

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144645.5 - phase: 0
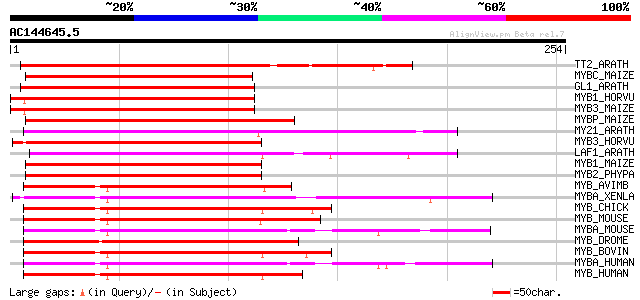
(254 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TT2_ARATH (Q9FJA2) TRANSPARENT TESTA 2 protein (Myb-related prot... 176 5e-44
MYBC_MAIZE (P10290) Anthocyanin regulatory C1 protein 172 9e-43
GL1_ARATH (P27900) Trichome differentiation protein GL1 (GLABROU... 164 2e-40
MYB1_HORVU (P20026) Myb-related protein Hv1 160 4e-39
MYB3_MAIZE (P20025) Myb-related protein Zm38 158 1e-38
MYBP_MAIZE (P27898) Myb-related protein P 157 2e-38
MY21_ARATH (Q9LK95) Transcription factor MYB21 (Myb-related prot... 145 9e-35
MYB3_HORVU (P20027) Myb-related protein Hv33 145 1e-34
LAF1_ARATH (Q9M0K4) Transcription factor LAF1 (Long after far-re... 140 2e-33
MYB1_MAIZE (P20024) Myb-related protein Zm1 139 7e-33
MYB2_PHYPA (P80073) Myb-related protein Pp2 133 4e-31
MYB_AVIMB (P01104) Transforming protein Myb 112 7e-25
MYBA_XENLA (Q05935) Myb-related protein A (A-Myb) (XAMYB) (MYB-r... 112 1e-24
MYB_CHICK (P01103) Myb proto-oncogene protein (C-myb) 110 3e-24
MYB_MOUSE (P06876) Myb proto-oncogene protein (C-myb) 110 3e-24
MYBA_MOUSE (P51960) Myb-related protein A (A-Myb) 108 1e-23
MYB_DROME (P04197) Myb protein 108 2e-23
MYB_BOVIN (P46200) Myb proto-oncogene protein (C-myb) 108 2e-23
MYBA_HUMAN (P10243) Myb-related protein A (A-Myb) 108 2e-23
MYB_HUMAN (P10242) Myb proto-oncogene protein (C-myb) 107 2e-23
>TT2_ARATH (Q9FJA2) TRANSPARENT TESTA 2 protein (Myb-related protein
123) (AtMYB123) (Myb-related transcription factor
LBM2-like)
Length = 258
Score = 176 bits (446), Expect = 5e-44
Identities = 93/186 (50%), Positives = 118/186 (63%), Gaps = 12/186 (6%)
Query: 6 KEVNKGAWSREEDDILSKYVVIHGEGKWQKVAQNAGLKRCGKSCRQRWLNYLKPGIKRGH 65
+E+N+GAW+ ED IL Y+ HGEGKW + AGLKRCGKSCR RW NYL+PGIKRG+
Sbjct: 12 EELNRGAWTDHEDKILRDYITTHGEGKWSTLPNQAGLKRCGKSCRLRWKNYLRPGIKRGN 71
Query: 66 ISTDEEDMIIRLHRLLGNRWSLIAKRLPGRTDNEIKNYWNTNLSKKLQKQPTSSSSLPSP 125
IS+DEE++IIRLH LLGNRWSLIA RLPGRTDNEIKN+WN+NL K+L K T P
Sbjct: 72 ISSDEEELIIRLHNLLGNRWSLIAGRLPGRTDNEIKNHWNSNLRKRLPKTQTKQ---PKR 128
Query: 126 SSVSLRHNHGKCGHVAPEAPKPRRLKAVHQYKILEKNSGS-------EYDQGSDETSIAD 178
S + + C + +A + + + +K+S S E DQG + + D
Sbjct: 129 IKHSTNNENNVC-VIRTKAIRCSKTLLFSDLSLQKKSSTSPLPLKEQEMDQGG-SSLMGD 186
Query: 179 FFIDFD 184
DFD
Sbjct: 187 LEFDFD 192
>MYBC_MAIZE (P10290) Anthocyanin regulatory C1 protein
