

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144657.9 + phase: 0
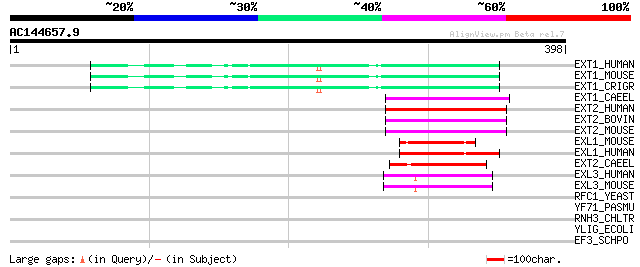
(398 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
EXT1_HUMAN (Q16394) Exostosin-1 (EC 2.4.1.224) (EC 2.4.1.225) (G... 67 6e-11
EXT1_MOUSE (P97464) Exostosin-1 (EC 2.4.1.224) (EC 2.4.1.225) (G... 67 8e-11
EXT1_CRIGR (Q9JK82) Exostosin-1 (EC 2.4.1.-) (Heparan sulfate co... 67 8e-11
EXT1_CAEEL (O01704) Multiple exostoses homolog 1 (Related to mam... 64 7e-10
EXT2_HUMAN (Q93063) Exostosin-2 (EC 2.4.1.224) (EC 2.4.1.225) (G... 63 1e-09
EXT2_BOVIN (O77783) Exostosin-2 (EC 2.4.1.224) (EC 2.4.1.225) (G... 63 2e-09
EXT2_MOUSE (P70428) Exostosin-2 (EC 2.4.1.224) (EC 2.4.1.225) (G... 61 4e-09
EXL1_MOUSE (Q9JKV7) Exostosin-like 1 (EC 2.4.1.224) (Glucuronosy... 52 3e-06
EXL1_HUMAN (Q92935) Exostosin-like 1 (EC 2.4.1.224) (Glucuronosy... 50 8e-06
EXT2_CAEEL (O01705) Exostosin-2 (EC 2.4.1.223) (EC 2.4.1.224) (G... 47 7e-05
EXL3_HUMAN (O43909) Exostosin-like 3 (EC 2.4.1.223) (Glucuronyl-... 44 6e-04
EXL3_MOUSE (Q9WVL6) Exostosin-like 3 (EC 2.4.1.-) (Multiple exos... 44 7e-04
RFC1_YEAST (P38630) Activator 1 95 kDa subunit (Replication fact... 33 1.3
YF71_PASMU (Q9CKN9) Hypothetical UPF0004 protein PM1571 33 1.7
RNH3_CHLTR (O84011) Ribonuclease HIII (EC 3.1.26.-) (RNase HIII) 32 3.7
YLIG_ECOLI (P75802) Hypothetical UPF0004 protein yliG 31 4.9
EF3_SCHPO (O94489) Elongation factor 3 (EF-3) 31 4.9
>EXT1_HUMAN (Q16394) Exostosin-1 (EC 2.4.1.224) (EC 2.4.1.225)
(Glucuronosyl-N-acetylglucosaminyl-proteoglycan/N-
acetylglucosaminyl-proteoglycan
4-alpha-N-acetylglucosaminyltransferase) (Putative tumor
suppressor protein EXT1) (Multiple exostoses protein
Length = 746
Score = 67.4 bits (163), Expect = 6e-11
Identities = 82/327 (25%), Positives = 127/327 (38%), Gaps = 70/327 (21%)
Query: 59 FKVWVYKEGEQPLVHDGPVNNKYSIEGQFIDEMDTSNKSPFKATHPELAHVFFLPFSVSK 118
FKV+VY + + + + N +IEG S F + P A +F L
Sbjct: 114 FKVYVYPQQKGEKIAESYQNILAAIEG-----------SRFYTSDPSQACLFVLSLDTLD 162
Query: 119 VIRYVYKPRKSRSDYNPHRLQLLVEDYIKIVANKYPYWNISQGADHFLLSCHDWGPRVSY 178
R S H L+ V+ WN G +H + + + G Y
Sbjct: 163 --------RDQLSPQYVHNLRSKVQSL--------HLWN--NGRNHLIFNLYS-GTWPDY 203
Query: 179 ANPKLFKHFIRALCNANTS-EGFWPNRDVSIPQLNLPVGKLG-----------PP----- 221
F L A+ S E F PN DVSIP + + G PP
Sbjct: 204 TEDVGFDIGQAMLAKASISTENFRPNFDVSIPLFSKDHPRTGGERGFLKFNTIPPLRKYM 263
