

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144657.4 + phase: 0
(776 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
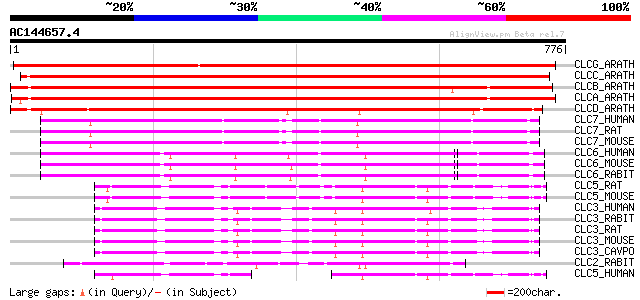
Score E
Sequences producing significant alignments: (bits) Value
CLCG_ARATH (P60300) Putative chloride channel-like protein CLC-G 1118 0.0
CLCC_ARATH (Q96282) Chloride channel protein CLC-c (AtCLC-c) 924 0.0
CLCB_ARATH (P92942) Chloride channel protein CLC-b (AtCLC-b) 823 0.0
CLCA_ARATH (P92941) Chloride channel protein CLC-a (AtCLC-a) 811 0.0
CLCD_ARATH (P92943) Chloride channel protein CLC-d (AtCLC-d) 614 e-175
CLC7_HUMAN (P51798) Chloride channel protein 7 (ClC-7) 387 e-107
CLC7_RAT (P51799) Chloride channel protein 7 (ClC-7) 386 e-106
CLC7_MOUSE (O70496) Chloride channel protein 7 (ClC-7) 386 e-106
CLC6_HUMAN (P51797) Chloride channel protein 6 (ClC-6) 326 1e-88
CLC6_MOUSE (O35454) Chloride channel protein 6 (ClC-6) 323 1e-87
CLC6_RABIT (Q9TT16) Chloride channel protein 6 (ClC-6) 321 5e-87
CLC5_RAT (P51796) Chloride channel protein 5 (ClC-5) 191 8e-48
CLC5_MOUSE (Q9WVD4) Chloride channel protein 5 (ClC-5) 191 8e-48
CLC3_HUMAN (P51790) Chloride channel protein 3 (ClC-3) 190 1e-47
CLC3_RABIT (O18894) Chloride channel protein 3 (ClC-3) 189 2e-47
CLC3_RAT (P51792) Chloride channel protein 3 (ClC-3) 189 3e-47
CLC3_MOUSE (P51791) Chloride channel protein 3 (ClC-3) 189 3e-47
CLC3_CAVPO (Q9R279) Chloride channel protein 3 (ClC-3) 189 3e-47
CLC2_RABIT (P51789) Chloride channel protein 2 (ClC-2) (PKA-acti... 169 3e-41
CLC5_HUMAN (P51795) Chloride channel protein 5 (ClC-5) 121 8e-27
>CLCG_ARATH (P60300) Putative chloride channel-like protein CLC-G
Length = 763
Score = 1118 bits (2891), Expect = 0.0
Identities = 549/758 (72%), Positives = 642/758 (84%), Gaps = 2/758 (0%)
Query: 6 LSNGESEPLLRRPLLSSQRSIINSTSQVAIVGANVCPIESLDYEIFENEFFKQDWRSRGV 65
+ N +E + PLL S R NSTSQVAIVGANVCPIESLDYEI EN+FFKQDWR R
Sbjct: 1 MPNSTTEDSVAVPLLPSLRRATNSTSQVAIVGANVCPIESLDYEIAENDFFKQDWRGRSK 60
Query: 66 VQILQYICMKWLLCFMIGLIVGFIGFCNNLAVENLAGIKFVTTSNMMLERRFMFAFFIFF 125
V+I QY+ MKWLLCF IG+IV IGF NNLAVENLAG+KFV TSNMM+ RF F +F
Sbjct: 61 VEIFQYVFMKWLLCFCIGIIVSLIGFANNLAVENLAGVKFVVTSNMMIAGRFAMGFVVFS 120
Query: 126 ASNLSLTLFASIITAFIAPTAAGSGISEVKAYLNGVDAPGIFTVRTLCVKIIGSITAVSG 185
+NL LTLFAS+ITAF+AP AAGSGI EVKAYLNGVDAP IF++RTL +KIIG+I+AVS
Sbjct: 121 VTNLILTLFASVITAFVAPAAAGSGIPEVKAYLNGVDAPEIFSLRTLIIKIIGNISAVSA 180
Query: 186 SLVIGKAGPMVHTGACVAALLGQGGSKRYGITWRWLRFFKNDRDRRDLIICGSAAGIAAA 245
SL+IGKAGPMVHTGACVA++LGQGGSKRY +TWRWLRFFKNDRDRRDL+ CG+AAGIAA+
Sbjct: 181 SLLIGKAGPMVHTGACVASILGQGGSKRYRLTWRWLRFFKNDRDRRDLVTCGAAAGIAAS 240
Query: 246 FRAPVGGVLFALEEMASWWRTALLWRAFFTTATVAIFLRAMIDVCLSDKCGLFGKGGLIM 305
FRAPVGGVLFALEEM+SW +ALLWR FF+TA VAI LRA+IDVCLS KCGLFGKGGLIM
Sbjct: 241 FRAPVGGVLFALEEMSSW--SALLWRIFFSTAVVAIVLRALIDVCLSGKCGLFGKGGLIM 298
Query: 306 FDAYSASISYHLVDVPPVFILAVVGGLLGSLFNFMTNKVLRIYNVINEKGTICRLFLACL 365
FD YS + SYHL DV PV +L VVGG+LGSL+NF+ +KVLR YN I EKG ++ LAC
Sbjct: 299 FDVYSENASYHLGDVLPVLLLGVVGGILGSLYNFLLDKVLRAYNYIYEKGVTWKILLACA 358
Query: 366 ISIFTSCLLFGLPWLAPCRPCPPDAVEPCPTIGRSGIYKKFQCPPNHYNGLASLIFNTND 425
ISIFTSCLLFGLP+LA C+PCP DA+E CPTIGRSG +KK+QCPP HYN LASLIFNTND
Sbjct: 359 ISIFTSCLLFGLPFLASCQPCPVDALEECPTIGRSGNFKKYQCPPGHYNDLASLIFNTND 418
Query: 426 DAIRNLFSMHTDNEFELSSMLVFFIICLFLSIFSCGIVAPAGLFVPIIVTGASYGRLVGI 485
DAI+NLFS +TD EF S+LVFF+ C FLSIFS GIVAPAGLFVP+IVTGASYGR VG+
Sbjct: 419 DAIKNLFSKNTDFEFHYFSVLVFFVTCFFLSIFSYGIVAPAGLFVPVIVTGASYGRFVGM 478
Query: 486 LVGERTNLSNGLYAVLGAASLLGGSMRTTVSLCVIMLELTNNLLLLPLIMMVLVVSKSVA 545
L+G +NL++GL+AVLGAAS LGG+MR TVS CVI+LELTNNLLLLP++M+VL++SK+VA
Sbjct: 479 LLGSNSNLNHGLFAVLGAASFLGGTMRMTVSTCVILLELTNNLLLLPMMMVVLLISKTVA 538
Query: 546 NVFNANVYDLIMKAKGLPYLETHAEPYMRQLTVGDVVTGPLQMFNGIEKVRNIVFILRTT 605
+ FNAN+Y+LIMK KG PYL +HAEPYMRQL VGDVVTGPLQ+FNGIEKV IV +L+TT
Sbjct: 539 DGFNANIYNLIMKLKGFPYLYSHAEPYMRQLLVGDVVTGPLQVFNGIEKVETIVHVLKTT 598
Query: 606 AHNGFPVIDEPPGSEAPILFGIILRHHLTTLLKKKAFLPSPVANSYDVVRKFSSDDFAKK 665
HNGFPV+D PP + AP+L G+ILR H+ TLLKK+ F+PSPVA + + +F +++FAKK
Sbjct: 599 NHNGFPVVDGPPLAAAPVLHGLILRAHILTLLKKRVFMPSPVACDSNTLSQFKAEEFAKK 658
Query: 666 YSVERVKIEDIQLTEEEMGMFVDLHPFTNASPYTVVETMSLAKALILFREVGLRHLLVIP 725
S KIED++L+EEE+ M++DLHPF+NASPYTVVETMSLAKALILFREVG+RHLLVIP
Sbjct: 659 GSGRSDKIEDVELSEEELNMYLDLHPFSNASPYTVVETMSLAKALILFREVGIRHLLVIP 718
Query: 726 KIPGRSPVVGILTRHDFTPEHILGMHPFLVKSRWKRLR 763
K R PVVGILTRHDF PEHILG+HP + +S+WKRLR
Sbjct: 719 KTSNRPPVVGILTRHDFMPEHILGLHPSVSRSKWKRLR 756
>CLCC_ARATH (Q96282) Chloride channel protein CLC-c (AtCLC-c)
Length = 779
Score = 924 bits (2387), Expect = 0.0
Identities = 452/740 (61%), Positives = 557/740 (75%), Gaps = 3/740 (0%)
Query: 16 RRPLLSSQRSIINSTSQVAIVGANVCPIESLDYEIFENEFFKQDWRSRGVVQILQYICMK 75
R+PLL+ R N+TSQ+AIVGAN CPIESLDYEIFEN+FFKQDWRSR ++ILQY +K
Sbjct: 38 RQPLLARNRK--NTTSQIAIVGANTCPIESLDYEIFENDFFKQDWRSRKKIEILQYTFLK 95
Query: 76 WLLCFMIGLIVGFIGFCNNLAVENLAGIKFVTTSNMMLERRFMFAFFIFFASNLSLTLFA 135
W L F+IGL G +GF NNL VEN+AG K + N+ML+ ++ AFF F NL L A
Sbjct: 96 WALAFLIGLATGLVGFLNNLGVENIAGFKLLLIGNLMLKEKYFQAFFAFAGCNLILATAA 155
Query: 136 SIITAFIAPTAAGSGISEVKAYLNGVDAPGIFTVRTLCVKIIGSITAVSGSLVIGKAGPM 195
+ + AFIAP AAGSGI EVKAYLNG+DA I TL VKI GSI V+ V+GK GPM
Sbjct: 156 ASLCAFIAPAAAGSGIPEVKAYLNGIDAYSILAPSTLFVKIFGSIFGVAAGFVVGKEGPM 215
Query: 196 VHTGACVAALLGQGGSKRYGITWRWLRFFKNDRDRRDLIICGSAAGIAAAFRAPVGGVLF 255
VHTGAC+A LLGQGGSK+Y +TW+WLRFFKNDRDRRDLI CG+AAG+AAAFRAPVGGVLF
Sbjct: 216 VHTGACIANLLGQGGSKKYRLTWKWLRFFKNDRDRRDLITCGAAAGVAAAFRAPVGGVLF 275
Query: 256 ALEEMASWWRTALLWRAFFTTATVAIFLRAMIDVCLSDKCGLFGKGGLIMFDAYSASISY 315
ALEE ASWWR ALLWR FFTTA VA+ LR++I+ C S +CGLFGKGGLIMFD S + Y
Sbjct: 276 ALEEAASWWRNALLWRTFFTTAVVAVVLRSLIEFCRSGRCGLFGKGGLIMFDVNSGPVLY 335
Query: 316 HLVDVPPVFILAVVGGLLGSLFNFMTNKVLRIYNVINEKGTICRLFLACLISIFTSCLLF 375
D+ + L V+GG+LGSL+N++ +KVLR Y++INEKG ++ L +SI +SC F
Sbjct: 336 STPDLLAIVFLGVIGGVLGSLYNYLVDKVLRTYSIINEKGPRFKIMLVMAVSILSSCCAF 395
Query: 376 GLPWLAPCRPCPPDAVE-PCPTIGRSGIYKKFQCPPNHYNGLASLIFNTNDDAIRNLFSM 434
GLPWL+ C PCP E CP++GRS IYK FQCPPNHYN L+SL+ NTNDDAIRNLF+
Sbjct: 396 GLPWLSQCTPCPIGIEEGKCPSVGRSSIYKSFQCPPNHYNDLSSLLLNTNDDAIRNLFTS 455
Query: 435 HTDNEFELSSMLVFFIICLFLSIFSCGIVAPAGLFVPIIVTGASYGRLVGILVGERTNLS 494
++NEF +S++ +FF+ L I + GI P+GLF+P+I+ GASYGRLVG L+G + L
Sbjct: 456 RSENEFHISTLAIFFVAVYCLGIITYGIAIPSGLFIPVILAGASYGRLVGRLLGPVSQLD 515
Query: 495 NGLYAVLGAASLLGGSMRTTVSLCVIMLELTNNLLLLPLIMMVLVVSKSVANVFNANVYD 554
GL+++LGAAS LGG+MR TVSLCVI+LELTNNLL+LPL+M+VL++SK+VA+ FN VYD
Sbjct: 516 VGLFSLLGAASFLGGTMRMTVSLCVILLELTNNLLMLPLVMLVLLISKTVADCFNRGVYD 575
Query: 555 LIMKAKGLPYLETHAEPYMRQLTVGDVVTGPLQMFNGIEKVRNIVFILRTTAHNGFPVID 614