Length = 273
Score = 172 bits (435), Expect = 9e-43
Identities = 76/104 (73%), Positives = 90/104 (86%)
Query: 8 VNKGAWSREEDDILSKYVVIHGEGKWQKVAQNAGLKRCGKSCRQRWLNYLKPGIKRGHIS 67
V +GAW+ +EDD L+ YV HGEGKW++V Q AGL+RCGKSCR RWLNYL+P I+RG+IS
Sbjct: 12 VKRGAWTSKEDDALAAYVKAHGEGKWREVPQKAGLRRCGKSCRLRWLNYLRPNIRRGNIS 71
Query: 68 TDEEDMIIRLHRLLGNRWSLIAKRLPGRTDNEIKNYWNTNLSKK 111
DEED+IIRLHRLLGNRWSLIA RLPGRTDNEIKNYWN+ L ++
Sbjct: 72 YDEEDLIIRLHRLLGNRWSLIAGRLPGRTDNEIKNYWNSTLGRR 115
>GL1_ARATH (P27900) Trichome differentiation protein GL1 (GLABROUS1
protein)
Length = 228
Score = 164 bits (414), Expect = 2e-40
Identities = 69/107 (64%), Positives = 88/107 (81%)
Query: 6 KEVNKGAWSREEDDILSKYVVIHGEGKWQKVAQNAGLKRCGKSCRQRWLNYLKPGIKRGH 65
+E KG W+ EED+IL YV+ HG G+W ++ + GLKRCGKSCR RW+NYL P + +G+
Sbjct: 12 QEYKKGLWTVEEDNILMDYVLNHGTGQWNRIVRKTGLKRCGKSCRLRWMNYLSPNVNKGN 71
Query: 66 ISTDEEDMIIRLHRLLGNRWSLIAKRLPGRTDNEIKNYWNTNLSKKL 112
+ EED+IIRLH+LLGNRWSLIAKR+PGRTDN++KNYWNT+LSKKL
Sbjct: 72 FTEQEEDLIIRLHKLLGNRWSLIAKRVPGRTDNQVKNYWNTHLSKKL 118
>MYB1_HORVU (P20026) Myb-related protein Hv1
Length = 267
Score = 160 bits (404), Expect = 4e-39
Identities = 72/116 (62%), Positives = 92/116 (79%), Gaps = 4/116 (3%)
Query: 1 MVRSP----KEVNKGAWSREEDDILSKYVVIHGEGKWQKVAQNAGLKRCGKSCRQRWLNY 56
M RSP NKGAW++EEDD L+ Y+ HGEG W+ + + AGL RCGKSCR RW+NY
Sbjct: 1 MGRSPCCEKAHTNKGAWTKEEDDRLTAYIKAHGEGCWRSLPKAAGLLRCGKSCRLRWINY 60
Query: 57 LKPGIKRGHISTDEEDMIIRLHRLLGNRWSLIAKRLPGRTDNEIKNYWNTNLSKKL 112
L+P +KRG+ S +E+++II+LH LLGN+WSLIA RLPGRTDNEIKNYWNT++ +KL
Sbjct: 61 LRPDLKRGNFSHEEDELIIKLHSLLGNKWSLIAGRLPGRTDNEIKNYWNTHIRRKL 116
>MYB3_MAIZE (P20025) Myb-related protein Zm38
Length = 255
Score = 158 bits (399), Expect = 1e-38
Identities = 70/116 (60%), Positives = 91/116 (78%), Gaps = 4/116 (3%)
Query: 1 MVRSP----KEVNKGAWSREEDDILSKYVVIHGEGKWQKVAQNAGLKRCGKSCRQRWLNY 56
M RSP N+GAW++EED+ L Y+ HGEG W+ + + AGL RCGKSCR RW+NY
Sbjct: 1 MGRSPCCEKAHTNRGAWTKEEDERLVAYIRAHGEGCWRSLPKAAGLLRCGKSCRLRWINY 60
Query: 57 LKPGIKRGHISTDEEDMIIRLHRLLGNRWSLIAKRLPGRTDNEIKNYWNTNLSKKL 112
L+P +KRG+ + DE+D+I++LH LLGN+WSLIA RLPGRTDNEIKNYWNT++ +KL
Sbjct: 61 LRPDLKRGNFTADEDDLIVKLHSLLGNKWSLIAARLPGRTDNEIKNYWNTHVRRKL 116
>MYBP_MAIZE (P27898) Myb-related protein P
Length = 399
Score = 157 bits (398), Expect = 2e-38
Identities = 66/123 (53%), Positives = 94/123 (75%)
Query: 8 VNKGAWSREEDDILSKYVVIHGEGKWQKVAQNAGLKRCGKSCRQRWLNYLKPGIKRGHIS 67
