

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144645.6 - phase: 0 /pseudo
(570 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
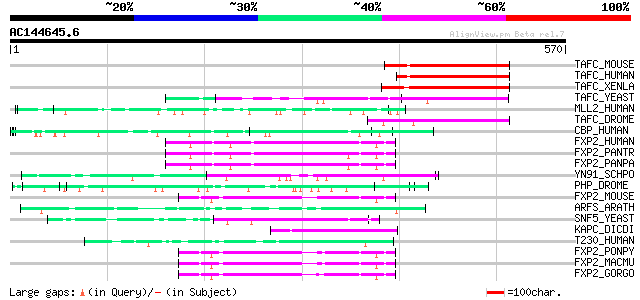
Score E
Sequences producing significant alignments: (bits) Value
TAFC_MOUSE (Q8VE65) Transcription initiation factor TFIID subuni... 121 4e-27
TAFC_HUMAN (Q16514) Transcription initiation factor TFIID subuni... 119 3e-26
TAFC_XENLA (Q91858) Transcription initiation factor TFIID subuni... 114 9e-25
TAFC_YEAST (Q03761) Transcription initiation factor TFIID subuni... 106 2e-22
MLL2_HUMAN (O14686) Myeloid/lymphoid or mixed-lineage leukemia p... 93 2e-18
TAFC_DROME (P49905) Transcription initiation factor TFIID subuni... 88 7e-17
CBP_HUMAN (Q92793) CREB-binding protein (EC 2.3.1.48) 80 1e-14
FXP2_HUMAN (O15409) Forkhead box protein P2 (CAG repeat protein ... 79 4e-14
FXP2_PANTR (Q8MJA0) Forkhead box protein P2 78 7e-14
FXP2_PANPA (Q8HZ00) Forkhead box protein P2 78 7e-14
YN91_SCHPO (Q9Y808) Hypothetical protein C146.01 in chromosome II 73 2e-12
PHP_DROME (P39769) Polyhomeotic-proximal chromatin protein 72 3e-12
FXP2_MOUSE (P58463) Forkhead box protein P2 72 5e-12
ARFS_ARATH (Q8RYC8) Auxin response factor 19 (Auxin-responsive p... 71 7e-12
SNF5_YEAST (P18480) Transcription regulatory protein SNF5 (SWI/S... 71 9e-12
KAPC_DICDI (P34099) cAMP-dependent protein kinase catalytic subu... 70 1e-11
T230_HUMAN (Q93074) Thyroid hormone receptor-associated protein ... 70 1e-11
FXP2_PONPY (Q8MJ98) Forkhead box protein P2 70 1e-11
FXP2_MACMU (Q8MJ97) Forkhead box protein P2 70 1e-11
FXP2_GORGO (Q8MJ99) Forkhead box protein P2 69 4e-11
>TAFC_MOUSE (Q8VE65) Transcription initiation factor TFIID subunit
12 (Transcription initiation factor TFIID 20 kDa
subunits) (TAFII-20) (TAFII20)
Length = 161
Score = 121 bits (304), Expect = 4e-27
Identities = 64/130 (49%), Positives = 87/130 (66%), Gaps = 4/130 (3%)
Query: 386 GSQPDATASG--ATTPGGSSSQGTEAATNQVLGKRKIQDLVAQVDPQGKLDPEVIDLLLE 443
GS ++T G A TPG E NQVL K+K+QDLV +VDP +LD +V ++LL+
Sbjct: 29 GSMANSTTVGKIAGTPGTGGRLSPE--NNQVLTKKKLQDLVREVDPNEQLDEDVEEMLLQ 86
Query: 444 FADDFIDSVTTHGCILAKHRKSSTLESKDLLLHLEKNWDLTIPGYSSEEKKYQSRPLSNE 503
ADDFI+SV T C LA+HRKSSTLE KD+ LHLE+ W++ IPG+ SEE + + + E
Sbjct: 87 IADDFIESVVTAACQLARHRKSSTLEVKDVQLHLERQWNMWIPGFGSEEIRPYKKACTTE 146
Query: 504 LHKRRLDAVR 513
HK+R+ +R
Sbjct: 147 AHKQRMALIR 156
>TAFC_HUMAN (Q16514) Transcription initiation factor TFIID subunit
12 (Transcription initiation factor TFIID 20/15 kDa
subunits) (TAFII-20/TAFII-15) (TAFII20/TAFII15)
Length = 161
Score = 119 bits (297), Expect = 3e-26
Identities = 59/116 (50%), Positives = 80/116 (68%), Gaps = 2/116 (1%)
Query: 398 TPGGSSSQGTEAATNQVLGKRKIQDLVAQVDPQGKLDPEVIDLLLEFADDFIDSVTTHGC 457
TPG E NQVL K+K+QDLV +VDP +LD +V ++LL+ ADDFI+SV T C
Sbjct: 43 TPGAGGRLSPE--NNQVLTKKKLQDLVREVDPNEQLDEDVEEMLLQIADDFIESVVTAAC 100
Query: 458 ILAKHRKSSTLESKDLLLHLEKNWDLTIPGYSSEEKKYQSRPLSNELHKRRLDAVR 513
LA+HRKSSTLE KD+ LHLE+ W++ IPG+ SEE + + + E HK+R+ +R
Sbjct: 101 QLARHRKSSTLEVKDVQLHLERQWNMWIPGFGSEEIRPYKKACTTEAHKQRMALIR 156
>TAFC_XENLA (Q91858) Transcription initiation factor TFIID subunit
12 (Transcription initiation factor TFIID 20/15 kDa
subunits) (TAFII-20/TAFII-15) (TAFII20/TAFII15) (TFIID
subunit p22)
Length = 164
Score = 114 bits (284), Expect = 9e-25
Identities = 57/131 (43%), Positives = 84/131 (63%), Gaps = 4/131 (3%)
Query: 383 SLTGSQPDATASGATTPGGSSSQGTEAATNQVLGKRKIQDLVAQVDPQGKLDPEVIDLLL 442
++ S P T GG +S NQVL K+K+ DLV +VDP +LD +V ++LL
Sbjct: 33 AMANSTPVGKVPVPTAGGGRASP----EANQVLSKKKLHDLVREVDPNEQLDEDVEEMLL 88
Query: 443 EFADDFIDSVTTHGCILAKHRKSSTLESKDLLLHLEKNWDLTIPGYSSEEKKYQSRPLSN 502
+ ADDFI+SV + C LA+HRKS+TLE KD+ LHLE+ W++ IPG+ SEE + + +
Sbjct: 89 QIADDFIESVVSAACQLARHRKSNTLEVKDVQLHLERQWNMWIPGFGSEEIRPYKKACTT 148
Query: 503 ELHKRRLDAVR 513
E HK+R+ ++
Sbjct: 149 EAHKQRMALIK 159
>TAFC_YEAST (Q03761) Transcription initiation factor TFIID subunit
12 (TBP-associated factor 12) (TBP-associated factor 61
kDa) (TAFII-61) (TAFII61) (TAFII-68) (TAFII68)
Length = 539
Score = 106 bits (264), Expect = 2e-22
Identities = 82/308 (26%), Positives = 140/308 (44%), Gaps = 40/308 (12%)
Query: 212 TQQQQIRLTPAQMRQQLSQQALNSQQVQGIPRSSSLAFMNSQLSGLSQNGQPGMMHNSLT 271
TQQQ+++ Q +QQ QQ QQ Q Q QN + NS
Sbjct: 243 TQQQRVQQQRVQQQQQQQQQQQQQQQQQ----------QQQQQQRQGQNQRKISSSNST- 291
Query: 272 QSQWLKQMPAMSGPASPLRLQQHQRQQQQLASSGQLQQNSMTLNQQQLSQFMQQQKSMGQ 331
++P+++GP + S Q QQN++T Q +
Sbjct: 292 ------EIPSVTGPDA--------------LKSQQQQQNTITATNNPRGNVNTSQTEQSK 331
Query: 332 PQLHQQQPSPQQQQQQQLLQPQQQSQLQASVHQQQHLHSPRVAGPTGQKSI--SLTGSQP 389
++ + + Q + ++ +++ S + P +S ++TG
Sbjct: 332 AKVTNVNATASMLNNISSSKSAIFKQTEPAIPISENI-STKTPAPVAYRSNRPTITGGSA 390
Query: 390 DATASGATTPGGSSSQGTEAATNQVLGKRKIQDLVAQV-----DPQGKLDPEVIDLLLEF 444
AS TP + E T +V+ KRK+++LV V D + +D +V +LLL+
Sbjct: 391 -MNASALNTPATTKLPPYEMDTQRVMSKRKLRELVKTVGIDEGDGETVIDGDVEELLLDL 449
Query: 445 ADDFIDSVTTHGCILAKHRKSSTLESKDLLLHLEKNWDLTIPGYSSEEKKYQSRPLSNEL 504
ADDF+ +VT C LAKHRKS LE++D+ LHLE+NW++ IPGYS++E + + ++
Sbjct: 450 ADDFVTNVTAFSCRLAKHRKSDNLEARDIQLHLERNWNIRIPGYSADEIRSTRKWNPSQN 509
Query: 505 HKRRLDAV 512
+ ++L ++
Sbjct: 510 YNQKLQSI 517
Score = 48.9 bits (115), Expect = 4e-05
Identities = 70/283 (24%), Positives = 101/283 (34%), Gaps = 60/283 (21%)
Query: 161 TSPRQKGGMVQSSQFSSANSAGQSLQGMQQAIGMMGSPNLASQMRTNGGLYTQQQQIRLT 220
T+PR K M Q+++ A + QQA N S ++ + + +
Sbjct: 58 TTPRGKELMFQAAKIKQVYDALTLNRRRQQAAQAY---NNTSNSNSSNPASIPTENVPNS 114
Query: 221 PAQMRQQLSQQALNSQQVQGIPRSSSLAFMNSQLSGLSQNGQPGMMHNSLTQSQ-WLKQM 279
Q +QQ Q NS + + + N + L QN Q + H S+ + + +LKQ
Sbjct: 115 SQQQQQQQQQTRNNSNKFSNMIKQVLTPEENQEYEKLWQNFQ--VRHTSIKEKETYLKQ- 171
Query: 280 PAMSGPASPLRLQQHQRQQQQLASSGQLQQNSMTL---------------NQQQLS---- 320
+ RL+Q +Q QLQ+ + L N Q S
Sbjct: 172 -------NIDRLEQEINKQTDEGPKQQLQEKKIELLNDWKVLKIEYTKLFNNYQNSKKTF 224
Query: 321 ---------------------QFMQQQKSMGQPQLHQQQPSPQQQQQQQLLQPQQQSQLQ 359
Q +QQQ+ Q Q QQQ QQQQQQQ Q Q+Q Q Q
Sbjct: 225 YVECARHNPALHKFLQESTQQQRVQQQRVQQQQQQQQQQQQQQQQQQQQ--QQQRQGQNQ 282
Query: 360 ASVHQQQHLHSPRVAGPTGQKSISLTGSQPDATASGATTPGGS 402
+ P V GP KS Q T + P G+
Sbjct: 283 RKISSSNSTEIPSVTGPDALKS----QQQQQNTITATNNPRGN 321
Score = 31.2 bits (69), Expect = 7.6
Identities = 38/163 (23%), Positives = 58/163 (35%), Gaps = 25/163 (15%)
Query: 65 NFQLQQTLQRSPSMSRLNQIQPQQQQQVVARQQAALYGGQMNFGGSAAVSAQQQQLSGGV 124
N L + LQ S R+ Q + QQQQQ +QQ QQQQ
Sbjct: 232 NPALHKFLQESTQQQRVQQQRVQQQQQQQQQQQ----------------QQQQQQQQQQQ 275
Query: 125 AAMGGSASNLSRSALIGQSGHFPMLSGAGA-----AQFNLLTSPRQKGGMVQSSQFSSAN 179
G + +S S S P ++G A Q N +T+ G V +SQ +
Sbjct: 276 QRQGQNQRKISSS----NSTEIPSVTGPDALKSQQQQQNTITATNNPRGNVNTSQTEQSK 331
Query: 180 SAGQSLQGMQQAIGMMGSPNLASQMRTNGGLYTQQQQIRLTPA 222
+ ++ + + S A +T + + TPA
Sbjct: 332 AKVTNVNATASMLNNISSSKSAIFKQTEPAIPISENISTKTPA 374
>MLL2_HUMAN (O14686) Myeloid/lymphoid or mixed-lineage leukemia
protein 2 (ALL1-related protein)
Length = 5262
Score = 92.8 bits (229), Expect = 2e-18
Identities = 122/416 (29%), Positives = 164/416 (39%), Gaps = 100/416 (24%)
Query: 46 SLSQDQQQLHT-------------------INTINPNSNFQLQQTLQRSPSMSRLNQIQP 86
+L+Q QQQ H+ + ++ P+S ++ LQ +L Q
Sbjct: 3401 ALAQQQQQQHSGGAGSLAGPSGGFFPGNLALRSLGPDSRLLQERQLQLQQQRMQLAQKLQ 3460
Query: 87 QQQQQVVARQQAALYGGQMNFGGSAAVSAQQQQLSGGVAAMGGSASNLSRSALIGQSGHF 146
QQQQQ +QQ Q + G A+ QQQQ G A L+ S H
Sbjct: 3461 QQQQQ---QQQ------QQHLLGQVAIQQQQQQ---GPGVQTNQALGPKPQGLMPPSSHQ 3508
Query: 147 PMLSGAGAAQFNLLTSPRQKGGMVQSSQFS--------SANSAGQSLQGMQQAIGMMGSP 198
+L + Q P+ GM+ +Q + + G Q + ++ SP