I+ KGLPY+E HAEPYMR L DVV+G L F+ +EKV I L+ T HNGFPVID
Sbjct: 576 QIVTMKGLPYMEDHAEPYMRNLVAKDVVSGALISFSRVEKVGVIWQALKMTRHNGFPVID 635
Query: 615 EPPGSEAPILFGIILRHHLTTLLKKKAFLPSPVANSYDVVRKFSSDDFAKKYSVERVKIE 674
EPP +EA L GI LR HL LL+ K F ++R + DF K + +KIE
Sbjct: 636 EPPFTEASELCGIALRSHLLVLLQGKKFSKQRTTFGSQILRSCKARDFGKAGLGKGLKIE 695
Query: 675 DIQLTEEEMGMFVDLHPFTNASPYTVVETMSLAKALILFREVGLRHLLVIPKIPGRSPVV 734
D+ L+EEEM M+VDLHP TN SPYTV+ET+SLAKA ILFR++GLRHL V+PK PGR P+V
Sbjct: 696 DLDLSEEEMEMYVDLHPITNTSPYTVLETLSLAKAAILFRQLGLRHLCVVPKTPGRPPIV 755
Query: 735 GILTRHDFTPEHILGMHPFL 754
GILTRHDF PEH+LG++P +
Sbjct: 756 GILTRHDFMPEHVLGLYPHI 775
>CLCB_ARATH (P92942) Chloride channel protein CLC-b (AtCLC-b)
Length = 780
Score = 823 bits (2126), Expect = 0.0
Identities = 408/769 (53%), Positives = 547/769 (71%), Gaps = 18/769 (2%)
Query: 2 SNNHLSNGESEP-LLRRPLLSSQRSIINSTSQVAIVGANVCPIESLDYEIFENEFFKQDW 60
SN + G+ E L +PL+ + R++ S++ +A+VGA V IESLDYEI EN+ FK DW
Sbjct: 13 SNYNGEGGDPESNTLNQPLVKANRTL--SSTPLALVGAKVSHIESLDYEINENDLFKHDW 70
Query: 61 RSRGVVQILQYICMKWLLCFMIGLIVGFIGFCNNLAVENLAGIKFVTTSNMMLERRFMFA 120
R R Q+LQY+ +KW L ++GL G I NLAVEN+AG K + + + + R++
Sbjct: 71 RKRSKAQVLQYVFLKWTLACLVGLFTGLIATLINLAVENIAGYKLLAVGHFLTQERYVTG 130
Query: 121 FFIFFASNLSLTLFASIITAFIAPTAAGSGISEVKAYLNGVDAPGIFTVRTLCVKIIGSI 180
+ +NL LTL AS++ APTAAG GI E+KAYLNGVD P +F T+ VKI+GSI
Sbjct: 131 LMVLVGANLGLTLVASVLCVCFAPTAAGPGIPEIKAYLNGVDTPNMFGATTMIVKIVGSI 190
Query: 181 TAVSGSLVIGKAGPMVHTGACVAALLGQGGSKRYGITWRWLRFFKNDRDRRDLIICGSAA 240
AV+ L +GK GP+VH G+C+A+LLGQGG+ + I WRWLR+F NDRDRRDLI CGSAA
Sbjct: 191 GAVAAGLDLGKEGPLVHIGSCIASLLGQGGTDNHRIKWRWLRYFNNDRDRRDLITCGSAA 250
Query: 241 GIAAAFRAPVGGVLFALEEMASWWRTALLWRAFFTTATVAIFLRAMIDVCLSDKCGLFGK 300
G+ AAFR+PVGGVLFALEE+A+WWR+ALLWR FF+TA V + LR I++C S KCGLFGK
Sbjct: 251 GVCAAFRSPVGGVLFALEEVATWWRSALLWRTFFSTAVVVVVLREFIEICNSGKCGLFGK 310
Query: 301 GGLIMFDAYSASISYHLVDVPPVFILAVVGGLLGSLFNFMTNKVLRIYNVINEKGTICRL 360
GGLIMFD + +YH+ D+ PV ++ V+GG+LGSL+N + +KVLR+YN+INEKG I ++
Sbjct: 311 GGLIMFDVSHVTYTYHVTDIIPVMLIGVIGGILGSLYNHLLHKVLRLYNLINEKGKIHKV 370
Query: 361 FLACLISIFTSCLLFGLPWLAPCRPCPPDAVEPCPTIGRSGIYKKFQCPPNHYNGLASLI 420
L+ +S+FTS L+GLP+LA C+PC P E CPT GRSG +K+F CP +YN LA+L+
Sbjct: 371 LLSLTVSLFTSVCLYGLPFLAKCKPCDPSIDEICPTNGRSGNFKQFHCPKGYYNDLATLL 430
Query: 421 FNTNDDAIRNLFSMHTDNEFELSSMLVFFIICLFLSIFSCGIVAPAGLFVPIIVTGASYG 480
TNDDA+RNLFS +T NEF + S+ +FF++ L +F+ GI P+GLF+PII+ GA+YG
Sbjct: 431 LTTNDDAVRNLFSSNTPNEFGMGSLWIFFVLYCILGLFTFGIATPSGLFLPIILMGAAYG 490
Query: 481 RLVGILVGERTNLSNGLYAVLGAASLLGGSMRTTVSLCVIMLELTNNLLLLPLIMMVLVV 540
R++G +G T++ GLYAVLGAA+L+ GSMR TVSLCVI LELTNNLLLLP+ M+VL++
Sbjct: 491 RMLGAAMGSYTSIDQGLYAVLGAAALMAGSMRMTVSLCVIFLELTNNLLLLPITMIVLLI 550
Query: 541 SKSVANVFNANVYDLIMKAKGLPYLETHAEPYMRQLTVGDV--VTGPLQMFNGIEKVRNI 598
+K+V + FN ++YD+I+ KGLP+LE + EP+MR LTVG++ P+ G+EKV NI
Sbjct: 551 AKTVGDSFNPSIYDIILHLKGLPFLEANPEPWMRNLTVGELGDAKPPVVTLQGVEKVSNI 610
Query: 599 VFILRTTAHNGFPVIDEPP------GSEAPILFGIILRHHLTTLLKKKAFLPSP-VANSY 651
V +L+ T HN FPV+DE + A L G+ILR HL +LKK+ FL +
Sbjct: 611 VDVLKNTTHNAFPVLDEAEVPQVGLATGATELHGLILRAHLVKVLKKRWFLTEKRRTEEW 670
Query: 652 DVVRKFSSDDFAKKYSVERVKIEDIQLTEEEMGMFVDLHPFTNASPYTVVETMSLAKALI 711
+V KF D+ A++ +D+ +T EM M+VDLHP TN +PYTV+E MS+AKAL+
Sbjct: 671 EVREKFPWDELAERED----NFDDVAITSAEMEMYVDLHPLTNTTPYTVMENMSVAKALV 726
Query: 712 LFREVGLRHLLVIPKI--PGRSPVVGILTRHDFTPEHILGMHPFLVKSR 758
LFR+VGLRHLL++PKI G PVVGILTR D +IL P L KS+
Sbjct: 727 LFRQVGLRHLLIVPKIQASGMCPVVGILTRQDLRAYNILQAFPLLEKSK 775
>CLCA_ARATH (P92941) Chloride channel protein CLC-a (AtCLC-a)
Length = 775
Score = 811 bits (2096), Expect = 0.0
Identities = 406/772 (52%), Positives = 540/772 (69%), Gaps = 17/772 (2%)
Query: 3 NNHLSNGESEP------LLRRPLLSSQRSIINSTSQVAIVGANVCPIESLDYEIFENEFF 56
+N NGE E L +PLL R++ S++ +A+VGA V IESLDYEI EN+ F
Sbjct: 10 SNSNYNGEEEGEDPENNTLNQPLLKRHRTL--SSTPLALVGAKVSHIESLDYEINENDLF 67
Query: 57 KQDWRSRGVVQILQYICMKWLLCFMIGLIVGFIGFCNNLAVENLAGIKFVTTSNMMLERR 116
K DWRSR Q+ QYI +KW L ++GL G I NLAVEN+AG K + + + R
Sbjct: 68 KHDWRSRSKAQVFQYIFLKWTLACLVGLFTGLIATLINLAVENIAGYKLLAVGYYIAQDR 127
Query: 117 FMFAFFIFFASNLSLTLFASIITAFIAPTAAGSGISEVKAYLNGVDAPGIFTVRTLCVKI 176
F +F +NL LTL A+++ + APTAAG GI E+KAYLNG+D P +F T+ VKI
Sbjct: 128 FWTGLMVFTGANLGLTLVATVLVVYFAPTAAGPGIPEIKAYLNGIDTPNMFGFTTMMVKI 187
Query: 177 IGSITAVSGSLVIGKAGPMVHTGACVAALLGQGGSKRYGITWRWLRFFKNDRDRRDLIIC 236
+GSI AV+ L +GK GP+VH G+C+A+LLGQGG + I WRWLR+F NDRDRRDLI C
Sbjct: 188 VGSIGAVAAGLDLGKEGPLVHIGSCIASLLGQGGPDNHRIKWRWLRYFNNDRDRRDLITC 247
Query: 237 GSAAGIAAAFRAPVGGVLFALEEMASWWRTALLWRAFFTTATVAIFLRAMIDVCLSDKCG 296
GSA+G+ AAFR+PVGGVLFALEE+A+WWR+ALLWR FF+TA V + LRA I++C S KCG
Sbjct: 248 GSASGVCAAFRSPVGGVLFALEEVATWWRSALLWRTFFSTAVVVVVLRAFIEICNSGKCG 307
Query: 297 LFGKGGLIMFDAYSASISYHLVDVPPVFILAVVGGLLGSLFNFMTNKVLRIYNVINEKGT 356
LFG GGLIMFD + YH D+ PV ++ V GG+LGSL+N + +KVLR+YN+IN+KG
Sbjct: 308 LFGSGGLIMFDVSHVEVRYHAADIIPVTLIGVFGGILGSLYNHLLHKVLRLYNLINQKGK 367
Query: 357 ICRLFLACLISIFTSCLLFGLPWLAPCRPCPPDAVEPCPTIGRSGIYKKFQCPPNHYNGL 416
I ++ L+ +S+FTS LFGLP+LA C+PC P E CPT GRSG +K+F CP +YN L
Sbjct: 368 IHKVLLSLGVSLFTSVCLFGLPFLAECKPCDPSIDEICPTNGRSGNFKQFNCPNGYYNDL 427
Query: 417 ASLIFNTNDDAIRNLFSMHTDNEFELSSMLVFFIICLFLSIFSCGIVAPAGLFVPIIVTG 476
++L+ TNDDA+RN+FS +T NEF + S+ +FF + L + + GI P+GLF+PII+ G
Sbjct: 428 STLLLTTNDDAVRNIFSSNTPNEFGMVSLWIFFGLYCILGLITFGIATPSGLFLPIILMG 487
Query: 477 ASYGRLVGILVGERTNLSNGLYAVLGAASLLGGSMRTTVSLCVIMLELTNNLLLLPLIMM 536
++YGR++G +G TN+ GLYAVLGAASL+ GSMR TVSLCVI LELTNNLLLLP+ M
Sbjct: 488 SAYGRMLGTAMGSYTNIDQGLYAVLGAASLMAGSMRMTVSLCVIFLELTNNLLLLPITMF 547
Query: 537 VLVVSKSVANVFNANVYDLIMKAKGLPYLETHAEPYMRQLTVGDV--VTGPLQMFNGIEK 594
VL+++K+V + FN ++Y++I+ KGLP+LE + EP+MR LTVG++ P+ NG+EK
Sbjct: 548 VLLIAKTVGDSFNLSIYEIILHLKGLPFLEANPEPWMRNLTVGELNDAKPPVVTLNGVEK 607
Query: 595 VRNIVFILRTTAHNGFPVIDEPPGSEAPILFGIILRHHLTTLLKKKAFL-PSPVANSYDV 653
V NIV +LR T HN FPV+D + L G+ILR HL +LKK+ FL ++V
Sbjct: 608 VANIVDVLRNTTHNAFPVLDGADQNTGTELHGLILRAHLVKVLKKRWFLNEKRRTEEWEV 667
Query: 654 VRKFSSDDFAKKYSVERVKIEDIQLTEEEMGMFVDLHPFTNASPYTVVETMSLAKALILF 713
KF+ + A++ +D+ +T EM ++VDLHP TN +PYTVV++MS+AKAL+LF
Sbjct: 668 REKFTPVELAERED----NFDDVAITSSEMQLYVDLHPLTNTTPYTVVQSMSVAKALVLF 723
Query: 714 REVGLRHLLVIPKI--PGRSPVVGILTRHDFTPEHILGMHPFLVKSRWKRLR 763
R VGLRHLLV+PKI G SPV+GILTR D +IL P L K + + R
Sbjct: 724 RSVGLRHLLVVPKIQASGMSPVIGILTRQDLRAYNILQAFPHLDKHKSGKAR 775
>CLCD_ARATH (P92943) Chloride channel protein CLC-d (AtCLC-d)
Length = 792
Score = 614 bits (1583), Expect = e-175
Identities = 344/763 (45%), Positives = 479/763 (62%), Gaps = 28/763 (3%)
Query: 1 MSNNHLSNG-ESEPLLRRPLLSSQRSIINSTSQVAIVGANVCP---IESLDYEIFENEFF 56
M +NHL NG ES+ LL + S + ST + ++ ++ + SLDYE+ EN +
