

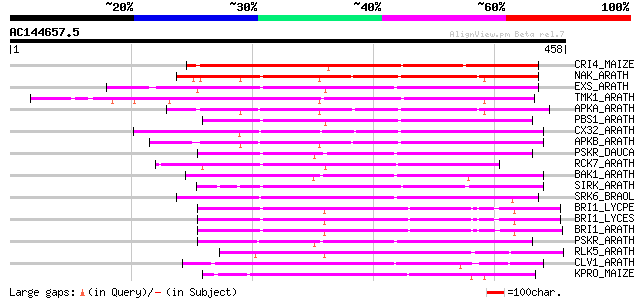
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144657.5 - phase: 0
(458 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
CRI4_MAIZE (O24585) Putative receptor protein kinase CRINKLY4 pr... 224 4e-58
NAK_ARATH (P43293) Probable serine/threonine-protein kinase NAK ... 213 6e-55
EXS_ARATH (Q9LYN8) Leucine-rich repeat receptor protein kinase E... 211 2e-54
TMK1_ARATH (P43298) Putative receptor protein kinase TMK1 precur... 209 9e-54
APKA_ARATH (Q06548) Protein kinase APK1A, chloroplast precursor ... 209 9e-54
PBS1_ARATH (Q9FE20) Serine/threonine-protein kinase PBS1 (EC 2.7... 201 4e-51
CX32_ARATH (P27450) Probable serine/threonine-protein kinase Cx3... 199 2e-50
APKB_ARATH (P46573) Protein kinase APK1B, chloroplast precursor ... 198 2e-50
PSKR_DAUCA (Q8LPB4) Phytosulfokine receptor precursor (EC 2.7.1.... 196 1e-49
RCK7_ARATH (Q9LQQ8) Putative serine/threonine-protein kinase RLC... 193 7e-49
BAK1_ARATH (Q94F62) BRASSINOSTEROID INSENSITIVE 1-associated rec... 193 7e-49
SIRK_ARATH (O64483) Senescence-induced receptor-like serine/thre... 188 3e-47
SRK6_BRAOL (Q09092) Putative serine/threonine-protein kinase rec... 187 6e-47
BRI1_LYCPE (Q8L899) Systemin receptor SR160 precursor (EC 2.7.1.... 182 2e-45
BRI1_LYCES (Q8GUQ5) Brassinosteroid LRR receptor kinase precurso... 182 2e-45
BRI1_ARATH (O22476) BRASSINOSTEROID INSENSITIVE 1 precursor (EC ... 182 2e-45
PSKR_ARATH (Q9ZVR7) Putative phytosulfokine receptor precursor (... 181 5e-45
RLK5_ARATH (P47735) Receptor-like protein kinase 5 precursor (EC... 175 2e-43
CLV1_ARATH (Q9SYQ8) Receptor protein kinase CLAVATA1 precursor (... 168 3e-41
KPRO_MAIZE (P17801) Putative receptor protein kinase ZmPK1 precu... 165 3e-40
>CRI4_MAIZE (O24585) Putative receptor protein kinase CRINKLY4
precursor (EC 2.7.1.-)
Length = 901
Score = 224 bits (571), Expect = 4e-58
Identities = 124/294 (42%), Positives = 185/294 (62%), Gaps = 9/294 (3%)
Query: 147 KLSRLQSLHLNLSQVSKATRNFSETLQIGEGGFGTVYKAHLDDGLVVAVKRA-KREHFES 205
K+ R Q + ++ +AT FSE Q+G+G F V+K L DG VVAVKRA K +
Sbjct: 486 KIRRAQEF--SYEELEQATGGFSEDSQVGKGSFSCVFKGILRDGTVVAVKRAIKASDVKK 543
Query: 206 LRTEFSSEVELLAKIDHRNLVKLLGYIDKGNERILITEFVANGTLREHLDGLRGKI---L 262
EF +E++LL++++H +L+ LLGY + G+ER+L+ EF+A+G+L +HL G + L
Sbjct: 544 SSKEFHNELDLLSRLNHAHLLNLLGYCEDGSERLLVYEFMAHGSLYQHLHGKDPNLKKRL 603
Query: 263 DFNQRLEIAIDVAHGLTYLHLYAEKQIIHRDVKSSNILLTESMRAKVADFGFAKLGPVNN 322
++ +R+ IA+ A G+ YLH YA +IHRD+KSSNIL+ E A+VADFG + LGP ++
Sbjct: 604 NWARRVTIAVQAARGIEYLHGYACPPVIHRDIKSSNILIDEDHNARVADFGLSILGPADS 663
Query: 323 DHTHISTKVKGTVGYLDPEYMKTYHLTPKSDVYSFGILLLEILTGRRPVELKKSAEERVT 382
T +S GT+GYLDPEY + ++LT KSDVYSFG++LLEIL+GR+ ++++ EE
Sbjct: 664 G-TPLSELPAGTLGYLDPEYYRLHYLTTKSDVYSFGVVLLEILSGRKAIDMQ--FEEGNI 720
Query: 383 LRWAFRKYNEGSVVALLDPLMQEAVKTDVAVKMFDLAFNCAAPVRSDRPDMKTV 436
+ WA G + A+LDP++ + K+ +A C DRP M V
Sbjct: 721 VEWAVPLIKAGDIFAILDPVLSPPSDLEALKKIASVACKCVRMRGKDRPSMDKV 774
>NAK_ARATH (P43293) Probable serine/threonine-protein kinase NAK (EC
2.7.1.37)
Length = 389
Score = 213 bits (543), Expect = 6e-55
Identities = 128/318 (40%), Positives = 194/318 (60%), Gaps = 23/318 (7%)
Query: 138 SPSRFSMSPKLSR--LQSLHL---NLSQVSKATRNFSETLQIGEGGFGTVYKAHLDD--- 189
S + FS P+ LQ+ +L +LS++ ATRNF +GEGGFG V+K +D+
Sbjct: 33 STASFSYMPRTEGEILQNANLKNFSLSELKSATRNFRPDSVVGEGGFGCVFKGWIDESSL 92
Query: 190 -------GLVVAVKRAKREHFESLRTEFSSEVELLAKIDHRNLVKLLGYIDKGNERILIT 242
G+V+AVKR +E F+ R E+ +E+ L ++DH NLVKL+GY + R+L+
Sbjct: 93 APSKPGTGIVIAVKRLNQEGFQGHR-EWLAEINYLGQLDHPNLVKLIGYCLEEEHRLLVY 151
