

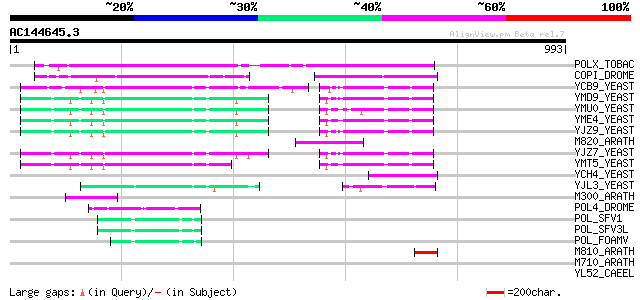
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144645.3 + phase: 0 /pseudo
(993 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
POLX_TOBAC (P10978) Retrovirus-related Pol polyprotein from tran... 342 4e-93
COPI_DROME (P04146) Copia protein (Gag-int-pol protein) [Contain... 162 4e-39
YCB9_YEAST (P25384) Transposon Ty2 protein B (Ty1-17 protein B) 109 3e-23
YMD9_YEAST (Q03434) Transposon Ty1 protein B 104 1e-21
YMU0_YEAST (Q04670) Transposon Ty1 protein B 103 2e-21
YME4_YEAST (Q04711) Transposon Ty1 protein B 103 2e-21
YJZ9_YEAST (P47100) Transposon Ty1 protein B 103 2e-21
M820_ARATH (P92520) Hypothetical mitochondrial protein AtMg00820... 102 5e-21
YJZ7_YEAST (P47098) Transposon Ty1 protein B 101 9e-21
YMT5_YEAST (Q04214) Transposon Ty1 protein B 101 1e-20
YCH4_YEAST (P25600) Transposon Ty5-1 34.5 kDa hypothetical protein 85 8e-16
YJL3_YEAST (P47024) Transposon Ty4 207.7 kDa hypothetical protein 73 3e-12
M300_ARATH (P93293) Hypothetical mitochondrial protein AtMg00300... 71 2e-11
POL4_DROME (P10394) Retrovirus-related Pol polyprotein from tran... 59 5e-08
POL_SFV1 (P23074) Pol polyprotein [Contains: Protease (EC 3.4.23... 58 1e-07
POL_SFV3L (P27401) Pol polyprotein [Contains: Protease (EC 3.4.2... 57 2e-07
POL_FOAMV (P14350) Pol polyprotein [Contains: Reverse transcript... 56 5e-07
M810_ARATH (P92519) Hypothetical mitochondrial protein AtMg00810... 50 4e-05
M710_ARATH (P92512) Hypothetical mitochondrial protein AtMg00710... 44 0.002
YL52_CAEEL (P34431) Hypothetical protein F44E2.2 in chromosome III 43 0.005
>POLX_TOBAC (P10978) Retrovirus-related Pol polyprotein from
transposon TNT 1-94 [Contains: Protease (EC 3.4.23.-);
Reverse transcriptase (EC 2.7.7.49); Endonuclease]
Length = 1328
Score = 342 bits (876), Expect = 4e-93
Identities = 229/729 (31%), Positives = 350/729 (47%), Gaps = 50/729 (6%)
Query: 45 LSLNNVLHAPKLIKNLIFVRRFTIDNNVTIEFDPFSFSVED-----IQTGIPLMRCDSTG 99
L L +V H P L NLI + + ++ D + + + + + + + G
Sbjct: 348 LVLKDVRHVPDLRMNLI--------SGIALDRDGYESYFANQKWRLTKGSLVIAKGVARG 399
Query: 100 DLYPLTTTTSHQSTSPATFAALSPTLWHNRLGHPGANVLSFLNKNKFIECKQISSPSICQ 159
LY T Q A +S LWH R+GH L L K I + ++ C
Sbjct: 400 TLYR-TNAEICQGELNAAQDEISVDLWHKRMGHMSEKGLQILAKKSLISYAKGTTVKPCD 458
Query: 160 SCIYGKHVKLPFSISNSTTSKPFDIIHSDLW-TSPILSSAGHKYYLFFLDDYTNFVWTFP 218
C++GK ++ F S+ D+++SD+ I S G+KY++ F+DD + +W +
Sbjct: 459 YCLFGKQHRVSFQTSSERKLNILDLVYSDVCGPMEIESMGGNKYFVTFIDDASRKLWVYI 518
Query: 219 IGRKSQVPSIFSSFHAFIKTQFGTTIKCFQCDNGTEFNNEYFTQFCHKNGMVFRFSCPHT 278
+ K QV +F FHA ++ + G +K + DNG E+ + F ++C +G+ + P T
Sbjct: 519 LKTKDQVFQVFQKFHALVERETGRKLKRLRSDNGGEYTSREFEEYCSSHGIRHEKTVPGT 578
Query: 279 SPQNGKAERKIRAINNFIRTSLAHSSMPPSFWHHALQITTYLQNILPSKILSHHSPTQYL 338
NG AER R I +R+ L + +P SFW A+Q YL N PS L+ P +
Sbjct: 579 PQHNGVAERMNRTIVEKVRSMLRMAKLPKSFWGEAVQTACYLINRSPSVPLAFEIPERVW 638