Query: 222 -----------------NTDQHPNNRTILTFFAGGAHGKIRKKLLKSWKDKDEEVQVHEY 264
N H +N + HGK +K S D+D EY
Sbjct: 264 LVFKGKRYLTGIGSDTRNALYHVHNGEDVVLLTTCKHGKDWQKHKDSRCDRDNT----EY 319
Query: 265 LPKGQDYTKLMGLSKFCLCPSGHEVASPRVVEAIYAGCVPVIICDNYSLPFSDVLNWSQF 324
+ DY +++ + FCL P G + S R +EA+ A CVPV++ + + LPFS+V+NW+Q
Sbjct: 320 --EKYDYREMLHNATFCLVPRGRRLGSFRFLEALQAACVPVMLSNGWELPFSEVINWNQA 377
Query: 325 SMEIAVDRIPEIKTILQNITETKYRVL 351
++ + +I + +++I + K L
Sbjct: 378 AVIGDERLLLQIPSTIRSIHQDKILAL 404
>EXT1_MOUSE (P97464) Exostosin-1 (EC 2.4.1.224) (EC 2.4.1.225)
(Glucuronosyl-N-acetylglucosaminyl-proteoglycan/N-
acetylglucosaminyl-proteoglycan
4-alpha-N-acetylglucosaminyltransferase) (Multiple
exostoses protein 1 homolog)
Length = 746
Score = 67.0 bits (162), Expect = 8e-11
Identities = 82/327 (25%), Positives = 127/327 (38%), Gaps = 70/327 (21%)
Query: 59 FKVWVYKEGEQPLVHDGPVNNKYSIEGQFIDEMDTSNKSPFKATHPELAHVFFLPFSVSK 118
FKV+VY + + + + N +IEG S F + P A +F L
Sbjct: 114 FKVYVYPQQKGEKIAESYQNILAAIEG-----------SRFYTSDPSQACLFVLSLDTLD 162
Query: 119 VIRYVYKPRKSRSDYNPHRLQLLVEDYIKIVANKYPYWNISQGADHFLLSCHDWGPRVSY 178
R S H L+ V+ WN G +H + + + G Y
Sbjct: 163 --------RDQLSPQYVHNLRSKVQSL--------HLWN--NGRNHLIFNLYS-GTWPDY 203
Query: 179 ANPKLFKHFIRALCNANTS-EGFWPNRDVSIPQLNLPVGKLG-----------PP----- 221
F L A+ S E F PN DVSIP + + G PP
Sbjct: 204 TEDVGFDIGQAMLAKASISTENFRPNFDVSIPLFSKDHPRTGGERGFLKFNTIPPLRKYM 263
Query: 222 -----------------NTDQHPNNRTILTFFAGGAHGKIRKKLLKSWKDKDEEVQVHEY 264
N H +N + HGK +K S D+D EY
Sbjct: 264 LVFKGKRYLTGIGSDTRNALYHVHNGEDVLLLTTCKHGKDWQKHKDSRCDRDNT----EY 319
Query: 265 LPKGQDYTKLMGLSKFCLCPSGHEVASPRVVEAIYAGCVPVIICDNYSLPFSDVLNWSQF 324
+ DY +++ + FCL P G + S R +EA+ A CVPV++ + + LPFS+V+NW+Q
Sbjct: 320 --EKYDYREMLHNATFCLVPRGRRLGSFRFLEALQAACVPVMLSNGWELPFSEVINWNQA 377
Query: 325 SMEIAVDRIPEIKTILQNITETKYRVL 351
++ + +I + +++I + K L
Sbjct: 378 AVIGDERLLLQIPSTIRSIHQDKILAL 404
>EXT1_CRIGR (Q9JK82) Exostosin-1 (EC 2.4.1.-) (Heparan sulfate
copolymerase) (Multiple exostoses protein 1 homolog)
Length = 746
Score = 67.0 bits (162), Expect = 8e-11
Identities = 82/327 (25%), Positives = 127/327 (38%), Gaps = 70/327 (21%)
Query: 59 FKVWVYKEGEQPLVHDGPVNNKYSIEGQFIDEMDTSNKSPFKATHPELAHVFFLPFSVSK 118
FKV+VY + + + + N +IEG S F + P A +F L
Sbjct: 114 FKVYVYPQQKGEKIAESYQNILAAIEG-----------SRFYTSDPSQACLFVLSLDTLD 162