Query: 243 EFVANGTLREHL--DGLRGKILDFNQRLEIAIDVAHGLTYLHLYAEKQIIHRDVKSSNIL 300
EF+ G+L HL G + L +N R+ +A+ A GL +LH A+ Q+I+RD K+SNIL
Sbjct: 152 EFMTRGSLENHLFRRGTFYQPLSWNTRVRMALGAARGLAFLH-NAQPQVIYRDFKASNIL 210
Query: 301 LTESMRAKVADFGFAKLGPVNNDHTHISTKVKGTVGYLDPEYMKTYHLTPKSDVYSFGIL 360
L + AK++DFG A+ GP+ D++H+ST+V GT GY PEY+ T HL+ KSDVYSFG++
Sbjct: 211 LDSNYNAKLSDFGLARDGPMG-DNSHVSTRVMGTQGYAAPEYLATGHLSVKSDVYSFGVV 269
Query: 361 LLEILTGRRPVELKKSAEERVTLRWAFRKY--NEGSVVALLDPLMQEAVKTDVAVKMFDL 418
LLE+L+GRR ++ + E + WA R Y N+ ++ ++DP +Q A+K+ L
Sbjct: 270 LLELLSGRRAIDKNQPVGEHNLVDWA-RPYLTNKRRLLRVMDPRLQGQYSLTRALKIAVL 328
Query: 419 AFNCAAPVRSDRPDMKTV 436
A +C + RP M +
Sbjct: 329 ALDCISIDAKSRPTMNEI 346
>EXS_ARATH (Q9LYN8) Leucine-rich repeat receptor protein kinase EXS
precursor (EC 2.7.1.37) (Extra sporogenous cells protein)
(EXCESS MICROSPOROCYTES1 protein)
Length = 1192
Score = 211 bits (538), Expect = 2e-54
Identities = 133/362 (36%), Positives = 202/362 (55%), Gaps = 14/362 (3%)
Query: 81 GALLVCCAVVCPCFYGKRRKATSHAVLEKD-PNSMELGSSFEPSVSDKIPASPLRVPPSP 139
G +L +V + RR A + V ++D P ME E + + + + S
Sbjct: 832 GLMLGFTIIVFVFVFSLRRWAMTKRVKQRDDPERME-----ESRLKGFVDQNLYFLSGSR 886
Query: 140 SRFSMSPKLSRLQS--LHLNLSQVSKATRNFSETLQIGEGGFGTVYKAHLDDGLVVAVKR 197
SR +S ++ + L + L + +AT +FS+ IG+GGFGTVYKA L VAVK+
Sbjct: 887 SREPLSINIAMFEQPLLKVRLGDIVEATDHFSKKNIIGDGGFGTVYKACLPGEKTVAVKK 946
Query: 198 AKREHFESLRTEFSSEVELLAKIDHRNLVKLLGYIDKGNERILITEFVANGTLREHLDGL 257
+ R EF +E+E L K+ H NLV LLGY E++L+ E++ NG+L L
Sbjct: 947 LSEAKTQGNR-EFMAEMETLGKVKHPNLVSLLGYCSFSEEKLLVYEYMVNGSLDHWLRNQ 1005
Query: 258 RG--KILDFNQRLEIAIDVAHGLTYLHLYAEKQIIHRDVKSSNILLTESMRAKVADFGFA 315
G ++LD+++RL+IA+ A GL +LH IIHRD+K+SNILL KVADFG A
Sbjct: 1006 TGMLEVLDWSKRLKIAVGAARGLAFLHHGFIPHIIHRDIKASNILLDGDFEPKVADFGLA 1065
Query: 316 KLGPVNNDHTHISTKVKGTVGYLDPEYMKTYHLTPKSDVYSFGILLLEILTGRRPVELK- 374
+L ++ +H+ST + GT GY+ PEY ++ T K DVYSFG++LLE++TG+ P
Sbjct: 1066 RL--ISACESHVSTVIAGTFGYIPPEYGQSARATTKGDVYSFGVILLELVTGKEPTGPDF 1123
Query: 375 KSAEERVTLRWAFRKYNEGSVVALLDPLMQEAVKTDVAVKMFDLAFNCAAPVRSDRPDMK 434
K +E + WA +K N+G V ++DPL+ + +++ +A C A + RP+M
Sbjct: 1124 KESEGGNLVGWAIQKINQGKAVDVIDPLLVSVALKNSQLRLLQIAMLCLAETPAKRPNML 1183
Query: 435 TV 436
V
Sbjct: 1184 DV 1185
>TMK1_ARATH (P43298) Putative receptor protein kinase TMK1 precursor
(EC 2.7.1.-)
Length = 942
Score = 209 bits (533), Expect = 9e-54
Identities = 154/439 (35%), Positives = 224/439 (50%), Gaps = 32/439 (7%)
Query: 18 FGSEVVLQTKGCGDNLVANSYSSHVNLPSTAGRKLLQKDLNNNSTSQGDPKQVSSSQVGI 77
F S VV+ T G D S S S +G +N + +G SS+ +GI
Sbjct: 429 FRSNVVVNTNGNPDIGKDKSSLSSPGSSSPSGGS--GSGINGDKDRRG---MKSSTFIGI 483
Query: 78 FAGGAL-----LVCCAVVCPCFYGKRRKA-----TSHAVLEKDPNSMELGSSFEPSVSDK 127
G L + ++ C+Y KR+K +S+AV+ +S S + +V+
Sbjct: 484 IVGSVLGGLLSIFLIGLLVFCWYKKRQKRFSGSESSNAVVVHPRHSGSDNESVKITVAGS 543
Query: 128 IPA----SPLRVPPSPSRFSMSPKLSRLQSLHLNLSQVSKATRNFSETLQIGEGGFGTVY 183
+ S P S + ++ ++ +++ + T NFS +G GGFG VY
Sbjct: 544 SVSVGGISDTYTLPGTSEVGDNIQMVEAGNMLISIQVLRSVTNNFSSDNILGSGGFGVVY 603
Query: 184 KAHLDDGLVVAVKRAKREHFESLR-TEFSSEVELLAKIDHRNLVKLLGYIDKGNERILIT 242
K L DG +AVKR + EF SE+ +L K+ HR+LV LLGY GNE++L+
Sbjct: 604 KGELHDGTKIAVKRMENGVIAGKGFAEFKSEIAVLTKVRHRHLVTLLGYCLDGNEKLLVY 663
Query: 243 EFVANGTLREHL-----DGLRGKILDFNQRLEIAIDVAHGLTYLHLYAEKQIIHRDVKSS 297
E++ GTL HL +GL K L + QRL +A+DVA G+ YLH A + IHRD+K S
Sbjct: 664 EYMPQGTLSRHLFEWSEEGL--KPLLWKQRLTLALDVARGVEYLHGLAHQSFIHRDLKPS 721
Query: 298 NILLTESMRAKVADFGFAKLGPVNNDHTHISTKVKGTVGYLDPEYMKTYHLTPKSDVYSF 357
NILL + MRAKVADFG +L P I T++ GT GYL PEY T +T K DVYSF
Sbjct: 722 NILLGDDMRAKVADFGLVRLAPEGKG--SIETRIAGTFGYLAPEYAVTGRVTTKVDVYSF 779