+ +G W+ EED +L+ Y+ HGEG W+ + +NAGL RCGKSCR RW+NYL+ +KRG+IS
Sbjct: 12 LKRGRWTAEEDQLLANYIAEHGEGSWRSLPKNAGLLRCGKSCRLRWINYLRADVKRGNIS 71
Query: 68 TDEEDMIIRLHRLLGNRWSLIAKRLPGRTDNEIKNYWNTNLSKKLQKQPTSSSSLPSPSS 127
+EED+II+LH LGNRWSLIA LPGRTDNEIKNYWN++LS+++ ++ P ++
Sbjct: 72 KEEEDIIIKLHATLGNRWSLIASHLPGRTDNEIKNYWNSHLSRQIHTYRRKYTAGPDDTA 131
Query: 128 VSL 130
+++
Sbjct: 132 IAI 134
>MY21_ARATH (Q9LK95) Transcription factor MYB21 (Myb-related protein
21) (AtMYB21) (Myb homolog 3) (AtMyb3)
Length = 226
Score = 145 bits (366), Expect = 9e-35
Identities = 76/200 (38%), Positives = 114/200 (57%), Gaps = 4/200 (2%)
Query: 7 EVNKGAWSREEDDILSKYVVIHGEGKWQKVAQNAGLKRCGKSCRQRWLNYLKPGIKRGHI 66
EV KG W+ EED IL Y+ HG+G W +A++AGLKR GKSCR RWLNYL+P ++RG+I
Sbjct: 19 EVRKGPWTMEEDLILINYIANHGDGVWNSLAKSAGLKRTGKSCRLRWLNYLRPDVRRGNI 78
Query: 67 STDEEDMIIRLHRLLGNRWSLIAKRLPGRTDNEIKNYWNTNLSKKL-QKQPTSSSSLPSP 125
+ +E+ +I+ LH GNRWS IAK LPGRTDNEIKN+W T + K + Q T++SS+ S
Sbjct: 79 TPEEQLIIMELHAKWGNRWSKIAKHLPGRTDNEIKNFWRTRIQKYIKQSDVTTTSSVGSH 138
Query: 126 SSVSLRHNHGKCGHVAPEAPKPRRLKAVHQYKILEKNSGSEYDQGSDETSIADFFIDFDH 185
S + + + ++ +++ E++ G+ + +D+
Sbjct: 139 HSSEINDQAASTSSHNVFCTQDQAMETYSPTPTSYQHTNMEFNYGNYSAAAVTATVDYPV 198
Query: 186 QDQLMVGDDESNSKIPQMED 205
M DD++ M+D
Sbjct: 199 P---MTVDDQTGENYWGMDD 215
>MYB3_HORVU (P20027) Myb-related protein Hv33
Length = 302
Score = 145 bits (365), Expect = 1e-34
Identities = 67/114 (58%), Positives = 83/114 (72%), Gaps = 1/114 (0%)
Query: 2 VRSPKEVNKGAWSREEDDILSKYVVIHGEGKWQKVAQNAGLKRCGKSCRQRWLNYLKPGI 61
V PK V KG WS EED+ L +++ HG G W V + A L RCGKSCR RW+NYL+P +
Sbjct: 9 VGQPK-VRKGLWSPEEDEKLYNHIIRHGVGCWSSVPRLAALNRCGKSCRLRWINYLRPDL 67
Query: 62 KRGHISTDEEDMIIRLHRLLGNRWSLIAKRLPGRTDNEIKNYWNTNLSKKLQKQ 115
KRG S EED I+ LH++LGNRWS IA LPGRTDNEIKN+WN+ + KKL++Q
Sbjct: 68 KRGCFSQQEEDHIVALHQILGNRWSQIASHLPGRTDNEIKNFWNSCIKKKLRQQ 121
>LAF1_ARATH (Q9M0K4) Transcription factor LAF1 (Long after far-red
light protein 1) (Myb-related protein 18) (AtMYB18)
Length = 283
Score = 140 bits (354), Expect = 2e-33
Identities = 86/212 (40%), Positives = 113/212 (52%), Gaps = 20/212 (9%)
Query: 10 KGAWSREEDDILSKYVVIHGEGKWQKVAQNAGLKRCGKSCRQRWLNYLKPGIKRGHISTD 69
KG WS EED+ L +++ +G W V AGL+R GKSCR RW+NYL+PG+KR IS +
Sbjct: 12 KGLWSPEEDEKLRSFILSYGHSCWTTVPIKAGLQRNGKSCRLRWINYLRPGLKRDMISAE 71
Query: 70 EEDMIIRLHRLLGNRWSLIAKRLPGRTDNEIKNYWNTNLSKKLQK----QPTSSSSLPSP 125