Sbjct: 3509 GLLVQQLSPQ-----PPQGPQGMLGPAQVAVLQQQHPGALGPQGPHRQVLMTQSRVLSSP 3563
Query: 199 NLASQMRTNGGLYTQQQQIRLTPAQMRQQLSQQALNSQQVQGIPRSSSLAFMNSQLSGLS 258
LA Q + GL RL AQ +QQ Q Q QG S + Q S +S
Sbjct: 3564 QLAQQGQ---GLMGH----RLVTAQQQQQQQQH-----QQQG----SMAGLSHLQQSLMS 3607
Query: 259 QNGQPGMMHNSLTQSQWLKQMPAMSGPASPLRLQQHQRQQQQLASSGQLQQNSMTLNQQQ 318
+GQP + + Q L+Q + + Q Q+QQQQL QLQQ L QQQ
Sbjct: 3608 HSGQPKLSAQPMGSLQQLQQQQQLQQ-----QQQLQQQQQQQLQQQQQLQQQQ--LQQQQ 3660
Query: 319 LSQFMQQQKSMGQPQLHQQQPSPQQQQQQQ-------------------LLQPQQQSQLQ 359
Q +QQQ+ Q Q QQQ QQQQQQQ LL PQQQ Q Q
Sbjct: 3661 QQQQLQQQQQQ-QLQQQQQQLQQQQQQQQQQFQQQQQQQQMGLLNQSRTLLSPQQQQQQQ 3719
Query: 360 ASV------HQQQHLHSPRVAGPTGQKSISLTGSQP---DATASGATTPGGSSSQG 406
++ QH SP GPT + LTG + D S T G S+ QG
Sbjct: 3720 VALGPGMPAKPLQHFSSPGALGPT----LLLTGKEQNTVDPAVSSEATEGPSTHQG 3771
Score = 54.3 bits (129), Expect = 8e-07
Identities = 104/402 (25%), Positives = 138/402 (33%), Gaps = 44/402 (10%)
Query: 7 GSPDSQIQNPSSSNPSIPSPSPISQQQQSPSIHMSNSSPSLSQDQQQLHTINTINPNSNF 66
G P Q P S + + QQQQ + Q Q Q +
Sbjct: 3610 GQPKLSAQ-PMGSLQQLQQQQQLQQQQQ---LQQQQQQQLQQQQQLQQQQLQQQQQQQQL 3665
Query: 67 QLQQTLQRSPSMSRLNQIQPQQQQQVVARQQAALYGGQMNFGGSAAVSAQQQQLSGGVAA 126
Q QQ Q +L Q Q QQQQQ +QQ G + +S QQQQ
Sbjct: 3666 QQQQQQQLQQQQQQLQQQQQQQQQQFQQQQQQQQMG--LLNQSRTLLSPQQQQQQQVALG 3723
Query: 127 MGGSASNLSRSALIGQSGHFPMLSG--------AGAAQFNLLTSPRQKG----GMVQSSQ 174
G A L + G G +L+G A +++ S Q G G S
Sbjct: 3724 PGMPAKPLQHFSSPGALGPTLLLTGKEQNTVDPAVSSEATEGPSTHQGGPLAIGTTPESM 3783
Query: 175 FSSANSAGQSLQGMQQAIGMMGSP-------NLASQMRTNGGLYTQQQQIRLTPAQMRQQ 227
+ SL G Q + + P L +R G QQQQ+ L
Sbjct: 3784 ATEPGEVKPSLSGDSQLLLVQPQPQPQPSSLQLQPPLRLPG---QQQQQVSLLHTAGGGS 3840
Query: 228 LSQQALNSQQVQGIPRSSSLAFMNSQLSGLSQNGQPGMMHNSLTQSQWLKQMPAMSGPAS 287
Q S +SS+ + +Q S +S QPG S+TQ+ Q P + P
Sbjct: 3841 HGQLGSGSSS-----EASSVPHLLAQPS-VSLGDQPG----SMTQNLLGPQQPMLERPMQ 3890
Query: 288 PLRLQQHQRQQQQLASSGQLQQNSMTLNQQQLSQFMQQQKSMGQPQLHQQQPSPQQQQQQ 347
Q + L S L + QL +Q + PQL + SPQQQQQ
Sbjct: 3891 NNTGPQPPKPGPVLQSGQGLPGVGIMPTVGQLRAQLQGVLAK-NPQL--RHLSPQQQQQL 3947
Query: 348 QLLQPQQQSQLQASVHQQQHLHSPRVAGPTGQKSISLTGSQP 389
Q L Q+Q Q +V Q P G L G QP
Sbjct: 3948 QALLMQRQLQQSQAVRQTPPYQEP---GTQTSPLQGLLGCQP 3986
Score = 46.6 bits (109), Expect = 2e-04
Identities = 104/441 (23%), Positives = 157/441 (35%), Gaps = 89/441 (20%)
Query: 9 PDSQIQNPSSSNPSIPSPSPISQQQQSPSIHMSNSSPSLSQDQQQLHTINTINPNSNFQL 68
P Q Q + +P+P P Q+ S+ SSPSL+ QQQL L
Sbjct: 2997 PAQQQQQQQQQHSLLPAPGPA----QAMSLPHEGSSPSLAGSQQQL------------SL 3040
Query: 69 QQTLQRSPSMSRLNQIQPQQQQQVVARQQAALYGGQMNFGGSAAVSAQQQQLSGGVAAMG 128
+ R P + + + P Q +Q+ A A VS Q LSG
Sbjct: 3041 GLAVARQPGLPQ--PLMPTQPPAHALQQRLA--------PSMAMVSNQGHMLSGQHGGQA 3090
Query: 129 GSASNLSRSALIGQSGHFPMLSGAGAAQFNLLTSPRQKGGMVQSSQFSSANSAGQSLQGM 188
G S ++ Q PM G ++ P+Q + Q ANS
Sbjct: 3091 GLVPQQSSQPVLSQK---PM----GTMPPSMCMKPQQ-----LAMQQQLANSFFPDTDLD 3138
Query: 189 QQAIGMMGSPNLASQMRTNGGL--YTQQQQIRLTPAQMRQQLSQQALNSQQVQGIPRS-- 244
+ A + P ++M G+ Q I + P RQQ+S L +Q++ G P S
Sbjct: 3139 KFAAEDIIGPIAKAKMVALKGIKKVMAQGSIGVAPGMNRQQVS---LLAQRLSGGPSSDL 3195
Query: 245 -SSLAFMNSQLSGLSQNGQPGMMHNSLTQ-----------SQWL---KQMPAMSGPASPL 289
+ +A + Q QP + Q +WL +Q+ M
Sbjct: 3196 QNHVAAGSGQERSAGDPSQPRPNPPTFAQGVINEADQRQYEEWLFHTQQLLQMQLKVLEE 3255
Query: 290 RLQQHQRQQQQL------------------ASSGQLQQNSMTLNQQQLSQFMQQQKSMGQ 331
++ H++ ++ L A +L + Q+QL Q +QQK
Sbjct: 3256 QIGVHRKSRKALCAKQRTAKKAGREFPEADAEKLKLVTEQQSKIQKQLDQVRKQQKE--H 3313
Query: 332 PQLHQQQPSPQQQQQQQLLQPQQQSQLQASVHQQQHLHSPRVAGPTGQKSISLTGSQPDA 391
L + + QQQQQQQ Q QQQ ++ Q SPR+ + + G QP
Sbjct: 3314 TNLMAEYRNKQQQQQQQQQQQQQQHSAVLALSPSQ---SPRLLTKLPGQLLPGHGLQPPQ 3370
Query: 392 TASGA------TTPGGSSSQG 406
G TPGG + G
Sbjct: 3371 GPPGGQAGGLRLTPGGMALPG 3391
Score = 36.6 bits (83), Expect = 0.18
Identities = 37/141 (26%), Positives = 54/141 (38%), Gaps = 16/141 (11%)
Query: 1 MAENVIGSPDSQIQNPSSSN--PSIPSPSPISQQQQS-PSIHMSNSSPSLSQDQQQLHTI 57
M +N++G ++ P +N P P P P+ Q Q P + + P++ Q + QL +
Sbjct: 3873 MTQNLLGPQQPMLERPMQNNTGPQPPKPGPVLQSGQGLPGVGI---MPTVGQLRAQLQGV 3929
Query: 58 NTINPNSNF---QLQQTLQRSPSMSRLNQIQ------PQQQQQVVARQQAALYGGQMNFG 108
NP Q QQ LQ +L Q Q P Q+ L G Q G
Sbjct: 3930 LAKNPQLRHLSPQQQQQLQALLMQRQLQQSQAVRQTPPYQEPGTQTSPLQGLLGCQPQLG 3989
Query: 109 G-SAAVSAQQQQLSGGVAAMG 128
G + Q+L G G
Sbjct: 3990 GFPGPQTGPLQELGAGPRPQG 4010
Score = 32.0 bits (71), Expect = 4.5
Identities = 31/120 (25%), Positives = 48/120 (39%), Gaps = 20/120 (16%)
Query: 282 MSGPASPLRLQQHQRQQQQL-------ASSGQLQQNSMTLNQQQLSQFMQ---------- 324
+SGP L++ + + L AS +L + +L +L ++
Sbjct: 2926 LSGPGGSSLLEKFELESGALTLPGGPAASGDELDKMESSLVASELPLLIEDLLEHEKKEL 2985
Query: 325 QQKSMGQPQLHQQQPSPQQQQQQQLLQPQQQSQLQASVHQQQHLHSPRVAGPTGQKSISL 384
Q+K QL Q QQQQQ LL +Q + H+ SP +AG Q S+ L
Sbjct: 2986 QKKQQLSAQLQPAQQQQQQQQQHSLLPAPGPAQAMSLPHEGS---SPSLAGSQQQLSLGL 3042
>TAFC_DROME (P49905) Transcription initiation factor TFIID subunit
12 (Transcription initiation factor TFIID 28-alpha
kDa/22 kDa subunits) (p28-alpha/p22) (TAFII30 alpha)
Length = 196
Score = 87.8 bits (216), Expect = 7e-17
Identities = 51/159 (32%), Positives = 85/159 (53%), Gaps = 13/159 (8%)
Query: 368 LHSPRVAGPTGQKSI---SLTGSQPDATASGATTPGGSSSQGTEAATNQ----------V 414
LH P +A P+ + S + SQ SG T G S G++++ + +
Sbjct: 32 LHDPPMASPSQHSPMTNNSNSSSQNGGPVSGLGTGTGPISGGSKSSNHTSSAAGSENTPM 91
Query: 415 LGKRKIQDLVAQVDPQGKLDPEVIDLLLEFADDFIDSVTTHGCILAKHRKSSTLESKDLL 474
L K ++ +LV +VD +LD +V +LLL+ DDF++ AKHRKS+ +E +D+
Sbjct: 92 LTKPRLTELVREVDTTTQLDEDVEELLLQIIDDFVEDTVKSTSAFAKHRKSNKIEVRDVQ 151
Query: 475 LHLEKNWDLTIPGYSSEEKKYQSRPLSNELHKRRLDAVR 513
LH E+ +++ IPG+ ++E + R E HK+RL +R
Sbjct: 152 LHFERKYNMWIPGFGTDELRPYKRAAVTEAHKQRLALIR 190
>CBP_HUMAN (Q92793) CREB-binding protein (EC 2.3.1.48)
Length = 2442
Score = 80.1 bits (196), Expect = 1e-14
Identities = 106/391 (27%), Positives = 143/391 (36%), Gaps = 59/391 (15%)
Query: 5 VIGSPDSQIQNPSSSNPSIPSP------SPISQQQQSPSIHMSNSSPSLSQDQQQLHTIN 58
V G +Q P S +PS SP S QQQ +++ S+P L + T
Sbjct: 2048 VAGPRMPSVQPPRSISPSALQDLLRTLKSPSSPQQQQQVLNILKSNPQLMAAFIKQRTAK 2107
Query: 59 TINPNSNFQLQQTLQRSPSMSRLNQIQPQQQQQVVARQQAALYGGQMNFGGSAAVSAQQQ 118
+ Q Q LQ P M Q QP QQ + A+ G G QQ
Sbjct: 2108 YVANQPGMQPQPGLQSQPGM----QPQPGMHQQPSLQNLNAMQAGVPRPG-----VPPQQ 2158
Query: 119 QLSGGVAAMGGSASNLSRSALIGQSGHFPMLSGAGAAQFNLLTSPRQKGGMVQSSQFSSA 178
Q GG+ G + + I GH P ++ +L RQ ++Q Q
Sbjct: 2159 QAMGGLNPQGQALN-------IMNPGHNPNMASMNPQYREMLR--RQ---LLQQQQ---- 2202
Query: 179 NSAGQSLQGMQQAIGMMGSPNLASQMRTNGGLYTQQQQIRLTPAQMRQQLSQQALNSQQV 238
Q Q QQ GS +A M +G Q PA +QQ QQ L Q
Sbjct: 2203 ---QQQQQQQQQQQQQQGSAGMAGGMAGHGQFQQPQGPGGYPPAMQQQQRMQQHLPLQ-- 2257
Query: 239 QGIPRSSSLAFMNSQLSGLSQNGQPGM-----------MHNSLTQSQWLKQMPAMSGPAS 287
SS+ M +Q+ L Q GQPG+ + + Q Q +KQ G +
Sbjct: 2258 -----GSSMGQMAAQMGQLGQMGQPGLGADSTPNIQQALQQRILQQQQMKQQIGSPGQPN 2312
Query: 288 PLRLQQHQRQQQQLASSGQLQQNSMTLNQQQLSQF-MQQQKSMGQPQLHQQQPSPQQQQQ 346
P+ QQH Q AS QQ + +L+ Q S +Q + QP P Q Q
Sbjct: 2313 PMSPQQHMLSGQPQASHLPGQQIATSLSNQVRSPAPVQSPRPQSQPPHSSPSPRIQPQPS 2372
Query: 347 QQLLQPQQQSQ------LQASVHQQQHLHSP 371
+ PQ S AS Q HL +P
Sbjct: 2373 PHHVSPQTGSPHPGLAVTMASSIDQGHLGNP 2403
Score = 60.5 bits (145), Expect = 1e-08
Identities = 106/427 (24%), Positives = 148/427 (33%), Gaps = 64/427 (14%)
Query: 7 GSPDSQIQNPSSSNPSIPSPSPIS----------QQQQSPSIHMSNSSP----SLSQDQQ 52
G P SQ+ P+ P+ P P+ + QQQ ++++NS P +
Sbjct: 1936 GKPTSQV--PAPPPPAQPPPAAVEAARQIEREAQQQQHLYRVNINNSMPPGRTGMGTPGS 1993
Query: 53 QLHTINTINPNSNFQLQQTLQRSPSMSRLNQIQPQQQ------QQVVARQQAALYGGQMN 106