Query: 358 GILLLEILTGRRPVELKKSAEERVTLRWAFRKY--NEGSVVALLDPLMQEAVKTDVAV-K 414
G++L+E++TGR+ ++ + E + W R Y E S +D + +T +V
Sbjct: 780 GVILMELITGRKSLDESQPEESIHLVSWFKRMYINKEASFKKAIDTTIDLDEETLASVHT 839
Query: 415 MFDLAFNCAAPVRSDRPDM 433
+ +LA +C A RPDM
Sbjct: 840 VAELAGHCCAREPYQRPDM 858
>APKA_ARATH (Q06548) Protein kinase APK1A, chloroplast precursor (EC
2.7.1.-)
Length = 410
Score = 209 bits (533), Expect = 9e-54
Identities = 128/330 (38%), Positives = 194/330 (58%), Gaps = 20/330 (6%)
Query: 130 ASPLRVPPSPSRFSMSPKLSRLQSLHLNLSQVSKATRNFSETLQIGEGGFGTVYKAHLDD 189
AS + V PSP + L+S + +++ ATRNF +GEGGFG V+K +D+
Sbjct: 32 ASSVSVRPSPRTEGEILQSPNLKSF--SFAELKSATRNFRPDSVLGEGGFGCVFKGWIDE 89
Query: 190 ----------GLVVAVKRAKREHFESLRTEFSSEVELLAKIDHRNLVKLLGYIDKGNERI 239
GLV+AVK+ ++ ++ E+ +EV L + HR+LVKL+GY + R+
Sbjct: 90 KSLTASRPGTGLVIAVKKLNQDGWQG-HQEWLAEVNYLGQFSHRHLVKLIGYCLEDEHRL 148
Query: 240 LITEFVANGTLREHL--DGLRGKILDFNQRLEIAIDVAHGLTYLHLYAEKQIIHRDVKSS 297
L+ EF+ G+L HL GL + L + RL++A+ A GL +LH +E ++I+RD K+S
Sbjct: 149 LVYEFMPRGSLENHLFRRGLYFQPLSWKLRLKVALGAAKGLAFLHS-SETRVIYRDFKTS 207
Query: 298 NILLTESMRAKVADFGFAKLGPVNNDHTHISTKVKGTVGYLDPEYMKTYHLTPKSDVYSF 357
NILL AK++DFG AK GP+ D +H+ST+V GT GY PEY+ T HLT KSDVYSF
Sbjct: 208 NILLDSEYNAKLSDFGLAKDGPIG-DKSHVSTRVMGTHGYAAPEYLATGHLTTKSDVYSF 266
Query: 358 GILLLEILTGRRPVELKKSAEERVTLRWAFRKY--NEGSVVALLDPLMQEAVKTDVAVKM 415
G++LLE+L+GRR V+ + + ER + WA + Y N+ + ++D +Q+ + A K+
Sbjct: 267 GVVLLELLSGRRAVDKNRPSGERNLVEWA-KPYLVNKRKIFRVIDNRLQDQYSMEEACKV 325
Query: 416 FDLAFNCAAPVRSDRPDMKTVGEQLWAIRA 445
L+ C RP+M V L I++
Sbjct: 326 ATLSLRCLTTEIKLRPNMSEVVSHLEHIQS 355
>PBS1_ARATH (Q9FE20) Serine/threonine-protein kinase PBS1 (EC
2.7.1.37) (AvrPphB susceptible protein 1)
Length = 456
Score = 201 bits (510), Expect = 4e-51
Identities = 117/276 (42%), Positives = 164/276 (59%), Gaps = 6/276 (2%)
Query: 160 QVSKATRNFSETLQIGEGGFGTVYKAHLDD-GLVVAVKRAKREHFESLRTEFSSEVELLA 218
+++ AT NF +GEGGFG VYK LD G VVAVK+ R + R EF EV +L+
Sbjct: 78 ELAAATMNFHPDTFLGEGGFGRVYKGRLDSTGQVVAVKQLDRNGLQGNR-EFLVEVLMLS 136
Query: 219 KIDHRNLVKLLGYIDKGNERILITEFVANGTLREHLDGLRG--KILDFNQRLEIAIDVAH 276
+ H NLV L+GY G++R+L+ EF+ G+L +HL L + LD+N R++IA A
Sbjct: 137 LLHHPNLVNLIGYCADGDQRLLVYEFMPLGSLEDHLHDLPPDKEALDWNMRMKIAAGAAK 196
Query: 277 GLTYLHLYAEKQIIHRDVKSSNILLTESMRAKVADFGFAKLGPVNNDHTHISTKVKGTVG 336
GL +LH A +I+RD KSSNILL E K++DFG AKLGP D +H+ST+V GT G
Sbjct: 197 GLEFLHDKANPPVIYRDFKSSNILLDEGFHPKLSDFGLAKLGPTG-DKSHVSTRVMGTYG 255
Query: 337 YLDPEYMKTYHLTPKSDVYSFGILLLEILTGRRPVELKKSAEERVTLRWAFRKYNE-GSV 395
Y PEY T LT KSDVYSFG++ LE++TGR+ ++ + E+ + WA +N+
Sbjct: 256 YCAPEYAMTGQLTVKSDVYSFGVVFLELITGRKAIDSEMPHGEQNLVAWARPLFNDRRKF 315
Query: 396 VALLDPLMQEAVKTDVAVKMFDLAFNCAAPVRSDRP 431
+ L DP ++ T + +A C + RP
Sbjct: 316 IKLADPRLKGRFPTRALYQALAVASMCIQEQAATRP 351
>CX32_ARATH (P27450) Probable serine/threonine-protein kinase Cx32,
chloroplast precursor (EC 2.7.1.37)
Length = 419
Score = 199 bits (505), Expect = 2e-50
Identities = 125/350 (35%), Positives = 187/350 (52%), Gaps = 16/350 (4%)
Query: 103 SHAVLEKDPNSMELGSSFEPSVSDKIPASPLRVPPSPSRFSMSPKLSRLQSLHL-NLSQV 161
SHA N E S+ + + + S S S KL +L + N +
Sbjct: 20 SHATTNNHSNGTEFSSTTGATTNSSVGQQSQFSDISTGIISDSGKLLESPNLKVYNFLDL 79
Query: 162 SKATRNFSETLQIGEGGFGTVYKAHLD----------DGLVVAVKRAKREHFESLRTEFS 211
AT+NF +G+GGFG VY+ +D G++VA+KR E + E+
Sbjct: 80 KTATKNFKPDSMLGQGGFGKVYRGWVDATTLAPSRVGSGMIVAIKRLNSESVQGF-AEWR 138
Query: 212 SEVELLAKIDHRNLVKLLGYIDKGNERILITEFVANGTLREHLDGLRGKILDFNQRLEIA 271
SEV L + HRNLVKLLGY + E +L+ EF+ G+L HL R ++ R++I