EE+ I+ H LGN+WS IAK LPGRTDNEIKNYW+++L KK K Q S S PS
Sbjct: 72 EEETILTFHSSLGNKWSQIAKFLPGRTDNEIKNYWHSHLKKKWLKSQSLQDAKSISPPSS 131
Query: 126 SSVSLRHNHGKCGHVAPEAP------KPRRLKAVHQYKILEKNSGSEYDQGSDETSIADF 179
SS SL CG PE +RL ++++G+ Q S I
Sbjct: 132 SSSSL----VACGKRNPETLISNHVFSFQRLLENKSSSPSQESNGNNSHQCSSAPEIPRL 187
Query: 180 FI------DFDHQDQLMVGDDESNSKIPQMED 205
F + H D D +S+ P +E+
Sbjct: 188 FFSEWLSSSYPHTDYSSEFTDSKHSQAPNVEE 219
>MYB1_MAIZE (P20024) Myb-related protein Zm1
Length = 340
Score = 139 bits (350), Expect = 7e-33
Identities = 61/108 (56%), Positives = 82/108 (75%)
Query: 8 VNKGAWSREEDDILSKYVVIHGEGKWQKVAQNAGLKRCGKSCRQRWLNYLKPGIKRGHIS 67
+N+G+W+ +ED L Y+ HG W+ + + AGL RCGKSCR RW+NYL+P +KRG+ +
Sbjct: 14 LNRGSWTPQEDMRLIAYIQKHGHTNWRALPKQAGLLRCGKSCRLRWINYLRPDLKRGNFT 73
Query: 68 TDEEDMIIRLHRLLGNRWSLIAKRLPGRTDNEIKNYWNTNLSKKLQKQ 115
+EE+ IIRLH LLGN+WS IA LPGRTDNEIKN WNT+L KK+ ++
Sbjct: 74 DEEEEAIIRLHGLLGNKWSKIAACLPGRTDNEIKNVWNTHLKKKVAQR 121
>MYB2_PHYPA (P80073) Myb-related protein Pp2
Length = 421
Score = 133 bits (335), Expect = 4e-31
Identities = 60/108 (55%), Positives = 77/108 (70%)
Query: 8 VNKGAWSREEDDILSKYVVIHGEGKWQKVAQNAGLKRCGKSCRQRWLNYLKPGIKRGHIS 67
+ +G W+ EED L ++ +G W+ + + AGL RCGKSCR RW NYL+P +KRG S
Sbjct: 12 LRRGPWTSEEDQKLVSHITNNGLSCWRAIPKLAGLLRCGKSCRLRWTNYLRPDLKRGIFS 71
Query: 68 TDEEDMIIRLHRLLGNRWSLIAKRLPGRTDNEIKNYWNTNLSKKLQKQ 115
EE++I+ LH LGNRWS IA +LPGRTDNEIKNYWNT L K+L+ Q
Sbjct: 72 EAEENLILDLHATLGNRWSRIAAQLPGRTDNEIKNYWNTRLKKRLRSQ 119
>MYB_AVIMB (P01104) Transforming protein Myb
Length = 382
Score = 112 bits (281), Expect = 7e-25
Identities = 53/126 (42%), Positives = 86/126 (68%), Gaps = 5/126 (3%)
Query: 7 EVNKGAWSREEDDILSKYVVIHGEGKWQKVAQNAGLK-RCGKSCRQRWLNYLKPGIKRGH 65
E+NKG W++EED + ++V +G +W +A++ LK R GK CR+RW N+L P +K+
Sbjct: 18 ELNKGPWTKEEDQRVIEHVQKYGPKRWSDIAKH--LKGRIGKQCRERWHNHLNPEVKKTS 75
Query: 66 ISTDEEDMIIRLHRLLGNRWSLIAKRLPGRTDNEIKNYWNTNLSKKLQKQ--PTSSSSLP 123
+ +E+ +I + H+ LGNRW+ IAK LPGRTDN +KN+WN+ + +K++++ P SS
Sbjct: 76 WTEEEDRIIYQAHKRLGNRWAEIAKLLPGRTDNAVKNHWNSTMRRKVEQEGYPQESSKAG 135
Query: 124 SPSSVS 129
PS+ +
Sbjct: 136 PPSATT 141
>MYBA_XENLA (Q05935) Myb-related protein A (A-Myb) (XAMYB)
(MYB-related protein 2) (XMYB2)
Length = 728
Score = 112 bits (279), Expect = 1e-24
Identities = 71/223 (31%), Positives = 113/223 (49%), Gaps = 15/223 (6%)
Query: 2 VRSPKEVNKGAWSREEDDILSKYVVIHGEGKWQKVAQNAGLK-RCGKSCRQRWLNYLKPG 60