Query: 339 YHRDPSYTHLRVFGCLCYPLFPSTAINKLQARSTPCAFLGYPQNHRGYKCYDLSSKKIII 398
+++ SY+HL+VFGC + P KL +S PC F+GY GY+ +D KK+I
Sbjct: 639 TNKEVSYSHLKVFGCRAFAHVPKEQRTKLDDKSIPCIFIGYGDEEFGYRLWDPVKKKVIR 698
Query: 399 SRHVIFDETQFPFAKTHTSSTHTYEFLNDSLHPLLHYHLQNDPKQDEPEPRKIESPQPAT 458
SR V+F E++ T++ + + N + P + P +
Sbjct: 699 SRDVVFRESE-----VRTAADMSEKVKNGII------------------PNFVTIPSTSN 735
Query: 459 TPASPINVTNQ-SILPPSPMSINQLPHPLVS--TELTSPTHTPQQIHQEPPRTIATHSMH 515
P S + T++ S P + + L E+ PT +Q +P R +
Sbjct: 736 NPTSAESTTDEVSEQGEQPGEVIEQGEQLDEGVEEVEHPTQGEEQ--HQPLRRSERPRVE 793
Query: 516 GIHKPKIQFNLTTSITSSPLPHNPKAALS---DSNWKAAMLDEFDALIKNKTWELVPRPQ 572
P ++ L I+ P + K LS + AM +E ++L KN T++LV P+
Sbjct: 794 SRRYPSTEYVL---ISDDREPESLKEVLSHPEKNQLMKAMQEEMESLQKNGTYKLVELPK 850
Query: 573 NVNFIRSMWIFRHKKKSDGSFERYKARLVGDGRSQ*VGVDCDETFSPVVKPTTIRIVLTI 632
++ W+F+ KK D RYKARLV G Q G+D DE FSPVVK T+IR +L++
Sbjct: 851 GKRPLKCKWVFKLKKDGDCKLVRYKARLVVKGFEQKKGIDFDEIFSPVVKMTSIRTILSL 910
Query: 633 ALSKSWPIHQLDVKNVFLHGELQETVYMHQPMGFRDPIHPDYVCLLKKSLYGLKQAPRAW 692
A S + QLDVK FLHG+L+E +YM QP GF VC L KSLYGLKQAPR W
Sbjct: 911 AASLDLEVEQLDVKTAFLHGDLEEEIYMEQPEGFEVAGKKHMVCKLNKSLYGLKQAPRQW 970
Query: 693 YQRFADFAFTIGFSHSKSDHSLFIYR-KGNDMTYILLYVDDIILTASSDVLRRSIMSLLA 751
Y +F F + + + SD ++ R N+ +LLYVDD+++ L + L+
Sbjct: 971 YMKFDSFMKSQTYLKTYSDPCVYFKRFSENNFIILLLYVDDMLIVGKDKGLIAKLKGDLS 1030
Query: 752 SEFAMKDLG 760
F MKDLG
Sbjct: 1031 KSFDMKDLG 1039
>COPI_DROME (P04146) Copia protein (Gag-int-pol protein) [Contains:
Copia VLP protein; Copia protease (EC 3.4.23.-)]
Length = 1409
Score = 162 bits (410), Expect = 4e-39
Identities = 119/395 (30%), Positives = 191/395 (48%), Gaps = 24/395 (6%)
Query: 45 LSLNNVLHAPKLIKNLIFVRRFTIDNNVTIEFDPFSFSVEDIQTGIPLMRCDSTGDLYPL 104
++L +VL + NL+ V+R + ++IEFD ++ + G+ +++ + P+
Sbjct: 343 ITLEDVLFCKEAAGNLMSVKRLQ-EAGMSIEFDKSGVTIS--KNGLMVVKNSGMLNNVPV 399
Query: 105 TTTTSHQSTSPATFAALSPTLWHNRLGH-PGANVLSFLNKNKFIECKQIS----SPSICQ 159
+ Q+ S + LWH R GH +L KN F + ++ S IC+
Sbjct: 400 I---NFQAYSINAKHKNNFRLWHERFGHISDGKLLEIKRKNMFSDQSLLNNLELSCEICE 456
Query: 160 SCIYGKHVKLPFSISNSTT--SKPFDIIHSDLWTSPI--LSSAGHKYYLFFLDDYTNFVW 215
C+ GK +LPF T +P ++HSD+ PI ++ Y++ F+D +T++
Sbjct: 457 PCLNGKQARLPFKQLKDKTHIKRPLFVVHSDV-CGPITPVTLDDKNYFVIFVDQFTHYCV 515
Query: 216 TFPIGRKSQVPSIFSSFHAFIKTQFGTTIKCFQCDNGTEFNNEYFTQFCHKNGMVFRFSC 275
T+ I KS V S+F F A + F + DNG E+ + QFC K G+ + +
Sbjct: 516 TYLIKYKSDVFSMFQDFVAKSEAHFNLKVVYLYIDNGREYLSNEMRQFCVKKGISYHLTV 575
Query: 276 PHTSPQNGKAERKIRAINNFIRTSLAHSSMPPSFWHHALQITTYLQNILPSKILSHHSPT 335
PHT NG +ER IR I RT ++ + + SFW A+ TYL N +PS+ L S T
Sbjct: 576 PHTPQLNGVSERMIRTITEKARTMVSGAKLDKSFWGEAVLTATYLINRIPSRALVDSSKT 635
Query: 336 QY--LYHRDPSYTHLRVFGCLCYPLFPSTAINKLQARSTPCAFLGYPQNHRGYKCYDLSS 393
Y +++ P HLRVFG Y + K +S F+GY N G+K +D +