Sbjct: 139 SEVNFLGMLSHRNLVKLLGYCREDKELLLVYEFMPKGSLESHLFR-RNDPFPWDLRIKIV 197
Query: 272 IDVAHGLTYLHLYAEKQIIHRDVKSSNILLTESMRAKVADFGFAKLGPVNNDHTHISTKV 331
I A GL +LH ++++I+RD K+SNILL + AK++DFG AKLGP + + +H++T++
Sbjct: 198 IGAARGLAFLHSL-QREVIYRDFKASNILLDSNYDAKLSDFGLAKLGPAD-EKSHVTTRI 255
Query: 332 KGTVGYLDPEYMKTYHLTPKSDVYSFGILLLEILTGRRPVELKKSAEERVTLRWAFRKY- 390
GT GY PEYM T HL KSDV++FG++LLEI+TG K+ + + W +
Sbjct: 256 MGTYGYAAPEYMATGHLYVKSDVFAFGVVLLEIMTGLTAHNTKRPRGQESLVDWLRPELS 315
Query: 391 NEGSVVALLDPLMQEAVKTDVAVKMFDLAFNCAAPVRSDRPDMKTVGEQL 440
N+ V ++D ++ T VA +M + +C P +RP MK V E L
Sbjct: 316 NKHRVKQIMDKGIKGQYTTKVATEMARITLSCIEPDPKNRPHMKEVVEVL 365
>APKB_ARATH (P46573) Protein kinase APK1B, chloroplast precursor (EC
2.7.1.-)
Length = 412
Score = 198 bits (504), Expect = 2e-50
Identities = 123/339 (36%), Positives = 186/339 (54%), Gaps = 22/339 (6%)
Query: 116 LGSSFEPSVSDKIPASPLRVPP-SPSRFSMSPKLSRLQSLHLNLSQVSKATRNFSETLQI 174
+ S S+ K + +R P + SP L +++ ATRNF +
Sbjct: 21 MSSEANDSLGSKSSSVSIRTNPRTEGEILQSPNLKSF-----TFAELKAATRNFRPDSVL 75
Query: 175 GEGGFGTVYKAHLDD----------GLVVAVKRAKREHFESLRTEFSSEVELLAKIDHRN 224
GEGGFG+V+K +D+ G+V+AVK+ ++ ++ E+ +EV L + H N
Sbjct: 76 GEGGFGSVFKGWIDEQTLTASKPGTGVVIAVKKLNQDGWQG-HQEWLAEVNYLGQFSHPN 134
Query: 225 LVKLLGYIDKGNERILITEFVANGTLREHL--DGLRGKILDFNQRLEIAIDVAHGLTYLH 282
LVKL+GY + R+L+ EF+ G+L HL G + L + RL++A+ A GL +LH
Sbjct: 135 LVKLIGYCLEDEHRLLVYEFMPRGSLENHLFRRGSYFQPLSWTLRLKVALGAAKGLAFLH 194
Query: 283 LYAEKQIIHRDVKSSNILLTESMRAKVADFGFAKLGPVNNDHTHISTKVKGTVGYLDPEY 342
AE +I+RD K+SNILL AK++DFG AK GP D +H+ST++ GT GY PEY
Sbjct: 195 -NAETSVIYRDFKTSNILLDSEYNAKLSDFGLAKDGPTG-DKSHVSTRIMGTYGYAAPEY 252
Query: 343 MKTYHLTPKSDVYSFGILLLEILTGRRPVELKKSAEERVTLRWAFRKY-NEGSVVALLDP 401
+ T HLT KSDVYS+G++LLE+L+GRR V+ + E+ + WA N+ + ++D
Sbjct: 253 LATGHLTTKSDVYSYGVVLLEVLSGRRAVDKNRPPGEQKLVEWARPLLANKRKLFRVIDN 312
Query: 402 LMQEAVKTDVAVKMFDLAFNCAAPVRSDRPDMKTVGEQL 440
+Q+ + A K+ LA C RP+M V L
Sbjct: 313 RLQDQYSMEEACKVATLALRCLTFEIKLRPNMNEVVSHL 351
>PSKR_DAUCA (Q8LPB4) Phytosulfokine receptor precursor (EC 2.7.1.37)
(Phytosulfokine LRR receptor kinase)
Length = 1021
Score = 196 bits (497), Expect = 1e-49
Identities = 114/280 (40%), Positives = 165/280 (58%), Gaps = 9/280 (3%)
Query: 156 LNLSQVSKATRNFSETLQIGEGGFGTVYKAHLDDGLVVAVKRAKREHFESLRTEFSSEVE 215
L+L + K+T +F++ IG GGFG VYKA L DG VA+KR + + R EF +EVE
Sbjct: 731 LSLDDILKSTSSFNQANIIGCGGFGLVYKATLPDGTKVAIKRLSGDTGQMDR-EFQAEVE 789
Query: 216 LLAKIDHRNLVKLLGYIDKGNERILITEFVANGTL----REHLDGLRGKILDFNQRLEIA 271
L++ H NLV LLGY + N+++LI ++ NG+L E +DG LD+ RL IA
Sbjct: 790 TLSRAQHPNLVHLLGYCNYKNDKLLIYSYMDNGSLDYWLHEKVDGPPS--LDWKTRLRIA 847
Query: 272 IDVAHGLTYLHLYAEKQIIHRDVKSSNILLTESMRAKVADFGFAKLGPVNNDHTHISTKV 331
A GL YLH E I+HRD+KSSNILL+++ A +ADFG A+L + TH++T +
Sbjct: 848 RGAAEGLAYLHQSCEPHILHRDIKSSNILLSDTFVAHLADFGLARL--ILPYDTHVTTDL 905
Query: 332 KGTVGYLDPEYMKTYHLTPKSDVYSFGILLLEILTGRRPVELKKSAEERVTLRWAFRKYN 391
GT+GY+ PEY + T K DVYSFG++LLE+LTGRRP+++ K R + W +
Sbjct: 906 VGTLGYIPPEYGQASVATYKGDVYSFGVVLLELLTGRRPMDVCKPRGSRDLISWVLQMKT 965
Query: 392 EGSVVALLDPLMQEAVKTDVAVKMFDLAFNCAAPVRSDRP 431
E + DP + + + + + ++A C RP
Sbjct: 966 EKRESEIFDPFIYDKDHAEEMLLVLEIACRCLGENPKTRP 1005
>RCK7_ARATH (Q9LQQ8) Putative serine/threonine-protein kinase
RLCKVII (EC 2.7.1.37)
Length = 423
Score = 193 bits (491), Expect = 7e-49
Identities = 118/301 (39%), Positives = 181/301 (59%), Gaps = 20/301 (6%)
Query: 121 EPSVSDKIPASPLRVPPSPSRFSMSPKLSR-LQSLHLN------------LSQVSKATRN 167
+PS SD SP R + +LS ++ L+LN ++++AT N
Sbjct: 44 QPS-SDSTKVSPYRDVNNEGGVGKEDQLSLDVKGLNLNDQVTGKKAQTFTFQELAEATGN 102