V SP E+ KG W++EED + + V +G KW +A++ LK R GK CR+RW N+L P
Sbjct: 79 VLSP-ELVKGPWTKEEDQRVIELVHKYGPKKWSIIAKH--LKGRIGKQCRERWHNHLNPD 135
Query: 61 IKRGHISTDEEDMIIRLHRLLGNRWSLIAKRLPGRTDNEIKNYWNTNLSKKLQKQPTSSS 120
+K+ + +E+ +I H+ +GNRW+ IAK LPGRTDN IKN+WN+ + +K++++
Sbjct: 136 VKKSSWTEEEDRIIYSAHKRMGNRWAEIAKLLPGRTDNSIKNHWNSTMKRKVEQEGYLQD 195
Query: 121 SLPSPSSVSLRHNHGKCGHVAPEAPKPRRLKAVHQYKILEKNSGSEYDQGSDETSIADFF 180
+ L+ A P L+A +Q+ I + Y S + + +
Sbjct: 196 LMNCDRPSKLQ---------AKSCAAPNHLQAQNQFYIPVQTQIPRYSSLSHDDNCIEIQ 246
Query: 181 IDFDHQDQLMV--GDDESNSKIPQMEDHKVSSTNSTHSSSSPS 221
F Q V D E +I ++E +S+ N PS
Sbjct: 247 NSFSFIQQPFVDADDPEKEKRIKELELLLMSAENEVRRKRVPS 289
Score = 33.1 bits (74), Expect = 0.66
Identities = 17/67 (25%), Positives = 33/67 (48%), Gaps = 2/67 (2%)
Query: 69 DEEDMIIRLHRLLGNRWSLIAKRLPGRTDNEIKNYWNTNLSKKLQKQPTSSSSLPSPSSV 128
DE+D + +L G W ++A+ R++ + ++ W+ LS +L K P + +
Sbjct: 41 DEDDKVKKLVEKHGEDWGVVARHFINRSEVQCQHRWHKVLSPELVKGPWTKEE--DQRVI 98
Query: 129 SLRHNHG 135
L H +G
Sbjct: 99 ELVHKYG 105
>MYB_CHICK (P01103) Myb proto-oncogene protein (C-myb)
Length = 641
Score = 110 bits (276), Expect = 3e-24
Identities = 58/147 (39%), Positives = 92/147 (62%), Gaps = 8/147 (5%)
Query: 7 EVNKGAWSREEDDILSKYVVIHGEGKWQKVAQNAGLK-RCGKSCRQRWLNYLKPGIKRGH 65
E+ KG W++EED + + V +G +W +A++ LK R GK CR+RW N+L P +K+
Sbjct: 89 ELIKGPWTKEEDQRVIELVQKYGPKRWSVIAKH--LKGRIGKQCRERWHNHLNPEVKKTS 146
Query: 66 ISTDEEDMIIRLHRLLGNRWSLIAKRLPGRTDNEIKNYWNTNLSKKLQK----QPTSSSS 121
+ +E+ +I + H+ LGNRW+ IAK LPGRTDN IKN+WN+ + +K+++ Q +S +
Sbjct: 147 WTEEEDRIIYQAHKRLGNRWAEIAKLLPGRTDNAIKNHWNSTMRRKVEQEGYLQESSKAG 206
Query: 122 LPSPSSVSLRHNHGKC-GHVAPEAPKP 147
LPS ++ + +H H P P P
Sbjct: 207 LPSATTGFQKSSHLMAFAHNPPAGPLP 233
Score = 30.8 bits (68), Expect = 3.3
Identities = 26/99 (26%), Positives = 41/99 (41%), Gaps = 21/99 (21%)
Query: 69 DEEDMIIRLHRLLGNR-WSLIAKRLPGRTDNEIKNYWNTNLSKKLQKQPTSSSS------ 121
+E++ + +L G W +IA LP RTD + ++ W L+ +L K P +
Sbjct: 46 EEDEKLKKLVEQNGTEDWKVIASFLPNRTDVQCQHRWQKVLNPELIKGPWTKEEDQRVIE 105
Query: 122 -----LPSPSSVSLRHNHGKCG---------HVAPEAPK 146
P SV +H G+ G H+ PE K
Sbjct: 106 LVQKYGPKRWSVIAKHLKGRIGKQCRERWHNHLNPEVKK 144
>MYB_MOUSE (P06876) Myb proto-oncogene protein (C-myb)
Length = 636
Score = 110 bits (275), Expect = 3e-24
Identities = 55/142 (38%), Positives = 90/142 (62%), Gaps = 8/142 (5%)