Sbjct: 636 PYEMWHNKKPYLKHLRVFGATVY-VHIKNKQGKFDDKSFKSIFVGYEPN--GFKLWDAVN 692
Query: 394 KKIIISRHVIFDETQFPFAKTHTSSTHTYEFLNDS 428
+K I++R V+ DET ++ T FL DS
Sbjct: 693 EKFIVARDVVVDETNMVNSRAVKFET---VFLKDS 724
Score = 159 bits (401), Expect = 5e-38
Identities = 82/222 (36%), Positives = 125/222 (55%), Gaps = 4/222 (1%)
Query: 546 SNWKAAMLDEFDALIKNKTWELVPRPQNVNFIRSMWIFRHKKKSDGSFERYKARLVGDGR 605
S+W+ A+ E +A N TW + RP+N N + S W+F K G+ RYKARLV G
Sbjct: 904 SSWEEAINTELNAHKINNTWTITKRPENKNIVDSRWVFSVKYNELGNPIRYKARLVARGF 963
Query: 606 SQ*VGVDCDETFSPVVKPTTIRIVLTIALSKSWPIHQLDVKNVFLHGELQETVYMHQPMG 665
+Q +D +ETF+PV + ++ R +L++ + + +HQ+DVK FL+G L+E +YM P G
Sbjct: 964 TQKYQIDYEETFAPVARISSFRFILSLVIQYNLKVHQMDVKTAFLNGTLKEEIYMRLPQG 1023
Query: 666 FRDPIHPDYVCLLKKSLYGLKQAPRAWYQRFADFAFTIGFSHSKSDHSLFIYRKG--NDM 723
+ D VC L K++YGLKQA R W++ F F +S D ++I KG N+
Sbjct: 1024 I--SCNSDNVCKLNKAIYGLKQAARCWFEVFEQALKECEFVNSSVDRCIYILDKGNINEN 1081
Query: 724 TYILLYVDDIILTASSDVLRRSIMSLLASEFAMKDLGTLSYF 765
Y+LLYVDD+++ + L +F M DL + +F
Sbjct: 1082 IYVLLYVDDVVIATGDMTRMNNFKRYLMEKFRMTDLNEIKHF 1123
>YCB9_YEAST (P25384) Transposon Ty2 protein B (Ty1-17 protein B)
Length = 1770
Score = 109 bits (273), Expect = 3e-23
Identities = 138/553 (24%), Positives = 231/553 (40%), Gaps = 55/553 (9%)
Query: 19 NIVVGSGQEIPIRGYGQTYISPPYPPLSLNNVLHAPKLIKNLIFVRRFTIDNNVTIEFDP 78
NIV Q+IPI G + + + LH P + +L+ + + N+T F
Sbjct: 481 NIVDAQKQDIPINAIGNLHFNFQNGTKTSIKALHTPNIAYDLLSLSELA-NQNITACFTR 539
Query: 79 FSFSVEDIQTGIPLMRCDSTGDLYPLTTT---TSHQSTSPATFAALSPT-------LWHN 128
+ D P+++ GD Y L+ SH S S + L H
Sbjct: 540 NTLERSDGTVLAPIVK---HGDFYWLSKKYLIPSHISKLTINNVNKSKSVNKYPYPLIHR 596
Query: 129 RLGHPG-ANVLSFLNKNKFIECKQI------SSPSICQSCIYGK-----HVKLPFSISNS 176
LGH ++ L KN K+ +S C C+ GK HVK +
Sbjct: 597 MLGHANFRSIQKSLKKNAVTYLKESDIEWSNASTYQCPDCLIGKSTKHRHVK-GSRLKYQ 655
Query: 177 TTSKPFDIIHSDLWTSPI--LSSAGHKYYLFFLDDYTNFVWTFPIG--RKSQVPSIFSSF 232
+ +PF +H+D++ P+ L + Y++ F D+ T F W +P+ R+ + ++F+S
Sbjct: 656 ESYEPFQYLHTDIF-GPVHHLPKSAPSYFISFTDEKTRFQWVYPLHDRREESILNVFTSI 714
Query: 233 HAFIKTQFGTTIKCFQCDNGTEFNNEYFTQFCHKNGMVFRFSCPHTSPQNGKAERKIRAI 292
AFIK QF + Q D G+E+ N+ +F G+ ++ S +G AER R +
Sbjct: 715 LAFIKNQFNARVLVIQMDRGSEYTNKTLHKFFTNRGITACYTTTADSRAHGVAERLNRTL 774
Query: 293 NNFIRTSLAHSSMPPSFWHHALQITTYLQNILPSKILSHHSPTQYLYHRDPSYTHLRVFG 352
N RT L S +P W A++ +T ++N L S + S Q+ T + FG
Sbjct: 775 LNDCRTLLHCSGLPNHLWFSAVEFSTIIRNSLVSP-KNDKSARQHAGLAGLDITTILPFG 833
Query: 353 CLCYPLFPS--TAINKLQARSTPCAFLGYPQNHRGYKCYDLSSKKII-ISRHVIFDETQF 409
P+ + +K+ R P L +N GY Y S KK + + +VI + Q
Sbjct: 834 ---QPVIVNNHNPDSKIHPRGIPGYALHPSRNSYGYIIYLPSLKKTVDTTNYVILQDKQ- 889
Query: 410 PFAKTHTSSTHTYEFLNDSLHPLLHYHLQNDPKQDEPEPRKIESPQPATTPASPINVTNQ 469
+K + T F +D L+ L H Q+ +Q+E E ++ + S I + +
Sbjct: 890 --SKLDQFNYDTLTF-DDDLNRLT-AHNQSFIEQNETEQSYDQNTESDHDYQSEIEINSD 945
Query: 470 SILPP-SPMSINQLPHPLVSTELTSPTHTPQQIHQE------PPRTIATHSMHGIHKPKI 522