Sbjct: 1 MLSNHLQNGIESDNLLWSRVPESDDT---STDDITLLNSHRDGDGGVNSLDYEVIENYAY 57
Query: 57 KQDWRSRGVVQILQYICMKWLLCFMIGLIVGFIGFCNNLAVENLAGIKFVTTSNMMLERR 116
+++ RG + + Y+ +KW +IG+ G NL+VEN AG KF T ++++
Sbjct: 58 REEQAHRGKLYVGYYVAVKWFFSLLIGIGTGLAAVFINLSVENFAGWKFALTF-AIIQKS 116
Query: 117 FMFAFFIFFASNLSLTLFASIITAFIAPTAAGSGISEVKAYLNGVDAPGIFTVRTLCVKI 176
+ F ++ NL L ++ I AP AAGSGI E+K YLNG+D PG RTL KI
Sbjct: 117 YFAGFIVYLLINLVLVFSSAYIITQFAPAAAGSGIPEIKGYLNGIDIPGTLLFRTLIGKI 176
Query: 177 IGSITAVSGSLVIGKAGPMVHTGACVAALLGQGGSKRYGITWRWLRFFKNDRDRRDLIIC 236
GSI +V G L +GK GP+VHTGAC+A+LLGQGGS +Y + RW + FK+DRDRRDL+ C
Sbjct: 177 FGSIGSVGGGLALGKEGPLVHTGACIASLLGQGGSTKYHLNSRWPQLFKSDRDRRDLVTC 236
Query: 237 GSAAGIAAAFRAPVGGVLFALEEMASWWRTALLWRAFFTTATVAIFLRAMIDVCLSDKCG 296
G AAG+AAAFRAPVGGVLFALEE+ SWWR+ L+WR FFT+A VA+ +R + C S CG
Sbjct: 237 GCAAGVAAAFRAPVGGVLFALEEVTSWWRSQLMWRVFFTSAIVAVVVRTAMGWCKSGICG 296
Query: 297 LFGKGGLIMFDAYSASISYHLVDVPPVFILAVVGGLLGSLFNFMTNKVLR-IYNVINEKG 355
FG GG I++D Y+ ++ P+ ++ V+GGLLG+LFN +T + N +++KG
Sbjct: 297 HFGGGGFIIWDVSDGQDDYYFKELLPMAVIGVIGGLLGALFNQLTLYMTSWRRNSLHKKG 356
Query: 356 TICRLFLACLISIFTSCLLFGLPWLAPCRPCP---PDAVEPCP-TIGRSGIYKKFQC-PP 410
++ AC+IS TS + FGLP L C PCP PD+ CP G G Y F C
Sbjct: 357 NRVKIIEACIISCITSAISFGLPLLRKCSPCPESVPDSGIECPRPPGMYGNYVNFFCKTD 416
Query: 411 NHYNGLASLIFNTNDDAIRNLFSMHTDNEFELSSMLVFFIICLFLSIFSCGIVAPAGLFV 470
N YN LA++ FNT DDAIRNLFS T EF S+L F + L++ + G PAG FV
Sbjct: 417 NEYNDLATIFFNTQDDAIRNLFSAKTMREFSAQSLLTFLAMFYTLAVVTFGTAVPAGQFV 476
Query: 471 PIIVTGASYGRLVGILV---GERTNLSNGLYAVLGAASLLGGSMRTTVSLCVIMLELTNN 527
P I+ G++YGRLVG+ V ++ N+ G YA+LGAAS LGGSMR TVSLCVIM+E+TNN
Sbjct: 477 PGIMIGSTYGRLVGMFVVRFYKKLNIEEGTYALLGAASFLGGSMRMTVSLCVIMVEITNN 536
Query: 528 LLLLPLIMMVLVVSKSVANVFNANVYDLIMKAKGLPYLETHAEPYMRQLTVGDVV-TGPL 586
L LLPLIM+VL++SK+V + FN +Y++ + KG+P LE+ + +MRQ+ + + +
Sbjct: 537 LKLLPLIMLVLLISKAVGDAFNEGLYEVQARLKGIPLLESRPKYHMRQMIAKEACQSQKV 596
Query: 587 QMFNGIEKVRNIVFILRTTAHNGFPVIDEPPGSEAPILFGIILRHHLTTLLKKKA-FLPS 645
+ +V ++ IL + HNGFPVID E ++ G++LR HL LL+ K F S
Sbjct: 597 ISLPRVIRVADVASILGSNKHNGFPVIDHTRSGET-LVIGLVLRSHLLVLLQSKVDFQHS 655
Query: 646 PV---ANSYDVVRKFSSDDFAKKYSVERVKIEDIQLTEEEMGMFVDLHPFTNASPYTVVE 702
P+ ++ ++ FS +FAK S + + IEDI LT +++ M++DL PF N SPY V E
Sbjct: 656 PLPCDPSARNIRHSFS--EFAKPVSSKGLCIEDIHLTSDDLEMYIDLAPFLNPSPYVVPE 713
Query: 703 TMSLAKALILFREVGLRHLLVIPKIPGRSPVVGILTRHDFTPE 745
MSL K LFR++GLRHL V+P+ P R V+G++TR D E
Sbjct: 714 DMSLTKVYNLFRQLGLRHLFVVPR-PSR--VIGLITRKDLLIE 753
>CLC7_HUMAN (P51798) Chloride channel protein 7 (ClC-7)
Length = 805
Score = 387 bits (994), Expect = e-107
Identities = 247/710 (34%), Positives = 391/710 (54%), Gaps = 30/710 (4%)
Query: 44 ESLDYEIFENEFFKQDWRSRGVVQILQYICMKWLLCFMIGLIVGFIGFCNNLAVENLAGI 103
ESLDY+ EN+ F ++ R +W++C +IG++ G + ++ VENLAG+
Sbjct: 95 ESLDYDNSENQLFLEEERRINHTAFRTVEIKRWVICALIGILTGLVACFIDIVVENLAGL 154
Query: 104 KF-VTTSNM---MLERRFMFAFFIFFASNLSLTLFASIITAFIAPTAAGSGISEVKAYLN 159
K+ V N+ + F+ ++ N + L S+I AFI P AAGSGI ++K +LN
Sbjct: 155 KYRVIKGNIDKFTEKGGLSFSLLLWATLNAAFVLVGSVIVAFIEPVAAGSGIPQIKCFLN 214
Query: 160 GVDAPGIFTVRTLCVKIIGSITAVSGSLVIGKAGPMVHTGACVAALLGQGGSKRYGITWR 219
GV P + ++TL +K+ G I +V G L +GK GPM+H+G+ +AA + QG S ++
Sbjct: 215 GVKIPHVVRLKTLVIKVSGVILSVVGGLAVGKEGPMIHSGSVIAAGISQGRSTSLKRDFK 274
Query: 220 WLRFFKNDRDRRDLIICGSAAGIAAAFRAPVGGVLFALEEMASWWRTALLWRAFFTTATV 279
+F+ D ++RD + G+AAG++AAF APVGGVLF+LEE AS+W L WR FF +
Sbjct: 275 IFEYFRRDTEKRDFVSAGAAAGVSAAFGAPVGGVLFSLEEGASFWNQFLTWRIFFASMIS 334
Query: 280 AIFLRAMIDVCLSDKCGLFGKGGLIMFDAY-SASISYHLVDVPPVFILAVVGGLLGSLFN 338
L ++ + + L GLI F + S ++Y + ++P + VVGG+LG++FN
Sbjct: 335 TFTLNFVLSIYHGNMWDL-SSPGLINFGRFDSEKMAYTIHEIPVFIAMGVVGGVLGAVFN 393
Query: 339 FMTNKVLRIYNVINEKGTICRLFLACLISIFTSCLLFGLPWLAPCRPCPPDAVEPCPTIG 398
+ N L ++ + ++ A L++ T+ + F L + R C P G
Sbjct: 394 AL-NYWLTMFRIRYIHRPCLQVIEAVLVAAVTATVAFVL--IYSSRDCQ-------PLQG 443
Query: 399 RSGIYK-KFQCPPNHYNGLASLIFNTNDDAIRNLFSMHTDNEFELSSMLVFFIICLFLSI 457
S Y + C YN +A+ FNT + ++ +LF + ++ +F ++ FL+
Sbjct: 444 GSMSYPLQLFCADGEYNSMAAAFFNTPEKSVVSLFH-DPPGSYNPLTLGLFTLVYFFLAC 502
Query: 458 FSCGIVAPAGLFVPIIVTGASYGRLVGI----LVGERTNLSNGLYAVLGAASLLGGSMRT 513
++ G+ AG+F+P ++ GA++GRL GI L G G YA++GAA+ LGG +R
Sbjct: 503 WTYGLTVSAGVFIPSLLIGAAWGRLFGISLSYLTGAAIWADPGKYALMGAAAQLGGIVRM 562
Query: 514 TVSLCVIMLELTNNLLLLPLIMMVLVVSKSVANVFNANVYDLIMKAKGLPYLETHAEPYM 573
T+SL VIM+E T+N+ IM+VL+ +K V +VF +YD+ ++ + +P+L A
Sbjct: 563 TLSLTVIMMEATSNVTYGFPIMLVLMTAKIVGDVFIEGLYDMHIQLQSVPFLHWEAPVTS 622
Query: 574 RQLTVGDVVTGPLQMFNGIEKVRNIVFILRTTA--HNGFPVIDEPPGSEAPILFGIILRH 631
LT +V++ P+ EKV IV +L TA HNGFPV++ ++ L G+ILR
Sbjct: 623 HSLTAREVMSTPVTCLRRREKVGVIVDVLSDTASNHNGFPVVEHADDTQPARLQGLILRS 682
Query: 632 HLTTLLKKKAFLPSPVANSYDVVRKFSSDDFAKKYSVERVKIEDIQLTEEEMGMFVDLHP 691
L LLK K F+ +N V R+ DF Y I+ I ++++E +DL
Sbjct: 683 QLIVLLKHKVFVER--SNLGLVQRRLRLKDFRDAYP-RFPPIQSIHVSQDERECTMDLSE 739
Query: 692 FTNASPYTVVETMSLAKALILFREVGLRHLLVIPKIPGRSPVVGILTRHD 741
F N SPYTV + SL + LFR +GLRHL+V + R+ VVG++TR D
Sbjct: 740 FMNPSPYTVPQEASLPRVFKLFRALGLRHLVV---VDNRNQVVGLVTRKD 786
>CLC7_RAT (P51799) Chloride channel protein 7 (ClC-7)
Length = 803
Score = 386 bits (992), Expect = e-106
Identities = 246/710 (34%), Positives = 391/710 (54%), Gaps = 30/710 (4%)
Query: 44 ESLDYEIFENEFFKQDWRSRGVVQILQYICMKWLLCFMIGLIVGFIGFCNNLAVENLAGI 103
ESLDY+ EN+ F ++ R +W++C +IG++ G + ++ VENLAG+
Sbjct: 93 ESLDYDNSENQLFLEEERRINHTAFRTVEIKRWVICALIGILTGLVACFIDIVVENLAGL 152
Query: 104 KF-VTTSNM---MLERRFMFAFFIFFASNLSLTLFASIITAFIAPTAAGSGISEVKAYLN 159
K+ V N+ + F+ ++ N + L S+I AFI P AAGSGI ++K +LN
Sbjct: 153 KYRVIKDNIDKFTEKGGLSFSLLLWATLNSAFVLVGSVIVAFIEPVAAGSGIPQIKCFLN 212
Query: 160 GVDAPGIFTVRTLCVKIIGSITAVSGSLVIGKAGPMVHTGACVAALLGQGGSKRYGITWR 219
GV P + ++TL +K+ G I +V G L +GK GPM+H+G+ +AA + QG S ++
Sbjct: 213 GVKIPHVVRLKTLVIKVSGVILSVVGGLAVGKEGPMIHSGSVIAAGISQGRSTSLKRDFK 272
Query: 220 WLRFFKNDRDRRDLIICGSAAGIAAAFRAPVGGVLFALEEMASWWRTALLWRAFFTTATV 279
+F+ D ++RD + G+AAG++AAF APVGGVLF+LEE AS+W L WR FF +
Sbjct: 273 IFEYFRRDTEKRDFVSAGAAAGVSAAFGAPVGGVLFSLEEGASFWNQFLTWRIFFASMIS 332
Query: 280 AIFLRAMIDVCLSDKCGLFGKGGLIMFDAY-SASISYHLVDVPPVFILAVVGGLLGSLFN 338
L ++ + + L GLI F + S ++Y + ++P + VVGG+LG++FN
Sbjct: 333 TFTLNFVLSIYHGNMWDL-SSPGLINFGRFDSEKMAYTIHEIPVFIAMGVVGGILGAVFN 391
Query: 339 FMTNKVLRIYNVINEKGTICRLFLACLISIFTSCLLFGLPWLAPCRPCPPDAVEPCPTIG 398
+ N L ++ + ++ A L++ T+ + F L + R C P G
Sbjct: 392 AL-NYWLTMFRIRYIHRPCLQVIEAMLVAAVTATVAFVL--IYSSRDCQ-------PLQG 441
Query: 399 RSGIYK-KFQCPPNHYNGLASLIFNTNDDAIRNLFSMHTDNEFELSSMLVFFIICLFLSI 457
S Y + C YN +A+ FNT + ++ +LF + ++ +F ++ FL+
Sbjct: 442 SSMSYPLQLFCADGEYNSMAAAFFNTPEKSVVSLFH-DPPGSYNPMTLGLFTLVYFFLAC 500