Query: 7 EVNKGAWSREEDDILSKYVVIHGEGKWQKVAQNAGLK-RCGKSCRQRWLNYLKPGIKRGH 65
E+ KG W++EED + + V +G +W +A++ LK R GK CR+RW N+L P +K+
Sbjct: 89 ELIKGPWTKEEDQRVIELVQKYGPKRWSVIAKH--LKGRIGKQCRERWHNHLNPEVKKTS 146
Query: 66 ISTDEEDMIIRLHRLLGNRWSLIAKRLPGRTDNEIKNYWNTNLSKKLQ-----KQPTSSS 120
+ +E+ +I + H+ LGNRW+ IAK LPGRTDN IKN+WN+ + +K++ ++P+ +S
Sbjct: 147 WTEEEDRIIYQAHKRLGNRWAEIAKLLPGRTDNAIKNHWNSTMRRKVEQEGYLQEPSKAS 206
Query: 121 SLPSPSSVSLRHNHGKCGHVAP 142
P +S ++ GH +P
Sbjct: 207 QTPVATSFQKNNHLMGFGHASP 228
Score = 30.8 bits (68), Expect = 3.3
Identities = 23/82 (28%), Positives = 33/82 (40%), Gaps = 20/82 (24%)
Query: 85 WSLIAKRLPGRTDNEIKNYWNTNLSKKLQKQPTSSSS-----------LPSPSSVSLRHN 133
W +IA LP RTD + ++ W L+ +L K P + P SV +H
Sbjct: 63 WKVIANYLPNRTDVQCQHRWQKVLNPELIKGPWTKEEDQRVIELVQKYGPKRWSVIAKHL 122
Query: 134 HGKCG---------HVAPEAPK 146
G+ G H+ PE K
Sbjct: 123 KGRIGKQCRERWHNHLNPEVKK 144
>MYBA_MOUSE (P51960) Myb-related protein A (A-Myb)
Length = 751
Score = 108 bits (270), Expect = 1e-23
Identities = 70/218 (32%), Positives = 112/218 (51%), Gaps = 18/218 (8%)
Query: 7 EVNKGAWSREEDDILSKYVVIHGEGKWQKVAQNAGLK-RCGKSCRQRWLNYLKPGIKRGH 65
E+ KG W++EED + + V +G +W +A++ LK R GK CR+RW N+L P +K+
Sbjct: 84 ELIKGPWTKEEDQRVIELVQKYGPKRWSLIAKH--LKGRIGKQCRERWHNHLNPEVKKSS 141
Query: 66 ISTDEEDMIIRLHRLLGNRWSLIAKRLPGRTDNEIKNYWNTNLSKKLQKQPTSSSSLPSP 125
+ +E+ +I H+ LGNRW+ IAK LPGRTDN IKN+WN+ + +K++++ + S
Sbjct: 142 WTEEEDRIIYEAHKRLGNRWAEIAKLLPGRTDNSIKNHWNSTMRRKVEQEGYLQDGIKSE 201
Query: 126 SSVSLRHNHGKCGHVAPEAPKPRRLKAVHQYKILEKNSGSEY---DQGSDETSIADFFID 182
S S + H C + L+ +Q+ I + G +Y D E FI
Sbjct: 202 RS-SSKLQHKPCATM-------DHLQTQNQFYIPVQIPGYQYVSPDGNCVEHVQTSAFIQ 253
Query: 183 FDHQDQLMVGDDESNSKIPQMEDHKVSSTNSTHSSSSP 220
D+ D + KI ++E +S+ N P
Sbjct: 254 QPFVDE----DPDKEKKIKELELLLMSAENEVRRKRLP 287
Score = 31.6 bits (70), Expect = 1.9
Identities = 30/108 (27%), Positives = 50/108 (45%), Gaps = 22/108 (20%)
Query: 10 KGAWSREEDDILSKYVVIHGEGKWQKVAQNAGLKRCGKSCRQRWLNYLKPGIKRGHISTD 69
K + S +EDD L + + ++ V Q GLK+ R +W + D
Sbjct: 3 KRSRSEDEDDDLQ-----YADHDYE-VPQQKGLKKLWN--RVKW-------------TRD 41
Query: 70 EEDMIIRLHRLLG-NRWSLIAKRLPGRTDNEIKNYWNTNLSKKLQKQP 116
E+D + +L G + W+LIA L R+D + ++ W L+ +L K P
Sbjct: 42 EDDKLKKLVEQHGTDDWTLIASHLQNRSDFQCQHRWQKVLNPELIKGP 89
>MYB_DROME (P04197) Myb protein
Length = 657
Score = 108 bits (269), Expect = 2e-23
Identities = 54/126 (42%), Positives = 78/126 (61%), Gaps = 1/126 (0%)