++ S SIN L + E P+++ + P + + + H I+K
Sbjct: 946 PLVNDFSSQSINPLQ---LDKEPVQKVRAPKEVDADISEYNILPSPVRSRTPHIINKEST 1002
Query: 523 QFNLTT-SITSSP 534
+ T S T+SP
Sbjct: 1003 EMGGTVESDTTSP 1015
Score = 68.9 bits (167), Expect = 6e-11
Identities = 61/212 (28%), Positives = 94/212 (43%), Gaps = 24/212 (11%)
Query: 555 EFDALIKNKTWELVPR-------PQNVNFIRSMWIFRHKKKSDGSFERYKARLVGDGRSQ 607
E L+K TW+ P+ V I SM+IF KK DG+ +KAR V G Q
Sbjct: 1272 EISQLLKMNTWDTNKYYDRNDIDPKKV--INSMFIFN--KKRDGT---HKARFVARGDIQ 1324
Query: 608 *VGVDCDETFSPVVKPTTIRIVLTIALSKSWPIHQLDVKNVFLHGELQETVYMHQP--MG 665
+ S V + L+IAL + I QLD+ + +L+ +++E +Y+ P +G
Sbjct: 1325 HPDTYDSDMQSNTVHHYALMTSLSIALDNDYYITQLDISSAYLYADIKEELYIRPPPHLG 1384
Query: 666 FRDPIHPDYVCLLKKSLYGLKQAPRAWYQRFADFAFTIGFSHSKSDHSLFIYRKGNDMTY 725
D + L+KSLYGLKQ+ WY+ + S N
Sbjct: 1385 LNDKL-----LRLRKSLYGLKQSGANWYETIKSYLINCCDMQEVRGWSCVF---KNSQVT 1436
Query: 726 ILLYVDDIILTASSDVLRRSIMSLLASEFAMK 757
I L+VDD+IL + + I++ L ++ K
Sbjct: 1437 ICLFVDDMILFSKDLNANKKIITTLKKQYDTK 1468
>YMD9_YEAST (Q03434) Transposon Ty1 protein B
Length = 1328
Score = 104 bits (259), Expect = 1e-21
Identities = 118/493 (23%), Positives = 200/493 (39%), Gaps = 59/493 (11%)
Query: 19 NIVVGSGQEIPIRGYGQTYISPPYPPLSLNNVLHAPKLIKNLIFVRRFTIDNNVTIEFDP 78
N+V + IPI G + VLH P + +L+ + +
Sbjct: 58 NVVDAQKRNIPINAIGDLQFHFQDNTKTSIKVLHTPNIAYDLLSLNELAAVDITAC---- 113
Query: 79 FSFSVEDIQTGIPLMRCDSTGDLYPLTT-----------TTSHQSTSPATFAALSPTLWH 127
F+ +V + G L GD Y ++ T ++ TS +T P + H
Sbjct: 114 FTKNVLERSDGTVLAPIVKYGDFYWVSKKYLLPSNISVPTINNVHTSESTRKYPYPFI-H 172
Query: 128 NRLGHPGANVLSFLNKNK----FIECKQISSPSI---CQSCIYGK-----HVKLPFSISN 175
L H A + + KN F E S +I C C+ GK H+K +
Sbjct: 173 RMLAHANAQTIRYSLKNNTITYFNESDVDRSSAIDYQCPDCLIGKSTKHRHIK-GSRLKY 231
Query: 176 STTSKPFDIIHSDLWTSPI--LSSAGHKYYLFFLDDYTNFVWTFPIG--RKSQVPSIFSS 231
+ +PF +H+D++ P+ L + Y++ F D+ T F W +P+ R+ + +F++
Sbjct: 232 QNSYEPFQYLHTDIF-GPVHNLPKSAPSYFISFTDETTKFRWVYPLHDRREDSILDVFTT 290
Query: 232 FHAFIKTQFGTTIKCFQCDNGTEFNNEYFTQFCHKNGMVFRFSCPHTSPQNGKAERKIRA 291
AFIK QF ++ Q D G+E+ N +F KNG+ ++ S +G AER R
Sbjct: 291 ILAFIKNQFQASVLVIQMDRGSEYTNRTLHKFLEKNGITPCYTTTADSRAHGVAERLNRT 350
Query: 292 INNFIRTSLAHSSMPPSFWHHALQITTYLQNILPSKILSHHSPTQYLYHRDPSYTHLRVF 351
+ + RT L S +P W A++ +T ++N L S S S Q+ + L F
Sbjct: 351 LLDDCRTQLQCSGLPNHLWFSAIEFSTIVRNSLASP-KSKKSARQHAGLAGLDISTLLPF 409
Query: 352 GCLCYPLFPS--TAINKLQARSTPCAFLGYPQNHRGYKCYDLSSKKIIISRHVI------ 403
G P+ + +K+ R P L +N GY Y S KK + + + +
Sbjct: 410 G---QPVIVNDHNPNSKIHPRGIPGYALHPSRNSYGYIIYLPSLKKTVDTTNYVILQGKE 466
Query: 404 ------------FDETQFPFAKTHTSSTHTYEF-LNDSLHPLLHYHLQNDPKQDEPEPRK 450
FDE ++ S + E +D L+ + Q+D + +PR
Sbjct: 467 SRLDQFNYDALTFDEDLNRLTASYQSFIASNEIQQSDDLNIESDHDFQSDIELHPEQPRN 526
Query: 451 IESPQPATTPASP 463
+ S + T ++P
Sbjct: 527 VLSKAVSPTDSTP 539
Score = 72.0 bits (175), Expect = 7e-12
