

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144720.3 + phase: 0 /pseudo
(949 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
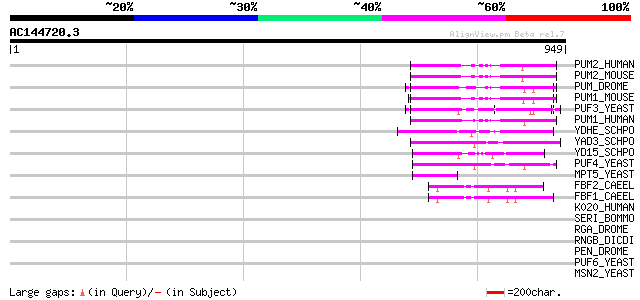
Score E
Sequences producing significant alignments: (bits) Value
PUM2_HUMAN (Q8TB72) Pumilio homolog 2 (Pumilio2) 139 3e-32
PUM2_MOUSE (Q80U58) Pumilio homolog 2 139 5e-32
PUM_DROME (P25822) Maternal pumilio protein 131 1e-29
PUM1_MOUSE (Q80U78) Pumilio homolog 1 128 6e-29
PUF3_YEAST (Q07807) PUF3 protein 127 2e-28
PUM1_HUMAN (Q14671) Pumilio homolog 1 (Pumilio1) (HsPUM) 125 5e-28
YDHE_SCHPO (Q92359) Hypothetical protein C6G9.14 in chromosome I 107 1e-22
YAD3_SCHPO (Q09829) Hypothetical protein C4G8.03c in chromosome I 89 7e-17
YD15_SCHPO (Q10238) Putative regulatory protein C4G9.05 82 5e-15
PUF4_YEAST (P25339) PUF4 protein 73 3e-12
MPT5_YEAST (P39016) Suppressor protein MPT5 (HTR1 protein) 52 7e-06
FBF2_CAEEL (Q09312) Fem-3 mRNA binding factor 2 51 1e-05
FBF1_CAEEL (Q9N5M6) Fem-3 mRNA binding factor 1 50 4e-05
K020_HUMAN (Q15397) Protein KIAA0020 40 0.023
SERI_BOMMO (P07856) Sericin precursor (Silk gum protein) 39 0.065
RGA_DROME (Q94547) Regulator of gene activity (Regena protein) 39 0.065
RNGB_DICDI (Q7M3S9) Ring finger protein B (Protein rngB) 39 0.086
PEN_DROME (O61345) Penguin protein 38 0.11
PUF6_YEAST (Q04373) PUF6 protein 37 0.25
MSN2_YEAST (P33748) Zinc finger protein MSN2 (Multicopy suppress... 37 0.25
>PUM2_HUMAN (Q8TB72) Pumilio homolog 2 (Pumilio2)
Length = 1066
Score = 139 bits (351), Expect = 3e-32
Identities = 98/286 (34%), Positives = 141/286 (49%), Gaps = 74/286 (25%)
Query: 685 YSSGFIQQKLETASVEEKTKIFPEILPHARALMTDVFGNYVIQKFFEHGTDSQRKELANQ 744
+ S FIQQKLE A+ E+ +F EIL A LMTDVFGNYVIQKFFE G+ Q+ LA +
Sbjct: 739 HGSRFIQQKLERATPAERQMVFNEILQAAYQLMTDVFGNYVIQKFFEFGSLDQKLALATR 798
Query: 745 LTGHVLPLSLQMYGCRVIQKLLLNLLTGLGGG*C*SAEPNGVRAQWCNNEMCT*PKWKPC 804
+ GHVLPL+LQMYGCRVIQK L ++ + Q +EM K
Sbjct: 799 IRGHVLPLALQMYGCRVIQKALESISSD----------------QQVISEMV-----KEL 837
Query: 805 YSEVYRVCPSE*NSVYHHVFLWASCCTFYSSLWVQSYSDCDDLKTQEIIMEEIMQSVCTL 864
V + C + N +HV VQ +C ++ + I++ V L
Sbjct: 838 DGHVLK-CVKDQNG--NHV--------------VQKCIECVQPQSLQFIIDAFKGQVFVL 880
Query: 865 AQDQYGNYVIQ------------------------------------HILEHGKPNERTI 888
+ YG VIQ H+LEHG+P +++
Sbjct: 881 STHPYGCRVIQRILEHCTAEQTLPILEELHQHTEQLVQDQYGNYVIQHVLEHGRPEDKSK 940
Query: 889 VISKLAGQIVKMSQQKFASNVIEKCLAFGSPEERQILVNEMLGTSD 934
++S++ G+++ +SQ KFASNV+EKC+ S ER +L++E+ +D
Sbjct: 941 IVSEIRGKVLALSQHKFASNVVEKCVTHASRAERALLIDEVCCQND 986
Score = 43.1 bits (100), Expect = 0.003
Identities = 21/78 (26%), Positives = 41/78 (51%)
Query: 854 MEEIMQSVCTLAQDQYGNYVIQHILEHGKPNERTIVISKLAGQIVKMSQQKFASNVIEKC 913
+ +++ + +QDQ+G+ IQ LE P ER +V +++ ++ F + VI+K
Sbjct: 724 LRDLIGHIVEFSQDQHGSRFIQQKLERATPAERQMVFNEILQAAYQLMTDVFGNYVIQKF 783
Query: 914 LAFGSPEERQILVNEMLG 931
FGS +++ L + G
Sbjct: 784 FEFGSLDQKLALATRIRG 801
Score = 42.4 bits (98), Expect = 0.006
Identities = 23/88 (26%), Positives = 45/88 (51%), Gaps = 7/88 (7%)
Query: 685 YSSGFIQQKLETASVEEKTKIFPEIL-----PHAR--ALMTDVFGNYVIQKFFEHGTDSQ 737
++S +++ + AS E+ + E+ PH+ +M D + NYV+QK + +Q
Sbjct: 957 FASNVVEKCVTHASRAERALLIDEVCCQNDGPHSALYTMMKDQYANYVVQKMIDMAEPAQ 1016
Query: 738 RKELANQLTGHVLPLSLQMYGCRVIQKL 765
RK + +++ H+ L YG ++ KL
Sbjct: 1017 RKIIMHKIRPHITTLRKYTYGKHILAKL 1044
>PUM2_MOUSE (Q80U58) Pumilio homolog 2
Length = 1066
Score = 139 bits (349), Expect = 5e-32
Identities = 98/286 (34%), Positives = 141/286 (49%), Gaps = 74/286 (25%)
Query: 685 YSSGFIQQKLETASVEEKTKIFPEILPHARALMTDVFGNYVIQKFFEHGTDSQRKELANQ 744
+ S FIQQKLE A+ E+ +F EIL A LMTDVFGNYVIQKFFE G+ Q+ LA +
Sbjct: 739 HGSRFIQQKLERATPAERQIVFNEILQAAYQLMTDVFGNYVIQKFFEFGSLDQKLALATR 798
Query: 745 LTGHVLPLSLQMYGCRVIQKLLLNLLTGLGGG*C*SAEPNGVRAQWCNNEMCT*PKWKPC 804
+ GHVLPL+LQMYGCRVIQK L ++ + Q +EM K
Sbjct: 799 IRGHVLPLALQMYGCRVIQKALESISSD----------------QQVISEMV-----KEL 837
Query: 805 YSEVYRVCPSE*NSVYHHVFLWASCCTFYSSLWVQSYSDCDDLKTQEIIMEEIMQSVCTL 864
V + C + N +HV VQ +C ++ + I++ V L
Sbjct: 838 DGHVLK-CVKDQNG--NHV--------------VQKCIECVQPQSLQFIIDAFKGQVFVL 880
Query: 865 AQDQYGNYVIQ------------------------------------HILEHGKPNERTI 888
+ YG VIQ H+LEHG+P +++