Query: 168 FSETLQIGEGGFGTVYKAHLDD-GLVVAVKRAKREHFESLRTEFSSEVELLAKIDHRNLV 226
F +GEGGFG V+K ++ VVA+K+ R + +R EF EV L+ DH NLV
Sbjct: 103 FRSDCFLGEGGFGKVFKGTIEKLDQVVAIKQLDRNGVQGIR-EFVVEVLTLSLADHPNLV 161
Query: 227 KLLGYIDKGNERILITEFVANGTLREHLDGLRG--KILDFNQRLEIAIDVAHGLTYLHLY 284
KL+G+ +G++R+L+ E++ G+L +HL L K LD+N R++IA A GL YLH
Sbjct: 162 KLIGFCAEGDQRLLVYEYMPQGSLEDHLHVLPSGKKPLDWNTRMKIAAGAARGLEYLHDR 221
Query: 285 AEKQIIHRDVKSSNILLTESMRAKVADFGFAKLGPVNNDHTHISTKVKGTVGYLDPEYMK 344
+I+RD+K SNILL E + K++DFG AK+GP + D TH+ST+V GT GY P+Y
Sbjct: 222 MTPPVIYRDLKCSNILLGEDYQPKLSDFGLAKVGP-SGDKTHVSTRVMGTYGYCAPDYAM 280
Query: 345 TYHLTPKSDVYSFGILLLEILTGRRPVELKKSAEERVTLRWAFRKYNE-GSVVALLDPLM 403
T LT KSD+YSFG++LLE++TGR+ ++ K+ +++ + WA + + + ++DPL+
Sbjct: 281 TGQLTFKSDIYSFGVVLLELITGRKAIDNTKTRKDQNLVGWARPLFKDRRNFPKMVDPLL 340
Query: 404 Q 404
Q
Sbjct: 341 Q 341
>BAK1_ARATH (Q94F62) BRASSINOSTEROID INSENSITIVE 1-associated
receptor kinase 1 precursor (EC 2.7.1.37)
(BRI1-associated receptor kinase 1) (Somatic
embryogenesis receptor-like kinase 3)
Length = 615
Score = 193 bits (491), Expect = 7e-49
Identities = 113/301 (37%), Positives = 175/301 (57%), Gaps = 10/301 (3%)
Query: 146 PKLSRLQSLHLNLSQVSKATRNFSETLQIGEGGFGTVYKAHLDDGLVVAVKRAKREHFES 205
P++ Q +L ++ A+ NFS +G GGFG VYK L DG +VAVKR K E +
Sbjct: 267 PEVHLGQLKRFSLRELQVASDNFSNKNILGRGGFGKVYKGRLADGTLVAVKRLKEERTQG 326
Query: 206 LRTEFSSEVELLAKIDHRNLVKLLGYIDKGNERILITEFVANGT----LREHLDGLRGKI 261
+F +EVE+++ HRNL++L G+ ER+L+ ++ANG+ LRE +
Sbjct: 327 GELQFQTEVEMISMAVHRNLLRLRGFCMTPTERLLVYPYMANGSVASCLRERPES--QPP 384
Query: 262 LDFNQRLEIAIDVAHGLTYLHLYAEKQIIHRDVKSSNILLTESMRAKVADFGFAKLGPVN 321
LD+ +R IA+ A GL YLH + + +IIHRDVK++NILL E A V DFG AKL ++
Sbjct: 385 LDWPKRQRIALGSARGLAYLHDHCDPKIIHRDVKAANILLDEEFEAVVGDFGLAKL--MD 442
Query: 322 NDHTHISTKVKGTVGYLDPEYMKTYHLTPKSDVYSFGILLLEILTGRRPVELKKSA--EE 379
TH++T V+GT+G++ PEY+ T + K+DV+ +G++LLE++TG+R +L + A ++
Sbjct: 443 YKDTHVTTAVRGTIGHIAPEYLSTGKSSEKTDVFGYGVMLLELITGQRAFDLARLANDDD 502
Query: 380 RVTLRWAFRKYNEGSVVALLDPLMQEAVKTDVAVKMFDLAFNCAAPVRSDRPDMKTVGEQ 439
+ L W E + AL+D +Q K + ++ +A C +RP M V
Sbjct: 503 VMLLDWVKGLLKEKKLEALVDVDLQGNYKDEEVEQLIQVALLCTQSSPMERPKMSEVVRM 562
Query: 440 L 440
L
Sbjct: 563 L 563
>SIRK_ARATH (O64483) Senescence-induced receptor-like
serine/threonine kinase precursor (FLG22-induced
receptor-like kinase 1)
Length = 876
Score = 188 bits (477), Expect = 3e-47
Identities = 113/287 (39%), Positives = 166/287 (57%), Gaps = 8/287 (2%)
Query: 155 HLNLSQVSKATRNFSETLQIGEGGFGTVYKAHLDDGLVVAVKRAKREHFESLRTEFSSEV 214
+ S+V T NF IG+GGFG VY ++ G VAVK E + + EF +EV
Sbjct: 563 YFKYSEVVNITNNFERV--IGKGGFGKVYHGVIN-GEQVAVKVLSEESAQGYK-EFRAEV 618
Query: 215 ELLAKIDHRNLVKLLGYIDKGNERILITEFVANGTLREHLDGLRGKILDFNQRLEIAIDV 274
+LL ++ H NL L+GY ++ N +LI E++AN L ++L G R IL + +RL+I++D
Sbjct: 619 DLLMRVHHTNLTSLVGYCNEINHMVLIYEYMANENLGDYLAGKRSFILSWEERLKISLDA 678
Query: 275 AHGLTYLHLYAEKQIIHRDVKSSNILLTESMRAKVADFGFAKLGPVNNDHTHISTKVKGT 334
A GL YLH + I+HRDVK +NILL E ++AK+ADFG ++ V IST V G+
Sbjct: 679 AQGLEYLHNGCKPPIVHRDVKPTNILLNEKLQAKMADFGLSRSFSVEGS-GQISTVVAGS 737
Query: 335 VGYLDPEYMKTYHLTPKSDVYSFGILLLEILTGRRPVELKKSAEERVTLRWAFRK-YNEG 393
+GYLDPEY T + KSDVYS G++LLE++TG+ + K+ E+V + R G
Sbjct: 738 IGYLDPEYYSTRQMNEKSDVYSLGVVLLEVITGQPAIASSKT--EKVHISDHVRSILANG 795
Query: 394 SVVALLDPLMQEAVKTDVAVKMFDLAFNCAAPVRSDRPDMKTVGEQL 440
+ ++D ++E A KM ++A C + RP M V +L
Sbjct: 796 DIRGIVDQRLRERYDVGSAWKMSEIALACTEHTSAQRPTMSQVVMEL 842
>SRK6_BRAOL (Q09092) Putative serine/threonine-protein kinase
receptor precursor (EC 2.7.1.37) (S-receptor kinase)
(SRK)
Length = 849
Score = 187 bits (474), Expect = 6e-47