Identities = 63/215 (29%), Positives = 99/215 (45%), Gaps = 30/215 (13%)
Query: 555 EFDALIKNKTW---------ELVPRPQNVNFIRSMWIFRHKKKSDGSFERYKARLVGDGR 605
E + L+K KTW E+ P+ I SM+IF KK DG+ +KAR V G
Sbjct: 830 EVNQLLKMKTWDTDEYYDRKEIDPK----RVINSMFIFN--KKRDGT---HKARFVARGD 880
Query: 606 SQ*VGVDCDETFSPVVKPTTIRIVLTIALSKSWPIHQLDVKNVFLHGELQETVYMHQP-- 663
Q S V + L++AL ++ I QLD+ + +L+ +++E +Y+ P
Sbjct: 881 IQHPDTYDSGMQSNTVHHYALMTSLSLALDNNYYITQLDISSAYLYADIKEELYIRPPPH 940
Query: 664 MGFRDPIHPDYVCLLKKSLYGLKQAPRAWYQRFADFAF-TIGFSHSKSDHSLFIYRKGND 722
+G D + LKKSLYGLKQ+ WY+ + G + +F N
Sbjct: 941 LGMNDKL-----IRLKKSLYGLKQSGANWYETIKSYLIKQCGMEEVRGWSCVF----KNS 991
Query: 723 MTYILLYVDDIILTASSDVLRRSIMSLLASEFAMK 757
I L+VDD+IL + + I++ L ++ K
Sbjct: 992 QVTICLFVDDMILFSKDLNANKKIITTLKKQYDTK 1026
>YMU0_YEAST (Q04670) Transposon Ty1 protein B
Length = 1328
Score = 103 bits (257), Expect = 2e-21
Identities = 118/493 (23%), Positives = 200/493 (39%), Gaps = 59/493 (11%)
Query: 19 NIVVGSGQEIPIRGYGQTYISPPYPPLSLNNVLHAPKLIKNLIFVRRFTIDNNVTIEFDP 78
N+V + IPI G + VLH P + +L+ + +
Sbjct: 58 NVVDAQKRNIPINAIGDLQFHFQDNTKTSIKVLHTPNIAYDLLSLNELAAVDITAC---- 113
Query: 79 FSFSVEDIQTGIPLMRCDSTGDLYPLTT-----------TTSHQSTSPATFAALSPTLWH 127
F+ +V + G L GD Y ++ T ++ TS +T P + H
Sbjct: 114 FTKNVLERSDGTVLAPIVKYGDFYWVSKKYLLPSNISVPTINNVHTSESTRKYPYPFI-H 172
Query: 128 NRLGHPGANVLSFLNKNK----FIECKQISSPSI---CQSCIYGK-----HVKLPFSISN 175
L H A + + KN F E S +I C C+ GK H+K +
Sbjct: 173 RMLAHANAQTIRYSLKNNTITYFNESDVDWSSAIDYQCPDCLIGKSTKHRHIK-GSRLKY 231
Query: 176 STTSKPFDIIHSDLWTSPI--LSSAGHKYYLFFLDDYTNFVWTFPIG--RKSQVPSIFSS 231
+ +PF +H+D++ P+ L + Y++ F D+ T F W +P+ R+ + +F++
Sbjct: 232 QNSYEPFQYLHTDIF-GPVHNLPKSAPSYFISFTDETTKFRWVYPLHDRREDSILDVFTT 290
Query: 232 FHAFIKTQFGTTIKCFQCDNGTEFNNEYFTQFCHKNGMVFRFSCPHTSPQNGKAERKIRA 291
AFIK QF ++ Q D G+E+ N +F KNG+ ++ S +G AER R
Sbjct: 291 ILAFIKNQFQASVLVIQMDRGSEYTNRTLHKFLEKNGITPCYTTTADSRAHGVAERLNRT 350
Query: 292 INNFIRTSLAHSSMPPSFWHHALQITTYLQNILPSKILSHHSPTQYLYHRDPSYTHLRVF 351
+ + RT L S +P W A++ +T ++N L S S S Q+ + L F
Sbjct: 351 LLDDCRTQLQCSGLPNHLWFSAIEFSTIVRNSLASP-KSKKSARQHAGLAGLDISTLLPF 409
Query: 352 GCLCYPLFPS--TAINKLQARSTPCAFLGYPQNHRGYKCYDLSSKKIIISRHVI------ 403
G P+ + +K+ R P L +N GY Y S KK + + + +
Sbjct: 410 G---QPVIVNDHNPNSKIHPRGIPGYALHPSRNSYGYIIYLPSLKKTVDTTNYVILQGKE 466
Query: 404 ------------FDETQFPFAKTHTSSTHTYEF-LNDSLHPLLHYHLQNDPKQDEPEPRK 450
FDE ++ S + E +D L+ + Q+D + +PR
Sbjct: 467 SRLDQFNYDALTFDEDLNRLTASYQSFIASNEIQQSDDLNIESDHDFQSDIELHPEQPRN 526
Query: 451 IESPQPATTPASP 463
+ S + T ++P
Sbjct: 527 VLSKAVSPTDSTP 539
Score = 73.2 bits (178), Expect = 3e-12
Identities = 62/220 (28%), Positives = 104/220 (47%), Gaps = 40/220 (18%)
Query: 555 EFDALIKNKTW---------ELVPRPQNVNFIRSMWIFRHKKKSDGSFERYKARLVGDGR 605
E + L+K KTW E+ P+ I SM+IF K+ DG+ +KAR V G
Sbjct: 830 EVNQLLKMKTWDTDRYYDRKEIDPK----RVINSMFIFNRKR--DGT---HKARFVARG- 879