Query: 458 FSCGIVAPAGLFVPIIVTGASYGRLVGI----LVGERTNLSNGLYAVLGAASLLGGSMRT 513
++ G+ AG+F+P ++ GA++GRL GI L G G YA++GAA+ LGG +R
Sbjct: 501 WTYGLTVSAGVFIPSLLIGAAWGRLFGISMSYLTGAAIWADPGKYALMGAAAQLGGIVRM 560
Query: 514 TVSLCVIMLELTNNLLLLPLIMMVLVVSKSVANVFNANVYDLIMKAKGLPYLETHAEPYM 573
T+SL VIM+E T+N+ IM+VL+ +K V +VF +YD+ ++ + +P+L A
Sbjct: 561 TLSLTVIMMEATSNVTYGFPIMLVLMTAKIVGDVFIEGLYDMHIQLQSVPFLHWEAPVTS 620
Query: 574 RQLTVGDVVTGPLQMFNGIEKVRNIVFILRTTA--HNGFPVIDEPPGSEAPILFGIILRH 631
LT +V++ P+ EKV IV +L TA HNGFPV+++ ++ L G+ILR
Sbjct: 621 HSLTAREVMSTPVTCLRRREKVGIIVDVLSDTASNHNGFPVVEDVGDTQPARLQGLILRS 680
Query: 632 HLTTLLKKKAFLPSPVANSYDVVRKFSSDDFAKKYSVERVKIEDIQLTEEEMGMFVDLHP 691
L LLK K F+ +N V R+ DF Y I+ I ++++E +DL
Sbjct: 681 QLIVLLKHKVFVER--SNMGLVQRRLRLKDFRDAYP-RFPPIQSIHVSQDERECTMDLSE 737
Query: 692 FTNASPYTVVETMSLAKALILFREVGLRHLLVIPKIPGRSPVVGILTRHD 741
F N SPYTV + SL + LFR +GLRHL+V + + VVG++TR D
Sbjct: 738 FMNPSPYTVPQEASLPRVFKLFRALGLRHLVV---VDNHNQVVGLVTRKD 784
>CLC7_MOUSE (O70496) Chloride channel protein 7 (ClC-7)
Length = 803
Score = 386 bits (992), Expect = e-106
Identities = 246/710 (34%), Positives = 391/710 (54%), Gaps = 30/710 (4%)
Query: 44 ESLDYEIFENEFFKQDWRSRGVVQILQYICMKWLLCFMIGLIVGFIGFCNNLAVENLAGI 103
ESLDY+ EN+ F ++ R +W++C +IG++ G + ++ VENLAG+
Sbjct: 93 ESLDYDNSENQLFLEEERRINHTAFRTVEIKRWVICALIGILTGLVACFIDIVVENLAGL 152
Query: 104 KF-VTTSNM---MLERRFMFAFFIFFASNLSLTLFASIITAFIAPTAAGSGISEVKAYLN 159
K+ V N+ + F+ ++ N + L S+I AFI P AAGSGI ++K +LN
Sbjct: 153 KYRVIKDNIDKFTEKGGLSFSLLLWATLNSAFVLVGSVIVAFIEPVAAGSGIPQIKCFLN 212
Query: 160 GVDAPGIFTVRTLCVKIIGSITAVSGSLVIGKAGPMVHTGACVAALLGQGGSKRYGITWR 219
GV P + ++TL +K+ G I +V G L +GK GPM+H+G+ +AA + QG S ++
Sbjct: 213 GVKIPHVVRLKTLVIKVSGVILSVVGGLAVGKEGPMIHSGSVIAAGISQGRSTSLKRDFK 272
Query: 220 WLRFFKNDRDRRDLIICGSAAGIAAAFRAPVGGVLFALEEMASWWRTALLWRAFFTTATV 279
+F+ D ++RD + G+AAG++AAF APVGGVLF+LEE AS+W L WR FF +
Sbjct: 273 IFEYFRRDTEKRDFVSAGAAAGVSAAFGAPVGGVLFSLEEGASFWNQFLTWRIFFASMIS 332
Query: 280 AIFLRAMIDVCLSDKCGLFGKGGLIMFDAY-SASISYHLVDVPPVFILAVVGGLLGSLFN 338
L ++ + + L GLI F + S ++Y + ++P + VVGG+LG++FN
Sbjct: 333 TFTLNFVLSIYHGNMWDL-SSPGLINFGRFDSEKMAYTIHEIPVFIAMGVVGGILGAVFN 391
Query: 339 FMTNKVLRIYNVINEKGTICRLFLACLISIFTSCLLFGLPWLAPCRPCPPDAVEPCPTIG 398
+ N L ++ + ++ A L++ T+ + F L + R C P G
Sbjct: 392 AL-NYWLTMFRIRYIHRPCLQVIEAMLVAAVTATVAFVL--IYSSRDCQ-------PLQG 441
Query: 399 RSGIYK-KFQCPPNHYNGLASLIFNTNDDAIRNLFSMHTDNEFELSSMLVFFIICLFLSI 457
S Y + C YN +A+ FNT + ++ +LF + ++ +F ++ FL+
Sbjct: 442 SSMSYPLQLFCADGEYNSMAAAFFNTPEKSVVSLFH-DPPGSYNPMTLGLFTLVYFFLAC 500
Query: 458 FSCGIVAPAGLFVPIIVTGASYGRLVGI----LVGERTNLSNGLYAVLGAASLLGGSMRT 513
++ G+ AG+F+P ++ GA++GRL GI L G G YA++GAA+ LGG +R
Sbjct: 501 WTYGLTVSAGVFIPSLLIGAAWGRLFGISLSYLTGAAIWADPGKYALMGAAAQLGGIVRM 560
Query: 514 TVSLCVIMLELTNNLLLLPLIMMVLVVSKSVANVFNANVYDLIMKAKGLPYLETHAEPYM 573
T+SL VIM+E T+N+ IM+VL+ +K V +VF +YD+ ++ + +P+L A
Sbjct: 561 TLSLTVIMMEATSNVTYGFPIMLVLMTAKIVGDVFIEGLYDMHIQLQSVPFLHWEAPVTS 620
Query: 574 RQLTVGDVVTGPLQMFNGIEKVRNIVFILRTTA--HNGFPVIDEPPGSEAPILFGIILRH 631
LT +V++ P+ EKV IV +L TA HNGFPV+++ ++ L G+ILR
Sbjct: 621 HSLTAREVMSTPVTCLRRREKVGIIVDVLSDTASNHNGFPVVEDVGDTQPARLQGLILRS 680
Query: 632 HLTTLLKKKAFLPSPVANSYDVVRKFSSDDFAKKYSVERVKIEDIQLTEEEMGMFVDLHP 691
L LLK K F+ +N V R+ DF Y I+ I ++++E +DL
Sbjct: 681 QLIVLLKHKVFVER--SNMGLVQRRLRLKDFRDAYP-RFPPIQSIHVSQDERECTMDLSE 737
Query: 692 FTNASPYTVVETMSLAKALILFREVGLRHLLVIPKIPGRSPVVGILTRHD 741
F N SPYTV + SL + LFR +GLRHL+V + + VVG++TR D
Sbjct: 738 FMNPSPYTVPQEASLPRVFKLFRALGLRHLVV---VDNHNQVVGLVTRKD 784
>CLC6_HUMAN (P51797) Chloride channel protein 6 (ClC-6)
Length = 869
Score = 326 bits (836), Expect = 1e-88
Identities = 210/608 (34%), Positives = 331/608 (53%), Gaps = 32/608 (5%)
Query: 44 ESLDYEIFENEFFKQDWRSRGVVQILQYICMKWLLCFMIGLIVGFIGFCNNLAVENLAGI 103
ESLDY+ N+ + + + + +Y +KW++ F IG+ G +G + V +
Sbjct: 49 ESLDYDRCINDPYLEVLETMDNKKGRRYEAVKWMVVFAIGVCTGLVGLFVDFFVRLFTQL 108
Query: 104 KF--VTTSNMMLERRFMFAFFIFFASNLSLT-LFASIITAFIAPTAAGSGISEVKAYLNG 160
KF V TS ++ A + +LT +F + + I P AAGSGI EVK YLNG
Sbjct: 109 KFGVVQTSVEECSQKGCLALSLLELLGFNLTFVFLASLLVLIEPVAAGSGIPEVKCYLNG 168
Query: 161 VDAPGIFTVRTLCVKIIGSITAVSGSLVIGKAGPMVHTGACVAALLGQGGSKRYGITWRW 220
V PGI +RTL K++G + +V+G L +GK GPM+H+G+ V A L Q S I+ R
Sbjct: 169 VKVPGIVRLRTLLCKVLGVLFSVAGGLFVGKEGPMIHSGSVVGAGLPQFQS----ISLRK 224
Query: 221 LRF----FKNDRDRRDLIICGSAAGIAAAFRAPVGGVLFALEEMASWWRTALLWRAFFTT 276
++F F++DRD+RD + G+AAG+AAAF AP+GG LF+LEE +S+W L W+ F +
Sbjct: 225 IQFNFPYFRSDRDKRDFVSAGAAAGVAAAFGAPIGGTLFSLEEGSSFWNQGLTWKVLFCS 284
Query: 277 ATVAIFLRAMIDVCLSDKCGLFGKGGLIMFDAYSASIS------YHLVDVPPVFILAVVG 330
+ L G F GL+ F + S S + +D+ ++ V+G
Sbjct: 285 MSATFTLNFFRSGIQFGSWGSFQLPGLLNFGEFKCSDSDKKCHLWTAMDLGFFVVMGVIG 344
Query: 331 GLLGSLFNFMTNKVLRIYNV--INEKGTICRLFLACLISIFTSCLLFGLPW-LAPCRPCP 387
GLLG+ FN + NK L Y + ++ K + R+ + L+S+ T+ ++F L CR
Sbjct: 345 GLLGATFNCL-NKRLAKYRMRNVHPKPKLVRVLESLLVSLVTTVVVFVASMVLGECRQMS 403
Query: 388 P------DAVEPCPTIGRSGIYKKFQCPPNHYNGLASLIFNTNDDAIRNLFSMHTDNEFE 441
D+ + T + K F CP + YN +A+L FN + AI LF H D F
Sbjct: 404 SSSQIGNDSFQLQVTEDVNSSIKTFFCPNDTYNDMATLFFNPQESAILQLF--HQDGTFS 461
Query: 442 LSSMLVFFIICLFLSIFSCGIVAPAGLFVPIIVTGASYGRLVGILVGERTNLSN---GLY 498
++ +FF++ L+ ++ GI P+GLFVP ++ GA++GRLV ++ L + G +
Sbjct: 462 PVTLALFFVLYFLLACWTYGISVPSGLFVPSLLCGAAFGRLVANVLKSYIGLGHIYSGTF 521
Query: 499 AVLGAASLLGGSMRTTVSLCVIMLELTNNLLLLPLIMMVLVVSKSVANVFNANVYDLIMK 558
A++GAA+ LGG +R T+SL VI++E TN + IM+ L+V+K + FN +YD+ +
Sbjct: 522 ALIGAAAFLGGVVRMTISLTVILIESTNEITYGLPIMVTLMVAKWTGDFFNKGIYDIHVG 581
Query: 559 AKGLPYLETHAEPYMRQLTVGDVVTGPLQMFNGIEKVRNIVFILRTTAHNGFPVIDEPPG 618
+G+P LE E M +L D++ L +++++V ILRTT H+ FPV+ E G
Sbjct: 582 LRGVPLLEWETEVEMDKLRASDIMEPNLTYVYPHTRIQSLVSILRTTVHHAFPVVTENRG 641
Query: 619 SEAPILFG 626
+E + G
Sbjct: 642 NEKEFMKG 649
Score = 61.2 bits (147), Expect = 1e-08
Identities = 43/127 (33%), Positives = 66/127 (51%), Gaps = 7/127 (5%)
Query: 622 PILF-GIILRHHLTTLLKKKAFLPSPVANSYDVVRKFSSDDFAKKYSVERVKIEDIQLTE 680
P+ F G+ILR L TLL + +++ + S + A+ Y I D+ LT
Sbjct: 738 PLTFHGLILRSQLVTLLVRGVCYSESQSSASQP--RLSYAEMAEDYP-RYPDIHDLDLTL 794
Query: 681 EEMGMFVDLHPFTNASPYTVVETMSLAKALILFREVGLRHLLVIPKIPGRSPVVGILTRH 740
M VD+ P+ N SP+TV +++ LFR +GLRHL V+ + +VGI+TRH
Sbjct: 795 LNPRMIVDVTPYMNPSPFTVSPNTHVSQVFNLFRTMGLRHLPVVNAV---GEIVGIITRH 851
Query: 741 DFTPEHI 747
+ T E +
Sbjct: 852 NLTYEFL 858
>CLC6_MOUSE (O35454) Chloride channel protein 6 (ClC-6)
Length = 870
Score = 323 bits (828), Expect = 1e-87
Identities = 208/609 (34%), Positives = 330/609 (54%), Gaps = 33/609 (5%)
Query: 44 ESLDYEIFENEFFKQDWRSRGVVQILQYICMKWLLCFMIGLIVGFIGFCNNLAVENLAGI 103
ESLDY+ N+ + + + + +Y +KW++ F IG+ G +G + +V +
