238) Putative regulatory protein C4G9.05
Length = 581
Score = 42.4 bits (98), Expect = 2e-04
Identities = 17/55 (30%), Positives = 33/55 (59%)
Query: 1 MQAMMKDPFGNYVVQKVLETCDDQSLELILSRIKVHLNALKRYTYGKHIVSRVEK 55
+Q + + +GNY+VQ +L+ + L+ +K H+ +L+ + YG+ I + VEK
Sbjct: 524 LQEIASNQYGNYIVQYLLQVATPSQINLMAEHLKKHMVSLRGHKYGQRIAALVEK 578
Score = 35.0 bits (79), Expect = 0.034
Identities = 14/49 (28%), Positives = 27/49 (54%)
Query: 4 MMKDPFGNYVVQKVLETCDDQSLELILSRIKVHLNALKRYTYGKHIVSR 52
+MK+ FGN+++QK E + L+ +K H+ L +G H++ +
Sbjct: 299 LMKNKFGNFLIQKCFEYSTEAQLQSFSYFLKKHVKELSIDAFGSHVLQK 347
>YDHE_SCHPO (Q92359) Hypothetical protein C6G9.14 in chromosome I
Length = 681
Score = 35.8 bits (81), Expect = 0.020
Identities = 15/52 (28%), Positives = 31/52 (58%)
Query: 4 MMKDPFGNYVVQKVLETCDDQSLELILSRIKVHLNALKRYTYGKHIVSRVEK 55
+++D F NYV+Q L+ + ++ RIK + ++K G+ I+S++E+
Sbjct: 606 LLRDSFANYVIQTALDNASVKQRAELVERIKPLIPSIKNTPCGRRILSKLER 657
Score = 32.7 bits (73), Expect = 0.17
Identities = 15/49 (30%), Positives = 27/49 (54%)
Query: 4 MMKDPFGNYVVQKVLETCDDQSLELILSRIKVHLNALKRYTYGKHIVSR 52
+++D FGNYV+Q VLE + E I+S + AL + +++ +
Sbjct: 532 LVQDAFGNYVLQYVLELNNPNHTEAIISYFLYKVRALSTQKFSSNVMEK 580
Score = 27.7 bits (60), Expect = 5.5
Identities = 13/59 (22%), Positives = 27/59 (45%)
Query: 1 MQAMMKDPFGNYVVQKVLETCDDQSLELILSRIKVHLNALKRYTYGKHIVSRVEKLITT 59
+ +M D FGNY+ QK+ E + + I L + +G + ++ L+++
Sbjct: 384 LAVLMVDAFGNYLCQKLFEHASEAQRSTFIQIIAPKLVPISFNMHGTRALQKIIDLVSS 442
>YAD3_SCHPO (Q09829) Hypothetical protein C4G8.03c in chromosome I
Length = 780
Score = 33.9 bits (76), Expect = 0.076
Identities = 14/53 (26%), Positives = 31/53 (58%)
Query: 1 MQAMMKDPFGNYVVQKVLETCDDQSLELILSRIKVHLNALKRYTYGKHIVSRV 53
++ ++ D + NYV+Q+ L D+ LIL I + ++ +G+HI++++
Sbjct: 722 IEQLLDDCYANYVMQRFLNVADESQKFLILRSISHVIPKIQNTRHGRHILAKL 774
Score = 29.6 bits (65), Expect = 1.4
Identities = 19/78 (24%), Positives = 35/78 (44%)
Query: 4 MMKDPFGNYVVQKVLETCDDQSLELILSRIKVHLNALKRYTYGKHIVSRVEKLITTGGNE 63
+M DPFGNY+ QK+ + +L+ I + + YG + + +T+
Sbjct: 506 LMIDPFGNYMCQKLFVYASREQKLSMLNGIGEGIVDICSNLYGTRSMQNIIDKLTSNEQI 565
Query: 64 CKLLRYMNSHLLSLNCSD 81
LL+ + L +L C +
Sbjct: 566 SLLLKIIIPSLTTLACDN 583
>MPT5_YEAST (P39016) Suppressor protein MPT5 (HTR1 protein)
Length = 834
Score = 33.1 bits (74), Expect = 0.13