Query: 606 SQ*VGVDCDETFSPVVKPTTIR-----IVLTIALSKSWPIHQLDVKNVFLHGELQETVYM 660
+ +T+ P ++ T+ L++AL ++ I QLD+ + +L+ +++E +Y+
Sbjct: 880 ----DIQHPDTYDPGMQSNTVHHYALMTSLSLALDNNYYITQLDISSAYLYADIKEELYI 935
Query: 661 HQP--MGFRDPIHPDYVCLLKKSLYGLKQAPRAWYQRFADFAF-TIGFSHSKSDHSLFIY 717
P +G D + LKKSLYGLKQ+ WY+ + G + +F
Sbjct: 936 RPPPHLGMNDKL-----IRLKKSLYGLKQSGANWYETIKSYLIKQCGMEEVRGWSCVF-- 988
Query: 718 RKGNDMTYILLYVDDIILTASSDVLRRSIMSLLASEFAMK 757
N I L+VDD+IL + + I++ L ++ K
Sbjct: 989 --KNSQVTICLFVDDMILFSKDLNANKKIITTLKKQYDTK 1026
>YME4_YEAST (Q04711) Transposon Ty1 protein B
Length = 1328
Score = 103 bits (257), Expect = 2e-21
Identities = 118/493 (23%), Positives = 200/493 (39%), Gaps = 59/493 (11%)
Query: 19 NIVVGSGQEIPIRGYGQTYISPPYPPLSLNNVLHAPKLIKNLIFVRRFTIDNNVTIEFDP 78
N+V + IPI G + VLH P + +L+ + +
Sbjct: 58 NVVDAQKRNIPINAIGDLQFHFQDNTKTSIKVLHTPNIAYDLLSLNELAAVDITAC---- 113
Query: 79 FSFSVEDIQTGIPLMRCDSTGDLYPLTT-----------TTSHQSTSPATFAALSPTLWH 127
F+ +V + G L GD Y ++ T ++ TS +T P + H
Sbjct: 114 FTKNVLERSDGTVLAPIVKYGDFYWVSKKYLLPSNISVPTINNVHTSESTRKYPYPFI-H 172
Query: 128 NRLGHPGANVLSFLNKNK----FIECKQISSPSI---CQSCIYGK-----HVKLPFSISN 175
L H A + + KN F E S +I C C+ GK H+K +
Sbjct: 173 RMLAHANAQTIRYSLKNNTITYFNESDVDWSSAIDYQCPDCLIGKSTKHRHIK-GSRLKY 231
Query: 176 STTSKPFDIIHSDLWTSPI--LSSAGHKYYLFFLDDYTNFVWTFPIG--RKSQVPSIFSS 231
+ +PF +H+D++ P+ L + Y++ F D+ T F W +P+ R+ + +F++
Sbjct: 232 QNSYEPFQYLHTDIF-GPVHNLPKSAPSYFISFTDETTKFRWVYPLHDRREDSILDVFTT 290
Query: 232 FHAFIKTQFGTTIKCFQCDNGTEFNNEYFTQFCHKNGMVFRFSCPHTSPQNGKAERKIRA 291
AFIK QF ++ Q D G+E+ N +F KNG+ ++ S +G AER R
Sbjct: 291 ILAFIKNQFQASVLVIQMDRGSEYTNRTLHKFLEKNGITPCYTTTADSRAHGVAERLNRT 350
Query: 292 INNFIRTSLAHSSMPPSFWHHALQITTYLQNILPSKILSHHSPTQYLYHRDPSYTHLRVF 351
+ + RT L S +P W A++ +T ++N L S S S Q+ + L F
Sbjct: 351 LLDDCRTQLQCSGLPNHLWFSAIEFSTIVRNSLASP-KSKKSARQHAGLAGLDISTLLPF 409
Query: 352 GCLCYPLFPS--TAINKLQARSTPCAFLGYPQNHRGYKCYDLSSKKIIISRHVI------ 403
G P+ + +K+ R P L +N GY Y S KK + + + +
Sbjct: 410 G---QPVIVNDHNPNSKIHPRGIPGYALHPSRNSYGYIIYLPSLKKTVDTTNYVILQGKE 466
Query: 404 ------------FDETQFPFAKTHTSSTHTYEF-LNDSLHPLLHYHLQNDPKQDEPEPRK 450
FDE ++ S + E +D L+ + Q+D + +PR
Sbjct: 467 SRLDQFNYDALTFDEDLNRLTASYQSFIASNEIQQSDDLNIESDHDFQSDIELHPEQPRN 526
Query: 451 IESPQPATTPASP 463
+ S + T ++P
Sbjct: 527 VLSKAVSPTDSTP 539
Score = 69.3 bits (168), Expect = 5e-11
Identities = 61/215 (28%), Positives = 98/215 (45%), Gaps = 30/215 (13%)
Query: 555 EFDALIKNKTW---------ELVPRPQNVNFIRSMWIFRHKKKSDGSFERYKARLVGDGR 605
E + L+K TW E+ P+ I SM+IF K+ DG+ +KAR V G
Sbjct: 830 EVNQLLKMNTWDTDKYYDRKEIDPK----RVINSMFIFNRKR--DGT---HKARFVARGD 880
Query: 606 SQ*VGVDCDETFSPVVKPTTIRIVLTIALSKSWPIHQLDVKNVFLHGELQETVYMHQP-- 663
Q S V + L++AL ++ I QLD+ + +L+ +++E +Y+ P
Sbjct: 881 IQHPDTYDSGMQSNTVHHYALMTSLSLALDNNYYITQLDISSAYLYADIKEELYIRPPPH 940