Query: 119 VIRYVYKPRKSRSDYNPHRLQLLVEDYIKIVANKYPYWNISQGADHFLLSCHDWGPRVSY 178
R S H L+ V+ WN G +H + + + G Y
Sbjct: 163 --------RDQLSPQYVHNLRSKVQSL--------HLWN--NGRNHLIFNLYS-GTWPDY 203
Query: 179 ANPKLFKHFIRALCNANTS-EGFWPNRDVSIPQLNLPVGKLG-----------PP----- 221
F L A+ S E F PN DVSIP + + G PP
Sbjct: 204 TEDVGFDIGQAMLAKASISTENFRPNFDVSIPLFSKDHPRTGGERGFLKFNTIPPLRKYM 263
Query: 222 -----------------NTDQHPNNRTILTFFAGGAHGKIRKKLLKSWKDKDEEVQVHEY 264
N H +N + HGK +K S D+D EY
Sbjct: 264 LVFKGKRYLTGIGSDTRNALYHVHNGEDVLLLTTCKHGKDWQKHKDSRCDRDNT----EY 319
Query: 265 LPKGQDYTKLMGLSKFCLCPSGHEVASPRVVEAIYAGCVPVIICDNYSLPFSDVLNWSQF 324
+ DY +++ + FCL P G + S R +EA+ A CVPV++ + + LPFS+V+NW+Q
Sbjct: 320 --EKYDYREMLHNATFCLVPRGRRLGSFRFLEALQAACVPVMLSNGWELPFSEVINWNQA 377
Query: 325 SMEIAVDRIPEIKTILQNITETKYRVL 351
++ + +I + +++I + K L
Sbjct: 378 AVIGDERLLLQIPSTIRSIHQDKILAL 404
>EXT1_CAEEL (O01704) Multiple exostoses homolog 1 (Related to
mammalian RIB protein 1)
Length = 378
Score = 63.9 bits (154), Expect = 7e-10
Identities = 29/89 (32%), Positives = 52/89 (57%)
Query: 270 DYTKLMGLSKFCLCPSGHEVASPRVVEAIYAGCVPVIICDNYSLPFSDVLNWSQFSMEIA 329
+Y +L+ S FCL P G + S R +E + +GCVPV+I D++ LPFS+ ++W+ ++ +A
Sbjct: 257 EYDELLANSTFCLVPRGRRLGSFRFLETLRSGCVPVVISDSWILPFSETIDWNSAAIVVA 316
Query: 330 VDRIPEIKTILQNITETKYRVLYSNVRRV 358
I +L + + + + L + R V
Sbjct: 317 ERDALSIPELLMSTSRRRVKELRESARNV 345
>EXT2_HUMAN (Q93063) Exostosin-2 (EC 2.4.1.224) (EC 2.4.1.225)
(Glucuronosyl-N-acetylglucosaminyl-proteoglycan/N-
acetylglucosaminyl-proteoglycan
4-alpha-N-acetylglucosaminyltransferase) (Putative tumor
suppressor protein EXT2) (Multiple exostoses protein
Length = 718
Score = 63.2 bits (152), Expect = 1e-09
Identities = 27/87 (31%), Positives = 53/87 (60%)
Query: 270 DYTKLMGLSKFCLCPSGHEVASPRVVEAIYAGCVPVIICDNYSLPFSDVLNWSQFSMEIA 329
DY +++ + FC+ G + + + + AGCVPV+I D+Y LPFS+VL+W + S+ +
Sbjct: 307 DYPQVLQEATFCVVLRGARLGQAVLSDVLQAGCVPVVIADSYILPFSEVLDWKRASVVVP 366
Query: 330 VDRIPEIKTILQNITETKYRVLYSNVR 356
+++ ++ +ILQ+I + + + R
Sbjct: 367 EEKMSDVYSILQSIPQRQIEEMQRQAR 393
>EXT2_BOVIN (O77783) Exostosin-2 (EC 2.4.1.224) (EC 2.4.1.225)
(Glucuronosyl-N-acetylglucosaminyl-proteoglycan/N-
acetylglucosaminyl-proteoglycan
4-alpha-N-acetylglucosaminyltransferase) (HS-polymerase)
(HS-POL) (Multiple exostoses protein 2 homolog)
Length = 718