Q+ ++ P N + P PQQQ + V++ Q A G
Sbjct: 1994 QMAPVSLNVPRPNQVSGPVMPSMPPGQWQQAPLPQQQPMPGLPRPVISMQAQAAVAGPRM 2053
Query: 107 FGGSAAVSAQQQQLSGGVAAMGGSASNLSRSALIGQSGHFPMLSGAGAAQFNLLTSPRQK 166
S L + + +S + ++ P L A Q Q
Sbjct: 2054 PSVQPPRSISPSALQDLLRTLKSPSSPQQQQQVLNILKSNPQLMAAFIKQRTAKYVANQP 2113
Query: 167 GGMVQSSQFSSANSAGQ-------SLQGMQQAIGMMGSPNLASQMRTNGGLYTQQQQIRL 219
G Q S Q SLQ + + P + Q + GGL Q Q + +
Sbjct: 2114 GMQPQPGLQSQPGMQPQPGMHQQPSLQNLNAMQAGVPRPGVPPQQQAMGGLNPQGQALNI 2173
Query: 220 TPA-----------QMRQQLSQQALNSQQVQGIPRSSSLAFMNSQLSGLSQNGQPGMMHN 268
Q R+ L +Q L QQ Q Q Q G GM
Sbjct: 2174 MNPGHNPNMASMNPQYREMLRRQLLQQQQQQQ---------QQQQQQQQQQQGSAGMAGG 2224
Query: 269 SLTQSQWLKQMPAMSGPAS-PLRLQQHQRQQQQLASSGQLQQNSMTLNQQQLSQFMQQQK 327
Q+ Q P GP P +QQ QR QQ L LQ +SM Q+ Q Q
Sbjct: 2225 MAGHGQF--QQP--QGPGGYPPAMQQQQRMQQHLP----LQGSSMGQMAAQMGQLGQ--- 2273
Query: 328 SMGQPQL-HQQQPSPQQQQQQQLLQPQQQSQLQASVHQQQHLHSPRVAGPTGQKSISLTG 386
MGQP L P+ QQ QQ++LQ QQ Q S Q + SP+ +GQ S
Sbjct: 2274 -MGQPGLGADSTPNIQQALQQRILQQQQMKQQIGSPGQPNPM-SPQQHMLSGQPQASHLP 2331
Query: 387 SQPDATA 393
Q AT+
Sbjct: 2332 GQQIATS 2338
Score = 53.1 bits (126), Expect = 2e-06
Identities = 105/465 (22%), Positives = 173/465 (36%), Gaps = 57/465 (12%)
Query: 4 NVIGSPDSQIQNPSSSNPSIPS--------PSPISQQQQSPSIHMSNSSPSLSQDQQQLH 55
N P + +P+S+ P P+ P P +Q Q SP PS+++ Q
Sbjct: 1873 NTRNVPQQSLPSPTSAPPGTPTQQPSTPQTPQPPAQPQPSPVSMSPAGFPSVARTQPPT- 1931
Query: 56 TINTINPNSNFQLQQTLQRSPSMSRLNQIQPQQQQQVVARQQAALYGGQMNFGGSAAVSA 115
T++T P S + P + ++ +Q + A+QQ LY ++N S
Sbjct: 1932 TVSTGKPTSQVPAPPPPAQPPPAA----VEAARQIEREAQQQQHLY--RVNINNSMPPGR 1985
Query: 116 QQQQLSGGVAAMGGSASNLSRSALIGQSGHFPMLSGAGAAQFNLLTSPRQKG--GMVQS- 172
G+ G + +S + P++ Q+ P+Q+ G+ +
Sbjct: 1986 T------GMGTPGSQMAPVSLNVPRPNQVSGPVMPSMPPGQWQQAPLPQQQPMPGLPRPV 2039
Query: 173 -SQFSSANSAGQSLQGMQQAIGMMGSPNLASQMRTNGGLYTQQQQIRLTPAQMRQQLSQQ 231
S + A AG + +Q + S L +RT + QQQ +Q L+
Sbjct: 2040 ISMQAQAAVAGPRMPSVQPPRSISPSA-LQDLLRTLKSPSSPQQQ--------QQVLNIL 2090
Query: 232 ALNSQQVQGIPRSSSLAFMNSQLSGLSQNG---QPGMMHN-SLTQSQWLKQMPAMSGPAS 287
N Q + + + ++ +Q Q G QPGM + Q L+ + AM
Sbjct: 2091 KSNPQLMAAFIKQRTAKYVANQPGMQPQPGLQSQPGMQPQPGMHQQPSLQNLNAMQAGVP 2150
Query: 288 PLRLQQHQRQQQQLASSGQLQQNSMTLNQQQLSQFMQQQKSMGQPQLHQQQPSPQQQQQQ 347
+ Q+ L GQ + ++ Q + M + QL QQQ QQQQQQ
Sbjct: 2151 RPGVPPQQQAMGGLNPQGQALNIMNPGHNPNMASMNPQYREMLRRQLLQQQQQQQQQQQQ 2210
Query: 348 QLLQPQQQSQLQASVHQQQHLHSPRVAGPTG-----------QKSISLTGSQPDATAS-- 394
Q Q QQQ + H + GP G Q+ + L GS A+
Sbjct: 2211 Q--QQQQQGSAGMAGGMAGHGQFQQPQGPGGYPPAMQQQQRMQQHLPLQGSSMGQMAAQM 2268
Query: 395 GATTPGGSSSQGTEAATN--QVLGKRKI--QDLVAQVDPQGKLDP 435
G G G ++ N Q L +R + Q + Q+ G+ +P
Sbjct: 2269 GQLGQMGQPGLGADSTPNIQQALQQRILQQQQMKQQIGSPGQPNP 2313
Score = 50.1 bits (118), Expect = 2e-05
Identities = 74/263 (28%), Positives = 96/263 (36%), Gaps = 30/263 (11%)
Query: 2 AENVIGSPDSQIQNPSSSNPSIPSPSPISQQQQSPSIHMSNSS------PSLSQDQQQL- 54
A+ V P Q Q S P + P P QQ PS+ N+ P + QQ +
Sbjct: 2106 AKYVANQPGMQPQPGLQSQPGM-QPQPGMHQQ--PSLQNLNAMQAGVPRPGVPPQQQAMG 2162
Query: 55 ------HTINTINPNSNFQLQQTLQRSPSMSRLNQIQPQQQQQVVARQQAALYGGQMNFG 108
+N +NP N + + M R +Q QQQQQ +QQ G
Sbjct: 2163 GLNPQGQALNIMNPGHNPNMASMNPQYREMLRRQLLQQQQQQQQQQQQQQQQQQGSAGMA 2222
Query: 109 GSAAVSAQQQQLSGGVAAMGGSASNLSRSALIGQSGHFPMLS---GAGAAQFNLLTSPRQ 165
G A Q QQ G GG + + + Q H P+ G AAQ L Q
Sbjct: 2223 GGMAGHGQFQQPQG----PGGYPPAMQQQQRMQQ--HLPLQGSSMGQMAAQMGQLGQMGQ 2276
Query: 166 KGGMVQSSQFSSANSAGQSL--QGMQQAIGMMGSPNLASQMRTNGGLYTQQQQIRLTPAQ 223
G S+ + L Q M+Q IG G PN S + L Q Q L Q
Sbjct: 2277 PGLGADSTPNIQQALQQRILQQQQMKQQIGSPGQPNPMSPQQHM--LSGQPQASHLPGQQ 2334
Query: 224 MRQQLSQQALNSQQVQGIPRSSS 246
+ LS Q + VQ PR S
Sbjct: 2335 IATSLSNQVRSPAPVQS-PRPQS 2356
Score = 42.4 bits (98), Expect = 0.003
Identities = 105/464 (22%), Positives = 168/464 (35%), Gaps = 99/464 (21%)
Query: 11 SQIQNPSSSNPSIPS-------PSPISQQQQSPS----IHMSNSSPSLSQDQQQLHTINT 59
S + SS PS+P P+P + Q +P + ++ SSP+ SQ +
Sbjct: 121 SPLSQGDSSAPSLPKQAASTSGPTPAASQALNPQAQKQVGLATSSPATSQTGPGI----C 176
Query: 60 INPNSNFQLQQTLQRSPSMSRLNQIQPQQQQQVVARQQAALYGGQMNF--------GGSA 111
+N N N L + S +NQ Q Q + AA G G S+
Sbjct: 177 MNANFNQTHPGLLNSNSGHSLINQASQGQAQVMNGSLGAAGRGRGAGMPYPTPAMQGASS 236
Query: 112 AVSAQ-----QQQLSGGVAAMGGSASNLSRSALIGQSGHFPM-LSGAGAAQFNLLTSPRQ 165
+V A+ Q++G A +++ + G + F S AG +
Sbjct: 237 SVLAETLTQVSPQMTGHAGLNTAQAGGMAKMGITGNTSPFGQPFSQAGG---------QP 287
Query: 166 KGGMVQSSQFSSANSAGQSLQGMQQAI---GMMGSPNLASQMRTNGGLY-TQQQQIRLTP 221
G + Q +S S SL I + PN+ SQM+T+ G+ TQ T
Sbjct: 288 MGATGVNPQLASKQSMVNSLPTFPTDIKNTSVTNVPNM-SQMQTSVGIVPTQAIATGPTA 346
Query: 222 AQMRQQLSQQAL----------NSQQVQGIPRSSSLAFMNSQLSGLS-----QNGQPGMM 266
+++L QQ L +Q G R+ SL + + L+ Q G+ +
Sbjct: 347 DPEKRKLIQQQLVLLLHAHKCQRREQANGEVRACSLPHCRTMKNVLNHMTHCQAGKACQV 406
Query: 267 HNSLTQSQWL---KQMPAMSGPAS-PLRLQQHQRQQQQLASS------------GQLQQN 310
+ + Q + K P PL+ +R QQ + S G QQN
Sbjct: 407 AHCASSRQIISHWKNCTRHDCPVCLPLKNASDKRNQQTILGSPASGIQNTIGSVGTGQQN 466
Query: 311 SMTLNQQQLSQ--------------FMQQQKSMGQPQLHQQQPS-PQQQQQQQLLQPQQQ 355
+ +L+ +M Q ++ QPQ+ QQP+ PQ QQ + L P
Sbjct: 467 ATSLSNPNPIDPSSMQRAYAALGLPYMNQPQTQLQPQVPGQQPAQPQTHQQMRTLNPLGN 526
Query: 356 SQLQASVHQQQHLHSPRVAGPTGQKSISLTGSQPDATASGATTP 399
+ ++ P T Q+ +L T+ GAT P
Sbjct: 527 NP----------MNIPAGGITTDQQPPNLISESALPTSLGATNP 560
>FXP2_HUMAN (O15409) Forkhead box protein P2 (CAG repeat protein 44)
(Trinucleotide repeat-containing gene 10 protein)
Length = 715
Score = 78.6 bits (192), Expect = 4e-14
Identities = 80/257 (31%), Positives = 113/257 (43%), Gaps = 26/257 (10%)
Query: 161 TSPRQKGGMVQSSQFSSANSAGQS------------LQGMQQAIGMMGSPNLASQMRTNG 208
+S Q G SSQ + + G+S L +QQ + + L Q +T+G
Sbjct: 13 SSMNQNGMSTLSSQLDAGSRDGRSSGDTSSEVSTVELLHLQQQQALQAARQLLLQQQTSG 72
Query: 209 GLYTQQQQIRLTPAQMR---QQLSQQALNSQQVQGIPRSSSLAFMNSQLSGLSQNGQPGM 265
L + + + P Q+ ++ Q + QQ+Q I + L+ QL L Q Q M
Sbjct: 73 -LKSPKSSDKQRPLQVPVSVAMMTPQVITPQQMQQILQQQVLS--PQQLQALLQQQQAVM 129
Query: 266 MHNSLTQSQWLKQMPAMSGPASPLRLQQHQRQQQQLASSGQLQQNSMTLNQQQLSQFMQQ 325
+ Q + KQ + + QQ Q+QQQQ Q QQ QQQ Q QQ
Sbjct: 130 LQQQQLQEFYKKQQEQLHLQLLQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQ 189
Query: 326 QKSMGQPQLHQQQPSPQQQQ--QQQLLQPQQQSQLQASVHQQQHLHSPRVAG----PTGQ 379
Q+ G+ QQQ QQQQ QQL+ QQ Q+Q + QQQHL S + G P GQ
Sbjct: 190 QQHPGKQAKEQQQQQQQQQQLAAQQLVFQQQLLQMQ-QLQQQQHLLSLQRQGLISIPPGQ 248
Query: 380 KSISLTGSQPDATASGA 396
++ + S P A S A
Sbjct: 249 AALPVQ-SLPQAGLSPA 264
Score = 38.1 bits (87), Expect = 0.062
Identities = 30/96 (31%), Positives = 39/96 (40%), Gaps = 6/96 (6%)
Query: 31 QQQQSPSIHMSNSSPSLSQDQQQLHTINTINPNSNFQLQQTLQRSPSMSRLNQIQPQQQQ 90
QQQQ+ + + Q+QLH Q QQ Q+ + Q Q QQQQ
Sbjct: 123 QQQQAVMLQQQQLQEFYKKQQEQLHLQLLQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQ 182
Query: 91 QVVARQQAALYGGQMNFGGSAAVSAQQQQLSGGVAA 126
Q +QQ Q + G A QQQQ +AA
Sbjct: 183 QQQQQQQ------QQHPGKQAKEQQQQQQQQQQLAA 212
Score = 33.1 bits (74), Expect = 2.0
Identities = 23/69 (33%), Positives = 26/69 (37%), Gaps = 5/69 (7%)
Query: 29 ISQQQQSPSIHMSNSSPSLSQDQQQLHTINTINPNSNFQLQQTLQRSPSMSRLNQIQPQQ 88
+ QQQQ Q QQQ Q QQ Q+ P Q Q QQ
Sbjct: 151 LQQQQQQQQQQQQQQQQQQQQQQQQQQ-----QQQQQQQQQQQQQQHPGKQAKEQQQQQQ 205
Query: 89 QQQVVARQQ 97
QQQ +A QQ
Sbjct: 206 QQQQLAAQQ 214
Score = 32.0 bits (71), Expect = 4.5
Identities = 32/119 (26%), Positives = 43/119 (35%), Gaps = 10/119 (8%)
Query: 11 SQIQNPSSSNPSIPSPSPISQQQQSPSIHMSNSSPSLSQDQ----QQLHTINTINPNSNF 66
S +++P SS+ P P+S +P + + Q Q QQL +
Sbjct: 71 SGLKSPKSSDKQRPLQVPVSVAMMTPQVITPQQMQQILQQQVLSPQQLQALLQQQQAVML 130