Identities = 112/308 (36%), Positives = 177/308 (57%), Gaps = 12/308 (3%)
Query: 138 SPSRFSMSPKLSRLQSLHLNLSQVSKATRNFSETLQIGEGGFGTVYKAHLDDGLVVAVKR 197
S FS K L+ + + V KAT NFS ++G+GGFG VYK L DG +AVKR
Sbjct: 498 SKREFSGEYKFEELELPLIEMETVVKATENFSSCNKLGQGGFGIVYKGRLLDGKEIAVKR 557
Query: 198 AKREHFESLRTEFSSEVELLAKIDHRNLVKLLGYIDKGNERILITEFVANGTLREHLDG- 256
+ + EF +EV L+A++ H NLV++LG +G+E++LI E++ N +L +L G
Sbjct: 558 LSKTSVQGT-DEFMNEVTLIARLQHINLVQVLGCCIEGDEKMLIYEYLENLSLDSYLFGK 616
Query: 257 LRGKILDFNQRLEIAIDVAHGLTYLHLYAEKQIIHRDVKSSNILLTESMRAKVADFGFAK 316
R L++N+R +I VA GL YLH + +IIHRD+K SNILL ++M K++DFG A+
Sbjct: 617 TRRSKLNWNERFDITNGVARGLLYLHQDSRFRIIHRDLKVSNILLDKNMIPKISDFGMAR 676
Query: 317 LGPVNNDHTHIST-KVKGTVGYLDPEYMKTYHLTPKSDVYSFGILLLEILTGRRPVELKK 375
+ D T +T KV GT GY+ PEY + KSDV+SFG+++LEI++G++
Sbjct: 677 I--FERDETEANTMKVVGTYGYMSPEYAMYGIFSEKSDVFSFGVIVLEIVSGKKNRGFYN 734
Query: 376 SAEERVTLRWAFRKYNEGSVVALLDPLMQEAVKTDVAV-------KMFDLAFNCAAPVRS 428
E L + + ++ EG + ++DP++ +++ + ++ K + C +
Sbjct: 735 LDYENDLLSYVWSRWKEGRALEIVDPVIVDSLSSQPSIFQPQEVLKCIQIGLLCVQELAE 794
Query: 429 DRPDMKTV 436
RP M +V
Sbjct: 795 HRPAMSSV 802
>BRI1_LYCPE (Q8L899) Systemin receptor SR160 precursor (EC 2.7.1.37)
(Brassinosteroid LRR receptor kinase)
Length = 1207
Score = 182 bits (461), Expect = 2e-45
Identities = 113/306 (36%), Positives = 175/306 (56%), Gaps = 14/306 (4%)
Query: 156 LNLSQVSKATRNFSETLQIGEGGFGTVYKAHLDDGLVVAVKRAKREHFESLRTEFSSEVE 215
L + + +AT F +G GGFG VYKA L DG VVA+K+ + R EF++E+E
Sbjct: 876 LTFADLLEATNGFHNDSLVGSGGFGDVYKAQLKDGSVVAIKKLIHVSGQGDR-EFTAEME 934
Query: 216 LLAKIDHRNLVKLLGYIDKGNERILITEFVANGTLREHLDGLR--GKILDFNQRLEIAID 273
+ KI HRNLV LLGY G ER+L+ E++ G+L + L + G L++ R +IAI
Sbjct: 935 TIGKIKHRNLVPLLGYCKVGEERLLVYEYMKYGSLEDVLHDRKKTGIKLNWPARRKIAIG 994
Query: 274 VAHGLTYLHLYAEKQIIHRDVKSSNILLTESMRAKVADFGFAKLGPVNNDHTHISTKVKG 333
A GL +LH IIHRD+KSSN+LL E++ A+V+DFG A+L + H +ST + G
Sbjct: 995 AARGLAFLHHNCIPHIIHRDMKSSNVLLDENLEARVSDFGMARLMSAMDTHLSVST-LAG 1053
Query: 334 TVGYLDPEYMKTYHLTPKSDVYSFGILLLEILTGRRPVELKKSAEERVTLRWAFRKYNEG 393
T GY+ PEY +++ + K DVYS+G++LLE+LTG++P + + + + W + + +G
Sbjct: 1054 TPGYVPPEYYQSFRCSTKGDVYSYGVVLLELLTGKQPTDSADFGDNNL-VGWV-KLHAKG 1111
Query: 394 SVVALLDPLMQEAVKTDVAVKM-----FDLAFNCAAPVRSDRPDMKTVGEQLWAIRADYL 448
+ + D +E +K D ++++ +A C RP M V I+A
Sbjct: 1112 KITDVFD---RELLKEDASIEIELLQHLKVACACLDDRHWKRPTMIQVMAMFKEIQAGSG 1168
Query: 449 KSSSTT 454
S++T
Sbjct: 1169 MDSTST 1174
>BRI1_LYCES (Q8GUQ5) Brassinosteroid LRR receptor kinase precursor (EC
2.7.1.37) (tBRI1) (Altered brassinolide sensitivity 1)
(Systemin receptor SR160)
Length = 1207
Score = 182 bits (461), Expect = 2e-45
Identities = 113/306 (36%), Positives = 175/306 (56%), Gaps = 14/306 (4%)
Query: 156 LNLSQVSKATRNFSETLQIGEGGFGTVYKAHLDDGLVVAVKRAKREHFESLRTEFSSEVE 215
L + + +AT F +G GGFG VYKA L DG VVA+K+ + R EF++E+E
Sbjct: 876 LTFADLLEATNGFHNDSLVGSGGFGDVYKAQLKDGSVVAIKKLIHVSGQGDR-EFTAEME 934
Query: 216 LLAKIDHRNLVKLLGYIDKGNERILITEFVANGTLREHLDGLR--GKILDFNQRLEIAID 273
+ KI HRNLV LLGY G ER+L+ E++ G+L + L + G L++ R +IAI
Sbjct: 935 TIGKIKHRNLVPLLGYCKVGEERLLVYEYMKYGSLEDVLHDRKKIGIKLNWPARRKIAIG 994
Query: 274 VAHGLTYLHLYAEKQIIHRDVKSSNILLTESMRAKVADFGFAKLGPVNNDHTHISTKVKG 333
A GL +LH IIHRD+KSSN+LL E++ A+V+DFG A+L + H +ST + G
Sbjct: 995 AARGLAFLHHNCIPHIIHRDMKSSNVLLDENLEARVSDFGMARLMSAMDTHLSVST-LAG 1053
Query: 334 TVGYLDPEYMKTYHLTPKSDVYSFGILLLEILTGRRPVELKKSAEERVTLRWAFRKYNEG 393
T GY+ PEY +++ + K DVYS+G++LLE+LTG++P + + + + W + + +G
Sbjct: 1054 TPGYVPPEYYQSFRCSTKGDVYSYGVVLLELLTGKQPTDSADFGDNNL-VGWV-KLHAKG 1111
Query: 394 SVVALLDPLMQEAVKTDVAVKM-----FDLAFNCAAPVRSDRPDMKTVGEQLWAIRADYL 448