Score = 62.8 bits (151), Expect = 2e-09
Identities = 28/87 (32%), Positives = 52/87 (59%)
Query: 270 DYTKLMGLSKFCLCPSGHEVASPRVVEAIYAGCVPVIICDNYSLPFSDVLNWSQFSMEIA 329
DY +++ + FC+ G + + + + AGCVPVII D+Y LPFS+VL+W + S+ +
Sbjct: 307 DYPQVLQEATFCMVLRGARLGQAVLSDVLRAGCVPVIIADSYVLPFSEVLDWKRASVVVP 366
Query: 330 VDRIPEIKTILQNITETKYRVLYSNVR 356
+++ ++ +ILQ+I + + R
Sbjct: 367 EEKMSDVYSILQSIPRRQIEEMQRQAR 393
>EXT2_MOUSE (P70428) Exostosin-2 (EC 2.4.1.224) (EC 2.4.1.225)
(Glucuronosyl-N-acetylglucosaminyl-proteoglycan/N-
acetylglucosaminyl-proteoglycan
4-alpha-N-acetylglucosaminyltransferase) (Multiple
exostoses protein 2 homolog)
Length = 718
Score = 61.2 bits (147), Expect = 4e-09
Identities = 27/87 (31%), Positives = 51/87 (58%)
Query: 270 DYTKLMGLSKFCLCPSGHEVASPRVVEAIYAGCVPVIICDNYSLPFSDVLNWSQFSMEIA 329
DY +++ + FC + + + + AGCVPV+I D+Y LPFS+VL+W + S+ +
Sbjct: 307 DYPQVLQEATFCTVLRRARLGQAVLSDVLQAGCVPVVIADSYILPFSEVLDWKKASVVVP 366
Query: 330 VDRIPEIKTILQNITETKYRVLYSNVR 356
+++ ++ +ILQNI + + + R
Sbjct: 367 EEKMSDVYSILQNIPQRQIEEMQRQAR 393
>EXL1_MOUSE (Q9JKV7) Exostosin-like 1 (EC 2.4.1.224)
(Glucuronosyl-N-acetylglucosaminyl-proteoglycan
4-alpha-N-acetylglucosaminyltransferase) (Exostosin-L)
(Multiple exostosis-like protein)
Length = 669
Score = 51.6 bits (122), Expect = 3e-06
Identities = 23/55 (41%), Positives = 40/55 (71%), Gaps = 2/55 (3%)
Query: 280 FCLCPSGHEVASPRVVEAIYAGCVPVIICDNYSLPFSDVLNWSQFSMEIAVDRIP 334
FCL P GH A+ ++A+ AGC+PV++ + LPFS+V++W++ ++ IA +R+P
Sbjct: 266 FCLIP-GHRSATSCFLQALQAGCIPVLLSPRWELPFSEVIDWTKAAI-IADERLP 318
>EXL1_HUMAN (Q92935) Exostosin-like 1 (EC 2.4.1.224)
(Glucuronosyl-N-acetylglucosaminyl-proteoglycan
4-alpha-N-acetylglucosaminyltransferase) (Exostosin-L)
(Multiple exostosis-like protein)
Length = 676
Score = 50.4 bits (119), Expect = 8e-06
Identities = 23/73 (31%), Positives = 46/73 (62%), Gaps = 2/73 (2%)
Query: 280 FCLCPSGHEVASPRVVEAIYAGCVPVIICDNYSLPFSDVLNWSQFSMEIAVDRIP-EIKT 338
FCL A+ R ++A+ AGC+PV++ + LPFS+V++W++ ++ +A +R+P ++
Sbjct: 272 FCLISGHRPEAASRFLQALQAGCIPVLLSPRWELPFSEVIDWTKAAI-VADERLPLQVLA 330
Query: 339 ILQNITETKYRVL 351
LQ ++ + L
Sbjct: 331 ALQEMSPARVLAL 343
>EXT2_CAEEL (O01705) Exostosin-2 (EC 2.4.1.223) (EC 2.4.1.224)
(Glucuronyl-galactosyl-proteoglycan/Glucuronosyl-N-
acetylglucosaminyl-proteoglycan
4-alpha-N-acetylglucosaminyltransferase) (Multiple
exostoses homolog 2)