Query: 67 QLQQTL------QRSPSMSRLNQIQPQQQQQVVARQQAALYGGQMNFGGSAAVSAQQQQ 119
Q QQ Q + L Q Q QQQQQ +QQ Q QQQQ
Sbjct: 131 QQQQLQEFYKKQQEQLHLQLLQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQ 189
>FXP2_PANTR (Q8MJA0) Forkhead box protein P2
Length = 716
Score = 77.8 bits (190), Expect = 7e-14
Identities = 80/258 (31%), Positives = 113/258 (43%), Gaps = 27/258 (10%)
Query: 161 TSPRQKGGMVQSSQFSSANSAGQS------------LQGMQQAIGMMGSPNLASQMRTNG 208
+S Q G SSQ + + G+S L +QQ + + L Q +T+G
Sbjct: 13 SSMNQNGMSTLSSQLDAGSRDGRSSGDTSSEVSTVELLHLQQQQALQAARQLLLQQQTSG 72
Query: 209 GLYTQQQQIRLTPAQMR---QQLSQQALNSQQVQGIPRSSSLAFMNSQLSGLSQNGQPGM 265
L + + + P Q+ ++ Q + QQ+Q I + L+ QL L Q Q M
Sbjct: 73 -LKSPKSSDKQRPLQVPVSVAMMTPQVITPQQMQQILQQQVLS--PQQLQALLQQQQAVM 129
Query: 266 MHNSLTQSQWLKQMPAMSGPASPLRLQQHQRQQQQLASSGQLQQNSMTLNQQQLSQFMQQ 325
+ Q + KQ + + QQ Q+QQQQ Q QQ QQQ Q QQ
Sbjct: 130 LQQQQLQEFYKKQQEQLHLQLLQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQ 189
Query: 326 QKSMGQPQLHQQQPSPQQQQQ---QQLLQPQQQSQLQASVHQQQHLHSPRVAG----PTG 378
Q+ Q +QQ QQQQQ QQL+ QQ Q+Q + QQQHL S + G P G
Sbjct: 190 QQQHPGKQAKEQQQQQQQQQQLAAQQLVFQQQLLQMQ-QLQQQQHLLSLQRQGLISIPPG 248
Query: 379 QKSISLTGSQPDATASGA 396
Q ++ + S P A S A
Sbjct: 249 QAALPVQ-SLPQAGLSPA 265
Score = 38.1 bits (87), Expect = 0.062
Identities = 30/96 (31%), Positives = 39/96 (40%), Gaps = 5/96 (5%)
Query: 31 QQQQSPSIHMSNSSPSLSQDQQQLHTINTINPNSNFQLQQTLQRSPSMSRLNQIQPQQQQ 90
QQQQ+ + + Q+QLH Q QQ Q+ + Q Q QQQQ
Sbjct: 123 QQQQAVMLQQQQLQEFYKKQQEQLHLQLLQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQ 182
Query: 91 QVVARQQAALYGGQMNFGGSAAVSAQQQQLSGGVAA 126
Q +QQ Q + G A QQQQ +AA
Sbjct: 183 QQQQQQQQ-----QQHPGKQAKEQQQQQQQQQQLAA 213
Score = 33.1 bits (74), Expect = 2.0
Identities = 23/69 (33%), Positives = 26/69 (37%), Gaps = 4/69 (5%)
Query: 29 ISQQQQSPSIHMSNSSPSLSQDQQQLHTINTINPNSNFQLQQTLQRSPSMSRLNQIQPQQ 88
+ QQQQ Q QQQ Q QQ Q+ P Q Q QQ
Sbjct: 151 LQQQQQQQQQQQQQQQQQQQQQQQQQQQ----QQQQQQQQQQQQQQHPGKQAKEQQQQQQ 206
Query: 89 QQQVVARQQ 97
QQQ +A QQ
Sbjct: 207 QQQQLAAQQ 215
Score = 32.0 bits (71), Expect = 4.5
Identities = 32/119 (26%), Positives = 43/119 (35%), Gaps = 10/119 (8%)
Query: 11 SQIQNPSSSNPSIPSPSPISQQQQSPSIHMSNSSPSLSQDQ----QQLHTINTINPNSNF 66
S +++P SS+ P P+S +P + + Q Q QQL +
Sbjct: 71 SGLKSPKSSDKQRPLQVPVSVAMMTPQVITPQQMQQILQQQVLSPQQLQALLQQQQAVML 130
Query: 67 QLQQTL------QRSPSMSRLNQIQPQQQQQVVARQQAALYGGQMNFGGSAAVSAQQQQ 119
Q QQ Q + L Q Q QQQQQ +QQ Q QQQQ
Sbjct: 131 QQQQLQEFYKKQQEQLHLQLLQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQ 189
>FXP2_PANPA (Q8HZ00) Forkhead box protein P2
Length = 716
Score = 77.8 bits (190), Expect = 7e-14
Identities = 80/258 (31%), Positives = 113/258 (43%), Gaps = 27/258 (10%)
Query: 161 TSPRQKGGMVQSSQFSSANSAGQS------------LQGMQQAIGMMGSPNLASQMRTNG 208
+S Q G SSQ + + G+S L +QQ + + L Q +T+G
Sbjct: 13 SSMNQNGMSTLSSQLDAGSRDGRSSGDTSSEVSTVELLHLQQQQALQAARQLLLQQQTSG 72
Query: 209 GLYTQQQQIRLTPAQMR---QQLSQQALNSQQVQGIPRSSSLAFMNSQLSGLSQNGQPGM 265
L + + + P Q+ ++ Q + QQ+Q I + L+ QL L Q Q M
Sbjct: 73 -LKSPKSSDKQRPLQVPVSVAMMTPQVITPQQMQQILQQQVLS--PQQLQALLQQQQAVM 129
Query: 266 MHNSLTQSQWLKQMPAMSGPASPLRLQQHQRQQQQLASSGQLQQNSMTLNQQQLSQFMQQ 325
+ Q + KQ + + QQ Q+QQQQ Q QQ QQQ Q QQ
Sbjct: 130 LQQQQLQEFYKKQQEQLHLQLLQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQ 189
Query: 326 QKSMGQPQLHQQQPSPQQQQQ---QQLLQPQQQSQLQASVHQQQHLHSPRVAG----PTG 378
Q+ Q +QQ QQQQQ QQL+ QQ Q+Q + QQQHL S + G P G
Sbjct: 190 QQQHPGKQAKEQQQQQQQQQQLAAQQLVFQQQLLQMQ-QLQQQQHLLSLQRQGLISIPPG 248
Query: 379 QKSISLTGSQPDATASGA 396
Q ++ + S P A S A
Sbjct: 249 QAALPVQ-SLPQAGLSPA 265
Score = 38.1 bits (87), Expect = 0.062
Identities = 30/96 (31%), Positives = 39/96 (40%), Gaps = 5/96 (5%)
Query: 31 QQQQSPSIHMSNSSPSLSQDQQQLHTINTINPNSNFQLQQTLQRSPSMSRLNQIQPQQQQ 90
QQQQ+ + + Q+QLH Q QQ Q+ + Q Q QQQQ
Sbjct: 123 QQQQAVMLQQQQLQEFYKKQQEQLHLQLLQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQ 182
Query: 91 QVVARQQAALYGGQMNFGGSAAVSAQQQQLSGGVAA 126
Q +QQ Q + G A QQQQ +AA
Sbjct: 183 QQQQQQQQ-----QQHPGKQAKEQQQQQQQQQQLAA 213
Score = 33.1 bits (74), Expect = 2.0
Identities = 23/69 (33%), Positives = 26/69 (37%), Gaps = 4/69 (5%)
Query: 29 ISQQQQSPSIHMSNSSPSLSQDQQQLHTINTINPNSNFQLQQTLQRSPSMSRLNQIQPQQ 88
+ QQQQ Q QQQ Q QQ Q+ P Q Q QQ
Sbjct: 151 LQQQQQQQQQQQQQQQQQQQQQQQQQQQ----QQQQQQQQQQQQQQHPGKQAKEQQQQQQ 206
Query: 89 QQQVVARQQ 97
QQQ +A QQ
Sbjct: 207 QQQQLAAQQ 215
Score = 32.0 bits (71), Expect = 4.5
Identities = 32/119 (26%), Positives = 43/119 (35%), Gaps = 10/119 (8%)
Query: 11 SQIQNPSSSNPSIPSPSPISQQQQSPSIHMSNSSPSLSQDQ----QQLHTINTINPNSNF 66
S +++P SS+ P P+S +P + + Q Q QQL +
Sbjct: 71 SGLKSPKSSDKQRPLQVPVSVAMMTPQVITPQQMQQILQQQVLSPQQLQALLQQQQAVML 130
Query: 67 QLQQTL------QRSPSMSRLNQIQPQQQQQVVARQQAALYGGQMNFGGSAAVSAQQQQ 119
Q QQ Q + L Q Q QQQQQ +QQ Q QQQQ
Sbjct: 131 QQQQLQEFYKKQQEQLHLQLLQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQ 189
>YN91_SCHPO (Q9Y808) Hypothetical protein C146.01 in chromosome II
Length = 1063
Score = 73.2 bits (178), Expect = 2e-12
Identities = 71/258 (27%), Positives = 109/258 (41%), Gaps = 31/258 (12%)
Query: 203 QMRTNGGLYTQQQQIRLTPAQMRQQLSQQALNSQQVQGI--PRSSSLAFMNSQLSGLSQN 260
Q N L QQ+Q L +Q++QQ QQ SQQ Q P++SS N++ +
Sbjct: 133 QFARNMKLTPQQRQFLLQQSQIQQQRQQQQQQSQQPQQTQQPQASSPTAPNTEANQQRSG 192
Query: 261 GQPGMMHNSLTQSQW---LKQMPAM----SGPASPLRLQQHQRQQQQLASSGQLQQNSMT 313
PG + +LTQ Q Q+ A+ P P+ Q+ Q +L ++
Sbjct: 193 SVPGRIVPALTQQQLNNLCNQITALLARNGNPPIPM-------QKLQSMPPARL----IS 241
Query: 314 LNQQQLSQFMQQQKSMGQPQLHQQQPSPQQQQQQQLLQPQQQSQLQASVHQQQHLHSPRV 373
+ Q Q+ +F Q Q Q QQQ QQQQQQQ Q QQQ Q Q Q+ P
Sbjct: 242 IYQNQIQKFRSLQHMQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQKQAPQNAFFPNP 301
Query: 374 AGPTGQKSI---------SLTGSQPDATASGATTPGGSSSQGTEAATN--QVLGKRKIQD 422
G G +S+ S QP TA+ P +++ + +++ D
Sbjct: 302 QGNVGAQSLQSMSPQDQPSTQQQQPQRTAAPPNNPNVNATNNNRINIEMLNIPVPKQLFD 361
Query: 423 LVAQVDPQGKLDPEVIDL 440
+ + P K+ +V++L
Sbjct: 362 QIPNLPPNVKIWRDVLEL 379
Score = 50.8 bits (120), Expect = 9e-06
Identities = 113/475 (23%), Positives = 165/475 (33%), Gaps = 90/475 (18%)
Query: 13 IQNPSSSNPSIPSPSPISQQQQSPSI--HMSNSSPSLSQDQQQLHTINTINPNSNFQLQQ 70
+ N + + ++P+ P ++ +P + + N + + QQ + P Q Q
Sbjct: 92 VYNNAGNVGALPTAGP-NRLANNPRVMPRLQNQNVPMQAGMQQFARNMKLTPQ---QRQF 147
Query: 71 TLQRSPSMSRLNQIQPQQQQQVVARQQAALYGGQMNFGGSAAVSAQQQQLSGGV------ 124
LQ+S QIQ Q+QQQ QQ +A + QQ SG V
Sbjct: 148 LLQQS-------QIQQQRQQQQQQSQQPQQTQQPQASSPTAPNTEANQQRSGSVPGRIVP 200
Query: 125 AAMGGSASNLSR--SALIGQSGHFPMLSGAGAAQFNLLTSPRQKGGMVQSSQFSSANSAG 182
A +NL +AL+ ++G+ P+ L + P + + +Q S
Sbjct: 201 ALTQQQLNNLCNQITALLARNGNPPI------PMQKLQSMPPARLISIYQNQIQKFRSLQ 254
Query: 183 QSLQGMQQAIGMMGSPNLASQMRTNGGLYTQQQQIRLTP-------------AQMRQQLS 229
Q QQ Q + QQQQ + P AQ Q +S
Sbjct: 255 HMQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQKQAPQNAFFPNPQGNVGAQSLQSMS 314
Query: 230 QQALNSQQVQGIPRSSSLAFMNSQLSGLSQNGQPGMMHNSLTQSQWLKQMPAMSGPAS-- 287
Q S Q Q P+ ++ N ++ + N M N Q Q+P +
Sbjct: 315 PQDQPSTQQQQ-PQRTAAPPNNPNVNATNNNRINIEMLNIPVPKQLFDQIPNLPPNVKIW 373
Query: 288 --PLRLQQHQR-QQQQLASSGQLQQNSMTL----NQQQLSQFMQ---------------- 324
L L Q QR +QL G L + + + QQQL++
Sbjct: 374 RDVLELGQSQRLPPEQLKLIGMLYRKHLQIILQHRQQQLNKIQNARMNSQNAPNTNKLGN 433
Query: 325 -QQKSMGQPQLHQQQPSPQQQQQQQLLQPQQQSQLQASVHQQQHLHSPRVAGPTGQKSIS 383
Q + G PQ QQ QQQQQ QQQ LH R + PT S
Sbjct: 434 PQPDNTGNPQAFSQQAFAQQQQQ-----------------QQQQLH--RTSNPTSASVTS 474
Query: 384 LTGSQPDATASGATTPGGS-SSQGTEAATNQVLGKRKIQDLVAQVDPQGKLDPEV 437
G QP T A + S T A N + Q V+QVD L P +
Sbjct: 475 QNGQQPINTKLSANAAKTNYQSYLTNKARN---ATQPTQPPVSQVDYSNNLPPNL 526