Sbjct: 49 ESLDYDRCINDPYLEVLETMDNKKGRRYEAVKWMVVFAIGVCTGLVGLFVDFSVRLFTQL 108
Query: 104 KF--VTTSNMMLERRFMFAFFIFFASNLSLT-LFASIITAFIAPTAAGSGISEVKAYLNG 160
KF V TS ++ A + +LT +F + + I P AAGSGI E+K YLNG
Sbjct: 109 KFGVVQTSVEECSQKGCLALSLLELLGFNLTFVFLASLLVLIEPVAAGSGIPEIKCYLNG 168
Query: 161 VDAPGIFTVRTLCVKIIGSITAVSGSLVIGKAGPMVHTGACVAALLGQGGSKRYGITWRW 220
V PGI +RTL K+ G + +VSG L +GK GPM+H+GA V A L Q S I+ R
Sbjct: 169 VKVPGIVRLRTLLCKVFGVLFSVSGGLFVGKEGPMIHSGAVVGAGLPQFQS----ISLRK 224
Query: 221 LRF----FKNDRDRRDLIICGSAAGIAAAFRAPVGGVLFALEEMASWWRTALLWRAFFTT 276
++F F++DRD+RD + G+AAG+AAAF AP+GG LF+LEE +S+W L W+ F +
Sbjct: 225 IQFNFPYFRSDRDKRDFVSAGAAAGVAAAFGAPIGGTLFSLEEGSSFWNQGLTWKVLFCS 284
Query: 277 ATVAIFLRAMIDVCLSDKCGLFGKGGLIMFDAYSASIS------YHLVDVPPVFILAVVG 330
+ L G F GL+ F + S S + +D+ ++ V+G
Sbjct: 285 MSATFTLNFFRSGIQFGSWGSFQLPGLLNFGEFKCSDSDKKCHLWTAMDLGFFVVMGVIG 344
Query: 331 GLLGSLFNFMTNKVLRIYNV--INEKGTICRLFLACLISIFTSCLLFGLPW-LAPCRPCP 387
GLLG+ FN + NK L Y + ++ K + R+ + L+S+ T+ ++F L CR
Sbjct: 345 GLLGATFNCL-NKRLAKYRMRNVHPKPKLVRVLESLLVSLVTTVVVFVASMVLGECRQMS 403
Query: 388 PDAVE-------PCPTIGRSGIYKKFQCPPNHYNGLASLIFNTNDDAIRNLFSMHTDNEF 440
+ + + K F CP + YN +A+L FN+ + AI LF H D F
Sbjct: 404 STSQTGNGSFQLQVTSEDVNSTIKAFFCPNDTYNDMATLFFNSQESAILQLF--HQDGTF 461
Query: 441 ELSSMLVFFIICLFLSIFSCGIVAPAGLFVPIIVTGASYGRLVGILVGERTNLSN---GL 497
++ +FFI+ L+ ++ G P+GLFVP ++ GA++GRLV ++ L + G
Sbjct: 462 SPVTLALFFILYFLLACWTFGTSVPSGLFVPSLLCGAAFGRLVANVLKSYIGLGHLYSGT 521
Query: 498 YAVLGAASLLGGSMRTTVSLCVIMLELTNNLLLLPLIMMVLVVSKSVANVFNANVYDLIM 557
+A++GAA+ LGG +R T+SL VI++E TN + IM+ L+V+K ++FN +YD+ +
Sbjct: 522 FALIGAAAFLGGVVRMTISLTVILIESTNEITYGLPIMVTLMVAKWTGDLFNKGIYDVHI 581
Query: 558 KAKGLPYLETHAEPYMRQLTVGDVVTGPLQMFNGIEKVRNIVFILRTTAHNGFPVIDEPP 617
+G+P LE + M +L D++ L +++++V ILRTT H+ FPV+ E
Sbjct: 582 GLRGVPLLEWETDVEMDKLRASDIMEPNLTYVYPHTRIQSLVSILRTTVHHAFPVVTENR 641
Query: 618 GSEAPILFG 626
G+E + G
Sbjct: 642 GNEKEFMKG 650
Score = 61.2 bits (147), Expect = 1e-08
Identities = 42/127 (33%), Positives = 66/127 (51%), Gaps = 7/127 (5%)
Query: 622 PILF-GIILRHHLTTLLKKKAFLPSPVANSYDVVRKFSSDDFAKKYSVERVKIEDIQLTE 680
P+ F G++LR L TLL + +++ + S + A+ Y I D+ LT
Sbjct: 739 PLTFHGLVLRSQLVTLLVRGVCYSESQSSASQP--RLSYAEMAEDYP-RYPDIHDLDLTL 795
Query: 681 EEMGMFVDLHPFTNASPYTVVETMSLAKALILFREVGLRHLLVIPKIPGRSPVVGILTRH 740
M VD+ P+ N SP+TV +++ LFR +GLRHL V+ + +VGI+TRH
Sbjct: 796 LNPRMIVDVTPYMNPSPFTVSPNTHVSQVFNLFRTMGLRHLPVVNAV---GEIVGIITRH 852
Query: 741 DFTPEHI 747
+ T E +
Sbjct: 853 NLTNEFL 859
>CLC6_RABIT (Q9TT16) Chloride channel protein 6 (ClC-6)
Length = 869
Score = 321 bits (822), Expect = 5e-87
Identities = 209/608 (34%), Positives = 329/608 (53%), Gaps = 32/608 (5%)
Query: 44 ESLDYEIFENEFFKQDWRSRGVVQILQYICMKWLLCFMIGLIVGFIGFCNNLAVENLAGI 103
ESLDY+ N+ + + + + Y +KW + F IG+ G +G + V+ +
Sbjct: 49 ESLDYDRCINDPYLEVLETMDHKKGRWYEVVKWTVVFAIGVCTGLVGLFVDFFVQLFTQL 108
Query: 104 KF--VTTSNMMLERRFMFAFFIFFASNLSLT-LFASIITAFIAPTAAGSGISEVKAYLNG 160
KF V S ++ A + +LT +F + + I P AAGSGI E+K YLNG
Sbjct: 109 KFGVVEASVEECSQKGCLALSLLELLGFNLTFVFLASLLVLIEPVAAGSGIPEIKCYLNG 168
Query: 161 VDAPGIFTVRTLCVKIIGSITAVSGSLVIGKAGPMVHTGACVAALLGQGGSKRYGITWRW 220
V PGI +RTL K+ G + +V+G L +GK GPM+H+GA V A L Q S I+ R
Sbjct: 169 VKVPGIVRLRTLLCKVFGVLFSVAGGLFVGKEGPMIHSGAVVGAGLPQFQS----ISLRK 224
Query: 221 LRF----FKNDRDRRDLIICGSAAGIAAAFRAPVGGVLFALEEMASWWRTALLWRAFFTT 276
++F F++DRD+RD + G+AAGIAAAF AP+G LF+LEE +S+W L W+ F +
Sbjct: 225 IQFNFPYFRSDRDKRDFVSAGAAAGIAAAFGAPIGATLFSLEEGSSFWNQGLTWKVLFCS 284
Query: 277 ATVAIFLRAMIDVCLSDKCGLFGKGGLIMFDAYSASIS------YHLVDVPPVFILAVVG 330
+ L G F GL+ F + S S + +D+ ++ V+G
Sbjct: 285 MSATFTLNFFRSGIQFGSWGSFQLPGLLNFGEFKCSDSDKKCHLWTAMDMGFFVVMGVIG 344
Query: 331 GLLGSLFNFMTNKVLRIYNV--INEKGTICRLFLACLISIFTSCLLFGLPW-LAPCRPCP 387
GLLG+ FN + NK L Y + ++ K + R+ + L+S+ T+ ++F L CR
Sbjct: 345 GLLGATFNCL-NKRLAKYRMRNVHPKPKLVRVLESLLVSLVTTLVVFVASMVLGECRQMS 403
Query: 388 PDA------VEPCPTIGRSGIYKKFQCPPNHYNGLASLIFNTNDDAIRNLFSMHTDNEFE 441
+ ++ T + K F CP + YN +A+L FN + AI LF H D F
Sbjct: 404 SSSQISNGSLKLQVTSDVNSSIKAFFCPNDTYNDMATLFFNPQESAILQLF--HQDGTFS 461
Query: 442 LSSMLVFFIICLFLSIFSCGIVAPAGLFVPIIVTGASYGRLVGILVGERTNLSN---GLY 498
++ +FF++ L+ ++ GI P+GLFVP ++ GA++GRLV ++ LS+ G +
Sbjct: 462 PITLALFFVLYFLLACWTYGISVPSGLFVPSLLCGAAFGRLVANVLKSYIGLSHIYSGTF 521
Query: 499 AVLGAASLLGGSMRTTVSLCVIMLELTNNLLLLPLIMMVLVVSKSVANVFNANVYDLIMK 558
+++GAA+LLGG +R T+SL VI++E TN + IM+ L+V+K + FN +YD+ +
Sbjct: 522 SLIGAAALLGGVVRMTISLTVILIESTNEITYGLPIMITLMVAKWTGDFFNKGIYDIHVG 581
Query: 559 AKGLPYLETHAEPYMRQLTVGDVVTGPLQMFNGIEKVRNIVFILRTTAHNGFPVIDEPPG 618
+G+P LE E M +L D++ L +++++V ILRTT H+ FPV+ E G
Sbjct: 582 LRGVPLLEWETEVEMDKLRASDIMEPNLTYVYPHTRIQSLVSILRTTVHHAFPVVTENRG 641
Query: 619 SEAPILFG 626
+E + G
Sbjct: 642 NEKEFMKG 649
Score = 60.5 bits (145), Expect = 2e-08
Identities = 43/127 (33%), Positives = 66/127 (51%), Gaps = 7/127 (5%)
Query: 622 PILF-GIILRHHLTTLLKKKAFLPSPVANSYDVVRKFSSDDFAKKYSVERVKIEDIQLTE 680
P+ F G+ILR L TLL + +++ + S + A+ Y I D+ LT
Sbjct: 738 PLTFHGLILRSQLVTLLVRGVCYSESQSSASQP--RLSYAEMAEDYP-RFPDIHDLDLTL 794
Query: 681 EEMGMFVDLHPFTNASPYTVVETMSLAKALILFREVGLRHLLVIPKIPGRSPVVGILTRH 740
M VD+ P+ N SP+TV +++ LFR +GLRHL V+ + +VGI+TRH
Sbjct: 795 LNPRMIVDVTPYMNPSPFTVSPNTHVSQVFNLFRTMGLRHLPVVNAV---GEIVGIVTRH 851
Query: 741 DFTPEHI 747
+ T E +
Sbjct: 852 NLTYEFL 858
>CLC5_RAT (P51796) Chloride channel protein 5 (ClC-5)
Length = 746
Score = 191 bits (484), Expect = 8e-48
Identities = 188/663 (28%), Positives = 300/663 (44%), Gaps = 92/663 (13%)
Query: 119 FAFFI-FFASNLSLTLFA----SIITAFIAPTAAGSGISEVKAYLNGVDAPGIFTVRTLC 173
FA+ + +F L LFA S++ AF AP A GSGI E+K L+G G TL
Sbjct: 134 FAYIVNYFMYVLWALLFAFLAVSLVKAF-APYACGSGIPEIKTILSGFIIRGYLGKWTLV 192
Query: 174 VKIIGSITAVSGSLVIGKAGPMVHTGACVAALLGQGGSKRYGITWRWLRFFKNDRDRRDL 233
+K I + AVS L +GK GP+VH C +L +K + KN+ RR++
Sbjct: 193 IKTITLVLAVSSGLSLGKEGPLVHVACCCGNILCHCFNK----------YRKNEAKRREV 242
Query: 234 IICGSAAGIAAAFRAPVGGVLFALEEMASWWRTALLWRAFFTTATVAIFLRAMIDVCLSD 293
+ +AAG++ AF AP+GGVLF+LEE++ ++ LWR+FF A LR++
Sbjct: 243 LSAAAAAGVSVAFGAPIGGVLFSLEEVSYYFPLKTLWRSFFAALVAAFTLRSI------- 295
Query: 294 KCGLFGKGGLIMFDAYSASISYHLVDVPPVFILAVVGGLLGSLFNFMTNKVLRIYNVINE 353
FG L++F +HL ++ P +L + GGL G+LF TN +
Sbjct: 296 --NPFGNSRLVLF-YVEFHTPWHLFELVPFIVLGIFGGLWGALF-IRTNIAWCRKRKTTQ 351
Query: 354 KGTICRLFLACLISIFTSCLLFGLPWL-APCRPCPPDAVEPCPTIGRSGIYKKFQCPPNH 412
G + +++ T+ L F + + C + S K NH
Sbjct: 352 LGKY-PVVEVLIVTAITAILAFPNEYTRMSTSELISELFNDCGLLDSS----KLCDYENH 406
Query: 413 YNGLASLIFNTNDDAIRNLFSMHTDNEFELSSMLVFFIICLFLSIFSCGIVAPAGLFVPI 472
+N + +M ++L+ L+ I+ ++IF+ G+ P+GLF+P
Sbjct: 407 FNTSKGGELPDRPAGVGVYSAM-----WQLALTLILKIV---ITIFTFGMKIPSGLFIPS 458