Length = 814
Score = 47.4 bits (111), Expect = 7e-05
Identities = 23/71 (32%), Positives = 44/71 (61%), Gaps = 3/71 (4%)
Query: 273 KLMGLSKFC-LCPSGHEVASPRVVEAIYAGCVPVIICDNYSLPFSDVLNWSQFSMEIAVD 331
+L+G S FC L PS E+ + ++ GC+P+I+ ++ LPF D+++W + + + +
Sbjct: 330 QLIGSSTFCFLLPS--EMFFQDFLSSLQLGCIPIILSNSQLLPFQDLIDWRRATYRLPLA 387
Query: 332 RIPEIKTILQN 342
R+PE I+Q+
Sbjct: 388 RLPEAHFIVQS 398
>EXL3_HUMAN (O43909) Exostosin-like 3 (EC 2.4.1.223)
(Glucuronyl-galactosyl-proteoglycan
4-alpha-N-acetylglucosaminyltransferase) (Putative tumor
suppressor protein EXTL3) (Multiple exostosis-like
protein 3) (Hereditary multiple exostoses gene isolog)
(EX
Length = 919
Score = 44.3 bits (103), Expect = 6e-04
Identities = 21/85 (24%), Positives = 48/85 (55%), Gaps = 7/85 (8%)
Query: 269 QDYTKLMGLSKFCLCPSGHEV-------ASPRVVEAIYAGCVPVIICDNYSLPFSDVLNW 321
+D +L+ LS F L + + + R+ EA+ G VPV++ + LP+ D+L W
Sbjct: 419 EDRLELLKLSTFALIITPGDPRLVISSGCATRLFEALEVGAVPVVLGEQVQLPYQDMLQW 478
Query: 322 SQFSMEIAVDRIPEIKTILQNITET 346
++ ++ + R+ E+ +L++++++
Sbjct: 479 NEAALVVPKPRVTEVHFLLRSLSDS 503
>EXL3_MOUSE (Q9WVL6) Exostosin-like 3 (EC 2.4.1.-) (Multiple
exostosis-like protein 3)
Length = 918
Score = 43.9 bits (102), Expect = 7e-04
Identities = 21/85 (24%), Positives = 48/85 (55%), Gaps = 7/85 (8%)
Query: 269 QDYTKLMGLSKFCLCPSGHEV-------ASPRVVEAIYAGCVPVIICDNYSLPFSDVLNW 321
+D +L+ LS F L + + + R+ EA+ G VPV++ + LP+ D+L W
Sbjct: 418 EDRLELLKLSTFALIITPGDPRLLISSGCATRLFEALEVGAVPVVLGEQVQLPYHDMLQW 477
Query: 322 SQFSMEIAVDRIPEIKTILQNITET 346
++ ++ + R+ E+ +L++++++
Sbjct: 478 NEAALVVPKPRVTEVHFLLRSLSDS 502
>RFC1_YEAST (P38630) Activator 1 95 kDa subunit (Replication factor
C subunit 1) (Replication factor C1) (Cell division
control protein 44)
Length = 861
Score = 33.1 bits (74), Expect = 1.3
Identities = 14/46 (30%), Positives = 25/46 (53%)
Query: 333 IPEIKTILQNITETKYRVLYSNVRRVRKHFEMNRPAKPFDLIHMIL 378
I ++ +L I+ T + + N+ + K +E N KPFD+ H +L
Sbjct: 514 IRQVINLLSTISTTTKTINHENINEISKAWEKNIALKPFDIAHKML 559
>YF71_PASMU (Q9CKN9) Hypothetical UPF0004 protein PM1571
Length = 446
Score = 32.7 bits (73), Expect = 1.7
Identities = 21/70 (30%), Positives = 35/70 (50%), Gaps = 3/70 (4%)
Query: 100 KATHPELAHVFFLPFSVSKVIRYVYKPRKSRSDYNPHRLQLLVEDYIKIVANKYPYWNIS 159
+ HP++ V P S V++ V+K + YNP+ + L+ + +K+ Y Y IS