Sbjct: 881 STHPYGCRVIQRILEHCTAEQTLPILEELHQHTEQLVQDQYGNYVIQHVLEHGRPEDKSK 940
Query: 889 VISKLAGQIVKMSQQKFASNVIEKCLAFGSPEERQILVNEMLGTSD 934
++S++ G+++ +SQ KFASNV+EKC+ S ER +L++E+ +D
Sbjct: 941 IVSEIRGKVLALSQHKFASNVVEKCVTHASRAERALLIDEVCCQND 986
Score = 44.3 bits (103), Expect = 0.002
Identities = 22/78 (28%), Positives = 41/78 (52%)
Query: 854 MEEIMQSVCTLAQDQYGNYVIQHILEHGKPNERTIVISKLAGQIVKMSQQKFASNVIEKC 913
+ +++ + +QDQ+G+ IQ LE P ER IV +++ ++ F + VI+K
Sbjct: 724 LRDLIGHIVEFSQDQHGSRFIQQKLERATPAERQIVFNEILQAAYQLMTDVFGNYVIQKF 783
Query: 914 LAFGSPEERQILVNEMLG 931
FGS +++ L + G
Sbjct: 784 FEFGSLDQKLALATRIRG 801
Score = 42.4 bits (98), Expect = 0.006
Identities = 23/88 (26%), Positives = 45/88 (51%), Gaps = 7/88 (7%)
Query: 685 YSSGFIQQKLETASVEEKTKIFPEIL-----PHAR--ALMTDVFGNYVIQKFFEHGTDSQ 737
++S +++ + AS E+ + E+ PH+ +M D + NYV+QK + +Q
Sbjct: 957 FASNVVEKCVTHASRAERALLIDEVCCQNDGPHSALYTMMKDQYANYVVQKMIDMAEPAQ 1016
Query: 738 RKELANQLTGHVLPLSLQMYGCRVIQKL 765
RK + +++ H+ L YG ++ KL
Sbjct: 1017 RKIIMHKIRPHITTLRKYTYGKHILAKL 1044
>PUM_DROME (P25822) Maternal pumilio protein
Length = 1533
Score = 131 bits (329), Expect = 1e-29
Identities = 97/286 (33%), Positives = 132/286 (45%), Gaps = 76/286 (26%)
Query: 685 YSSGFIQQKLETASVEEKTKIFPEILPHARALMTDVFGNYVIQKFFEHGTDSQRKELANQ 744
+ S FIQQKLE A+ EK +F EIL A +LMTDVFGNYVIQKFFE GT Q+ L Q
Sbjct: 1124 HGSRFIQQKLERATAAEKQMVFSEILAAAYSLMTDVFGNYVIQKFFEFGTPEQKNTLGMQ 1183
Query: 745 LTGHVLPLSLQMYGCRVIQKLLLNLLTGLGGG*C*SAEPNGVRAQWCNNEMCT*PKWKPC 804
+ GHVL L+LQMYGCRVIQK L ++ S E + +
Sbjct: 1184 VKGHVLQLALQMYGCRVIQKALESI----------SPEQQQEIVHELDGHVLK------- 1226
Query: 805 YSEVYRVCPSE*NSVYHHVFLWASCCTFYSSLWVQSYSDCDDLKTQEIIMEEIMQSVCTL 864
C + N +HV VQ +C D + I+ V +L
Sbjct: 1227 -------CVKDQNG--NHV--------------VQKCIECVDPVALQFIINAFKGQVYSL 1263
Query: 865 AQDQYGNYVIQHIL------------------------------------EHGKPNERTI 888
+ YG VIQ IL EHGK +++I
Sbjct: 1264 STHPYGCRVIQRILEHCTAEQTTPILDELHEHTEQLIQDQYGNYVIQHVLEHGKQEDKSI 1323
Query: 889 VISKLAGQIVKMSQQKFASNVIEKCLAFGSPEERQILVNEMLGTSD 934
+I+ + G+++ +SQ KFASNV+EKC+ + ER L++E+ +D
Sbjct: 1324 LINSVRGKVLVLSQHKFASNVVEKCVTHATRGERTGLIDEVCTFND 1369
Score = 60.8 bits (146), Expect = 2e-08
Identities = 57/256 (22%), Positives = 105/256 (40%), Gaps = 45/256 (17%)
Query: 678 QISLIMWYSSGFIQQKLETASVEEKTKIFPEILPHARALMTDVFGNYVIQKFFEHGTDSQ 737
Q++L M Y IQ+ LE+ S E++ +I E+ H + D GN+V+QK E
Sbjct: 1190 QLALQM-YGCRVIQKALESISPEQQQEIVHELDGHVLKCVKDQNGNHVVQKCIECVDPVA 1248
Query: 738 RKELANQLTGHVLPLSLQMYGCRVIQKLLLNLLTGLGGG*C*SAEPNGVRAQWCNNEMCT 797
+ + N G V LS YGCRVIQ++L E CT
Sbjct: 1249 LQFIINAFKGQVYSLSTHPYGCRVIQRIL---------------------------EHCT 1281
Query: 798 *PKWKPCYSEVYRVCPSE*NSVYHHVFLWASCCTFYSSLWVQSYSDCDDLKTQEIIMEEI 857
+ P E+ H Y + +Q + + + I++ +
Sbjct: 1282 AEQTTPILDEL-------------HEHTEQLIQDQYGNYVIQHVLEHGKQEDKSILINSV 1328
Query: 858 MQSVCTLAQDQYGNYVIQHILEHGKPNERTIVISKLA----GQIVKMSQQKFASNVIEKC 913
V L+Q ++ + V++ + H ERT +I ++ + M + ++A+ V++K
Sbjct: 1329 RGKVLVLSQHKFASNVVEKCVTHATRGERTGLIDEVCTFNDNALHVMMKDQYANYVVQKM 1388
Query: 914 LAFGSPEERQILVNEM 929
+ P + + L+ ++
Sbjct: 1389 IDVSEPTQLKKLMTKI 1404
Score = 43.9 bits (102), Expect = 0.002
Identities = 21/78 (26%), Positives = 40/78 (50%)
Query: 854 MEEIMQSVCTLAQDQYGNYVIQHILEHGKPNERTIVISKLAGQIVKMSQQKFASNVIEKC 913
+ ++ + +QDQ+G+ IQ LE E+ +V S++ + F + VI+K
Sbjct: 1109 LRDLANHIVEFSQDQHGSRFIQQKLERATAAEKQMVFSEILAAAYSLMTDVFGNYVIQKF 1168
Query: 914 LAFGSPEERQILVNEMLG 931
FG+PE++ L ++ G
Sbjct: 1169 FEFGTPEQKNTLGMQVKG 1186
Score = 37.4 bits (85), Expect = 0.19
Identities = 21/85 (24%), Positives = 41/85 (47%), Gaps = 4/85 (4%)
Query: 685 YSSGFIQQKLETASVEEKTKIFPEILPH----ARALMTDVFGNYVIQKFFEHGTDSQRKE 740
++S +++ + A+ E+T + E+ +M D + NYV+QK + +Q K+
Sbjct: 1340 FASNVVEKCVTHATRGERTGLIDEVCTFNDNALHVMMKDQYANYVVQKMIDVSEPTQLKK 1399
Query: 741 LANQLTGHVLPLSLQMYGCRVIQKL 765
L ++ H+ L YG + KL
Sbjct: 1400 LMTKIRPHMAALRKYTYGKHINAKL 1424
>PUM1_MOUSE (Q80U78) Pumilio homolog 1
Length = 1189
Score = 128 bits (322), Expect = 6e-29
Identities = 94/286 (32%), Positives = 135/286 (46%), Gaps = 74/286 (25%)
Query: 685 YSSGFIQQKLETASVEEKTKIFPEILPHARALMTDVFGNYVIQKFFEHGTDSQRKELANQ 744
+ S FIQ KLE A+ E+ +F EIL A LM DVFGNYVIQKFFE G+ Q+ LA +