Score = 38.5 bits (88), Expect = 0.048
Identities = 32/99 (32%), Positives = 44/99 (44%), Gaps = 12/99 (12%)
Query: 272 QSQWLKQMPAMSG---------PASPLRLQQHQRQQQQLASSGQLQQNSMTLNQQQLSQF 322
Q+ +KQMP++ A P RL + R +L + Q M + +
Sbjct: 82 QATQMKQMPSVYNNAGNVGALPTAGPNRLANNPRVMPRLQNQNVPMQAGMQQFARNMKLT 141
Query: 323 MQQQKSMGQPQLHQQQPSPQQQQQQQLLQPQQQSQLQAS 361
QQ++ + Q QQQ QQQQ Q QPQQ Q QAS
Sbjct: 142 PQQRQFLLQQSQIQQQRQQQQQQSQ---QPQQTQQPQAS 177
Score = 34.3 bits (77), Expect = 0.90
Identities = 88/437 (20%), Positives = 155/437 (35%), Gaps = 60/437 (13%)
Query: 4 NVIGSPDSQIQNPSSSNPSIPSPSPISQQQQSPSIHMSNSSPSLSQDQQQLHTINTINPN 63
N +G+P Q ++ NP S +QQQQ Q QQQLH T NP
Sbjct: 429 NKLGNP----QPDNTGNPQAFSQQAFAQQQQ--------------QQQQQLH--RTSNPT 468
Query: 64 SNFQLQQTLQRSPSMSRLNQIQPQQQQQVVARQQAALYGGQMNFGGSAAVSAQQQQLSGG 123
S Q Q+ P ++L+ A Y + A Q +S
Sbjct: 469 SASVTSQNGQQ-PINTKLS-----------ANAAKTNYQSYLTNKARNATQPTQPPVSQ- 515
Query: 124 VAAMGGSASNLSRSALIGQSGHFPM-LSGAGAAQFNLLTSPRQKGGMVQSSQFSSANSAG 182
V NL S+ S P + AG ++ S + + + ++ NS+
Sbjct: 516 VDYSNNLPPNLDTSSTFRSSASPPSAFTKAGNEALSVPLSGARNTAASRPTNLAAGNSSA 575
Query: 183 QSLQGMQQAIGMMGSPNLASQMRTNGGLYTQQQQIRLTPAQMRQQLSQQALNSQQVQG-- 240
+Q + + G MG + S++R L + + P + Q+ + + ++
Sbjct: 576 SIVQQLLECGGTMGQQKMTSRIRE---LTERVMHSLMRPVPLDLPHDQKVMIASLIKSAY 632
Query: 241 --IPRSSSLAFMNSQLSGLSQNGQP--GMMHNSLTQSQWLKQMPAMSGPASPLRLQQHQR 296
R++ L + L+G + M H Q + L+Q P + ++++
Sbjct: 633 PMFSRTNQLICLFYCLTGNEEATIQLIQMRHIFKLQLEGLQQGVFTCAPQTLAKIKEKTS 692
Query: 297 QQQQLASSGQLQ-QNSMTLNQQQLSQFMQQQKSMGQPQLHQQQPSPQQQQQQQLLQPQQQ 355
+ + L+ + + N + + S+ ++QQQ Q QQQQ
Sbjct: 693 RYFAFVKAQLLRLHHEVNNNNMSIQNALAHISSLRTASINQQQQPQPQSQQQQ------- 745
Query: 356 SQLQASVHQQQHLHSPRVAGPTGQKSISLTGSQPDATASGATTPGGSSSQGTEAATNQVL 415
QAS Q +P V+ + S+ +QP +S T + L
Sbjct: 746 ---QAS----QFPQAPSVSSNVRPINGSIPNAQPSVPGQAQPAAKANSVGNTSFPVDSKL 798
Query: 416 GKRKIQDLVAQVDPQGK 432
+ + V+Q D Q K
Sbjct: 799 AFQSLD--VSQPDLQAK 813
>PHP_DROME (P39769) Polyhomeotic-proximal chromatin protein
Length = 1589
Score = 72.4 bits (176), Expect = 3e-12
Identities = 99/428 (23%), Positives = 162/428 (37%), Gaps = 75/428 (17%)
Query: 14 QNPSSSNPSIPSPSPISQQQQ--------SPSIHMSNSSPSLSQDQQQL----------- 54
Q P+ + + + +P QQQQ + ++ ++ L Q QQQL
Sbjct: 603 QTPTKARTQLDALAPKQQQQQQQVGTTNQTQQQQLAVATAQLQQQQQQLTAAALQRPGAP 662
Query: 55 ---HTINTINPNSNFQLQQTLQRSPSMSRL-NQIQPQQQQQVVARQQAALYGGQMNFGGS 110
H + P S+ Q +S +++ N+ QP + + + GQ G
Sbjct: 663 VMPHNGTQVRPASSVSTQTAQNQSLLKAKMRNKQQPVRPALATLKTEIGQVAGQNKVVGH 722
Query: 111 AAVSAQQQQLSGGVAAMGGSASNLSRSALIGQSGHFPMLSGAGAAQFNLLTSPRQKGGMV 170
QQQQ A+NL + ++ +G+ ++ L Q G +
Sbjct: 723 LTTVQQQQQ-----------ATNLQQ--VVNAAGNKMVVMSTTGTPITL-----QNGQTL 764
Query: 171 QSSQFSSANSAGQSLQGMQQAIGMMGSPNLASQMRTNGGLYTQQQQIRLTPAQMRQQL-- 228
++ + + Q LQ Q+ + L Q+ + QQQQ + Q +QQ
Sbjct: 765 HAATAAGVDKQQQQLQLFQKQQILQQQQMLQQQI---AAIQMQQQQAAVQAQQQQQQQVS 821
Query: 229 SQQALNSQQVQGIPRSSSLAFMNSQLSGLSQNGQPGMMHNSLTQSQWLKQMPAMSGPASP 288
QQ +N+QQ Q + + Q Q + Q+Q Q +
Sbjct: 822 QQQQVNAQQQQAVAQQQQAVAQAQQ--------QQREQQQQVAQAQAQHQQALANATQQI 873
Query: 289 LR------LQQHQRQQQQLASSGQLQQNSMTLNQQQLSQFMQQQKSMGQPQLHQQQPSPQ 342
L+ + HQ+QQQQ + +QQ QQ +Q Q + Q Q QQQ Q
Sbjct: 874 LQVAPNQFITSHQQQQQQQLHNQLIQQQL-----QQQAQAQVQAQVQAQAQQQQQQREQQ 928
Query: 343 QQQQQQLLQPQQQSQLQASVHQQQHLHSPRVAGPTGQKSISLTGSQPDATASGATTPGGS 402
Q QQ++ Q + Q + QQQH S GQ +S S P + +S T G +
Sbjct: 929 QNIIQQIVVQQSGATSQQTSQQQQHHQS-------GQLQLS---SVPFSVSSSTTPAGIA 978
Query: 403 SSQGTEAA 410
+S +AA
Sbjct: 979 TSSALQAA 986
Score = 63.9 bits (154), Expect = 1e-09
Identities = 121/489 (24%), Positives = 172/489 (34%), Gaps = 97/489 (19%)
Query: 4 NVIGSPDSQIQNPSSSNPSIPSPSPISQQQQSPSIHMSNSSPSLSQDQQQLHT-INTINP 62
NVI SP Q QN S + ++QQQQ + L+Q QQQL + +
Sbjct: 406 NVI-SPQQQ-QNLLQSMAAAAQQQQLTQQQQQFN---QQQQQQLTQQQQQLTAALAKVGV 460
Query: 63 NSNFQLQQ-----------TLQRSPSMSRLNQIQPQQQQQVVARQQAALYGGQMNFGGSA 111
++ +L Q +Q + Q QQ QQV +QQ Q
Sbjct: 461 DAQGKLAQKVVQKVTTTSSAVQAATGPGSTGSTQTQQVQQVQQQQQQTTQTTQQ------ 514
Query: 112 AVSAQQQQLSGGVAAMGGSASNLSRSALIGQSGHFPM---------LSGAGAAQFNLLTS 162
V Q L GV GG + ++ GQ+ + L G Q L
Sbjct: 515 CVQVSQSTLPVGV---GGQSVQTAQLLNAGQAQQMQIPWFLQNAAGLQPFGPNQIILRNQ 571
Query: 163 PRQKGGMVQSSQFSSANSAGQSLQGMQQAI---GMMGSPNLA-------------SQMRT 206
P GM F A Q+LQ Q I + +P A Q +
Sbjct: 572 PDGTQGM-----FIQQQPATQTLQTQQNQIIQCNVTQTPTKARTQLDALAPKQQQQQQQV 626
Query: 207 NGGLYTQQQQIRLTPAQMRQQLSQQALNSQQVQGIPR--------------SSSLAFMNS 252
TQQQQ+ + AQ++QQ Q + Q G P S+ A S
Sbjct: 627 GTTNQTQQQQLAVATAQLQQQQQQLTAAALQRPGAPVMPHNGTQVRPASSVSTQTAQNQS 686
Query: 253 QLSGLSQNGQPGMMHNSLTQSQWLKQMPAMSGPASPLRLQQHQRQ----QQQLASSGQLQ 308
L +N Q + T + Q+ + L Q Q+Q QQ + ++G
Sbjct: 687 LLKAKMRNKQQPVRPALATLKTEIGQVAGQNKVVGHLTTVQQQQQATNLQQVVNAAGNKM 746
Query: 309 ------------QNSMTLN---------QQQLSQFMQQQKSMGQPQLHQQQ-PSPQQQQQ 346
QN TL+ QQQ Q Q+Q+ + Q Q+ QQQ + Q QQQ
Sbjct: 747 VVMSTTGTPITLQNGQTLHAATAAGVDKQQQQLQLFQKQQILQQQQMLQQQIAAIQMQQQ 806
Query: 347 QQLLQPQQQSQLQASVHQQQHLHSPRVAGPTGQKSISLTGSQPDATASGATTPGGSSSQG 406
Q +Q QQQ Q Q S QQ + + Q++++ Q Q
Sbjct: 807 QAAVQAQQQQQQQVSQQQQVNAQQQQAVAQQ-QQAVAQAQQQQREQQQQVAQAQAQHQQA 865
Query: 407 TEAATNQVL 415
AT Q+L
Sbjct: 866 LANATQQIL 874
Score = 63.2 bits (152), Expect = 2e-09
Identities = 102/402 (25%), Positives = 151/402 (37%), Gaps = 92/402 (22%)
Query: 52 QQLHTINTINPNSNFQLQQTLQRSPSMSR--LNQIQPQQQQQVVARQQAALYGGQMNFGG 109
QQ T+ N +Q + ++P+ +R L+ + P+QQQQ +QQ G N
Sbjct: 581 QQQPATQTLQTQQNQIIQCNVTQTPTKARTQLDALAPKQQQQ---QQQV----GTTN--- 630
Query: 110 SAAVSAQQQQLSGGVAAMGGSASNLSRSALIGQSGHFPMLSGAGAA---QFNLLTSPRQK 166
QQQQL+ A + L+ +AL Q P++ G ++ T Q
Sbjct: 631 ----QTQQQQLAVATAQLQQQQQQLTAAAL--QRPGAPVMPHNGTQVRPASSVSTQTAQN 684
Query: 167 GGMVQSSQFSSANSAGQSLQGMQQAIGMMGSPNLASQMRTNGGLYTQQQQIRLTPAQMRQ 226
++++ + +L ++ IG + A Q + G L T QQQ
Sbjct: 685 QSLLKAKMRNKQQPVRPALATLKTEIGQV-----AGQNKVVGHLTTVQQQ---------- 729
Query: 227 QLSQQALNSQQVQGIPRSSSLAFMNSQLSGLSQNGQPGMMHNSLTQSQWLKQMPAMSGPA 286
QQA N QQV + +M MS
Sbjct: 730 ---QQATNLQQVVNAAGN---------------------------------KMVVMSTTG 753
Query: 287 SPLRLQ-------------QHQRQQQQLASSGQLQQNSMTLNQQQLSQFMQQQKSMGQPQ 333
+P+ LQ Q+QQ QL Q+ Q L QQ + MQQQ++ Q Q
Sbjct: 754 TPITLQNGQTLHAATAAGVDKQQQQLQLFQKQQILQQQQMLQQQIAAIQMQQQQAAVQAQ 813
Query: 334 LHQQQPSPQQQ----QQQQLLQPQQQSQLQASVHQQQHLHSPRVAGPTGQKSISL-TGSQ 388
QQQ QQQ QQQQ + QQQ+ QA QQQ +VA Q +L +Q
Sbjct: 814 QQQQQQVSQQQQVNAQQQQAVAQQQQAVAQA--QQQQREQQQQVAQAQAQHQQALANATQ 871
Query: 389 PDATASGATTPGGSSSQGTEAATNQVLGKRKIQDLVAQVDPQ 430
+ Q + NQ++ ++ Q AQV Q
Sbjct: 872 QILQVAPNQFITSHQQQQQQQLHNQLIQQQLQQQAQAQVQAQ 913
Score = 56.6 bits (135), Expect = 2e-07
Identities = 77/331 (23%), Positives = 129/331 (38%), Gaps = 28/331 (8%)
Query: 59 TINPNSNFQLQQTLQRSPSMSRLNQIQPQQQQQVV-----ARQQAALYGGQMNFG--GSA 111
T P ++ Q QT+ SP +N I PQQQQ ++ A QQ L Q F
Sbjct: 387 TCIPTNHNQSPQTVLFSP----MNVISPQQQQNLLQSMAAAAQQQQLTQQQQQFNQQQQQ 442
Query: 112 AVSAQQQQLSGGVAAMGGSASNLSRSALIGQSGHFPMLSGAGAAQFNLLTSPRQKGGMVQ 171
++ QQQQL+ +A +G A ++ + A + ++ Q+ VQ
Sbjct: 443 QLTQQQQQLTAALAKVGVDAQGKLAQKVVQKVTTTSSAVQAATGPGSTGSTQTQQVQQVQ 502
Query: 172 SSQFSSANSAGQSLQGMQQAIGM-MGSPNLASQMRTNGGLYTQQQQIRLTPAQMRQQLSQ 230
Q + + Q +Q Q + + +G ++ + N G QQ QI P ++
Sbjct: 503 QQQQQTTQTTQQCVQVSQSTLPVGVGGQSVQTAQLLNAG-QAQQMQI---PWFLQNAAGL 558
Query: 231 QALNSQQV--QGIPRSSSLAFMNSQLSGLSQNGQPGMMHNSLTQSQWLKQMPAMSGPASP 288
Q Q+ + P + F+ Q + + Q N + Q + Q P +
Sbjct: 559 QPFGPNQIILRNQPDGTQGMFIQQQPATQTLQTQ----QNQIIQCN-VTQTPTKARTQLD 613
Query: 289 LRLQQHQRQQQQLASSGQLQQNSMTLNQQQLSQFMQQQKSM-----GQPQLHQQQPSPQQ 343