Query: 7 EVNKGAWSREEDDILSKYVVIHGEGKWQKVAQNAGLKRCGKSCRQRWLNYLKPGIKRGHI 66
E+ KG W+R+EDD++ K V G KW +A+ R GK CR+RW N+L P IK+
Sbjct: 133 ELIKGPWTRDEDDMVIKLVRNFGPKKWTLIARYLN-GRIGKQCRERWHNHLNPNIKKTAW 191
Query: 67 STDEEDMIIRLHRLLGNRWSLIAKRLPGRTDNEIKNYWNTNLSKKLQKQPTSSSSLPSPS 126
+ E+++I + H LGN+W+ IAKRLPGRTDN IKN+WN+ + +K + S ++ S
Sbjct: 192 TEKEDEIIYQAHLELGNQWAKIAKRLPGRTDNAIKNHWNSTMRRKYDVERRSVNASGSDL 251
Query: 127 SVSLRH 132
S H
Sbjct: 252 KSSRTH 257
>MYB_BOVIN (P46200) Myb proto-oncogene protein (C-myb)
Length = 640
Score = 108 bits (269), Expect = 2e-23
Identities = 57/147 (38%), Positives = 93/147 (62%), Gaps = 8/147 (5%)
Query: 7 EVNKGAWSREEDDILSKYVVIHGEGKWQKVAQNAGLK-RCGKSCRQRWLNYLKPGIKRGH 65
E+ KG W++EED + + V +G +W +A++ LK R GK CR+RW N+L P +K+
Sbjct: 89 ELIKGPWTKEEDQRVIELVQKYGPKRWSVIAKH--LKGRIGKQCRERWHNHLNPEVKKTS 146
Query: 66 ISTDEEDMIIRLHRLLGNRWSLIAKRLPGRTDNEIKNYWNTNLSKKLQK----QPTSSSS 121
+ +E+ +I + H+ LGNRW+ IAK LPGRTDN IKN+WN+ + +K+++ Q +S +S
Sbjct: 147 WTEEEDRIIYQAHKRLGNRWAEIAKLLPGRTDNAIKNHWNSTMRRKVEQEGYLQESSKAS 206
Query: 122 LPSPSSVSLRHNH-GKCGHVAPEAPKP 147
P+ ++ +++H H P A P
Sbjct: 207 QPAVTTSFQKNSHLMGFTHAPPSAQLP 233
Score = 30.8 bits (68), Expect = 3.3
Identities = 23/82 (28%), Positives = 33/82 (40%), Gaps = 20/82 (24%)
Query: 85 WSLIAKRLPGRTDNEIKNYWNTNLSKKLQKQPTSSSS-----------LPSPSSVSLRHN 133
W +IA LP RTD + ++ W L+ +L K P + P SV +H
Sbjct: 63 WKVIANYLPNRTDVQCQHRWQKVLNPELIKGPWTKEEDQRVIELVQKYGPKRWSVIAKHL 122
Query: 134 HGKCG---------HVAPEAPK 146
G+ G H+ PE K
Sbjct: 123 KGRIGKQCRERWHNHLNPEVKK 144
>MYBA_HUMAN (P10243) Myb-related protein A (A-Myb)
Length = 752
Score = 108 bits (269), Expect = 2e-23
Identities = 69/220 (31%), Positives = 115/220 (51%), Gaps = 19/220 (8%)
Query: 7 EVNKGAWSREEDDILSKYVVIHGEGKWQKVAQNAGLK-RCGKSCRQRWLNYLKPGIKRGH 65
E+ KG W++EED + + V +G +W +A++ LK R GK CR+RW N+L P +K+
Sbjct: 84 ELIKGPWTKEEDQRVIELVQKYGPKRWSLIAKH--LKGRIGKQCRERWHNHLNPEVKKSS 141
Query: 66 ISTDEEDMIIRLHRLLGNRWSLIAKRLPGRTDNEIKNYWNTNLSKKLQKQPTSSSSLPSP 125
+ +E+ +I H+ LGNRW+ IAK LPGRTDN IKN+WN+ + +K++++ + S
Sbjct: 142 WTEEEDRIIYEAHKRLGNRWAEIAKLLPGRTDNSIKNHWNSTMRRKVEQEGYLQDGIKSE 201
Query: 126 SSVSLRHNHGKCGHVAPEAPKPRRLKAVHQYKILEKNSGSEY--DQGS--DETSIADFFI 181
S S + H C + ++ +Q+ I + G +Y +G+ + FI
Sbjct: 202 RS-SSKLQHKPCAAM-------DHMQTQNQFYIPVQIPGYQYVSPEGNCIEHVQPTSAFI 253
Query: 182 DFDHQDQLMVGDDESNSKIPQMEDHKVSSTNSTHSSSSPS 221
Q + D + KI ++E +S+ N PS