Query: 473 IVTGASYGRLVGILVGERTN------------------LSNGLYAVLGAASLLGGSMRTT 514
+ GA GRL+G+ + + ++ GLYA++GAA+ LGG R T
Sbjct: 459 MAVGAIAGRLLGVGMEQLAYYHHDWGIFNSWCSQGADCITPGLYAMVGAAACLGGVTRMT 518
Query: 515 VSLCVIMLELTNNLLLLPLIMMVLVVSKSVANVF-NANVYDLIMKAKGLPYLETHAEPYM 573
VSL VIM ELT L + +M + SK VA+ +YD ++ G P+LE E +
Sbjct: 519 VSLVVIMFELTGGLEYIVPLMAAAMTSKWVADALGREGIYDAHIRLNGYPFLEA-KEEFA 577
Query: 574 RQLTVGDVV----TGPL--QMFNGIEKVRNIVFILRTTAHNGFPVIDEPPGSEAPILFGI 627
+ DV+ PL + V ++ I+ T ++GFPV+ E+ L G
Sbjct: 578 HKTLAMDVMKPRRNDPLLTVLTQDSMTVEDVETIISETTYSGFPVV---VSRESQRLVGF 634
Query: 628 ILRHHLTTLLKKKAFLPSPVANSYDVVRKFSSDDFAKKYSVERVKIEDIQLTEEEMGMFV 687
+LR L ++ V S ++ Y+ +K+ +I +
Sbjct: 635 VLRRDLIISIENARKKQDGVV-STSIIYFTEHSPPMPPYTPPTLKLRNI----------L 683
Query: 688 DLHPFTNASPYTVVETMSLAKALILFREVGLRHLLVIPKIPGRSPVVGILTRHDFTPEHI 747
DL SP+TV + + + +FR++GLR LV GR ++GI+T+ D +HI
Sbjct: 684 DL------SPFTVTDLTPMEIVVDIFRKLGLRQCLVTHN--GR--LLGIITKKDVL-KHI 732
Query: 748 LGM 750
M
Sbjct: 733 AQM 735
>CLC5_MOUSE (Q9WVD4) Chloride channel protein 5 (ClC-5)
Length = 746
Score = 191 bits (484), Expect = 8e-48
Identities = 188/663 (28%), Positives = 300/663 (44%), Gaps = 92/663 (13%)
Query: 119 FAFFI-FFASNLSLTLFA----SIITAFIAPTAAGSGISEVKAYLNGVDAPGIFTVRTLC 173
FA+ + +F L LFA S++ AF AP A GSGI E+K L+G G TL
Sbjct: 134 FAYIVNYFMYVLWALLFAFLAVSLVKAF-APYACGSGIPEIKTILSGFIIRGYLGKWTLV 192
Query: 174 VKIIGSITAVSGSLVIGKAGPMVHTGACVAALLGQGGSKRYGITWRWLRFFKNDRDRRDL 233
+K I + AVS L +GK GP+VH C +L +K + KN+ RR++
Sbjct: 193 IKTITLVLAVSSGLSLGKEGPLVHVACCCGNILCHCFNK----------YRKNEAKRREV 242
Query: 234 IICGSAAGIAAAFRAPVGGVLFALEEMASWWRTALLWRAFFTTATVAIFLRAMIDVCLSD 293
+ +AAG++ AF AP+GGVLF+LEE++ ++ LWR+FF A LR++
Sbjct: 243 LSAAAAAGVSVAFGAPIGGVLFSLEEVSYYFPLKTLWRSFFAALVAAFTLRSI------- 295
Query: 294 KCGLFGKGGLIMFDAYSASISYHLVDVPPVFILAVVGGLLGSLFNFMTNKVLRIYNVINE 353
FG L++F +HL ++ P +L + GGL G+LF TN +
Sbjct: 296 --NPFGNSRLVLF-YVEFHTPWHLFELVPFIVLGIFGGLWGALF-IRTNIAWCRKRKTTQ 351
Query: 354 KGTICRLFLACLISIFTSCLLFGLPWL-APCRPCPPDAVEPCPTIGRSGIYKKFQCPPNH 412
G + +++ T+ L F + + C + S K NH
Sbjct: 352 LGKY-PVVEVLIVTAITAILAFPNEYTRMSTSELISELFNDCGLLDSS----KLCDYENH 406
Query: 413 YNGLASLIFNTNDDAIRNLFSMHTDNEFELSSMLVFFIICLFLSIFSCGIVAPAGLFVPI 472
+N + +M ++L+ L+ I+ ++IF+ G+ P+GLF+P
Sbjct: 407 FNTSKGGELPDRPAGVGIYSAM-----WQLALTLILKIV---ITIFTFGMKIPSGLFIPS 458
Query: 473 IVTGASYGRLVGILVGERTN------------------LSNGLYAVLGAASLLGGSMRTT 514
+ GA GRL+G+ + + ++ GLYA++GAA+ LGG R T
Sbjct: 459 MAVGAIAGRLLGVGMEQLAYYHHDWGIFNSWCSQGADCITPGLYAMVGAAACLGGVTRMT 518
Query: 515 VSLCVIMLELTNNLLLLPLIMMVLVVSKSVANVF-NANVYDLIMKAKGLPYLETHAEPYM 573
VSL VIM ELT L + +M + SK VA+ +YD ++ G P+LE E +
Sbjct: 519 VSLVVIMFELTGGLEYIVPLMAAAMTSKWVADALGREGIYDAHIRLNGYPFLEA-KEEFA 577
Query: 574 RQLTVGDVV----TGPL--QMFNGIEKVRNIVFILRTTAHNGFPVIDEPPGSEAPILFGI 627
+ DV+ PL + V ++ I+ T ++GFPV+ E+ L G
Sbjct: 578 HKTLAMDVMKPRRNDPLLTVLTQDSMTVEDVETIISETTYSGFPVV---VSRESQRLVGF 634
Query: 628 ILRHHLTTLLKKKAFLPSPVANSYDVVRKFSSDDFAKKYSVERVKIEDIQLTEEEMGMFV 687
+LR L ++ V S ++ Y+ +K+ +I +
Sbjct: 635 VLRRDLIISIENARKKQDGVV-STSIIYFTEHSPPMPPYTPPTLKLRNI----------L 683
Query: 688 DLHPFTNASPYTVVETMSLAKALILFREVGLRHLLVIPKIPGRSPVVGILTRHDFTPEHI 747
DL SP+TV + + + +FR++GLR LV GR ++GI+T+ D +HI
Sbjct: 684 DL------SPFTVTDLTPMEIVVDIFRKLGLRQCLVTHN--GR--LLGIITKKDVL-KHI 732
Query: 748 LGM 750
M
Sbjct: 733 AQM 735
>CLC3_HUMAN (P51790) Chloride channel protein 3 (ClC-3)
Length = 762
Score = 190 bits (483), Expect = 1e-47
Identities = 176/661 (26%), Positives = 286/661 (42%), Gaps = 108/661 (16%)
Query: 119 FAFFIFFASNLSLTLFASIITAFIAPTAAGSGISEVKAYLNGVDAPGIFTVRTLCVKIIG 178
+ +IF+A LS A + AP A GSGI E+K L+G G TL +K I
Sbjct: 153 YIMYIFWA--LSFAFLAVSLVKVFAPYACGSGIPEIKTILSGFIIRGYLGKWTLMIKTIT 210
Query: 179 SITAVSGSLVIGKAGPMVHTGACVAALLGQGGSKRYGITWRWLRFFKNDRDRRDLIICGS 238
+ AV+ L +GK GP+VH C + ++ + ++ N+ +R+++ S
Sbjct: 211 LVLAVASGLSLGKEGPLVHVACCCGNIF----------SYLFPKYSTNEAKKREVLSAAS 260
Query: 239 AAGIAAAFRAPVGGVLFALEEMASWWRTALLWRAFFTTATVAIFLRAMIDVCLSDKCGLF 298
AAG++ AF AP+GGVLF+LEE++ ++ LWR+FF A LR++ F
Sbjct: 261 AAGVSVAFGAPIGGVLFSLEEVSYYFPLKTLWRSFFAALVAAFVLRSI---------NPF 311
Query: 299 GKGGLIMFDAYSASISYH----LVDVPPVFILAVVGGLLGSLFNFMTNKVLRIYNVINEK 354
G L++F + YH L ++ P +L V GGL G+ F+ + + K
Sbjct: 312 GNSRLVLF-----YVEYHTPWYLFELFPFILLGVFGGLWGAF--FIRANIAWCRRRKSTK 364
Query: 355 GTICRLFLACLISIFTSCLLFGLPWLAPCRPCPPDAVEPCPTIGRSGIYKKF--QCPPNH 412
+ +++ T+ + F P+ + S + K+ C P
Sbjct: 365 FGKYPVLEVIIVAAITAVIAFPNPY---------------TRLNTSELIKELFTDCGPLE 409
Query: 413 YNGLASLIFNTNDDAIRNLFSMHTDNEFELSSMLVFFIICL------FLSIFSCGIVAPA 466
+ L ND + D + + +CL +++F+ GI P+
Sbjct: 410 SSSLCDY---RNDMNASKIVDDIPDRPAGIGVYSAIWQLCLALIFKIIMTVFTFGIKVPS 466
Query: 467 GLFVPIIVTGASYGRLVGILVGERTN------------------LSNGLYAVLGAASLLG 508
GLF+P + GA GR+VGI V + ++ GLYA++GAA+ LG
Sbjct: 467 GLFIPSMAIGAIAGRIVGIAVEQLAYYHHDWFIFKEWCEVGADCITPGLYAMVGAAACLG 526
Query: 509 GSMRTTVSLCVIMLELTNNLLLLPLIMMVLVVSKSVANVF-NANVYDLIMKAKGLPYLET 567
G R TVSL VI+ ELT L + +M ++ SK V + F +Y+ ++ G P+L+
Sbjct: 527 GVTRMTVSLVVIVFELTGGLEYIVPLMAAVMTSKWVGDAFGREGIYEAHIRLNGYPFLDA 586
Query: 568 HAEPYMRQLTVGDVVTGPLQ-------MFNGIEKVRNIVFILRTTAHNGFPVIDEPPGSE 620
E T+ V P + + V +I ++ T++NGFPVI E
Sbjct: 587 KEEFEFTHTTLAADVMRPRRNDPPLAVLTQDNMTVDDIENMINETSYNGFPVI---MSKE 643
Query: 621 APILFGIILRHHLTTLLKKKAFLPSPVANSYDVVRKFSSDDFAKKYSVERVKIEDIQLTE 680
+ L G LR LT ++ + S V FA +
Sbjct: 644 SQRLVGFALRRDLTIAIESARKKQEGIVGSSRVC-------FA----------QHTPSLP 686
Query: 681 EEMGMFVDLHPFTNASPYTVVETMSLAKALILFREVGLRHLLVIPKIPGRSPVVGILTRH 740
E + L + SP+TV + + + +FR++GLR LV GR ++GI+T+
Sbjct: 687 AESPRPLKLRSILDMSPFTVTDHTPMEIVVDIFRKLGLRQCLVTHN--GR--LLGIITKK 742
Query: 741 D 741
D
Sbjct: 743 D 743
>CLC3_RABIT (O18894) Chloride channel protein 3 (ClC-3)
Length = 760
Score = 189 bits (480), Expect = 2e-47
Identities = 177/660 (26%), Positives = 287/660 (42%), Gaps = 108/660 (16%)
Query: 119 FAFFIFFASNLSLTLFASIITAFIAPTAAGSGISEVKAYLNGVDAPGIFTVRTLCVKIIG 178
+ +IF+A LS A + AP A GSGI E+K L+G G TL +K I
Sbjct: 153 YIMYIFWA--LSFAFLAVSLVKVFAPYACGSGIPEIKTILSGFIIRGYLGKWTLMIKTIT 210
Query: 179 SITAVSGSLVIGKAGPMVHTGACVAALLGQGGSKRYGITWRWLRFFKNDRDRRDLIICGS 238
+ AV+ L +GK GP+VH C + ++ + ++ N+ +R+++ S
Sbjct: 211 LVLAVASGLSLGKEGPLVHVACCCGNIF----------SYLFPKYSTNEAKKREVLSAAS 260
Query: 239 AAGIAAAFRAPVGGVLFALEEMASWWRTALLWRAFFTTATVAIFLRAMIDVCLSDKCGLF 298
AAG++ AF AP+GGVLF+LEE++ ++ LWR+FF A LR++ F
Sbjct: 261 AAGVSVAFGAPIGGVLFSLEEVSYYFPLKTLWRSFFAALVAAFVLRSI---------NPF 311
Query: 299 GKGGLIMFDAYSASISYH----LVDVPPVFILAVVGGLLGSLFNFMTNKVLRIYNVINEK 354
G L++F + YH L ++ P +L V GGL G+ F+ + + K
Sbjct: 312 GNSRLVLF-----YVEYHTPWYLFELFPFILLGVFGGLWGAF--FIRANIAWCRRRKSTK 364