Query: 664 MGFRDPIHPDYVCLLKKSLYGLKQAPRAWYQRFADFAF-TIGFSHSKSDHSLFIYRKGND 722
+G D + LKKSLYGLKQ+ WY+ + G + +F N
Sbjct: 941 LGMNDKL-----IRLKKSLYGLKQSGANWYETIKSYLIKQCGMEEVRGWSCVF----KNS 991
Query: 723 MTYILLYVDDIILTASSDVLRRSIMSLLASEFAMK 757
I L+VDD+IL + + I++ L ++ K
Sbjct: 992 QVTICLFVDDMILFSKDLNANKKIITTLKKQYDTK 1026
>YJZ9_YEAST (P47100) Transposon Ty1 protein B
Length = 1755
Score = 103 bits (257), Expect = 2e-21
Identities = 118/493 (23%), Positives = 200/493 (39%), Gaps = 59/493 (11%)
Query: 19 NIVVGSGQEIPIRGYGQTYISPPYPPLSLNNVLHAPKLIKNLIFVRRFTIDNNVTIEFDP 78
N+V + IPI G + VLH P + +L+ + +
Sbjct: 485 NVVDAQKRNIPINAIGDLQFHFQDNTKTSIKVLHTPNIAYDLLSLNELAAVDITAC---- 540
Query: 79 FSFSVEDIQTGIPLMRCDSTGDLYPLTT-----------TTSHQSTSPATFAALSPTLWH 127
F+ +V + G L GD Y ++ T ++ TS +T P + H
Sbjct: 541 FTKNVLERSDGTVLAPIVKYGDFYWVSKKYLLPSNISVPTINNVHTSESTRKYPYPFI-H 599
Query: 128 NRLGHPGANVLSFLNKNK----FIECKQISSPSI---CQSCIYGK-----HVKLPFSISN 175
L H A + + KN F E S +I C C+ GK H+K +
Sbjct: 600 RMLAHANAQTIRYSLKNNTITYFNESDVDWSSAIDYQCPDCLIGKSTKHRHIK-GSRLKY 658
Query: 176 STTSKPFDIIHSDLWTSPI--LSSAGHKYYLFFLDDYTNFVWTFPIG--RKSQVPSIFSS 231
+ +PF +H+D++ P+ L + Y++ F D+ T F W +P+ R+ + +F++
Sbjct: 659 QNSYEPFQYLHTDIF-GPVHNLPKSAPSYFISFTDETTKFRWVYPLHDRREDSILDVFTT 717
Query: 232 FHAFIKTQFGTTIKCFQCDNGTEFNNEYFTQFCHKNGMVFRFSCPHTSPQNGKAERKIRA 291
AFIK QF ++ Q D G+E+ N +F KNG+ ++ S +G AER R
Sbjct: 718 ILAFIKNQFQASVLVIQMDRGSEYTNRTLHKFLEKNGITPCYTTTADSRAHGVAERLNRT 777
Query: 292 INNFIRTSLAHSSMPPSFWHHALQITTYLQNILPSKILSHHSPTQYLYHRDPSYTHLRVF 351
+ + RT L S +P W A++ +T ++N L S S S Q+ + L F
Sbjct: 778 LLDDCRTQLQCSGLPNHLWFSAIEFSTIVRNSLASP-KSKKSARQHAGLAGLDISTLLPF 836
Query: 352 GCLCYPLFPS--TAINKLQARSTPCAFLGYPQNHRGYKCYDLSSKKIIISRHVI------ 403
G P+ + +K+ R P L +N GY Y S KK + + + +
Sbjct: 837 G---QPVIVNDHNPNSKIHPRGIPGYALHPSRNSYGYIIYLPSLKKTVDTTNYVILQGKE 893
Query: 404 ------------FDETQFPFAKTHTSSTHTYEF-LNDSLHPLLHYHLQNDPKQDEPEPRK 450
FDE ++ S + E +D L+ + Q+D + +PR
Sbjct: 894 SRLDQFNYDALTFDEDLNRLTASYQSFIASNEIQQSDDLNIESDHDFQSDIELHPEQPRN 953
Query: 451 IESPQPATTPASP 463
+ S + T ++P
Sbjct: 954 VLSKAVSPTDSTP 966
Score = 70.1 bits (170), Expect = 3e-11
Identities = 62/215 (28%), Positives = 99/215 (45%), Gaps = 30/215 (13%)
Query: 555 EFDALIKNKTW---------ELVPRPQNVNFIRSMWIFRHKKKSDGSFERYKARLVGDGR 605
E + L+K KTW E+ P+ I SM+IF KK DG+ +KAR V G
Sbjct: 1257 EVNQLLKMKTWDTDEYYDRKEIDPK----RVINSMFIFN--KKRDGT---HKARFVARGD 1307
Query: 606 SQ*VGVDCDETFSPVVKPTTIRIVLTIALSKSWPIHQLDVKNVFLHGELQETVYMHQP-- 663
Q S V + L++AL ++ I QLD+ + +L+ +++E +Y+ P
Sbjct: 1308 IQHPDTYDSGMQSNTVHHYALMTSLSLALDNNYYITQLDISSAYLYADIKEELYIRPPPH 1367
Query: 664 MGFRDPIHPDYVCLLKKSLYGLKQAPRAWYQRFADFAF-TIGFSHSKSDHSLFIYRKGND 722
+G D + LKKSLYGLKQ+ WY+ + G + +F N
Sbjct: 1368 LGMNDKL-----IRLKKSLYGLKQSGANWYETIKSYLIQQCGMEEVRGWSCVF----KNS 1418
Query: 723 MTYILLYVDDIILTASSDVLRRSIMSLLASEFAMK 757
I L+VDD++L + + + I+ L ++ K