Sbjct: 862 HGSRFIQLKLERATAAERQLVFNEILQAAYQLMVDVFGNYVIQKFFEFGSHEQKLALAER 921
Query: 745 LTGHVLPLSLQMYGCRVIQKLLLNLLTGLGGG*C*SAEPNGVRAQWCNNEMCT*PKWKPC 804
+ GHVL L+LQMYGCRVIQK L + + Q NEM +
Sbjct: 922 IRGHVLSLALQMYGCRVIQKALEFIPSD----------------QQVINEMV-----REL 960
Query: 805 YSEVYRVCPSE*NSVYHHVFLWASCCTFYSSLWVQSYSDCDDLKTQEIIMEEIMQSVCTL 864
V + C + N +HV VQ +C ++ + I++ V L
Sbjct: 961 DGHVLK-CVKDQNG--NHV--------------VQKCIECVQPQSLQFIIDAFKGQVFAL 1003
Query: 865 AQDQYGNYVIQHI------------------------------------LEHGKPNERTI 888
+ YG VIQ I LEHG+P +++
Sbjct: 1004 STHPYGCRVIQRILEHCLPDQTLPILEELHQHTEQLVQDQYGNYVIQHVLEHGRPEDKSK 1063
Query: 889 VISKLAGQIVKMSQQKFASNVIEKCLAFGSPEERQILVNEMLGTSD 934
+++++ G ++ +SQ KFASNV+EKC+ S ER +L++E+ +D
Sbjct: 1064 IVAEIRGNVLVLSQHKFASNVVEKCVTHASRTERAVLIDEVCTMND 1109
Score = 52.4 bits (124), Expect = 6e-06
Identities = 51/257 (19%), Positives = 102/257 (38%), Gaps = 49/257 (19%)
Query: 682 IMWYSSGFIQQKLETASVEEKT--KIFPEILPHARALMTDVFGNYVIQKFFEHGTDSQRK 739
+ Y IQ+ LE +++ ++ E+ H + D GN+V+QK E +
Sbjct: 931 LQMYGCRVIQKALEFIPSDQQVINEMVRELDGHVLKCVKDQNGNHVVQKCIECVQPQSLQ 990
Query: 740 ELANQLTGHVLPLSLQMYGCRVIQKLLLNLLTGLGGG*C*SAEPNGVRAQWCNNEMCT*P 799
+ + G V LS YGCRVIQ++L E C
Sbjct: 991 FIIDAFKGQVFALSTHPYGCRVIQRIL---------------------------EHCLPD 1023
Query: 800 KWKPCYSEVYRVCPSE*NSVYHHVFLWASCCTFYSSLWVQSYSDCDDLKTQEIIMEEIMQ 859
+ P E+ H Y + +Q + + + I+ EI
Sbjct: 1024 QTLPILEEL-------------HQHTEQLVQDQYGNYVIQHVLEHGRPEDKSKIVAEIRG 1070
Query: 860 SVCTLAQDQYGNYVIQHILEHGKPNERTIVISKLA-------GQIVKMSQQKFASNVIEK 912
+V L+Q ++ + V++ + H ER ++I ++ + M + ++A+ V++K
Sbjct: 1071 NVLVLSQHKFASNVVEKCVTHASRTERAVLIDEVCTMNDGPHSALYTMMKDQYANYVVQK 1130
Query: 913 CLAFGSPEERQILVNEM 929
+ P +R+I+++++
Sbjct: 1131 MIDVAEPGQRKIVMHKI 1147
Score = 41.6 bits (96), Expect = 0.010
Identities = 23/78 (29%), Positives = 39/78 (49%)
Query: 854 MEEIMQSVCTLAQDQYGNYVIQHILEHGKPNERTIVISKLAGQIVKMSQQKFASNVIEKC 913
+ EI + +QDQ+G+ IQ LE ER +V +++ ++ F + VI+K
Sbjct: 847 LREIAGHIMEFSQDQHGSRFIQLKLERATAAERQLVFNEILQAAYQLMVDVFGNYVIQKF 906
Query: 914 LAFGSPEERQILVNEMLG 931
FGS E++ L + G
Sbjct: 907 FEFGSHEQKLALAERIRG 924
Score = 41.2 bits (95), Expect = 0.013
Identities = 23/88 (26%), Positives = 44/88 (49%), Gaps = 7/88 (7%)
Query: 685 YSSGFIQQKLETASVEEKTKIFPEIL-----PHAR--ALMTDVFGNYVIQKFFEHGTDSQ 737
++S +++ + AS E+ + E+ PH+ +M D + NYV+QK + Q
Sbjct: 1080 FASNVVEKCVTHASRTERAVLIDEVCTMNDGPHSALYTMMKDQYANYVVQKMIDVAEPGQ 1139
Query: 738 RKELANQLTGHVLPLSLQMYGCRVIQKL 765
RK + +++ H+ L YG ++ KL
Sbjct: 1140 RKIVMHKIRPHIATLRKYTYGKHILAKL 1167
>PUF3_YEAST (Q07807) PUF3 protein
Length = 879
Score = 127 bits (318), Expect = 2e-28
Identities = 90/271 (33%), Positives = 142/271 (52%), Gaps = 27/271 (9%)
Query: 685 YSSGFIQQKLETASVEEKTKIFPEILPHARALMTDVFGNYVIQKFFEHGTDSQRKELANQ 744
+ S FIQ++L T+ EK IF EI A L DVFGNYVIQKFFE G+ Q+ L +Q
Sbjct: 551 HGSRFIQRELATSPASEKEVIFNEIRDDAIELSNDVFGNYVIQKFFEFGSKIQKNTLVDQ 610
Query: 745 LTGHVLPLSLQMYGCRVIQKLL--------LNLLTGLGGG*C*SAEPNGVRAQWCNNEMC 796
G++ LSLQMY CRVIQK L + L+ L + ++ Q N+ +
Sbjct: 611 FKGNMKQLSLQMYACRVIQKALEYIDSNQRIELVLELS-----DSVLQMIKDQNGNHVI- 664
Query: 797 T*PKWKPCYSEVYRVCPSE*NSVYHHVFLWASCCTFYSSLWVQSYSDCDDLKTQEIIMEE 856
K + P +S+ H++ ++ Y +Q + + QE I+ E
Sbjct: 665 ----QKAIETIPIEKLPFILSSLTGHIYHLST--HSYGCRVIQRLLEFGSSEDQESILNE 718
Query: 857 IMQSVCTLAQDQYGNYVIQHILEHGKPNERTIV------ISKLAGQIVKMSQQKFASNVI 910
+ + L QDQYGNYVIQ++L+ + + +V I +A +V+ S+ KFASNV+
Sbjct: 719 LKDFIPYLIQDQYGNYVIQYVLQQDQFTNKEMVDIKQEIIETVANNVVEYSKHKFASNVV 778
Query: 911 EKCLAFGSPEERQILVNEMLGTSDENEPLQV 941
EK + +GS ++ ++++++L D+N L +
Sbjct: 779 EKSILYGSKNQKDLIISKIL-PRDKNHALNL 808
Score = 57.0 bits (136), Expect = 2e-07
Identities = 60/262 (22%), Positives = 117/262 (43%), Gaps = 48/262 (18%)
Query: 678 QISLIMWYSSGFIQQKLETASVEEKTKIFPEILPHARALMTDVFGNYVIQKFFEHGTDSQ 737
Q+SL M Y+ IQ+ LE ++ ++ E+ ++ D GN+VIQK E +
Sbjct: 617 QLSLQM-YACRVIQKALEYIDSNQRIELVLELSDSVLQMIKDQNGNHVIQKAIETIPIEK 675
Query: 738 RKELANQLTGHVLPLSLQMYGCRVIQKLLLNLLTGLGGG*C*SAEPNGVRAQWCNNEMCT 797
+ + LTGH+ LS YGCRVIQ+LL G S + + NE+
Sbjct: 676 LPFILSSLTGHIYHLSTHSYGCRVIQRLL-----EFGS----SEDQESIL-----NEL-- 719
Query: 798 *PKWKPCYSEVYRVCPSE*NSVYHHVFLWASCCTFYSSLWVQSYSDCDDLKTQEIIMEEI 857
+ P Y + N V +V L +++ + + ++ I+E +