+ Q+QQQQ+ ++ Q QQ + + QL Q QQ + G P + +
Sbjct: 614 ALAPKQQQQQQQVGTTNQTQQQQLAVATAQLQQQQQQLTAAALQRPGAPVMPHNGTQVRP 673
Query: 344 QQQQQLLQPQQQSQLQASVHQQQHLHSPRVA 374
Q QS L+A + +Q P +A
Sbjct: 674 ASSVSTQTAQNQSLLKAKMRNKQQPVRPALA 704
>FXP2_MOUSE (P58463) Forkhead box protein P2
Length = 714
Score = 71.6 bits (174), Expect = 5e-12
Identities = 73/232 (31%), Positives = 98/232 (41%), Gaps = 30/232 (12%)
Query: 174 QFSSANSAGQSLQGMQQAIGMMGSPNLASQMR-----TNGGLYTQQQQIRLTPAQMRQQL 228
Q A A + L QQ G+ SP + + R + + T Q +TP QM+Q L
Sbjct: 53 QQQQALQAARQLLLQQQTSGLK-SPKSSEKQRPLQVPVSVAMMTPQV---ITPQQMQQIL 108
Query: 229 SQQALNSQQVQGIPRSSSLAFMNSQLSGLSQNGQPGMMHNSLTQSQWLKQMPAMSGPASP 288
QQ L+ QQ+Q + + + Q Q +H L Q Q +Q
Sbjct: 109 QQQVLSPQQLQALLQQQQAVMLQQQQLQEFYKKQQEQLHLQLLQQQQQQQQQQQQQQQQQ 168
Query: 289 LRLQQHQRQQQQLASSGQLQQNSMTLNQQQLSQFMQQQKSMGQPQLHQQQPSPQQQQQQQ 348
+ QQ Q+QQQQ QQQ Q QQQ Q + QQQ QQ QQ
Sbjct: 169 QQQQQQQQQQQQ---------------QQQQQQQQQQQHPGKQAKEQQQQQQQQQLAAQQ 213
Query: 349 LLQPQQQSQLQASVHQQQHLHSPRVAG----PTGQKSISLTGSQPDATASGA 396
L+ QQ Q+Q + QQQHL S + G P GQ ++ + S P A S A
Sbjct: 214 LVFQQQLLQMQ-QLQQQQHLLSLQRQGLISIPPGQAALPVQ-SLPQAGLSPA 263
Score = 39.3 bits (90), Expect = 0.028
Identities = 30/96 (31%), Positives = 38/96 (39%), Gaps = 7/96 (7%)
Query: 31 QQQQSPSIHMSNSSPSLSQDQQQLHTINTINPNSNFQLQQTLQRSPSMSRLNQIQPQQQQ 90
QQQQ+ + + Q+QLH Q QQ Q+ + Q Q QQQQ
Sbjct: 123 QQQQAVMLQQQQLQEFYKKQQEQLHL-------QLLQQQQQQQQQQQQQQQQQQQQQQQQ 175
Query: 91 QVVARQQAALYGGQMNFGGSAAVSAQQQQLSGGVAA 126
Q +QQ Q G A QQQQ +AA
Sbjct: 176 QQQQQQQQQQQQQQQQHPGKQAKEQQQQQQQQQLAA 211
Score = 32.7 bits (73), Expect = 2.6
Identities = 22/68 (32%), Positives = 26/68 (37%), Gaps = 5/68 (7%)
Query: 29 ISQQQQSPSIHMSNSSPSLSQDQQQLHTINTINPNSNFQLQQTLQRSPSMSRLNQIQPQQ 88
+ QQQQ Q QQQ Q QQ Q+ P Q Q QQ
Sbjct: 151 LQQQQQQQQQQQQQQQQQQQQQQQQQQ-----QQQQQQQQQQQQQQHPGKQAKEQQQQQQ 205
Query: 89 QQQVVARQ 96
QQQ+ A+Q
Sbjct: 206 QQQLAAQQ 213
Score = 31.6 bits (70), Expect = 5.8
Identities = 32/119 (26%), Positives = 42/119 (34%), Gaps = 10/119 (8%)
Query: 11 SQIQNPSSSNPSIPSPSPISQQQQSPSIHMSNSSPSLSQDQ----QQLHTINTINPNSNF 66
S +++P SS P P+S +P + + Q Q QQL +
Sbjct: 71 SGLKSPKSSEKQRPLQVPVSVAMMTPQVITPQQMQQILQQQVLSPQQLQALLQQQQAVML 130
Query: 67 QLQQTL------QRSPSMSRLNQIQPQQQQQVVARQQAALYGGQMNFGGSAAVSAQQQQ 119
Q QQ Q + L Q Q QQQQQ +QQ Q QQQQ
Sbjct: 131 QQQQLQEFYKKQQEQLHLQLLQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQ 189
>ARFS_ARATH (Q8RYC8) Auxin response factor 19 (Auxin-responsive
protein IAA22)
Length = 1086
Score = 71.2 bits (173), Expect = 7e-12
Identities = 104/428 (24%), Positives = 154/428 (35%), Gaps = 82/428 (19%)
Query: 12 QIQNPSSSNPSIPSPSPISQ-------QQQSPSIHMSNSSPSLSQDQQQLHTINTINPNS 64
Q NP S + + PS +S PS ++ SP+LS Q + NT+N
Sbjct: 423 QQNNPLSGSATPQLPSALSSFNLPNNFASNDPSKLLNFQSPNLSSANSQFNKPNTVN--- 479
Query: 65 NFQLQQTLQRSPSMSRLNQIQPQQQQQVVARQQAALYGGQMNFGGSAAVSAQQQQLSGGV 124
+ Q +Q P+M + Q Q QQQQQ +QQ Q+ S QQ + G
Sbjct: 480 --HISQQMQAQPAMVKSQQQQQQQQQQHQHQQQQLQQQQQLQM--SQQQVQQQGIYNNGT 535
Query: 125 AAMGGSASNLSRSALIGQSGHFPMLSGAGAAQFNLLTSPRQKGGMVQSSQFSSANSAGQS 184
A+ S SP Q G QS Q +
Sbjct: 536 IAVANQVS---------------------------CQSPNQPTGFSQS-QLQQQSMLPTG 567
Query: 185 LQGMQQAIGMMGSPNLASQMRTNGGLYTQQQQIRLTPAQMRQQLSQQALNSQQVQGIPRS 244
+ Q I MG+ L SQM + + Q+ Q + Q+ S Q L +QQ Q
Sbjct: 568 AKMTHQNINSMGNKGL-SQMTS----FAQEMQFQ---QQLEMHNSSQLLRNQQEQ----- 614
Query: 245 SSLAFMNSQLSGLSQNGQPGMMHNSLTQSQWLKQMPAMSGPASPLRLQQHQRQQQQLASS 304
+ ++S LSQN Q M +Q + P+ L+LQ Q+ QQQ
Sbjct: 615 ---SSLHSLQQNLSQNPQQLQM----------QQQSSKPSPSQQLQLQLLQKLQQQ---- 657
Query: 305 GQLQQNSMTLNQQQLSQFMQQQKSMGQPQLHQQQPSPQQQQQQQLLQPQQQSQLQASVHQ 364
Q QQ+ ++ S Q ++ Q Q HQ Q Q QQ L +
Sbjct: 658 -QQQQSIPPVS----SSLQPQLSALQQTQSHQLQQLLSSQNQQPLAHGNNSFPASTFMQP 712
Query: 365 QQHLHSPRVAGPTGQKSISLTGSQPDATASG-----ATTPGGSSSQGTEAATNQVLGKRK 419
Q SP+ G K++ G G +T+P +++ + L + +
Sbjct: 713 PQIQVSPQQQGQMSNKNLVAAGRSHSGHTDGEAPSCSTSPSANNTGHDNVSPTNFLSRNQ 772
Query: 420 IQDLVAQV 427
Q A V
Sbjct: 773 QQGQAASV 780
Score = 36.2 bits (82), Expect = 0.24
Identities = 40/152 (26%), Positives = 54/152 (35%), Gaps = 21/152 (13%)
Query: 263 PGMMHNSLTQSQWLKQMPAMSGPASPLRLQQHQRQQQQLASSGQLQQNSMTLNQQQLSQF 322
PG+ SL Q ++Q +SG A+P Q SS L N + + +L F
Sbjct: 412 PGL---SLVQWMSMQQNNPLSGSATP--------QLPSALSSFNLPNNFASNDPSKLLNF 460
Query: 323 MQQQKSMGQPQLHQQQPSPQQQQQQQLLQPQQQSQLQASVHQQQHLHSPRVAGPTGQKSI 382
S Q ++ QQ Q +SQ Q QQQH H + Q +
Sbjct: 461 QSPNLSSANSQFNKPNTVNHISQQMQAQPAMVKSQQQQQQQQQQHQHQQQQLQQQQQLQM 520
Query: 383 SLTGSQPDATASGATTPGGSSSQGTEAATNQV 414
S Q G + GT A NQV
Sbjct: 521 SQQQVQQQ----------GIYNNGTIAVANQV 542
>SNF5_YEAST (P18480) Transcription regulatory protein SNF5 (SWI/SNF
complex component SNF5) (Transcription factor TYE4)
Length = 905
Score = 70.9 bits (172), Expect = 9e-12
Identities = 60/182 (32%), Positives = 80/182 (42%), Gaps = 15/182 (8%)
Query: 210 LYTQQQQIRLTPA---------QMRQQLSQQALNSQQVQGIPRSSSL---AFMNSQLSGL 257
L+ Q Q+ L PA Q+ QQ LN + Q I +++ A +Q
Sbjct: 108 LHPQIGQVPLAPAPINLPPQIAQLPLATQQQVLNKLRQQAIAKNNPQVVNAITVAQQQVQ 167
Query: 258 SQNGQPGMMHNSLTQSQWLKQMPAMSGPASPLRLQQHQRQQQQLASSGQLQQNSMTLNQQ 317
Q Q + TQ + +Q+ LR Q ++QQQQ Q+QQ QQ
Sbjct: 168 RQIEQQKGQQTAQTQLEQQRQLLVQQQQQQQLRNQIQRQQQQQFRHHVQIQQQQQKQQQQ 227
Query: 318 QLSQFMQQQKSMGQPQLHQQQPSPQQQQQQQLLQPQQQSQLQASVHQQQHLHSPRVAGPT 377
Q QQQ+ Q Q QQQ QQQQQQQ Q QQQ Q Q + Q Q + P+V +
Sbjct: 228 QQQHQQQQQQQQQQQQQQQQQQQQQQQQQQQ-QQQQQQQQQQGQIPQSQQV--PQVRSMS 284
Query: 378 GQ 379
GQ
Sbjct: 285 GQ 286
Score = 54.7 bits (130), Expect = 6e-07
Identities = 88/332 (26%), Positives = 122/332 (36%), Gaps = 53/332 (15%)
Query: 39 HMSNSSPSLSQDQQQLHTINTINPNSNFQLQQTLQRSPSM--SRLNQIQPQQQQQVVARQ 96
++ S +++Q QQL+ ++ P QLQ QR + SRL Q Q QQQQ Q
Sbjct: 21 NIGTPSFNMAQIPQQLY--QSLTPQ---QLQMIQQRHQQLLRSRLQQQQQQQQQTSPPPQ 75
Query: 97 QAALYGGQMNFGGSAAVSAQQQQLSGGVAAMGGSASNLSRSALIGQSGHFPMLSGAGAAQ 156
S QQ Q +A + L Q G P+ A
Sbjct: 76 THQ----------SPPPPPQQSQPIANQSATSTPPPPPAPHNLHPQIGQVPL----APAP 121
Query: 157 FNLLTSPRQKGGMVQSSQFSSANSAGQSLQGMQQAIGMMGSPNLASQMRTNGGLYTQQQQ 216
NL P Q + ++Q N QQAI +P + + + QQQ
Sbjct: 122 INL---PPQIAQLPLATQQQVLNKL------RQQAIAK-NNPQVVNAITV-----AQQQV 166
Query: 217 IRLTPAQMRQQLSQQALNSQQVQGIPRSSSLAFMNSQLSGLSQNGQPGMMHNSLTQSQWL 276
R Q QQ +Q L Q+ + + N + + Q H+ Q Q
Sbjct: 167 QRQIEQQKGQQTAQTQLEQQRQLLVQQQQQQQLRNQ----IQRQQQQQFRHHVQIQQQQQ 222
Query: 277 KQMPAMSGPASPLRLQQHQRQQQQLASSGQLQQNSMTLNQQQLSQFMQQQKSMGQPQLHQ 336
KQ QQHQ+QQQQ Q QQ QQQ Q QQQ+ Q Q
Sbjct: 223 KQQQQQ---------QQHQQQQQQQQQQQQQQQQQ----QQQQQQQQQQQQQQQQQQQQG 269
Query: 337 QQPSPQQQQQQQLLQPQQQSQLQASVHQQQHL 368
Q P QQ Q + + Q + +Q ++ Q L
Sbjct: 270 QIPQSQQVPQVRSMSGQPPTNVQPTIGQLPQL 301
Score = 39.3 bits (90), Expect = 0.028
Identities = 36/100 (36%), Positives = 49/100 (49%), Gaps = 15/100 (15%)
Query: 250 MNSQLSGLSQ--NGQPGMMHNSLTQSQWLKQMPA-MSGPASPLRLQQHQRQQQQLASSGQ 306
MN+Q G + N + N T S + Q+P + +P +LQ Q++ QQL S +
Sbjct: 1 MNNQPQGTNSVPNSIGNIFSNIGTPSFNMAQIPQQLYQSLTPQQLQMIQQRHQQLLRS-R 59
Query: 307 LQQNSMTLNQQQLSQFMQQQKSMGQPQLHQQQPSPQQQQQ 346
LQQ QQQ QQQ++ PQ HQ P P QQ Q
Sbjct: 60 LQQ------QQQ-----QQQQTSPPPQTHQSPPPPPQQSQ 88
Score = 35.0 bits (79), Expect = 0.53
Identities = 30/87 (34%), Positives = 39/87 (44%), Gaps = 13/87 (14%)
Query: 313 TLNQQQLSQFMQQQKSMGQPQLHQQQPSPQQQQQQQLLQPQQQSQLQASVHQQQHLHSPR 372
+ N Q+ Q + Q + Q Q+ QQ+ QQ + LQ QQQ Q Q S Q H P
Sbjct: 26 SFNMAQIPQQLYQSLTPQQLQMIQQR---HQQLLRSRLQQQQQQQQQTSPPPQTHQSPP- 81
Query: 373 VAGPTGQKSISLTGSQPDATASGATTP 399
P Q+ SQP A S +TP
Sbjct: 82 ---PPPQQ------SQPIANQSATSTP 99