Sbjct: 254 ----QQPFIDEDPDKEKKIKELEMLLMSAENEVRRKRIPS 289
Score = 31.6 bits (70), Expect = 1.9
Identities = 30/108 (27%), Positives = 50/108 (45%), Gaps = 22/108 (20%)
Query: 10 KGAWSREEDDILSKYVVIHGEGKWQKVAQNAGLKRCGKSCRQRWLNYLKPGIKRGHISTD 69
K + S +EDD L + + ++ V Q GLK+ R +W + D
Sbjct: 3 KRSRSEDEDDDLQ-----YADHDYE-VPQQKGLKKLWN--RVKW-------------TRD 41
Query: 70 EEDMIIRLHRLLG-NRWSLIAKRLPGRTDNEIKNYWNTNLSKKLQKQP 116
E+D + +L G + W+LIA L R+D + ++ W L+ +L K P
Sbjct: 42 EDDKLKKLVEQHGTDDWTLIASHLQNRSDFQCQHRWQKVLNPELIKGP 89
>MYB_HUMAN (P10242) Myb proto-oncogene protein (C-myb)
Length = 640
Score = 107 bits (268), Expect = 2e-23
Identities = 53/133 (39%), Positives = 89/133 (66%), Gaps = 7/133 (5%)
Query: 7 EVNKGAWSREEDDILSKYVVIHGEGKWQKVAQNAGLK-RCGKSCRQRWLNYLKPGIKRGH 65
E+ KG W++EED + + V +G +W +A++ LK R GK CR+RW N+L P +K+
Sbjct: 89 ELIKGPWTKEEDQRVIELVQKYGPKRWSVIAKH--LKGRIGKQCRERWHNHLNPEVKKTS 146
Query: 66 ISTDEEDMIIRLHRLLGNRWSLIAKRLPGRTDNEIKNYWNTNLSKKLQK----QPTSSSS 121
+ +E+ +I + H+ LGNRW+ IAK LPGRTDN IKN+WN+ + +K+++ Q +S +S
Sbjct: 147 WTEEEDRIIYQAHKRLGNRWAEIAKLLPGRTDNAIKNHWNSTMRRKVEQEGYLQESSKAS 206
Query: 122 LPSPSSVSLRHNH 134
P+ ++ +++H
Sbjct: 207 QPAVATSFQKNSH 219
Score = 30.8 bits (68), Expect = 3.3
Identities = 23/82 (28%), Positives = 33/82 (40%), Gaps = 20/82 (24%)
Query: 85 WSLIAKRLPGRTDNEIKNYWNTNLSKKLQKQPTSSSS-----------LPSPSSVSLRHN 133
W +IA LP RTD + ++ W L+ +L K P + P SV +H
Sbjct: 63 WKVIANYLPNRTDVQCQHRWQKVLNPELIKGPWTKEEDQRVIELVQKYGPKRWSVIAKHL 122
Query: 134 HGKCG---------HVAPEAPK 146
G+ G H+ PE K
Sbjct: 123 KGRIGKQCRERWHNHLNPEVKK 144
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.313 0.131 0.387
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 32,935,685
Number of Sequences: 164201
Number of extensions: 1422412
Number of successful extensions: 3339
Number of sequences better than 10.0: 62
Number of HSP's better than 10.0 without gapping: 39
Number of HSP's successfully gapped in prelim test: 23
Number of HSP's that attempted gapping in prelim test: 3187
Number of HSP's gapped (non-prelim): 114
length of query: 254
length of database: 59,974,054
effective HSP length: 108
effective length of query: 146
effective length of database: 42,240,346
effective search space: 6167090516
effective search space used: 6167090516
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 64 (29.3 bits)
Medicago: description of AC144645.5