Sbjct: 720 -KDFIP-----YLIQDQYGNYVIQYV------------LQQDQFTNKEMVDIKQEIIETV 761
Query: 858 MQSVCTLAQDQYGNYVIQHILEHGKPNERTIVISKLA-------------GQIVKMSQQK 904
+V ++ ++ + V++ + +G N++ ++ISK+ ++ M + +
Sbjct: 762 ANNVVEYSKHKFASNVVEKSILYGSKNQKDLIISKILPRDKNHALNLEDDSPMILMIKDQ 821
Query: 905 FASNVIEKCLAFGSPEERQILV 926
FA+ VI+K + E ++++V
Sbjct: 822 FANYVIQKLVNVSEGEGKKLIV 843
Score = 53.9 bits (128), Expect = 2e-06
Identities = 29/101 (28%), Positives = 49/101 (47%)
Query: 829 CCTFYSSLWVQSYSDCDDLKTQEIIMEEIMQSVCTLAQDQYGNYVIQHILEHGKPNERTI 888
C + S ++Q +E+I EI L+ D +GNYVIQ E G ++
Sbjct: 547 CKDQHGSRFIQRELATSPASEKEVIFNEIRDDAIELSNDVFGNYVIQKFFEFGSKIQKNT 606
Query: 889 VISKLAGQIVKMSQQKFASNVIEKCLAFGSPEERQILVNEM 929
++ + G + ++S Q +A VI+K L + +R LV E+
Sbjct: 607 LVDQFKGNMKQLSLQMYACRVIQKALEYIDSNQRIELVLEL 647
Score = 42.4 bits (98), Expect = 0.006
Identities = 21/90 (23%), Positives = 44/90 (48%)
Query: 842 SDCDDLKTQEIIMEEIMQSVCTLAQDQYGNYVIQHILEHGKPNERTIVISKLAGQIVKMS 901
S D + +++I +DQ+G+ IQ L +E+ ++ +++ +++S
Sbjct: 524 SSSDKNSNSNMSLKDIFGHSLEFCKDQHGSRFIQRELATSPASEKEVIFNEIRDDAIELS 583
Query: 902 QQKFASNVIEKCLAFGSPEERQILVNEMLG 931
F + VI+K FGS ++ LV++ G
Sbjct: 584 NDVFGNYVIQKFFEFGSKIQKNTLVDQFKG 613
>PUM1_HUMAN (Q14671) Pumilio homolog 1 (Pumilio1) (HsPUM)
Length = 1186
Score = 125 bits (314), Expect = 5e-28
Identities = 93/286 (32%), Positives = 134/286 (46%), Gaps = 76/286 (26%)
Query: 685 YSSGFIQQKLETASVEEKTKIFPEILPHARALMTDVFGNYVIQKFFEHGTDSQRKELANQ 744
+ S FIQ KLE A+ E+ +F EIL A LM DVFGNYVIQKFFE G+ Q+ LA +
Sbjct: 861 HGSRFIQLKLERATPAERQLVFNEILQAAYQLMVDVFGNYVIQKFFEFGSLEQKLALAER 920
Query: 745 LTGHVLPLSLQMYGCRVIQKLLLNLLTGLGGG*C*SAEPNGVRAQWCNNEMCT*PKWKPC 804
+ GHVL L+LQMYGCRVIQK L + + NEM +
Sbjct: 921 IRGHVLSLALQMYGCRVIQKALEFIPSDQ------------------QNEMV-----REL 957
Query: 805 YSEVYRVCPSE*NSVYHHVFLWASCCTFYSSLWVQSYSDCDDLKTQEIIMEEIMQSVCTL 864
V + C + N +HV VQ +C ++ + I++ V L
Sbjct: 958 DGHVLK-CVKDQNG--NHV--------------VQKCIECVQPQSLQFIIDAFKGQVFAL 1000
Query: 865 AQDQYGNYVIQHIL------------------------------------EHGKPNERTI 888
+ YG VIQ IL EHG+P +++
Sbjct: 1001 STHPYGCRVIQRILEHCLPDQTLPILEELHQHTEQLVQDQYGNYVIQHVLEHGRPEDKSK 1060
Query: 889 VISKLAGQIVKMSQQKFASNVIEKCLAFGSPEERQILVNEMLGTSD 934
+++++ G ++ +SQ KFASNV+EKC+ S ER +L++E+ +D
Sbjct: 1061 IVAEIRGNVLVLSQHKFASNVVEKCVTHASRTERAVLIDEVCTMND 1106
Score = 44.3 bits (103), Expect = 0.002
Identities = 24/78 (30%), Positives = 40/78 (50%)
Query: 854 MEEIMQSVCTLAQDQYGNYVIQHILEHGKPNERTIVISKLAGQIVKMSQQKFASNVIEKC 913
+ EI + +QDQ+G+ IQ LE P ER +V +++ ++ F + VI+K
Sbjct: 846 LREIAGHIMEFSQDQHGSRFIQLKLERATPAERQLVFNEILQAAYQLMVDVFGNYVIQKF 905
Query: 914 LAFGSPEERQILVNEMLG 931
FGS E++ L + G
Sbjct: 906 FEFGSLEQKLALAERIRG 923
Score = 41.2 bits (95), Expect = 0.013
Identities = 23/88 (26%), Positives = 44/88 (49%), Gaps = 7/88 (7%)
Query: 685 YSSGFIQQKLETASVEEKTKIFPEIL-----PHAR--ALMTDVFGNYVIQKFFEHGTDSQ 737
++S +++ + AS E+ + E+ PH+ +M D + NYV+QK + Q
Sbjct: 1077 FASNVVEKCVTHASRTERAVLIDEVCTMNDGPHSALYTMMKDQYANYVVQKMIDVAEPGQ 1136
Query: 738 RKELANQLTGHVLPLSLQMYGCRVIQKL 765
RK + +++ H+ L YG ++ KL
Sbjct: 1137 RKIVMHKIRPHIATLRKYTYGKHILAKL 1164
>YDHE_SCHPO (Q92359) Hypothetical protein C6G9.14 in chromosome I
Length = 681
Score = 107 bits (267), Expect = 1e-22
Identities = 82/282 (29%), Positives = 131/282 (46%), Gaps = 31/282 (10%)
Query: 664 SMSSRTTKPNLLNFQISLIMWYSSG--FIQQKLETASVEEKTKIFPEILPHARALMTDVF 721
S+SS +L N I G ++Q+ LE + +F E P+ LM D F
Sbjct: 333 SLSSSVASLSLQNSNILSFCKDQHGCRYLQRLLEKKNQSHIDAVFAETHPYLAVLMVDAF 392
Query: 722 GNYVIQKFFEHGTDSQRKELANQLTGHVLPLSLQMYGCRVIQKLLLNLLTGLGGG*C*-- 779
GNY+ QK FEH +++QR + ++P+S M+G R +QK++ +L++ C
Sbjct: 393 GNYLCQKLFEHASEAQRSTFIQIIAPKLVPISFNMHGTRALQKII-DLVSSPDQISCIVN 451
Query: 780 SAEPNGVRA----------QWCNNEMCT*PKWKPCYSEVYRVCPSE*N-SVYHHVFLWAS 828
+ PN V Q C N+ + C +C + S + H
Sbjct: 452 ALRPNVVLLTKDLNGNHVIQKCLNKFSQ----EDCQFIFDAICEDPLDVSTHRH-----G 502
Query: 829 CCTFYSSLWVQSYSDCDDLKTQEIIMEEIMQSVCTLAQDQYGNYVIQHILEHGKPNERTI 888
CC VQ D E ++E I+ TL QD +GNYV+Q++LE PN
Sbjct: 503 CCV------VQRCFDHASPAQIEQLVEHIVPHALTLVQDAFGNYVLQYVLELNNPNHTEA 556
Query: 889 VISKLAGQIVKMSQQKFASNVIEKCLAFGSPEERQILVNEML 930
+IS ++ +S QKF+SNV+EKC+ F ++ L++E++
Sbjct: 557 IISYFLYKVRALSTQKFSSNVMEKCIFFAPAAIKEKLISELM 598