Query: 355 GTICRLFLACLISIFTSCLLFGLPWLAPCRPCPPDAVEPCPTIGRSGIYKKF--QCPPNH 412
+ +++ T+ + F P+ + S + K+ C P
Sbjct: 365 FGKYPVLEVIIVAAITAVIAFPNPY---------------TRLNTSELIKELFTDCGPLE 409
Query: 413 YNGLASLIFNTNDDAIRNLFSMHTDNEFELSSMLVFFIICL------FLSIFSCGIVAPA 466
+ L ND + D + + +CL +++F+ GI P+
Sbjct: 410 SSSLCDY---RNDMNASKIVDDIPDRPAGIGVYSAIWQLCLALIFKIIMTVFTFGIKVPS 466
Query: 467 GLFVPIIVTGASYGRLVGILVGERTN------------------LSNGLYAVLGAASLLG 508
GLF+P + GA GR+VGI V + ++ GLYA++GAA+ LG
Sbjct: 467 GLFIPSMAIGAIAGRIVGIAVEQLAYYHHDWFIFKEWCEVGADCITPGLYAMVGAAACLG 526
Query: 509 GSMRTTVSLCVIMLELTNNLLLLPLIMMVLVVSKSVANVF-NANVYDLIMKAKGLPYLET 567
G R TVSL VI+ ELT L + +M ++ SK V + F +Y+ ++ G P+L+
Sbjct: 527 GVTRMTVSLVVIVFELTGGLEYIVPLMAAVMTSKWVGDAFGREGIYEAHIRLNGYPFLDA 586
Query: 568 HAEPYMRQLTVGDVV-----TGPLQMFNGIE-KVRNIVFILRTTAHNGFPVIDEPPGSEA 621
E + DV+ PL + V +I ++ T++NGFPVI E+
Sbjct: 587 -KEEFTHTTLAADVMRPRRNDPPLAVLTQDNMTVDDIENMINETSYNGFPVI---MSKES 642
Query: 622 PILFGIILRHHLTTLLKKKAFLPSPVANSYDVVRKFSSDDFAKKYSVERVKIEDIQLTEE 681
L G LR LT ++ + S V FA +
Sbjct: 643 QRLVGFALRRDLTIAIESARKKQEGIVGSSRVC-------FA----------QHTPSLPA 685
Query: 682 EMGMFVDLHPFTNASPYTVVETMSLAKALILFREVGLRHLLVIPKIPGRSPVVGILTRHD 741
E + L + SP+TV + + + +FR++GLR LV GR ++GI+T+ D
Sbjct: 686 ESPRPLKLRSILDMSPFTVTDHTPMEIVVDIFRKLGLRQCLVTHN--GR--LLGIITKKD 741
>CLC3_RAT (P51792) Chloride channel protein 3 (ClC-3)
Length = 760
Score = 189 bits (479), Expect = 3e-47
Identities = 178/660 (26%), Positives = 287/660 (42%), Gaps = 108/660 (16%)
Query: 119 FAFFIFFASNLSLTLFASIITAFIAPTAAGSGISEVKAYLNGVDAPGIFTVRTLCVKIIG 178
+ +IF+A LS A + AP A GSGI E+K L+G G TL +K I
Sbjct: 153 YIMYIFWA--LSFAFLAVSLVKVFAPYACGSGIPEIKTILSGFIIRGYLGKWTLMIKTIT 210
Query: 179 SITAVSGSLVIGKAGPMVHTGACVAALLGQGGSKRYGITWRWLRFFKNDRDRRDLIICGS 238
+ AV+ L +GK GP+VH C + ++ + ++ N+ +R+++ S
Sbjct: 211 LVLAVASGLSLGKEGPLVHVACCCGNIF----------SYLFPKYSTNEAKKREVLSAAS 260
Query: 239 AAGIAAAFRAPVGGVLFALEEMASWWRTALLWRAFFTTATVAIFLRAMIDVCLSDKCGLF 298
AAG++ AF AP+GGVLF+LEE++ ++ LWR+FF A LR++ F
Sbjct: 261 AAGVSVAFGAPIGGVLFSLEEVSYYFPLKTLWRSFFAALVAAFVLRSI---------NPF 311
Query: 299 GKGGLIMFDAYSASISYH----LVDVPPVFILAVVGGLLGSLFNFMTNKVLRIYNVINEK 354
G L++F + YH L ++ P +L V GGL G+ F+ + + K
Sbjct: 312 GNSRLVLF-----YVEYHTPWYLFELFPFILLGVFGGLWGAF--FIRANIAWCRRRKSTK 364
Query: 355 GTICRLFLACLISIFTSCLLFGLPWLAPCRPCPPDAVEPCPTIGRSGIYKKF--QCPPNH 412
+ +++ T+ + F P+ + S + K+ C P
Sbjct: 365 FGKYPVLEVIIVAAITAVIAFPNPY---------------TRLNTSELIKELFTDCGPLE 409
Query: 413 YNGLASLIFNTNDDAIRNLFSMHTDNEFELSSMLVFFIICL------FLSIFSCGIVAPA 466
+ L ND + D + + +CL +++F+ GI P+
Sbjct: 410 SSSLCDY---RNDMNASKIVDDIPDRPAGVGVYSAIWQLCLALIFKIIMTVFTFGIKVPS 466
Query: 467 GLFVPIIVTGASYGRLVGILVGERTN------------------LSNGLYAVLGAASLLG 508
GLF+P + GA GR+VGI V + ++ GLYA++GAA+ LG
Sbjct: 467 GLFIPSMAIGAIAGRIVGIAVEQLAYYHHDWFIFKEWCEVGADCITPGLYAMVGAAACLG 526
Query: 509 GSMRTTVSLCVIMLELTNNLLLLPLIMMVLVVSKSVANVF-NANVYDLIMKAKGLPYLET 567
G R TVSL VI+ ELT L + +M ++ SK V + F +Y+ ++ G P+L+
Sbjct: 527 GVTRMTVSLVVIVFELTGGLEYIVPLMAAVMTSKWVGDAFGREGIYEAHIRLNGYPFLDA 586
Query: 568 HAEPYMRQLTVGDVV-----TGPLQMFNGIE-KVRNIVFILRTTAHNGFPVIDEPPGSEA 621
E + DV+ PL + V +I ++ T++NGFPVI E+
Sbjct: 587 -KEEFTHTTLAADVMRPRRSDPPLAVLTQDNMTVDDIENMINETSYNGFPVI---MSKES 642
Query: 622 PILFGIILRHHLTTLLKKKAFLPSPVANSYDVVRKFSSDDFAKKYSVERVKIEDIQLTEE 681
L G LR LT ++ V S V FA +
Sbjct: 643 QRLVGFALRRDLTIAIESARKKQEGVVGSSRVC-------FA----------QHTPSLPA 685
Query: 682 EMGMFVDLHPFTNASPYTVVETMSLAKALILFREVGLRHLLVIPKIPGRSPVVGILTRHD 741
E + L + SP+TV + + + +FR++GLR LV GR ++GI+T+ D
Sbjct: 686 ESPRPLKLRSILDMSPFTVTDHTPMEIVVDVFRKLGLRQCLVTHN--GR--LLGIITKKD 741
>CLC3_MOUSE (P51791) Chloride channel protein 3 (ClC-3)
Length = 760
Score = 189 bits (479), Expect = 3e-47
Identities = 177/660 (26%), Positives = 287/660 (42%), Gaps = 108/660 (16%)
Query: 119 FAFFIFFASNLSLTLFASIITAFIAPTAAGSGISEVKAYLNGVDAPGIFTVRTLCVKIIG 178
+ +IF+A LS A + AP A GSGI E+K L+G G TL +K I
Sbjct: 153 YIMYIFWA--LSFAFLAVSLVKVFAPYACGSGIPEIKTILSGFIIRGYLGKWTLMIKTIT 210
Query: 179 SITAVSGSLVIGKAGPMVHTGACVAALLGQGGSKRYGITWRWLRFFKNDRDRRDLIICGS 238
+ AV+ L +GK GP+VH C + ++ + ++ N+ +R+++ S
Sbjct: 211 LVLAVASGLSLGKEGPLVHVACCCGNIF----------SYLFPKYSTNEAKKREVLSAAS 260
Query: 239 AAGIAAAFRAPVGGVLFALEEMASWWRTALLWRAFFTTATVAIFLRAMIDVCLSDKCGLF 298
AAG++ AF AP+GGVLF+LEE++ ++ LWR+FF A LR++ F
Sbjct: 261 AAGVSVAFGAPIGGVLFSLEEVSYYFPLKTLWRSFFAALVAAFVLRSI---------NPF 311
Query: 299 GKGGLIMFDAYSASISYH----LVDVPPVFILAVVGGLLGSLFNFMTNKVLRIYNVINEK 354
G L++F + YH L ++ P +L V GGL G+ F+ + + K
Sbjct: 312 GNSRLVLF-----YVEYHTPWYLFELFPFILLGVFGGLWGAF--FIRANIAWCRRRKSTK 364
Query: 355 GTICRLFLACLISIFTSCLLFGLPWLAPCRPCPPDAVEPCPTIGRSGIYKKF--QCPPNH 412
+ +++ T+ + F P+ + S + K+ C P
Sbjct: 365 FGKYPVLEVIIVAAITAVIAFPNPY---------------TRLNTSELIKELFTDCGPLE 409
Query: 413 YNGLASLIFNTNDDAIRNLFSMHTDNEFELSSMLVFFIICL------FLSIFSCGIVAPA 466
+ L ND + D + + +CL +++F+ GI P+
Sbjct: 410 SSSLCDY---RNDMNASKIVDDIPDRPAGVGVYSAIWQLCLALIFKIIMTVFTFGIKVPS 466
Query: 467 GLFVPIIVTGASYGRLVGILVGERTN------------------LSNGLYAVLGAASLLG 508
GLF+P + GA GR+VGI V + ++ GLYA++GAA+ LG
Sbjct: 467 GLFIPSMAIGAIAGRIVGIAVEQLAYYHHDWFIFKEWCEVGADCITPGLYAMVGAAACLG 526
Query: 509 GSMRTTVSLCVIMLELTNNLLLLPLIMMVLVVSKSVANVF-NANVYDLIMKAKGLPYLET 567
G R TVSL VI+ ELT L + +M ++ SK V + F +Y+ ++ G P+L+
Sbjct: 527 GVTRMTVSLVVIVFELTGGLEYIVPLMAAVMTSKWVGDAFGREGIYEAHIRLNGYPFLDA 586
Query: 568 HAEPYMRQLTVGDVV-----TGPLQMFNGIE-KVRNIVFILRTTAHNGFPVIDEPPGSEA 621
E + DV+ PL + V +I ++ T++NGFPVI E+
Sbjct: 587 -KEEFTHTTLAADVMRPRRSDPPLAVLTQDNMTVDDIENMINETSYNGFPVI---MSKES 642
Query: 622 PILFGIILRHHLTTLLKKKAFLPSPVANSYDVVRKFSSDDFAKKYSVERVKIEDIQLTEE 681
L G LR LT ++ + S V FA +
Sbjct: 643 QRLVGFALRRDLTIAIESARKKQEGIVGSSRVC-------FA----------QHTPSLPA 685
Query: 682 EMGMFVDLHPFTNASPYTVVETMSLAKALILFREVGLRHLLVIPKIPGRSPVVGILTRHD 741
E + L + SP+TV + + + +FR++GLR LV GR ++GI+T+ D
Sbjct: 686 ESPRPLKLRSILDMSPFTVTDHTPMEIVVDIFRKLGLRQCLVTHN--GR--LLGIITKKD 741
>CLC3_CAVPO (Q9R279) Chloride channel protein 3 (ClC-3)
Length = 760
Score = 189 bits (479), Expect = 3e-47
Identities = 177/660 (26%), Positives = 287/660 (42%), Gaps = 108/660 (16%)
Query: 119 FAFFIFFASNLSLTLFASIITAFIAPTAAGSGISEVKAYLNGVDAPGIFTVRTLCVKIIG 178
+ +IF+A LS A + AP A GSGI E+K L+G G TL +K I
Sbjct: 153 YIMYIFWA--LSFAFLAVSLVKVFAPYACGSGIPEIKTILSGFIIRGYLGKWTLMIKTIT 210
Query: 179 SITAVSGSLVIGKAGPMVHTGACVAALLGQGGSKRYGITWRWLRFFKNDRDRRDLIICGS 238
+ AV+ L +GK GP+VH C + ++ + ++ N+ +R+++ S
Sbjct: 211 LVLAVASGLSLGKEGPLVHVACCCGNIF----------SYLFPKYSTNEAKKREVLSAAS 260
Query: 239 AAGIAAAFRAPVGGVLFALEEMASWWRTALLWRAFFTTATVAIFLRAMIDVCLSDKCGLF 298
AAG++ AF AP+GGVLF+LEE++ ++ LWR+FF A LR++ F