+ + D +E +K D ++++ +A C RP M V I+A
Sbjct: 1112 KITDVFD---RELLKEDASIEIELLQHLKVACACLDDRHWKRPTMIQVMAMFKEIQAGSG 1168
Query: 449 KSSSTT 454
S++T
Sbjct: 1169 MDSTST 1174
>BRI1_ARATH (O22476) BRASSINOSTEROID INSENSITIVE 1 precursor (EC
2.7.1.37) (AtBRI1) (Brassinosteroid LRR receptor kinase)
Length = 1196
Score = 182 bits (461), Expect = 2e-45
Identities = 117/308 (37%), Positives = 175/308 (55%), Gaps = 14/308 (4%)
Query: 156 LNLSQVSKATRNFSETLQIGEGGFGTVYKAHLDDGLVVAVKRAKREHFESLRTEFSSEVE 215
L + + +AT F IG GGFG VYKA L DG VA+K+ + R EF +E+E
Sbjct: 871 LTFADLLQATNGFHNDSLIGSGGFGDVYKAILKDGSAVAIKKLIHVSGQGDR-EFMAEME 929
Query: 216 LLAKIDHRNLVKLLGYIDKGNERILITEFVANGTLREHLDGLR--GKILDFNQRLEIAID 273
+ KI HRNLV LLGY G+ER+L+ EF+ G+L + L + G L+++ R +IAI
Sbjct: 930 TIGKIKHRNLVPLLGYCKVGDERLLVYEFMKYGSLEDVLHDPKKAGVKLNWSTRRKIAIG 989
Query: 274 VAHGLTYLHLYAEKQIIHRDVKSSNILLTESMRAKVADFGFAKLGPVNNDHTHISTKVKG 333
A GL +LH IIHRD+KSSN+LL E++ A+V+DFG A+L + H +ST + G
Sbjct: 990 SARGLAFLHHNCSPHIIHRDMKSSNVLLDENLEARVSDFGMARLMSAMDTHLSVST-LAG 1048
Query: 334 TVGYLDPEYMKTYHLTPKSDVYSFGILLLEILTGRRPVELKKSAEERVTLRWAFRKYNEG 393
T GY+ PEY +++ + K DVYS+G++LLE+LTG+RP + + + + W +++ +
Sbjct: 1049 TPGYVPPEYYQSFRCSTKGDVYSYGVVLLELLTGKRPTDSPDFGDNNL-VGWV-KQHAKL 1106
Query: 394 SVVALLDPLMQEAVKTDVAVKM-----FDLAFNCAAPVRSDRPDMKTVGEQLWAIRADYL 448
+ + DP E +K D A+++ +A C RP M V I+A
Sbjct: 1107 RISDVFDP---ELMKEDPALEIELLQHLKVAVACLDDRAWRRPTMVQVMAMFKEIQAGSG 1163
Query: 449 KSSSTTAR 456
S +T R
Sbjct: 1164 IDSQSTIR 1171
>PSKR_ARATH (Q9ZVR7) Putative phytosulfokine receptor precursor (EC
2.7.1.37) (Phytosulfokine LRR receptor kinase)
Length = 1008
Score = 181 bits (458), Expect = 5e-45
Identities = 108/280 (38%), Positives = 159/280 (56%), Gaps = 9/280 (3%)
Query: 156 LNLSQVSKATRNFSETLQIGEGGFGTVYKAHLDDGLVVAVKRAKREHFESLRTEFSSEVE 215
L+ + +T +F + IG GGFG VYKA L DG VA+K+ + + + EF +EVE
Sbjct: 722 LSYDDLLDSTNSFDQANIIGCGGFGMVYKATLPDGKKVAIKKLSGDCGQ-IEREFEAEVE 780
Query: 216 LLAKIDHRNLVKLLGYIDKGNERILITEFVANGTL----REHLDGLRGKILDFNQRLEIA 271
L++ H NLV L G+ N+R+LI ++ NG+L E DG +L + RL IA
Sbjct: 781 TLSRAQHPNLVLLRGFCFYKNDRLLIYSYMENGSLDYWLHERNDG--PALLKWKTRLRIA 838
Query: 272 IDVAHGLTYLHLYAEKQIIHRDVKSSNILLTESMRAKVADFGFAKLGPVNNDHTHISTKV 331
A GL YLH + I+HRD+KSSNILL E+ + +ADFG A+L ++ TH+ST +
Sbjct: 839 QGAAKGLLYLHEGCDPHILHRDIKSSNILLDENFNSHLADFGLARL--MSPYETHVSTDL 896
Query: 332 KGTVGYLDPEYMKTYHLTPKSDVYSFGILLLEILTGRRPVELKKSAEERVTLRWAFRKYN 391
GT+GY+ PEY + T K DVYSFG++LLE+LT +RPV++ K R + W + +
Sbjct: 897 VGTLGYIPPEYGQASVATYKGDVYSFGVVLLELLTDKRPVDMCKPKGCRDLISWVVKMKH 956
Query: 392 EGSVVALLDPLMQEAVKTDVAVKMFDLAFNCAAPVRSDRP 431
E + DPL+ ++ ++A C + RP
Sbjct: 957 ESRASEVFDPLIYSKENDKEMFRVLEIACLCLSENPKQRP 996
>RLK5_ARATH (P47735) Receptor-like protein kinase 5 precursor (EC
2.7.1.37)
Length = 999
Score = 175 bits (443), Expect = 2e-43
Identities = 107/296 (36%), Positives = 169/296 (56%), Gaps = 15/296 (5%)
Query: 174 IGEGGFGTVYKAHLDDGLVVAVKRAKRE--------HFESL-RTEFSSEVELLAKIDHRN 224
IG G G VYK L G VVAVK+ + +SL R F++EVE L I H++
Sbjct: 689 IGFGSSGKVYKVELRGGEVVAVKKLNKSVKGGDDEYSSDSLNRDVFAAEVETLGTIRHKS 748
Query: 225 LVKLLGYIDKGNERILITEFVANGTLREHLDGLR--GKILDFNQRLEIAIDVAHGLTYLH 282
+V+L G+ ++L+ E++ NG+L + L G R G +L + +RL IA+D A GL+YLH
Sbjct: 749 IVRLWCCCSSGDCKLLVYEYMPNGSLADVLHGDRKGGVVLGWPERLRIALDAAEGLSYLH 808
Query: 283 LYAEKQIIHRDVKSSNILLTESMRAKVADFGFAKLGPVNNDHT-HISTKVKGTVGYLDPE 341
I+HRDVKSSNILL AKVADFG AK+G ++ T + + G+ GY+ PE
Sbjct: 809 HDCVPPIVHRDVKSSNILLDSDYGAKVADFGIAKVGQMSGSKTPEAMSGIAGSCGYIAPE 868
Query: 342 YMKTYHLTPKSDVYSFGILLLEILTGRRPVELKKSAEERVTLRWAFRKYNEGSVVALLDP 401