Score = 37.4 bits (85), Expect = 0.19
Identities = 23/83 (27%), Positives = 41/83 (48%), Gaps = 2/83 (2%)
Query: 685 YSSGFIQQKLETASVEEKTKIFPEILP--HARALMTDVFGNYVIQKFFEHGTDSQRKELA 742
+SS +++ + A K K+ E++ H L+ D F NYVIQ ++ + QR EL
Sbjct: 573 FSSNVMEKCIFFAPAAIKEKLISELMDEKHLPKLLRDSFANYVIQTALDNASVKQRAELV 632
Query: 743 NQLTGHVLPLSLQMYGCRVIQKL 765
++ + + G R++ KL
Sbjct: 633 ERIKPLIPSIKNTPCGRRILSKL 655
>YAD3_SCHPO (Q09829) Hypothetical protein C4G8.03c in chromosome I
Length = 780
Score = 88.6 bits (218), Expect = 7e-17
Identities = 70/264 (26%), Positives = 115/264 (43%), Gaps = 15/264 (5%)
Query: 685 YSSGFIQQKLETASVEEKTKIFPEILPHARALMTDVFGNYVIQKFFEHGTDSQRKELANQ 744
Y ++Q+ L+ + FPEI LM D FGNY+ QK F + + Q+ + N
Sbjct: 475 YGCRYLQKLLDENPKVNASLFFPEIRQSVVQLMIDPFGNYMCQKLFVYASREQKLSMLNG 534
Query: 745 LTGHVLPLSLQMYGCRVIQKLLLNLLTGLGGG*C*SAEPNGVRAQWCNN------EMCT* 798
+ ++ + +YG R +Q ++ L + + C+N + C
Sbjct: 535 IGEGIVDICSNLYGTRSMQNIIDKLTSNEQISLLLKIIIPSLTTLACDNNGTHVLQKCI- 593
Query: 799 PKWKPCYSEVYRVCPSE*NSVYHHVFLWASCCTFYSSLWVQSYSDCDDLKTQEIIMEEIM 858
K+ P E + E ++ CC L D + QE ++ I+
Sbjct: 594 AKFPPEKLEPLFLSMEE--NLITLATNRHGCCILQRCL------DRTNGDIQERLVNSII 645
Query: 859 QSVCTLAQDQYGNYVIQHILEHGKPNERTIVISKLAGQIVKMSQQKFASNVIEKCLAFGS 918
+S L Q+ YGNY++QH+LE +I K G I K+S QKF+SN IE+C+ S
Sbjct: 646 KSCLLLVQNAYGNYLVQHVLELNIQPYTERIIEKFFGNICKLSLQKFSSNAIEQCIRTAS 705
Query: 919 PEERQILVNEMLGTSDENEPLQVC 942
P R+ ++ E L + + L C
Sbjct: 706 PSTREQMLQEFLSFPNIEQLLDDC 729
Score = 41.6 bits (96), Expect = 0.010
Identities = 27/84 (32%), Positives = 45/84 (53%), Gaps = 4/84 (4%)
Query: 685 YSSGFIQQKLETASVEEKTKIFPEIL--PHARALMTDVFGNYVIQKFFEHGTDSQRKELA 742
+SS I+Q + TAS + ++ E L P+ L+ D + NYV+Q+F +SQ K L
Sbjct: 692 FSSNAIEQCIRTASPSTREQMLQEFLSFPNIEQLLDDCYANYVMQRFLNVADESQ-KFLI 750
Query: 743 NQLTGHVLP-LSLQMYGCRVIQKL 765
+ HV+P + +G ++ KL
Sbjct: 751 LRSISHVIPKIQNTRHGRHILAKL 774
Score = 33.1 bits (74), Expect = 3.6
Identities = 20/88 (22%), Positives = 39/88 (43%), Gaps = 4/88 (4%)
Query: 829 CCTFYSSLWVQSYSDCDDLKTQEIIMEEIMQSVCTLAQDQYGNYVIQHILEHGKPNERTI 888
C Y ++Q D + + EI QSV L D +GNY+ Q + + ++
Sbjct: 471 CKDQYGCRYLQKLLDENPKVNASLFFPEIRQSVVQLMIDPFGNYMCQKLFVYASREQKLS 530
Query: 889 VISKLAGQIVKMSQQKFAS----NVIEK 912
+++ + IV + + + N+I+K
Sbjct: 531 MLNGIGEGIVDICSNLYGTRSMQNIIDK 558
>YD15_SCHPO (Q10238) Putative regulatory protein C4G9.05
Length = 581
Score = 82.4 bits (202), Expect = 5e-15
Identities = 64/240 (26%), Positives = 115/240 (47%), Gaps = 30/240 (12%)
Query: 690 IQQKLETASVEEKTKIFPEILPHARALMTDVFGNYVIQKFFEHGTDSQRKELANQLTGHV 749
+QQ+L+ +E+ + ILP + LM + FGN++IQK FE+ T++Q + + L HV
Sbjct: 273 LQQQLKNEDIEDNINLINTILPFSVTLMKNKFGNFLIQKCFEYSTEAQLQSFSYFLKKHV 332
Query: 750 LPLSLQMYGCRVIQKLL--------LNLLTGLGGG*C*SAEPNGVRAQWCNNEMCT*PKW 801
LS+ +G V+QK L NL+ L + A C W
Sbjct: 333 KELSIDAFGSHVLQKSLEIYPERFTNNLIEEL---------IECLPATLMQRHSC--HVW 381
Query: 802 KPCYSEVYRVCPSE*NSVYHHV-------FLWASCCTFYSSLWVQSYSDCDDLKTQEIIM 854
+ + E R S + ++ H +L S S + + +C + K + +
Sbjct: 382 QK-FFETRR--KSLVDGIFDHFNKKMQGKWLQVSVSEMGSLVVQTIFENCKE-KDKRTCL 437
Query: 855 EEIMQSVCTLAQDQYGNYVIQHILEHGKPNERTIVISKLAGQIVKMSQQKFASNVIEKCL 914
+EI+ ++ + Q+GN+VIQHI+EHG ++ +++ L ++ S ++AS V+E+ L
Sbjct: 438 DEIINNMDQIICGQWGNWVIQHIIEHGSEPDKQRILNSLLKEVESYSTNRYASKVVERAL 497
Score = 35.0 bits (79), Expect = 0.95
Identities = 12/46 (26%), Positives = 27/46 (58%)
Query: 864 LAQDQYGNYVIQHILEHGKPNERTIVISKLAGQIVKMSQQKFASNV 909
+A +QYGNY++Q++L+ P++ ++ L +V + K+ +
Sbjct: 527 IASNQYGNYIVQYLLQVATPSQINLMAEHLKKHMVSLRGHKYGQRI 572
Score = 32.0 bits (71), Expect = 8.0
Identities = 22/85 (25%), Positives = 36/85 (41%), Gaps = 8/85 (9%)
Query: 685 YSSGFIQQKLETASVEEKTKIFPEILPHARALMT--------DVFGNYVIQKFFEHGTDS 736
Y+S +++ L V + EI L T + +GNY++Q + T S
Sbjct: 488 YASKVVERALRVCHVTFFDRYVKEITTPQNELPTIFLQEIASNQYGNYIVQYLLQVATPS 547
Query: 737 QRKELANQLTGHVLPLSLQMYGCRV 761
Q +A L H++ L YG R+
Sbjct: 548 QINLMAEHLKKHMVSLRGHKYGQRI 572
>PUF4_YEAST (P25339) PUF4 protein
Length = 888
Score = 73.2 bits (178), Expect = 3e-12
Identities = 71/257 (27%), Positives = 109/257 (41%), Gaps = 21/257 (8%)
Query: 689 FIQQKLETASVEEKTKIFPEILPHARALMTDVFGNYVIQKFFEHGTDSQRKELANQLTGH 748
F+Q++L+ + IF E + LMTD FGNY+IQK E T QR L + H