Sbjct: 261 AAGVSVAFGAPIGGVLFSLEEVSYYFPLKTLWRSFFAALVAAFVLRSI---------NPF 311
Query: 299 GKGGLIMFDAYSASISYH----LVDVPPVFILAVVGGLLGSLFNFMTNKVLRIYNVINEK 354
G L++F + YH L ++ P +L V GGL G+ F+ + + K
Sbjct: 312 GNSRLVLF-----YVEYHTPWYLFELFPFILLGVFGGLWGAF--FIRANIAWCRRRKSTK 364
Query: 355 GTICRLFLACLISIFTSCLLFGLPWLAPCRPCPPDAVEPCPTIGRSGIYKKF--QCPPNH 412
+ +++ T+ + F P+ + S + K+ C P
Sbjct: 365 FGKYPVLEVIIVAAITAVIAFPNPY---------------TRLNTSELIKELFTDCGPLE 409
Query: 413 YNGLASLIFNTNDDAIRNLFSMHTDNEFELSSMLVFFIICL------FLSIFSCGIVAPA 466
+ L ND + D + + +CL +++F+ GI P+
Sbjct: 410 SSSLCDY---RNDMNASKIVDDIPDRPAGVGVYSAIWQLCLALIFKIIMTVFTFGIKVPS 466
Query: 467 GLFVPIIVTGASYGRLVGILVGERTN------------------LSNGLYAVLGAASLLG 508
GLF+P + GA GR+VGI V + ++ GLYA++GAA+ LG
Sbjct: 467 GLFIPSMAIGAIAGRIVGIAVEQLAYFHHDWFIFKEWCEVGADCITPGLYAMVGAAACLG 526
Query: 509 GSMRTTVSLCVIMLELTNNLLLLPLIMMVLVVSKSVANVF-NANVYDLIMKAKGLPYLET 567
G R TVSL VI+ ELT L + +M ++ SK V + F +Y+ ++ G P+L+
Sbjct: 527 GVTRMTVSLVVIVFELTGGLEYIVPLMAAVMTSKWVGDAFGREGIYEAHIRLNGYPFLDA 586
Query: 568 HAEPYMRQLTVGDVV-----TGPLQMFNGIE-KVRNIVFILRTTAHNGFPVIDEPPGSEA 621
E + DV+ PL + V +I ++ T++NGFPVI E+
Sbjct: 587 -KEEFTHTTLAADVMRPRRNDPPLAVLTQDNMTVDDIENMINETSYNGFPVI---MSKES 642
Query: 622 PILFGIILRHHLTTLLKKKAFLPSPVANSYDVVRKFSSDDFAKKYSVERVKIEDIQLTEE 681
L G LR LT ++ + S V FA +
Sbjct: 643 QRLVGFALRRDLTIAIESARKKQEGIVGSSRVC-------FA----------QHTPSLPA 685
Query: 682 EMGMFVDLHPFTNASPYTVVETMSLAKALILFREVGLRHLLVIPKIPGRSPVVGILTRHD 741
E + L + SP+TV + + + +FR++GLR LV GR ++GI+T+ D
Sbjct: 686 ESPRPLKLRSILDMSPFTVTDHTPMEIVVDIFRKLGLRQCLVTHN--GR--LLGIITKKD 741
>CLC2_RABIT (P51789) Chloride channel protein 2 (ClC-2)
(PKA-activated chloride channel)
Length = 898
Score = 169 bits (428), Expect = 3e-41
Identities = 145/578 (25%), Positives = 260/578 (44%), Gaps = 48/578 (8%)
Query: 76 WLLCFMIGLIVGFIGFCNNLAVENLAGIKFVTTSNMMLERRFMFAFFIFFASNLSLTLFA 135
W+ ++GL++ + + + A+ A ++ + L + + + + L F+
Sbjct: 94 WIFLVLLGLLMALVSWAMDYAIA--ACLQAQQWMSRGLNTNLLLQYLAWVTYPVVLITFS 151
Query: 136 SIITAFIAPTAAGSGISEVKAYLNGVDAPGIFTVRTLCVKIIGSITAVSGSLVIGKAGPM 195
+ T +AP A GSGI E+K L GV T++T K+IG A+ + +GK GP
Sbjct: 152 AGFTQILAPQAVGSGIPEMKTILRGVVLKEYLTLKTFVAKVIGLTCALGSGMPLGKEGPF 211
Query: 196 VHTGACVAALLGQGGSKRYGITWRWLRFFKNDRDRRDLIICGSAAGIAAAFRAPVGGVLF 255
VH + AALL + S GI ++N+ +++ A G+ F AP+GGVLF
Sbjct: 212 VHIASMCAALLSKFLSLFGGI-------YENESRNTEMLAAACAVGVGCCFAAPIGGVLF 264
Query: 256 ALEEMASWWRTALLWRAFFTTATVAIFLRAMIDVCLSDKCGLFGKGGLIMFDAYSASISY 315
++E ++++ WR FF AT + F+ ++ V D+ + + + +
Sbjct: 265 SIEVTSTFFAVRNYWRGFF-AATFSAFIFRVLAVWNRDEETITA----LFKTRFRLDFPF 319
Query: 316 HLVDVPPVFILAVVGGLLGSLFNFMTNK---VLRIYNVINEKGTICRLFLACLISIFTSC 372
L ++P ++ + G G+LF ++ K V+R IN RL L+++ S
Sbjct: 320 DLQELPAFAVIGIASGFGGALFVYLNRKIVQVMRKQKTINRFLMRKRLLFPALVTLLIST 379
Query: 373 LLFGLPWLAPCRPCPPDAVEPCPTIGRSGIYKKFQCPPNHYNGLASLIFNTNDDAIRNLF 432
L F P E T+ + + + L+ + +
Sbjct: 380 LTFP-PGFGQFMAGQLSQKETLVTLFDNRTWVR-----------QGLVEELEPPSTSQAW 427
Query: 433 SMHTDNEFELSSMLVFFIICLFLSIFSCGIVAPAGLFVPIIVTGASYGRLVGILV----- 487
S N F ++++F ++ ++S + I P G F+P+ V GA++GRLVG +
Sbjct: 428 SPPRANVF--LTLVIFILMKFWMSALATTIPVPCGAFMPVFVIGAAFGRLVGESMAAWFP 485
Query: 488 -GERTNLSN-----GLYAVLGAASLLGGSMRTTVSLCVIMLELTNNLLLLPLIMMVLVVS 541
G T+ S G YAV+GAA+ L G++ TVS VI+ ELT + + +M+ ++++
Sbjct: 486 DGIHTDSSTYRIVPGGYAVVGAAA-LAGAVTHTVSTAVIVFELTGQIAHILPVMIAVILA 544
Query: 542 KSVANVFNANVYDLIMKAKGLPYLET--HAEPYMRQLTVGDVVTGPLQMFNGIEKVRNIV 599
+VA ++YD I++ K LPYL ++ V D++ + R++
Sbjct: 545 NAVAQSLQPSLYDSIIRIKKLPYLPELGWGRHQQYRVRVEDIMVRDVPHVALSCTFRDLR 604
Query: 600 FILRTTAHNGFPVIDEPPGSEAPILFGIILRHHLTTLL 637
L T +++ P E+ IL G I R + LL
Sbjct: 605 LALHRTKGRTLALVESP---ESMILLGSIERTQVVALL 639
>CLC5_HUMAN (P51795) Chloride channel protein 5 (ClC-5)
Length = 746
Score = 121 bits (303), Expect = 8e-27
Identities = 77/223 (34%), Positives = 116/223 (51%), Gaps = 24/223 (10%)
Query: 119 FAFFI-FFASNLSLTLFASIITAFI---APTAAGSGISEVKAYLNGVDAPGIFTVRTLCV 174
FA+ + +F L LFA + + + AP A GSGI E+K L+G G TL +
Sbjct: 134 FAYIVNYFMYVLWALLFAFLAVSLVKVFAPYACGSGIPEIKTILSGFIIRGYLGKWTLVI 193
Query: 175 KIIGSITAVSGSLVIGKAGPMVHTGACVAALLGQGGSKRYGITWRWLRFFKNDRDRRDLI 234
K I + AVS L +GK GP+VH C +L +K + KN+ RR+++
Sbjct: 194 KTITLVLAVSSGLSLGKEGPLVHVACCCGNILCHCFNK----------YRKNEAKRREVL 243
Query: 235 ICGSAAGIAAAFRAPVGGVLFALEEMASWWRTALLWRAFFTTATVAIFLRAMIDVCLSDK 294
+AAG++ AF AP+GGVLF+LEE++ ++ LWR+FF A LR++
Sbjct: 244 SAAAAAGVSVAFGAPIGGVLFSLEEVSYYFPLKTLWRSFFAALVAAFTLRSI-------- 295
Query: 295 CGLFGKGGLIMFDAYSASISYHLVDVPPVFILAVVGGLLGSLF 337
FG L++F +HL ++ P +L + GGL G+LF
Sbjct: 296 -NPFGNSRLVLF-YVEFHTPWHLFELVPFILLGIFGGLWGALF 336
Score = 97.4 bits (241), Expect = 1e-19
Identities = 93/326 (28%), Positives = 152/326 (46%), Gaps = 51/326 (15%)
Query: 450 IICLFLSIFSCGIVAPAGLFVPIIVTGASYGRLVGILVGERTN----------------- 492
I+ + ++IF+ G+ P+GLF+P + GA GRL+G+ + +
Sbjct: 436 ILKIVITIFTFGMKIPSGLFIPSMAVGAIAGRLLGVGMEQLAYYHQEWTVFNSWCSQGAD 495
Query: 493 -LSNGLYAVLGAASLLGGSMRTTVSLCVIMLELTNNLLLLPLIMMVLVVSKSVANVF-NA 550
++ GLYA++GAA+ LGG R TVSL VIM ELT L + +M + SK VA+
Sbjct: 496 CITPGLYAMVGAAACLGGVTRMTVSLVVIMFELTGGLEYIVPLMAAAMTSKWVADALGRE 555
Query: 551 NVYDLIMKAKGLPYLETHAEPYMRQLTVGDVV----TGPL--QMFNGIEKVRNIVFILRT 604
+YD ++ G P+LE E + + DV+ PL + V ++ I+
Sbjct: 556 GIYDAHIRLNGYPFLEA-KEEFAHKTLAMDVMKPRRNDPLLTVLTQDSMTVEDVETIISE 614
Query: 605 TAHNGFPVIDEPPGSEAPILFGIILRHHLTTLLKKKAFLPSPVANSYDVVRKFSSDDFAK 664
T ++GFPV+ E+ L G +LR L ++ V S ++
Sbjct: 615 TTYSGFPVV---VSRESQRLVGFVLRRDLIISIENARKKQDGVV-STSIIYFTEHSPPLP 670
Query: 665 KYSVERVKIEDIQLTEEEMGMFVDLHPFTNASPYTVVETMSLAKALILFREVGLRHLLVI 724
Y+ +K+ +I +DL SP+TV + + + +FR++GLR LV
Sbjct: 671 PYTPPTLKLRNI----------LDL------SPFTVTDLTPMEIVVDIFRKLGLRQCLVT 714
Query: 725 PKIPGRSPVVGILTRHDFTPEHILGM 750
GR ++GI+T+ D +HI M
Sbjct: 715 HN--GR--LLGIITKKDVL-KHIAQM 735
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.328 0.143 0.436
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 90,013,741
Number of Sequences: 164201
Number of extensions: 3877644
Number of successful extensions: 12256
Number of sequences better than 10.0: 70
Number of HSP's better than 10.0 without gapping: 60
Number of HSP's successfully gapped in prelim test: 10
Number of HSP's that attempted gapping in prelim test: 11873
Number of HSP's gapped (non-prelim): 174
length of query: 776
length of database: 59,974,054
effective HSP length: 118
effective length of query: 658
effective length of database: 40,598,336
effective search space: 26713705088
effective search space used: 26713705088
T: 11
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.7 bits)
S2: 70 (31.6 bits)
Medicago: description of AC144657.4