Score = 33.9 bits (76), Expect = 1.2
Identities = 46/192 (23%), Positives = 68/192 (34%), Gaps = 7/192 (3%)
Query: 236 QQVQGIPRSSSLAFMNSQLSGLSQNGQPGMMHNSLTQSQWLKQMPAMSGPASPLRLQQHQ 295
Q +P S F N + P ++ SLT Q L+ + RLQQ Q
Sbjct: 6 QGTNSVPNSIGNIFSNIGTPSFNMAQIPQQLYQSLTPQQ-LQMIQQRHQQLLRSRLQQQQ 64
Query: 296 RQQQQLASSGQLQQNSMTLNQQQLSQFMQQQKSMGQPQLHQQQPSPQQQQ----QQQLLQ 351
+QQQQ + Q Q+ QQ Q S P PQ Q +
Sbjct: 65 QQQQQTSPPPQTHQSPPPPPQQSQPIANQSATSTPPPPPAPHNLHPQIGQVPLAPAPINL 124
Query: 352 PQQQSQLQASVHQQ--QHLHSPRVAGPTGQKSISLTGSQPDATASGATTPGGSSSQGTEA 409
P Q +QL + QQ L +A Q ++T +Q G ++Q
Sbjct: 125 PPQIAQLPLATQQQVLNKLRQQAIAKNNPQVVNAITVAQQQVQRQIEQQKGQQTAQTQLE 184
Query: 410 ATNQVLGKRKIQ 421
Q+L +++ Q
Sbjct: 185 QQRQLLVQQQQQ 196
Score = 33.1 bits (74), Expect = 2.0
Identities = 28/114 (24%), Positives = 40/114 (34%), Gaps = 11/114 (9%)
Query: 17 SSSNPSIPSPSPISQQQ-----------QSPSIHMSNSSPSLSQDQQQLHTINTINPNSN 65
+ +NP + + ++QQQ Q+ + L Q QQQ N I
Sbjct: 149 AKNNPQVVNAITVAQQQVQRQIEQQKGQQTAQTQLEQQRQLLVQQQQQQQLRNQIQRQQQ 208
Query: 66 FQLQQTLQRSPSMSRLNQIQPQQQQQVVARQQAALYGGQMNFGGSAAVSAQQQQ 119
Q + +Q + Q Q Q QQQ +QQ Q QQQQ
Sbjct: 209 QQFRHHVQIQQQQQKQQQQQQQHQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQ 262
Score = 31.2 bits (69), Expect = 7.6
Identities = 54/243 (22%), Positives = 77/243 (31%), Gaps = 26/243 (10%)
Query: 31 QQQQSPSIHMSNSSPSLSQDQQ----QLHTINTINPNSNFQLQQTLQRSPSMSRLNQIQP 86
QQQ SP S P Q Q Q T P + L + + P + P
Sbjct: 67 QQQTSPPPQTHQSPPPPPQQSQPIANQSATSTPPPPPAPHNLHPQIGQVPLAPAPINLPP 126
Query: 87 QQQQQVVARQQAALYG------GQMNFGGSAAVSAQQQQLSGGVAAMGGSASNLSRSALI 140
Q Q +A QQ L + N A++ QQQ+ + G + ++
Sbjct: 127 QIAQLPLATQQQVLNKLRQQAIAKNNPQVVNAITVAQQQVQRQIEQQKGQQTAQTQLE-- 184
Query: 141 GQSGHFPMLSGAGAAQFNLLTSPRQKGGMVQSSQFSSANSAGQSLQGMQQAIGMMGSPNL 200
Q LL +Q+ + Q +Q QQ
Sbjct: 185 --------------QQRQLLVQQQQQQQLRNQIQRQQQQQFRHHVQIQQQQQKQQQQQQQ 230
Query: 201 ASQMRTNGGLYTQQQQIRLTPAQMRQQLSQQALNSQQVQGIPRSSSLAFMNSQLSGLSQN 260
Q + QQQQ + Q +QQ QQ QQ IP+S + + S N
Sbjct: 231 HQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQGQIPQSQQVPQVRSMSGQPPTN 290
Query: 261 GQP 263
QP
Sbjct: 291 VQP 293
>KAPC_DICDI (P34099) cAMP-dependent protein kinase catalytic subunit
(EC 2.7.1.37)
Length = 648
Score = 70.5 bits (171), Expect = 1e-11
Identities = 50/130 (38%), Positives = 58/130 (44%), Gaps = 1/130 (0%)
Query: 269 SLTQSQWLKQMPAMSGPASPLRLQQHQRQQQQLASSGQLQQNSMTLNQQQLSQFMQQQKS 328
SLT + KQ P + QQ Q QQQQ Q Q QQQL Q QQQ+
Sbjct: 127 SLTPPEPNKQQQPQQQPQQQ-QPQQQQPQQQQPQQQQQQQPQQQQQPQQQLQQNNQQQQQ 185
Query: 329 MGQPQLHQQQPSPQQQQQQQLLQPQQQSQLQASVHQQQHLHSPRVAGPTGQKSISLTGSQ 388
Q Q QQQ QQQQQQQ Q QQQ Q + QQQHLH + S T +
Sbjct: 186 QLQQQQLQQQLQQQQQQQQQQQQQQQQKQQKQQQQQQQHLHQDGIVNTPSTTQTSTTTTT 245
Query: 389 PDATASGATT 398
T + T+
Sbjct: 246 TTTTTNPHTS 255
Score = 43.1 bits (100), Expect = 0.002
Identities = 56/234 (23%), Positives = 80/234 (33%), Gaps = 50/234 (21%)
Query: 9 PDSQIQNPSSSNPSIPSPSPISQQQQSPSIHMSNSSPSLSQDQQQLHTINTINPNSNFQL 68
P+ Q P P QQQ P Q QQQL +N Q
Sbjct: 131 PEPNKQQQPQQQPQQQQPQQQQPQQQQPQQQQQQQPQQQQQPQQQLQ-------QNNQQQ 183
Query: 69 QQTLQRSPSMSRLNQIQPQQQQQVVARQQAALYGGQMNFGGSAAVSAQQQQLSGGVAAMG 128
QQ LQ+ +L Q Q QQQQQ +QQ QQQQ
Sbjct: 184 QQQLQQQQLQQQLQQQQQQQQQQQQQQQQKQ--------------QKQQQQ--------- 220
Query: 129 GSASNLSRSALIGQSGHFPMLSGAGAAQFNLLTSPRQKGGMVQSSQFSSANSAGQSLQGM 188
+L + ++ N ++ + ++ ++ +++G SLQ
Sbjct: 221 -QQQHLHQDGIV-----------------NTPSTTQTSTTTTTTTTTTNPHTSGLSLQHA 262
Query: 189 QQAIGMMGSPNLASQMRTNGGLYTQQQQIRLTPAQMRQQLSQQALNSQQVQGIP 242
+ S L S L T+ +T +QQ SQQ L QQ+Q IP
Sbjct: 263 HSSY--TPSNVLHSPTHFQSSLPTRLDTNPITTPIRQQQQSQQQLQQQQLQQIP 314
Score = 39.7 bits (91), Expect = 0.021
Identities = 34/119 (28%), Positives = 46/119 (38%), Gaps = 3/119 (2%)
Query: 200 LASQMRTNGGLYTQQQQIRLTPAQ--MRQQLSQQALNSQQVQGIPRSSSLAFMNSQLSGL 257
LAS + + G + LTP + +QQ QQ Q Q P+ Q
Sbjct: 110 LASNLNSFGN-FKVPSTFSLTPPEPNKQQQPQQQPQQQQPQQQQPQQQQPQQQQQQQPQQ 168
Query: 258 SQNGQPGMMHNSLTQSQWLKQMPAMSGPASPLRLQQHQRQQQQLASSGQLQQNSMTLNQ 316
Q Q + N+ Q Q L+Q + QQ Q+QQQQ Q QQ L+Q
Sbjct: 169 QQQPQQQLQQNNQQQQQQLQQQQLQQQLQQQQQQQQQQQQQQQQKQQKQQQQQQQHLHQ 227
Score = 36.6 bits (83), Expect = 0.18
Identities = 49/193 (25%), Positives = 63/193 (32%), Gaps = 9/193 (4%)
Query: 160 LTSPRQKGGMVQSSQFSSANSAGQSLQGMQQAIGMMGSPNLASQMRTNGGLYTQQQQIRL 219
LT P Q Q Q Q P Q + QQQQ +L
Sbjct: 128 LTPPEPNKQQQPQQQPQQQQPQQQQPQQQQPQQQQQQQPQQQQQPQQQLQQNNQQQQQQL 187
Query: 220 TPAQMRQQLSQQALNSQQVQGIPRSSSLAFMNSQLSGLSQNGQPGMMHNSLTQSQWLKQM 279
Q++QQL QQ QQ Q + Q L Q+G + T +
Sbjct: 188 QQQQLQQQLQQQQQQQQQQQQQQQQKQQKQQQQQQQHLHQDGIVNTPSTTQTSTTTTTTT 247
Query: 280 PAMSGPASPLRLQQHQRQQQQLASSGQLQQNSM-TLNQQQLSQFMQQQKSMGQPQLHQQQ 338
+ S L LQ A S N + + Q S + + + QQQ
Sbjct: 248 TTTNPHTSGLSLQH--------AHSSYTPSNVLHSPTHFQSSLPTRLDTNPITTPIRQQQ 299
Query: 339 PSPQQQQQQQLLQ 351
S QQ QQQQL Q
Sbjct: 300 QSQQQLQQQQLQQ 312
Score = 35.4 bits (80), Expect = 0.40
Identities = 21/47 (44%), Positives = 22/47 (46%)
Query: 320 SQFMQQQKSMGQPQLHQQQPSPQQQQQQQLLQPQQQSQLQASVHQQQ 366
S F + Q QQQP QQ QQQQ Q Q Q Q Q QQQ
Sbjct: 124 STFSLTPPEPNKQQQPQQQPQQQQPQQQQPQQQQPQQQQQQQPQQQQ 170
>T230_HUMAN (Q93074) Thyroid hormone receptor-associated protein
complex 230 kDa component (Trap230) (Activator-recruited
cofactor 240 KDa component) (ARC240) (CAG repeat protein
45) (OPA-containing protein) (Trinucleotide repeat
containing 11)
Length = 2212
Score = 70.1 bits (170), Expect = 1e-11
Identities = 92/326 (28%), Positives = 128/326 (39%), Gaps = 42/326 (12%)
Query: 77 SMSRLNQIQPQQQQQVVARQQA-ALYGGQMNFGGSAAVSAQQQQLSGGVAAMGGSASNLS 135
+M+ + ++P + V RQQ A+ GQ QQ Q S G+ S ++
Sbjct: 1916 TMTGVMGLEPSSYKTSVYRQQQPAVPQGQR--------LRQQLQQSQGMLGQS-SVHQMT 1966
Query: 136 RSALIG---QSGHFPMLSGAGAAQFNLLTSPRQKGGMVQSSQFSSANSAGQSLQGMQQAI 192
S+ G G+ P +S G Q T P G MV S S Q Q +
Sbjct: 1967 PSSSYGLQTSQGYTPYVSHVGLQQH---TGPA--GTMVPPSYSS------QPYQSTHPST 2015
Query: 193 G-MMGSPNLASQMRTNGGLYTQQQQIRLTPAQMRQQLSQQALNSQQVQGIPRSSSLAFMN 251
+ P Q R +G Y QQ P S Q + Q +Q P S++ M+
Sbjct: 2016 NPTLVDPTRHLQQRPSG--YVHQQ----APTYGHGLTSTQRFSHQTLQQTPMISTMTPMS 2069
Query: 252 SQLSGLSQNGQPGMMHNSLTQSQWLKQMPAMSGPASPLRLQQHQRQQQQLASSGQLQQNS 311
+Q Q G+ ++ Q +Q + QQ Q+QQQQ Q QQ
Sbjct: 2070 AQ------GVQAGVRSTAILPEQ--QQQQQQQQQQQQQQQQQQQQQQQQQYHIRQQQQQQ 2121
Query: 312 MTLNQQQLSQFMQQQKSMGQPQLHQQQPSPQQQQQQQLLQPQQQSQLQASVHQ---QQHL 368
+ QQQ Q QQQ+ Q Q QQQ QQQQQQQ PQ Q Q Q + QQ
Sbjct: 2122 ILRQQQQQQQQQQQQQQQQQQQQQQQQQQHQQQQQQQAAPPQPQPQSQPQFQRQGLQQTQ 2181
Query: 369 HSPRVAGPTGQKSISLTGSQPDATAS 394
+ A Q L+ +QP + +
Sbjct: 2182 QQQQTAALVRQLQQQLSNTQPQPSTN 2207
Score = 38.1 bits (87), Expect = 0.062
Identities = 53/228 (23%), Positives = 78/228 (33%), Gaps = 49/228 (21%)
Query: 18 SSNPSIPSPSPISQQQQSPSIH---------MSNSSPSLSQDQQQLHTINTINPNSNFQL 68
S+NP++ P+ QQ+ S +H ++++ Q QQ I+T+ P S +
Sbjct: 2014 STNPTLVDPTRHLQQRPSGYVHQQAPTYGHGLTSTQRFSHQTLQQTPMISTMTPMSAQGV 2073
Query: 69 QQTLQRSPSMSRLNQIQPQQQQQVVARQQAALYGGQMNFGGSAAVSAQQQQLSGGVAAMG 128
Q ++ + + Q Q QQQQQ +QQ QQQQ
Sbjct: 2074 QAGVRSTAILPEQQQQQQQQQQQQQQQQQQ---------------QQQQQQ--------- 2109
Query: 129 GSASNLSRSALIGQSGHFPMLSGAGAAQFNLLTSPRQKGGMVQSSQFSSANSAGQSLQGM 188
Q H + +Q+ Q Q Q
Sbjct: 2110 -------------QQYHIRQQQQQQILRQQQQQQQQQQQQQQQQQQQQQQQQQQHQQQQQ 2156
Query: 189 QQAIGMMGSPNLASQMRTNGGLYTQQQQIRLTPAQMRQQLSQQALNSQ 236
QQA P Q + G TQQQQ A + +QL QQ N+Q
Sbjct: 2157 QQAAPPQPQPQSQPQFQRQGLQQTQQQQ---QTAALVRQLQQQLSNTQ 2201
>FXP2_PONPY (Q8MJ98) Forkhead box protein P2
Length = 714
Score = 70.1 bits (170), Expect = 1e-11
Identities = 75/234 (32%), Positives = 101/234 (43%), Gaps = 34/234 (14%)