Sbjct: 580 FLQKQLDILGSKAADAIFEETKDYTVELMTDSFGNYLIQKLLEEVTTEQRIVLTKISSPH 639
Query: 749 VLPLSLQMYGCRVIQKLLLNLLTGLGGG-*C*SAEPNGVRAQWCNN-----EMCT*PKWK 802
+ +SL +G R +QKL+ + T S P V+ N + C
Sbjct: 640 FVEISLNPHGTRALQKLIECIKTDEEAQIVVDSLRPYTVQLSKDLNGNHVIQKCLQRLKP 699
Query: 803 PCYSEVYRVCPSE*NSVYHHVFLWASCCTFYSSLWVQSYSDCDDLKTQEIIMEEIMQSVC 862
+ ++ + H CC L + CD+L ++++ V
Sbjct: 700 ENFQFIFDAISDSCIDIATHRH---GCCVLQRCLDHGTTEQCDNL------CDKLLALVD 750
Query: 863 TLAQDQYGNYVIQHIL----EHGKPNERTIVISKLAGQIVKMSQQKFASNVIEKCLAFGS 918
L D +GNYV+Q+I+ E K + ++ L + +++S KF SNVIEK L
Sbjct: 751 KLTLDPFGNYVVQYIITKEAEKNKYDYTHKIVHLLKPRAIELSIHKFGSNVIEKILKTAI 810
Query: 919 PEERQILVNEMLGTSDE 935
E IL E+L E
Sbjct: 811 VSEPMIL--EILNNGGE 825
Score = 40.4 bits (93), Expect = 0.023
Identities = 25/102 (24%), Positives = 46/102 (44%), Gaps = 1/102 (0%)
Query: 829 CCTFYSSLWVQSYSDCDDLKTQEIIMEEIMQSVCTLAQDQYGNYVIQHILEHGKPNERTI 888
C + ++Q D K + I EE L D +GNY+IQ +LE +R +
Sbjct: 572 CKDQHGCRFLQKQLDILGSKAADAIFEETKDYTVELMTDSFGNYLIQKLLEEVTTEQRIV 631
Query: 889 VISKLAGQIVKMSQQKFASNVIEKCL-AFGSPEERQILVNEM 929
+ + V++S + ++K + + EE QI+V+ +
Sbjct: 632 LTKISSPHFVEISLNPHGTRALQKLIECIKTDEEAQIVVDSL 673
>MPT5_YEAST (P39016) Suppressor protein MPT5 (HTR1 protein)
Length = 834
Score = 52.0 bits (123), Expect = 7e-06
Identities = 27/80 (33%), Positives = 46/80 (56%), Gaps = 2/80 (2%)
Query: 689 FIQQKLETASVEEKTK--IFPEILPHARALMTDVFGNYVIQKFFEHGTDSQRKELANQLT 746
F+Q+KLET S + ++ +I P L+ D FGNY++QK ++ T Q+ L +
Sbjct: 202 FLQKKLETPSESNMVRDLMYEQIKPFFLDLILDPFGNYLVQKLCDYLTAEQKTLLIQTIY 261
Query: 747 GHVLPLSLQMYGCRVIQKLL 766
+V +S+ YG R +QK++
Sbjct: 262 PNVFQISINQYGTRSLQKII 281
Score = 39.7 bits (91), Expect = 0.038
Identities = 21/62 (33%), Positives = 41/62 (65%), Gaps = 2/62 (3%)
Query: 853 IMEEIMQSVCTLAQDQYGNYVIQHILEHGKPNERTI--VISKLAGQIVKMSQQKFASNVI 910
I +I+Q + L DQ+GNY+IQ +L+ + + + + ++L+ ++ ++S KF+SNV+
Sbjct: 373 ISVKIVQFLPGLINDQFGNYIIQFLLDIKELDFYLLAELFNRLSNELCQLSCLKFSSNVV 432
Query: 911 EK 912
EK
Sbjct: 433 EK 434
Score = 34.3 bits (77), Expect = 1.6
Identities = 24/88 (27%), Positives = 43/88 (48%), Gaps = 5/88 (5%)
Query: 690 IQQKLETASVEEKTKIFPEILPHARALMTDVFGNYVIQKFFE--HGTDSQRKELANQLTG 747
+Q+ L ++++ KI +I+ L+ D FGNY+IQ + EL N+L+
Sbjct: 358 LQKLLSVCTLQQIFKISVKIVQFLPGLINDQFGNYIIQFLLDIKELDFYLLAELFNRLSN 417
Query: 748 HVLPLSLQMYGCRVIQKL---LLNLLTG 772
+ LS + V++K L ++TG
Sbjct: 418 ELCQLSCLKFSSNVVEKFIKKLFRIITG 445
Score = 33.5 bits (75), Expect = 2.8
Identities = 12/65 (18%), Positives = 37/65 (56%)
Query: 850 QEIIMEEIMQSVCTLAQDQYGNYVIQHILEHGKPNERTIVISKLAGQIVKMSQQKFASNV 909
++++ E+I L D +GNY++Q + ++ ++T++I + + ++S ++ +
Sbjct: 217 RDLMYEQIKPFFLDLILDPFGNYLVQKLCDYLTAEQKTLLIQTIYPNVFQISINQYGTRS 276
Query: 910 IEKCL 914
++K +
Sbjct: 277 LQKII 281
Score = 32.3 bits (72), Expect = 6.1
Identities = 29/108 (26%), Positives = 54/108 (49%), Gaps = 12/108 (11%)
Query: 668 RTTKPNLLNFQISLIMWYSSGFIQQKLETASVEEKTKIFPE-------ILPHARALMTDV 720
+T PN+ FQIS I Y + +Q+ ++T E + + + + L+ D+
Sbjct: 258 QTIYPNV--FQIS-INQYGTRSLQKIIDTVDNEVQIDLIIKGFSQEFTSIEQVVTLINDL 314
Query: 721 FGNYVIQKFFEHGTDSQRKELANQLT--GHVLPLSLQMYGCRVIQKLL 766
GN+VIQK + S+ + + + +++ +S +GC V+QKLL
Sbjct: 315 NGNHVIQKCIFKFSPSKFGFIIDAIVEQNNIITISTHKHGCCVLQKLL 362
>FBF2_CAEEL (Q09312) Fem-3 mRNA binding factor 2
Length = 632
Score = 51.2 bits (121), Expect = 1e-05
Identities = 59/227 (25%), Positives = 97/227 (41%), Gaps = 35/227 (15%)
Query: 716 LMTDVFGNYVIQKFF-------EHGTDSQRKELANQLTGHVLPLSLQMYGCRVIQKLLLN 768
L T++FGNY++Q + G ++++L N ++ + + L + CRVIQ L N
Sbjct: 237 LSTNIFGNYLVQSVIGISLATNDDGYTKRQEKLKNFISSQMTDMCLDKFACRVIQSSLQN 296
Query: 769 LLTGLGGG*C*SAEPNGVRAQWCNNEMCT*PKWKPCYSEVYRVCPSE*-----NSVYHHV 823
+ L A P R +C +V V P + + V
Sbjct: 297 MDLSLACKLV-QALPRDARLI----AICVDQNANHVIQKVVAVIPLKNWEFIVDFVATPE 351
Query: 824 FLWASCCTFYSSLWVQSYSD---CDDLKT---------QEIIMEEIMQSVCT----LAQD 867
L C Y VQ+ + D + +E ++ +M SV LA +
Sbjct: 352 HLRQICSDKYGCRVVQTIIEKLTADSMNVDLTSAAQNLRERALQRLMTSVTNRCQELATN 411
Query: 868 QYGNYVIQHILEHGK-PNERTIVISK-LAGQIVKMSQQKFASNVIEK 912
+Y NY+IQHI+ + R +I K L ++ +SQ+KFAS+V+EK
Sbjct: 412 EYANYIIQHIVSNDDLAVYRECIIEKCLMRNLLSLSQEKFASHVVEK 458
>FBF1_CAEEL (Q9N5M6) Fem-3 mRNA binding factor 1
Length = 614