Y+ T + KSD+YSFG++LLE++TG++P + + ++ +W ++ + ++DP
Sbjct: 869 YVYTLRVNEKSDIYSFGVVLLELVTGKQPTDSELGDKDMA--KWVCTALDKCGLEPVIDP 926
Query: 402 LMQEAVKTDVAVKMFDLAFNCAAPVRSDRPDMKTVGEQLWAIRADYLKSSSTTARR 457
+ K +++ K+ + C +P+ +RP M+ V L + SS T++R
Sbjct: 927 KLDLKFKEEIS-KVIHIGLLCTSPLPLNRPSMRKVVIMLQEVSGAVPCSSPNTSKR 981
>CLV1_ARATH (Q9SYQ8) Receptor protein kinase CLAVATA1 precursor (EC
2.7.1.-)
Length = 980
Score = 168 bits (425), Expect = 3e-41
Identities = 106/308 (34%), Positives = 165/308 (53%), Gaps = 20/308 (6%)
Query: 143 SMSPKLSRLQSLHLNLSQVSKATRNFSETLQIGEGGFGTVYKAHLDDGLVVAVKRAKREH 202
S++ KL+ Q L V + + E IG+GG G VY+ + + + VA+KR
Sbjct: 670 SLAWKLTAFQKLDFKSEDVLECLK---EENIIGKGGAGIVYRGSMPNNVDVAIKRLVGRG 726
Query: 203 FESLRTEFSSEVELLAKIDHRNLVKLLGYIDKGNERILITEFVANGTLREHLDGLRGKIL 262
F++E++ L +I HR++V+LLGY+ + +L+ E++ NG+L E L G +G L
Sbjct: 727 TGRSDHGFTAEIQTLGRIRHRHIVRLLGYVANKDTNLLLYEYMPNGSLGELLHGSKGGHL 786
Query: 263 DFNQRLEIAIDVAHGLTYLHLYAEKQIIHRDVKSSNILLTESMRAKVADFGFAKLGPVNN 322
+ R +A++ A GL YLH I+HRDVKS+NILL A VADFG AK V+
Sbjct: 787 QWETRHRVAVEAAKGLCYLHHDCSPLILHRDVKSNNILLDSDFEAHVADFGLAKF-LVDG 845
Query: 323 DHTHISTKVKGTVGYLDPEYMKTYHLTPKSDVYSFGILLLEILTGRRP----------VE 372
+ + + G+ GY+ PEY T + KSDVYSFG++LLE++ G++P V
Sbjct: 846 AASECMSSIAGSYGYIAPEYAYTLKVDEKSDVYSFGVVLLELIAGKKPVGEFGEGVDIVR 905
Query: 373 LKKSAEERVTLRWAFRKYNEGSVVALLDPLMQEAVKTDVAVKMFDLAFNCAAPVRSDRPD 432
++ EE +T + + VVA++DP + T V + +F +A C + RP
Sbjct: 906 WVRNTEEEIT-----QPSDAAIVVAIVDPRLTGYPLTSV-IHVFKIAMMCVEEEAAARPT 959
Query: 433 MKTVGEQL 440
M+ V L
Sbjct: 960 MREVVHML 967
>KPRO_MAIZE (P17801) Putative receptor protein kinase ZmPK1
precursor (EC 2.7.1.37)
Length = 817
Score = 165 bits (417), Expect = 3e-40
Identities = 106/284 (37%), Positives = 158/284 (55%), Gaps = 14/284 (4%)
Query: 160 QVSKATRNFSETLQIGEGGFGTVYKAHLDDGLVVAVKRAKREHFESLRTEFSSEVELLAK 219
++ KATR F +++G G GTVYK L+D VAVK K E+ + F +E+ ++ +
Sbjct: 528 ELVKATRKFK--VELGRGESGTVYKGVLEDDRHVAVK--KLENVRQGKEVFQAELSVIGR 583
Query: 220 IDHRNLVKLLGYIDKGNERILITEFVANGTLREHLDGLRGKIL-DFNQRLEIAIDVAHGL 278
I+H NLV++ G+ +G+ R+L++E+V NG+L L G IL D+ R IA+ VA GL
Sbjct: 584 INHMNLVRIWGFCSEGSHRLLVSEYVENGSLANILFSEGGNILLDWEGRFNIALGVAKGL 643
Query: 279 TYLHLYAEKQIIHRDVKSSNILLTESMRAKVADFGFAKLGPVNNDHTHISTKVKGTVGYL 338
YLH + +IH DVK NILL ++ K+ DFG KL ++S V+GT+GY+
Sbjct: 644 AYLHHECLEWVIHCDVKPENILLDQAFEPKITDFGLVKLLNRGGSTQNVS-HVRGTLGYI 702
Query: 339 DPEYMKTYHLTPKSDVYSFGILLLEILTGRRPVELKKSAEE-----RVTLRWAFRKY--- 390
PE++ + +T K DVYS+G++LLE+LTG R EL +E R +R K
Sbjct: 703 APEWVSSLPITAKVDVYSYGVVLLELLTGTRVSELVGGTDEVHSMLRKLVRMLSAKLEGE 762
Query: 391 NEGSVVALLDPLMQEAVKTDVAVKMFDLAFNCAAPVRSDRPDMK 434
+ + LD + V A + LA +C RS RP M+
Sbjct: 763 EQSWIDGYLDSKLNRPVNYVQARTLIKLAVSCLEEDRSKRPTME 806
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.318 0.134 0.381
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 50,651,945
Number of Sequences: 164201
Number of extensions: 2087470
Number of successful extensions: 9871
Number of sequences better than 10.0: 1672
Number of HSP's better than 10.0 without gapping: 1155
Number of HSP's successfully gapped in prelim test: 517
Number of HSP's that attempted gapping in prelim test: 6164
Number of HSP's gapped (non-prelim): 1896
length of query: 458
length of database: 59,974,054
effective HSP length: 114
effective length of query: 344
effective length of database: 41,255,140
effective search space: 14191768160
effective search space used: 14191768160
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 67 (30.4 bits)
Medicago: description of AC144657.5