Query: 174 QFSSANSAGQSLQGMQQAIGMMGSPNLASQMR-----TNGGLYTQQQQIRLTPAQMRQQL 228
Q A A + L QQ G+ SP + + R + + T Q +TP QM+Q L
Sbjct: 53 QQQQALQAARQLLLQQQTSGLK-SPKSSDKQRPLQVPVSVAMMTPQV---ITPQQMQQIL 108
Query: 229 SQQALNSQQVQGIPRSSSLAFMNSQLSGLSQNGQPGMMHNSLTQSQWLKQMPAMSGPASP 288
QQ L+ QQ+Q + + + Q Q +H L Q Q +Q
Sbjct: 109 QQQVLSPQQLQALLQQQQAVMLQQQQLQEFYKKQQEQLHLQLLQQQQQQQQQQQQQQQQQ 168
Query: 289 LRLQQHQRQQQQLASSGQLQQNSMTLNQQQLSQFMQQQKSMGQPQLHQQQPSPQQQQ--Q 346
+ QQ Q+QQQQ QQQ Q QQQ+ G+ QQQ QQQQ
Sbjct: 169 QQQQQQQQQQQQ---------------QQQQQQ--QQQQHPGKQAKEQQQQQQQQQQLAA 211
Query: 347 QQLLQPQQQSQLQASVHQQQHLHSPRVAG----PTGQKSISLTGSQPDATASGA 396
QQL+ QQ Q+Q + QQQHL S + G P GQ ++ + S P A S A
Sbjct: 212 QQLVFQQQLLQMQ-QLQQQQHLLSLQRQGLISIPPGQAALPVQ-SLPQAGLSPA 263
Score = 35.8 bits (81), Expect = 0.31
Identities = 29/96 (30%), Positives = 38/96 (39%), Gaps = 7/96 (7%)
Query: 31 QQQQSPSIHMSNSSPSLSQDQQQLHTINTINPNSNFQLQQTLQRSPSMSRLNQIQPQQQQ 90
QQQQ+ + + Q+QLH Q QQ Q+ + Q Q QQQQ
Sbjct: 123 QQQQAVMLQQQQLQEFYKKQQEQLHLQLLQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQ 182
Query: 91 QVVARQQAALYGGQMNFGGSAAVSAQQQQLSGGVAA 126
Q +Q Q + G A QQQQ +AA
Sbjct: 183 QQQQQQ-------QQHPGKQAKEQQQQQQQQQQLAA 211
Score = 32.7 bits (73), Expect = 2.6
Identities = 34/126 (26%), Positives = 45/126 (34%), Gaps = 10/126 (7%)
Query: 11 SQIQNPSSSNPSIPSPSPISQQQQSPSIHMSNSSPSLSQDQ----QQLHTINTINPNSNF 66
S +++P SS+ P P+S +P + + Q Q QQL +
Sbjct: 71 SGLKSPKSSDKQRPLQVPVSVAMMTPQVITPQQMQQILQQQVLSPQQLQALLQQQQAVML 130
Query: 67 QLQQTL------QRSPSMSRLNQIQPQQQQQVVARQQAALYGGQMNFGGSAAVSAQQQQL 120
Q QQ Q + L Q Q QQQQQ +QQ Q QQQQ
Sbjct: 131 QQQQLQEFYKKQQEQLHLQLLQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQ 190
Query: 121 SGGVAA 126
G A
Sbjct: 191 HPGKQA 196
Score = 32.7 bits (73), Expect = 2.6
Identities = 23/69 (33%), Positives = 26/69 (37%), Gaps = 6/69 (8%)
Query: 29 ISQQQQSPSIHMSNSSPSLSQDQQQLHTINTINPNSNFQLQQTLQRSPSMSRLNQIQPQQ 88
+ QQQQ Q QQQ Q QQ Q+ P Q Q QQ
Sbjct: 151 LQQQQQQQQQQQQQQQQQQQQQQQQQQQ------QQQQQQQQQQQQHPGKQAKEQQQQQQ 204
Query: 89 QQQVVARQQ 97
QQQ +A QQ
Sbjct: 205 QQQQLAAQQ 213
>FXP2_MACMU (Q8MJ97) Forkhead box protein P2
Length = 714
Score = 70.1 bits (170), Expect = 1e-11
Identities = 75/234 (32%), Positives = 101/234 (43%), Gaps = 34/234 (14%)
Query: 174 QFSSANSAGQSLQGMQQAIGMMGSPNLASQMR-----TNGGLYTQQQQIRLTPAQMRQQL 228
Q A A + L QQ G+ SP + + R + + T Q +TP QM+Q L
Sbjct: 53 QQQQALQAARQLLLQQQTSGLK-SPKSSDKQRPLQVPVSVAMMTPQV---ITPQQMQQIL 108
Query: 229 SQQALNSQQVQGIPRSSSLAFMNSQLSGLSQNGQPGMMHNSLTQSQWLKQMPAMSGPASP 288
QQ L+ QQ+Q + + + Q Q +H L Q Q +Q
Sbjct: 109 QQQVLSPQQLQALLQQQQAVMLQQQQLQEFYKKQQEQLHLQLLQQQQQQQQQQQQQQQQQ 168
Query: 289 LRLQQHQRQQQQLASSGQLQQNSMTLNQQQLSQFMQQQKSMGQPQLHQQQPSPQQQQ--Q 346
+ QQ Q+QQQQ QQQ Q QQQ+ G+ QQQ QQQQ
Sbjct: 169 QQQQQQQQQQQQ---------------QQQQQQ--QQQQHPGKQAKEQQQQQQQQQQLAA 211
Query: 347 QQLLQPQQQSQLQASVHQQQHLHSPRVAG----PTGQKSISLTGSQPDATASGA 396
QQL+ QQ Q+Q + QQQHL S + G P GQ ++ + S P A S A
Sbjct: 212 QQLVFQQQLLQMQ-QLQQQQHLLSLQRQGLISIPPGQAALPVQ-SLPQAGLSPA 263
Score = 35.8 bits (81), Expect = 0.31
Identities = 29/96 (30%), Positives = 38/96 (39%), Gaps = 7/96 (7%)
Query: 31 QQQQSPSIHMSNSSPSLSQDQQQLHTINTINPNSNFQLQQTLQRSPSMSRLNQIQPQQQQ 90
QQQQ+ + + Q+QLH Q QQ Q+ + Q Q QQQQ
Sbjct: 123 QQQQAVMLQQQQLQEFYKKQQEQLHLQLLQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQ 182
Query: 91 QVVARQQAALYGGQMNFGGSAAVSAQQQQLSGGVAA 126
Q +Q Q + G A QQQQ +AA
Sbjct: 183 QQQQQQ-------QQHPGKQAKEQQQQQQQQQQLAA 211
Score = 32.7 bits (73), Expect = 2.6
Identities = 34/126 (26%), Positives = 45/126 (34%), Gaps = 10/126 (7%)
Query: 11 SQIQNPSSSNPSIPSPSPISQQQQSPSIHMSNSSPSLSQDQ----QQLHTINTINPNSNF 66
S +++P SS+ P P+S +P + + Q Q QQL +
Sbjct: 71 SGLKSPKSSDKQRPLQVPVSVAMMTPQVITPQQMQQILQQQVLSPQQLQALLQQQQAVML 130
Query: 67 QLQQTL------QRSPSMSRLNQIQPQQQQQVVARQQAALYGGQMNFGGSAAVSAQQQQL 120
Q QQ Q + L Q Q QQQQQ +QQ Q QQQQ
Sbjct: 131 QQQQLQEFYKKQQEQLHLQLLQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQ 190
Query: 121 SGGVAA 126
G A
Sbjct: 191 HPGKQA 196
Score = 32.7 bits (73), Expect = 2.6
Identities = 23/69 (33%), Positives = 26/69 (37%), Gaps = 6/69 (8%)
Query: 29 ISQQQQSPSIHMSNSSPSLSQDQQQLHTINTINPNSNFQLQQTLQRSPSMSRLNQIQPQQ 88
+ QQQQ Q QQQ Q QQ Q+ P Q Q QQ
Sbjct: 151 LQQQQQQQQQQQQQQQQQQQQQQQQQQQ------QQQQQQQQQQQQHPGKQAKEQQQQQQ 204
Query: 89 QQQVVARQQ 97
QQQ +A QQ
Sbjct: 205 QQQQLAAQQ 213
>FXP2_GORGO (Q8MJ99) Forkhead box protein P2
Length = 713
Score = 68.6 bits (166), Expect = 4e-11
Identities = 74/234 (31%), Positives = 100/234 (42%), Gaps = 35/234 (14%)
Query: 174 QFSSANSAGQSLQGMQQAIGMMGSPNLASQMR-----TNGGLYTQQQQIRLTPAQMRQQL 228
Q A A + L QQ G+ SP + + R + + T Q +TP QM+Q L
Sbjct: 53 QQQQALQAARQLLLQQQTSGLK-SPKSSDKQRPLQVPVSVAMMTPQV---ITPQQMQQIL 108
Query: 229 SQQALNSQQVQGIPRSSSLAFMNSQLSGLSQNGQPGMMHNSLTQSQWLKQMPAMSGPASP 288
QQ L+ QQ+Q + + + Q Q +H L Q Q +Q
Sbjct: 109 QQQVLSPQQLQALLQQQQAVMLQQQQLQEFYKKQQEQLHLQLLQQQQQQQQQQQQQQQQQ 168
Query: 289 LRLQQHQRQQQQLASSGQLQQNSMTLNQQQLSQFMQQQKSMGQPQLHQQQPSPQQQQ--Q 346
+ QQ Q+QQQQ QQQ QQQ+ G+ QQQ QQQQ
Sbjct: 169 QQQQQQQQQQQQ--------------QQQQ----QQQQQHPGKQAKEQQQQQQQQQQLAA 210
Query: 347 QQLLQPQQQSQLQASVHQQQHLHSPRVAG----PTGQKSISLTGSQPDATASGA 396
QQL+ QQ Q+Q + QQQHL S + G P GQ ++ + S P A S A
Sbjct: 211 QQLVFQQQLLQMQ-QLQQQQHLLSLQRQGLISIPPGQAALPVQ-SLPQAGLSPA 262
Score = 35.4 bits (80), Expect = 0.40
Identities = 29/96 (30%), Positives = 38/96 (39%), Gaps = 8/96 (8%)
Query: 31 QQQQSPSIHMSNSSPSLSQDQQQLHTINTINPNSNFQLQQTLQRSPSMSRLNQIQPQQQQ 90
QQQQ+ + + Q+QLH Q QQ Q+ + Q Q QQQQ
Sbjct: 123 QQQQAVMLQQQQLQEFYKKQQEQLHLQLLQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQ 182
Query: 91 QVVARQQAALYGGQMNFGGSAAVSAQQQQLSGGVAA 126
Q +QQ + G A QQQQ +AA
Sbjct: 183 QQQQQQQ--------HPGKQAKEQQQQQQQQQQLAA 210
Score = 33.1 bits (74), Expect = 2.0
Identities = 23/69 (33%), Positives = 26/69 (37%), Gaps = 7/69 (10%)
Query: 29 ISQQQQSPSIHMSNSSPSLSQDQQQLHTINTINPNSNFQLQQTLQRSPSMSRLNQIQPQQ 88
+ QQQQ Q QQQ Q QQ Q+ P Q Q QQ
Sbjct: 151 LQQQQQQQQQQQQQQQQQQQQQQQQ-------QQQQQQQQQQQQQQHPGKQAKEQQQQQQ 203
Query: 89 QQQVVARQQ 97
QQQ +A QQ
Sbjct: 204 QQQQLAAQQ 212
Score = 32.7 bits (73), Expect = 2.6
Identities = 33/122 (27%), Positives = 44/122 (36%), Gaps = 10/122 (8%)
Query: 11 SQIQNPSSSNPSIPSPSPISQQQQSPSIHMSNSSPSLSQDQ----QQLHTINTINPNSNF 66
S +++P SS+ P P+S +P + + Q Q QQL +
Sbjct: 71 SGLKSPKSSDKQRPLQVPVSVAMMTPQVITPQQMQQILQQQVLSPQQLQALLQQQQAVML 130
Query: 67 QLQQTL------QRSPSMSRLNQIQPQQQQQVVARQQAALYGGQMNFGGSAAVSAQQQQL 120
Q QQ Q + L Q Q QQQQQ +QQ Q QQQQ
Sbjct: 131 QQQQLQEFYKKQQEQLHLQLLQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQH 190
Query: 121 SG 122
G
Sbjct: 191 PG 192
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.309 0.121 0.328
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 61,809,495
Number of Sequences: 164201
Number of extensions: 2597346
Number of successful extensions: 41365
Number of sequences better than 10.0: 881
Number of HSP's better than 10.0 without gapping: 403
Number of HSP's successfully gapped in prelim test: 495
Number of HSP's that attempted gapping in prelim test: 12962
Number of HSP's gapped (non-prelim): 6480
length of query: 570
length of database: 59,974,054
effective HSP length: 116
effective length of query: 454
effective length of database: 40,926,738
effective search space: 18580739052
effective search space used: 18580739052
T: 11
A: 40
X1: 16 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.7 bits)
S2: 68 (30.8 bits)
Medicago: description of AC144645.6