Score = 49.7 bits (117), Expect = 4e-05
Identities = 60/244 (24%), Positives = 103/244 (41%), Gaps = 35/244 (14%)
Query: 716 LMTDVFGNYVIQKF-------FEHGTDSQRKELANQLTGHVLPLSLQMYGCRVIQKLLLN 768
L T++FGNY +Q+ ++ ++++L N ++ + + L + CRVIQ L N
Sbjct: 235 LSTNIFGNYFVQEIIGMSLTTYDDDNIKRQEKLKNFISSQMTDMCLDKFACRVIQSSLQN 294
Query: 769 LLTGLGGG*C*SAEPNGVRAQWCNNEMCT*PKWKPCYSEVYRVCPSE*-----NSVYHHV 823
+ L A P R +C +V V P + + V
Sbjct: 295 MDLSLACKLV-QALPRDARLI----AICVDQNANHVIQKVVAVIPLKNWEFIVDFVATPE 349
Query: 824 FLWASCCTFYSSLWVQSYSD---CDDLKT---------QEIIMEEIMQSVCT----LAQD 867
L C Y VQ+ + D + +E ++ +M SV LA +
Sbjct: 350 HLRQICFDKYGCRVVQTIIEKLTADSINVDLTSAAQHLRERALQRLMTSVTNRCQELATN 409
Query: 868 QYGNYVIQHILEHGK-PNERTIVISK-LAGQIVKMSQQKFASNVIEKCLAFGSPEERQIL 925
+Y NY+IQHI+ + R +I K L ++ +SQ+KFAS+V+EK E +
Sbjct: 410 EYANYIIQHIVSNDDLAVYRECIIEKCLMRNLLSLSQEKFASHVVEKAFLHAPMELLAEM 469
Query: 926 VNEM 929
++E+
Sbjct: 470 MDEI 473
>K020_HUMAN (Q15397) Protein KIAA0020
Length = 508
Score = 40.4 bits (93), Expect = 0.023
Identities = 22/92 (23%), Positives = 50/92 (53%), Gaps = 3/92 (3%)
Query: 843 DCDDLKTQEII--MEEIMQS-VCTLAQDQYGNYVIQHILEHGKPNERTIVISKLAGQIVK 899
DCD K +++ +++++Q + T+A VIQ +++G +R +L +V+
Sbjct: 9 DCDKEKRVKLMSDLQKLIQGKIKTIAFAHDSTRVIQCYIQYGNEEQRKQAFEELRDDLVE 68
Query: 900 MSQQKFASNVIEKCLAFGSPEERQILVNEMLG 931
+S+ K++ N+++K L +GS + ++ G
Sbjct: 69 LSKAKYSRNIVKKFLMYGSKPQIAEIIRSFKG 100
Score = 36.6 bits (83), Expect = 0.33
Identities = 24/88 (27%), Positives = 44/88 (49%), Gaps = 12/88 (13%)
Query: 851 EIIMEEIMQSVCTLAQDQYGNYVIQHILEH---------GKPNERTIVISKLAGQIVKMS 901
E+IM+E+ Q + +AQ + VI+H L H P R+ +I + +V ++
Sbjct: 163 ELIMDEMKQILTPMAQKEA---VIKHSLVHKVFLDFFTYAPPKLRSEMIEAIREAVVYLA 219
Query: 902 QQKFASNVIEKCLAFGSPEERQILVNEM 929
+ V CL G+P++R+++V M
Sbjct: 220 HTHDGARVAMHCLWHGTPKDRKVIVKTM 247
Score = 36.2 bits (82), Expect = 0.42
Identities = 21/89 (23%), Positives = 47/89 (52%), Gaps = 6/89 (6%)
Query: 848 KTQEIIMEEIMQSVCTLAQDQYGNYVIQHILEHGKPNERTIVISKLAGQIVKMSQQKFAS 907
K + ++E I ++V LA G V H L HG P +R +++ + + K++ +++
Sbjct: 202 KLRSEMIEAIREAVVYLAHTHDGARVAMHCLWHGTPKDRKVIVKTMKTYVEKVANGQYSH 261
Query: 908 NVIEKCLAFGSPEE----RQILVNEMLGT 932
V+ AF ++ +QI+++E++ +
Sbjct: 262 LVL--LAAFDCIDDTKLVKQIIISEIISS 288
Score = 33.5 bits (75), Expect = 2.8
Identities = 21/99 (21%), Positives = 46/99 (46%), Gaps = 5/99 (5%)
Query: 838 VQSYSDCDDLKTQEIIMEEIMQSVCTLAQDQYGNYVIQHILEHGKPNERTIVISKLAGQI 897
+Q Y + + ++ EE+ + L++ +Y +++ L +G + +I G +
Sbjct: 43 IQCYIQYGNEEQRKQAFEELRDDLVELSKAKYSRNIVKKFLMYGSKPQIAEIIRSFKGHV 102
Query: 898 VKMSQQKFASNVIEKCLAFGSP---EERQILVNEMLGTS 933
KM + AS ++E A+ E+R +L E+ G +
Sbjct: 103 RKMLRHAEASAIVE--YAYNDKAILEQRNMLTEELYGNT 139
>SERI_BOMMO (P07856) Sericin precursor (Silk gum protein)
Length = 389
Score = 38.9 bits (89), Expect = 0.065
Identities = 79/318 (24%), Positives = 115/318 (35%), Gaps = 41/318 (12%)
Query: 127 QRFQAGGGSSSIERFGDWRKNATSNGDSSSLFSMQPGFSVQQAENDLMELRKASGRNLPR 186
Q +GG SS K+A+ SSS S Q S + + E +S R
Sbjct: 93 QAIISGGTKSSNSNVQSDEKSASQ---SSSSRSSQESASYSSSSSSSTEESSSSSS---R 146
Query: 187 QSSTQLLDRHMDGMTRMPGTSL-GVRRTC-YSDILQDGFDQPTLSSNMSRPASHNAFVDI 244
+S+ + D + G+S G RRT YS +DG T SS+ + S NA
Sbjct: 147 AASSTDASSNTDSNSNSAGSSTSGGRRTYGYSSNSRDGSVSSTGSSSNTDSNSSNA---- 202
Query: 245 RDSTGIVDREPLEGLRSSASTPGLVGLQNHGVNSHNFSSVVGSSLSRSTTPESHVIGRPV 304
+ST G + NS + S S S + + + V R
Sbjct: 203 -----------------GSSTSGGSSTYGYSSNSRDGSVSTTGSSSNTDSNSNSVGSRRS 245
Query: 305 GSGVPQMGSKVFSAENIGLGNHNGHSSNMTDLADMVSSLSGLNLSGARRAEQDNLLKSKL 364
G S EN+ G SSN D S+ G + SG RR +
Sbjct: 246 GGSSSHEDSSKSRDENVST---TGSSSN----TDSNSNSVGSSTSGGRRTYGYSSNSRDG 298
Query: 365 QVEVDNHANVMLSTPNNVNLPKHNELATDLNTFSLNERVNLLKKTASYANLRSNAHSTGN 424
V ++ S N+V +T +S N R + T S +N SN++S G+
Sbjct: 299 SVSSTGSSSNTDSNSNSVGSSTSGGSST--YGYSSNSRDGSVSSTGSSSNTDSNSNSAGS 356
Query: 425 LTSIDFAGQVPSAYPANT 442
TS G Y +N+
Sbjct: 357 STS---GGSSTYGYSSNS 371
>RGA_DROME (Q94547) Regulator of gene activity (Regena protein)
Length = 585
Score = 38.9 bits (89), Expect = 0.065
Identities = 36/132 (27%), Positives = 56/132 (42%), Gaps = 12/132 (9%)
Query: 224 DQPTLSSNMSRPASHNAFVDIRDSTGIVDR---EPLEGLRSSASTPGLVGLQNHGVNSHN 280
DQ SN +P + + S G+V + E E S+ P L G QN ++