Identities = 18/63 (28%), Positives = 34/63 (53%), Gaps = 3/63 (4%)
Query: 4 MMKDPFGNYVVQKVLETCDDQSLELILSRIKVHLNALKRYTYGKHIVSRVEKLITTGGNE 63
++ DPFGNY+VQK+ + + L++ I ++ + YG ++K+I T NE
Sbjct: 231 LILDPFGNYLVQKLCDYLTAEQKTLLIQTIYPNVFQISINQYG---TRSLQKIIDTVDNE 287
Query: 64 CKL 66
++
Sbjct: 288 VQI 290
>PUF4_YEAST (P25339) PUF4 protein
Length = 888
Score = 29.3 bits (64), Expect = 1.9
Identities = 14/56 (25%), Positives = 27/56 (48%)
Query: 4 MMKDPFGNYVVQKVLETCDDQSLELILSRIKVHLNALKRYTYGKHIVSRVEKLITT 59
+M D FGNY++QK+LE + ++ H + +G + ++ + I T
Sbjct: 607 LMTDSFGNYLIQKLLEEVTTEQRIVLTKISSPHFVEISLNPHGTRALQKLIECIKT 662
Score = 27.7 bits (60), Expect = 5.5
Identities = 18/70 (25%), Positives = 36/70 (50%), Gaps = 6/70 (8%)
Query: 7 DPFGNYVVQKVLETCDDQS----LELILSRIKVHLNALKRYTYGKHIVSRVEKLITTGGN 62
DPFGNYVVQ ++ +++ I+ +K L + +G +++ ++ K T +
Sbjct: 755 DPFGNYVVQYIITKEAEKNKYDYTHKIVHLLKPRAIELSIHKFGSNVIEKILK--TAIVS 812
Query: 63 ECKLLRYMNS 72
E +L +N+
Sbjct: 813 EPMILEILNN 822
Score = 26.9 bits (58), Expect = 9.3
Identities = 17/54 (31%), Positives = 29/54 (53%), Gaps = 4/54 (7%)
Query: 1 MQAMMKDPFGNYVVQKVLETCDDQS--LELILSRIKVHL--NALKRYTYGKHIV 50
+Q+++ D +GNYV+Q L+ Q+ L LS I L ++ +GK I+
Sbjct: 828 IQSLLNDSYGNYVLQTALDISHKQNDYLYKRLSEIVAPLLVGPIRNTPHGKRII 881
>FBF2_CAEEL (Q09312) Fem-3 mRNA binding factor 2
Length = 632
Score = 28.5 bits (62), Expect = 3.2
Identities = 21/79 (26%), Positives = 33/79 (41%), Gaps = 20/79 (25%)
Query: 1 MQAMMKDPFGNYVVQKVLETCDD--------------------QSLELILSRIKVHLNAL 40
+ MM FGNYVVQ +L C D L+ + SR+ + L
Sbjct: 490 LDIMMFHQFGNYVVQCMLTICCDAVSGRRQTKEGGYDHAISFQDWLKKLHSRVTKERHRL 549
Query: 41 KRYTYGKHIVSRVEKLITT 59
R++ GK ++ + L +T
Sbjct: 550 SRFSSGKKMIETLANLRST 568
>YOJN_ECOLI (P39838) Putative sensor-like histidine kinase yojN (EC
2.7.3.-)
Length = 890
Score = 28.1 bits (61), Expect = 4.2
Identities = 24/79 (30%), Positives = 42/79 (52%), Gaps = 7/79 (8%)
Query: 1 MQAMMKDPFGNYVVQKVLETCDDQSLELILSRIKVHLNALKRYTYGKHIVSRVEKLI--T 58
M+ + + P+G + L+T + L L+L+ I + AL YT +H SR + + T
Sbjct: 290 MRLVWQVPYGTLL----LDTLQNILLPLLLN-IGLLALALFGYTTFRHFSSRSTENVPST 344
Query: 59 TGGNECKLLRYMNSHLLSL 77
NE ++LR +N ++SL
Sbjct: 345 AVNNELRILRAINEEIVSL 363
>FBF1_CAEEL (Q9N5M6) Fem-3 mRNA binding factor 1
Length = 614
Score = 28.1 bits (61), Expect = 4.2
Identities = 21/79 (26%), Positives = 33/79 (41%), Gaps = 20/79 (25%)