Sbjct: 89 REVHPKVLEVTG-PHSYEAVMQQVHK-YVPKPAYNPY-VNLVPKQGVKLTPKHYAYLKIS 145
Query: 160 QGADHFLLSC 169
+G DH C
Sbjct: 146 EGCDHRCTFC 155
>RNH3_CHLTR (O84011) Ribonuclease HIII (EC 3.1.26.-) (RNase HIII)
Length = 300
Score = 31.6 bits (70), Expect = 3.7
Identities = 19/77 (24%), Positives = 37/77 (47%), Gaps = 11/77 (14%)
Query: 282 LCPSGHEVASPRVVEAIYAGCVPVIICDNYSLPFSDVLNWSQFSMEIAVDRIPEIKTILQ 341
LC +G +SP+ +EA+Y ICD+ +P + +L+ +Q + ++
Sbjct: 103 LCTAGVYASSPQAIEALY----KTSICDSKLIPDAKILSLAQNIRSLCACKV-------I 151
Query: 342 NITETKYRVLYSNVRRV 358
+ KY LY+N + +
Sbjct: 152 TLFPEKYNALYANFQNL 168
>YLIG_ECOLI (P75802) Hypothetical UPF0004 protein yliG
Length = 441
Score = 31.2 bits (69), Expect = 4.9
Identities = 19/78 (24%), Positives = 36/78 (45%), Gaps = 7/78 (8%)
Query: 94 SNKSPFKATHPELAHVFFLPFSVSKVIRYV--YKPRKSRSDYNPHRLQLLVEDYIKIVAN 151
+ + + HP++ + P S +V+ +V Y P+ + + L L+ E +K+
Sbjct: 85 AKEDQIREVHPKVLEITG-PHSYEQVLEHVHHYVPKPKHNPF----LSLVPEQGVKLTPR 139
Query: 152 KYPYWNISQGADHFLLSC 169
Y Y IS+G +H C
Sbjct: 140 HYAYLKISEGCNHRCTFC 157
>EF3_SCHPO (O94489) Elongation factor 3 (EF-3)
Length = 1047
Score = 31.2 bits (69), Expect = 4.9
Identities = 26/105 (24%), Positives = 47/105 (44%), Gaps = 18/105 (17%)
Query: 13 QTRALIQRAIRSKKSTTNMKQSFVPKGSIYLNPHAFHQSHKEMVKRFKVWVYKEGEQPLV 72
+ R+++QRAI + + N+ +P+ S NP ++ K ++ KV +E
Sbjct: 310 ECRSVVQRAITTLERVGNVVDGKIPEVSTAANPEVCLETLKAVLGEIKVPTNEE------ 363
Query: 73 HDGPVNNKY--SIEGQFIDEMDTSNKS------PFKATHPELAHV 109
V KY +I Q ++E D N+S P+ + AH+
Sbjct: 364 ----VIAKYVANIAAQLVEEKDNENESWVLNITPYLTAFIDEAHI 404
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.321 0.137 0.426
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 50,049,854
Number of Sequences: 164201
Number of extensions: 2214613
Number of successful extensions: 4619
Number of sequences better than 10.0: 17
Number of HSP's better than 10.0 without gapping: 13
Number of HSP's successfully gapped in prelim test: 4
Number of HSP's that attempted gapping in prelim test: 4603
Number of HSP's gapped (non-prelim): 20
length of query: 398
length of database: 59,974,054
effective HSP length: 112
effective length of query: 286
effective length of database: 41,583,542
effective search space: 11892893012
effective search space used: 11892893012
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 67 (30.4 bits)
Medicago: description of AC144657.9