Sbjct: 176 DQTLPQSNPLQPPGSKPYGNFFTSFGMVKQPTSEQSEFTMSNEDFPALPGTQNSDGTTNA 235
Query: 281 FSSVVGSSLSRSTTPESHVIG--RPVGSGVPQMGSKVFSAENIGLGNHNGHSSNMTDLAD 338
SV G+ S + E+H+ G +P+ S V + S N+G+ NG L
Sbjct: 236 VGSVAGTGGSGGASTENHLDGTEKPMNSIVVSGSASGSSGSNVGVVGGNG-------LGA 288
Query: 339 MVSSLSGLNLSG 350
+ S + GL + G
Sbjct: 289 VGSGIGGLAVGG 300
>RNGB_DICDI (Q7M3S9) Ring finger protein B (Protein rngB)
Length = 943
Score = 38.5 bits (88), Expect = 0.086
Identities = 48/226 (21%), Positives = 91/226 (40%), Gaps = 16/226 (7%)
Query: 242 VDIRDSTGIVDREPLEGLRSSASTPGLVGLQNHGVNSHNFSSVVGSSLSRSTTPESHVIG 301
+ I +S IV P + S G G++ H S++ SS S S S +
Sbjct: 334 IPITNSNSIVG-SPNTSISCGVSNSGASSSSGGGISGH--PSILSSSSSSSYLSTSPLST 390
Query: 302 RPVGSGVPQMGSKVFSA---ENIGLGNHNGHSSNMTDLADMVSSLSGLNLSGARRAEQDN 358
+ S S F+ +N N+N +++N+ ++ + N + +N
Sbjct: 391 SSLASSYQSSQSLQFNQNQNQNNNNNNNNNNNNNIQTTTTTTTNNNNNNNNNNNNNNNNN 450
Query: 359 LLKSKLQVEVDNHA----NVMLSTPNNVNLPKHNELATDLNTFSLNERVNLLKKTASYAN 414
++S Q + H +V+ + +N++L N ++ + + T S+A+
Sbjct: 451 NVESNQQQQQIQHQTSPMSVLSRSNSNISLNSLNSSSSSILSTPSTLSTTTTTTTTSHAS 510
Query: 415 LRSNAH-----STGNLTSID-FAGQVPSAYPANTTLNNVYNNHLET 454
S+ S G + SI F G+ + N + +N YNNH +T
Sbjct: 511 HTSHTSNRSNGSRGGIPSIPPFNGRSSNHNNNNNSNSNNYNNHQQT 556
>PEN_DROME (O61345) Penguin protein
Length = 737
Score = 38.1 bits (87), Expect = 0.11
Identities = 19/59 (32%), Positives = 33/59 (55%)
Query: 873 VIQHILEHGKPNERTIVISKLAGQIVKMSQQKFASNVIEKCLAFGSPEERQILVNEMLG 931
VIQ +L++ P R + KL V+M Q K+A +++ L +G+P + LV+ + G
Sbjct: 184 VIQSMLKYASPALRAEISEKLLPFTVEMCQSKYAQFCVQRMLKYGAPATKAKLVDSLYG 242
Score = 35.0 bits (79), Expect = 0.95
Identities = 16/76 (21%), Positives = 38/76 (49%)
Query: 690 IQQKLETASVEEKTKIFPEILPHARALMTDVFGNYVIQKFFEHGTDSQRKELANQLTGHV 749
IQ L+ AS + +I ++LP + + + +Q+ ++G + + +L + L GH+
Sbjct: 185 IQSMLKYASPALRAEISEKLLPFTVEMCQSKYAQFCVQRMLKYGAPATKAKLVDSLYGHI 244
Query: 750 LPLSLQMYGCRVIQKL 765
+ L+ G ++ +
Sbjct: 245 VRLAGHSIGSGLLDSM 260
>PUF6_YEAST (Q04373) PUF6 protein
Length = 656
Score = 37.0 bits (84), Expect = 0.25
Identities = 19/82 (23%), Positives = 40/82 (48%)
Query: 686 SSGFIQQKLETASVEEKTKIFPEILPHARALMTDVFGNYVIQKFFEHGTDSQRKELANQL 745
+S +Q ++ +S + + +I + L T +G Y++ K +G+ S R+ + N+L
Sbjct: 170 ASRIVQTLVKYSSKDRREQIVDALKGKFYVLATSAYGKYLLVKLLHYGSRSSRQTIINEL 229
Query: 746 TGHVLPLSLQMYGCRVIQKLLL 767
G + L G V++ L +
Sbjct: 230 HGSLRKLMRHREGAYVVEDLFV 251
Score = 37.0 bits (84), Expect = 0.25
Identities = 18/88 (20%), Positives = 44/88 (49%), Gaps = 8/88 (9%)
Query: 853 IMEEIMQSVCTLAQDQYGNYVIQH--------ILEHGKPNERTIVISKLAGQIVKMSQQK 904
I E++ + L++D + V++H ++++ + R ++ L G+ ++
Sbjct: 145 IREKLSNEIWELSKDCISDLVLKHDASRIVQTLVKYSSKDRREQIVDALKGKFYVLATSA 204
Query: 905 FASNVIEKCLAFGSPEERQILVNEMLGT 932
+ ++ K L +GS RQ ++NE+ G+
Sbjct: 205 YGKYLLVKLLHYGSRSSRQTIINELHGS 232
>MSN2_YEAST (P33748) Zinc finger protein MSN2 (Multicopy suppressor
of SNF1 protein 2)
Length = 704
Score = 37.0 bits (84), Expect = 0.25
Identities = 33/103 (32%), Positives = 56/103 (54%), Gaps = 17/103 (16%)
Query: 330 SSNMTDLADMVSSLSGLNLSGARRAEQDNLLK-SKLQVEVDNHANVMLST-PNNVNLPKH 387
+SN T+L+ S+ + +S RA+Q +K ++L + ++N+ + T PNN+N
Sbjct: 167 ASNETNLSPQTSNGNETLISP--RAQQHTSIKDNRLSLPNGANSNLFIDTNPNNLNEKLR 224
Query: 388 NELATDLNTFSLNERVNLLKKTASYANLRSNAHSTGNLTSIDF 430
N+L +D N++S S +N SN++STGNL S F
Sbjct: 225 NQLNSDTNSYS-----------NSISN--SNSNSTGNLNSSYF 254
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.326 0.139 0.423
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 104,607,264
Number of Sequences: 164201
Number of extensions: 4250139
Number of successful extensions: 13685
Number of sequences better than 10.0: 38
Number of HSP's better than 10.0 without gapping: 16
Number of HSP's successfully gapped in prelim test: 22
Number of HSP's that attempted gapping in prelim test: 13478
Number of HSP's gapped (non-prelim): 204
length of query: 949
length of database: 59,974,054
effective HSP length: 120
effective length of query: 829
effective length of database: 40,269,934
effective search space: 33383775286
effective search space used: 33383775286
T: 11
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.7 bits)
S2: 71 (32.0 bits)
Medicago: description of AC144720.3