Query: 1 MQAMMKDPFGNYVVQKVLETCDDQS--------------------LELILSRIKVHLNAL 40
+ MM FGNYVVQ +L C D L+ + SR+ + L
Sbjct: 488 LDIMMFHQFGNYVVQCMLTICCDAVSGRRQTKEGSYDHANSFQVWLKKLHSRVTKERHRL 547
Query: 41 KRYTYGKHIVSRVEKLITT 59
R++ GK ++ + L +T
Sbjct: 548 SRFSSGKKMIETLAHLRST 566
>RL24_PROMM (Q7V534) 50S ribosomal protein L24
Length = 118
Score = 27.3 bits (59), Expect = 7.1
Identities = 15/58 (25%), Positives = 29/58 (49%)
Query: 11 NYVVQKVLETCDDQSLELILSRIKVHLNALKRYTYGKHIVSRVEKLITTGGNECKLLR 68
N + V T + +S ++ VH + + Y+ K I SRVE ++ G++ + L+
Sbjct: 54 NIRTRHVKPTQEGESGRIVTEEASVHASNVMLYSNKKKIASRVELVVEKDGSKKRRLK 111
>DHOM_PSEAE (P29365) Homoserine dehydrogenase (EC 1.1.1.3) (HDH)
Length = 434
Score = 27.3 bits (59), Expect = 7.1
Identities = 16/41 (39%), Positives = 20/41 (48%)
Query: 23 DQSLELILSRIKVHLNALKRYTYGKHIVSRVEKLITTGGNE 63
D +ELI H LK GKH+V+ + LI GNE
Sbjct: 72 DVVVELIGGYTLAHELVLKAIENGKHVVTANKALIAVHGNE 112
>YCF2_EPIVI (P30072) Protein ycf2
Length = 2216
Score = 26.9 bits (58), Expect = 9.3
Identities = 18/39 (46%), Positives = 22/39 (56%), Gaps = 1/39 (2%)
Query: 23 DQSLELILSRIKVHLNALKRYTYGKHIVSRVEKLITTGG 61
+Q LE I +I VHL K + G H SR KL+ TGG
Sbjct: 861 NQFLEPIDLKI-VHLKKRKPFLLGYHGTSRKLKLLITGG 898
>YCF2_AMBTC (P61241) Protein ycf2
Length = 2304
Score = 26.9 bits (58), Expect = 9.3
Identities = 17/39 (43%), Positives = 22/39 (55%), Gaps = 1/39 (2%)
Query: 23 DQSLELILSRIKVHLNALKRYTYGKHIVSRVEKLITTGG 61
+Q LE I +I VHLN LK + H S+ KL+ GG
Sbjct: 913 NQLLESIGVQI-VHLNKLKPFLLDDHDTSQRSKLLINGG 950
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.322 0.136 0.393
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 9,033,603
Number of Sequences: 164201
Number of extensions: 274812
Number of successful extensions: 675
Number of sequences better than 10.0: 18
Number of HSP's better than 10.0 without gapping: 13
Number of HSP's successfully gapped in prelim test: 5
Number of HSP's that attempted gapping in prelim test: 621
Number of HSP's gapped (non-prelim): 59
length of query: 83
length of database: 59,974,054
effective HSP length: 59
effective length of query: 24
effective length of database: 50,286,195
effective search space: 1206868680
effective search space used: 1206868680
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 58 (26.9 bits)
Medicago: description of AC144720.4