Sbjct: 1419 QVTICLFVDDMVLFSKNLNSNKRIIEKLKMQYDTK 1453
>M820_ARATH (P92520) Hypothetical mitochondrial protein AtMg00820
(ORF170)
Length = 170
Score = 102 bits (254), Expect = 5e-21
Identities = 55/123 (44%), Positives = 73/123 (58%)
Query: 511 THSMHGIHKPKIQFNLTTSITSSPLPHNPKAALSDSNWKAAMLDEFDALIKNKTWELVPR 570
T S GI+K +++LT + T P + AL D W AM +E DAL +NKTW LVP
Sbjct: 3 TRSKAGINKLNPKYSLTITTTIKKEPKSVIFALKDPGWCQAMQEELDALSRNKTWILVPP 62
Query: 571 PQNVNFIRSMWIFRHKKKSDGSFERYKARLVGDGRSQ*VGVDCDETFSPVVKPTTIRIVL 630
P N N + W+F+ K SDG+ +R KARLV G Q G+ ET+SPVV+ TIR +L
Sbjct: 63 PVNQNILGCKWVFKTKLHSDGTLDRLKARLVAKGFHQEEGIYFVETYSPVVRTATIRTIL 122
Query: 631 TIA 633
+A
Sbjct: 123 NVA 125
>YJZ7_YEAST (P47098) Transposon Ty1 protein B
Length = 1755
Score = 101 bits (252), Expect = 9e-21
Identities = 116/493 (23%), Positives = 200/493 (40%), Gaps = 59/493 (11%)
Query: 19 NIVVGSGQEIPIRGYGQTYISPPYPPLSLNNVLHAPKLIKNLIFVRRFTIDNNVTIEFDP 78
N+V + IPI G + VLH P + +L+ + +
Sbjct: 485 NVVDAQKRNIPINAIGDLQFHFQDNTKTSIKVLHTPNIAYDLLSLNELAAVDITAC---- 540
Query: 79 FSFSVEDIQTGIPLMRCDSTGDLYPLTT-----------TTSHQSTSPATFAALSPTLWH 127
F+ +V + G L GD Y ++ T ++ TS +T P + H
Sbjct: 541 FTKNVLERSDGTVLAPIVQYGDFYWVSKRYLLPSNISVPTINNVHTSESTRKYPYPFI-H 599
Query: 128 NRLGHPGANVLSFLNKNK----FIECKQISSPSI---CQSCIYGK-----HVKLPFSISN 175
L H A + + KN F E S +I C C+ GK H+K +
Sbjct: 600 RMLAHANAQTIRYSLKNNTITYFNESDVDWSSAIDYQCPDCLIGKSTKHRHIK-GSRLKY 658
Query: 176 STTSKPFDIIHSDLWTSPI--LSSAGHKYYLFFLDDYTNFVWTFPIG--RKSQVPSIFSS 231
+ +PF +H+D++ P+ L + Y++ F D+ T F W +P+ R+ + +F++
Sbjct: 659 QNSYEPFQYLHTDIF-GPVHNLPKSAPSYFISFTDETTKFRWVYPLHDRREDSILDVFTT 717
Query: 232 FHAFIKTQFGTTIKCFQCDNGTEFNNEYFTQFCHKNGMVFRFSCPHTSPQNGKAERKIRA 291
AFIK QF ++ Q D G+E+ N +F KNG+ ++ S +G AER R
Sbjct: 718 ILAFIKNQFQASVLVIQMDRGSEYTNRTLHKFLEKNGITPCYTTTADSRAHGVAERLNRT 777
Query: 292 INNFIRTSLAHSSMPPSFWHHALQITTYLQNILPSKILSHHSPTQYLYHRDPSYTHLRVF 351
+ + RT L S +P W A++ +T ++N L S S S Q+ + L F
Sbjct: 778 LLDDCRTQLQCSGLPNHLWFSAIEFSTIVRNSLASP-KSKKSARQHAGLAGLDISTLLPF 836
Query: 352 GCLCYPLFPS--TAINKLQARSTPCAFLGYPQNHRGYKCYDLSSKKIIISRHVI------ 403
G P+ + +K+ R P L +N GY Y S KK + + + +
Sbjct: 837 G---QPVIVNDHNPNSKIHPRGIPGYALHPSRNSYGYIIYLPSLKKTVDTTNYVILQGKE 893
Query: 404 -----FDETQFPFAKTHTSSTHTYEFL--------NDSLHPLLHYHLQNDPKQDEPEPRK 450
F+ F + T +Y+ ++ L+ + Q+D + +PR
Sbjct: 894 SRLDQFNYDALTFDEDLNRLTASYQSFIASNEIQESNDLNIESDHDFQSDIELHPEQPRN 953
Query: 451 IESPQPATTPASP 463
+ S + T ++P
Sbjct: 954 VLSKAVSPTDSTP 966
Score = 68.9 bits (167), Expect = 6e-11
Identities = 62/215 (28%), Positives = 98/215 (44%), Gaps = 30/215 (13%)
Query: 555 EFDALIKNKTW---------ELVPRPQNVNFIRSMWIFRHKKKSDGSFERYKARLVGDGR 605
E + L+K KTW E+ P+ I SM+IF KK DG+ +KAR V G
Sbjct: 1257 EVNQLLKMKTWDTDEYYDRKEIDPK----RVINSMFIFN--KKRDGT---HKARFVARGD 1307
Query: 606 SQ*VGVDCDETFSPVVKPTTIRIVLTIALSKSWPIHQLDVKNVFLHGELQETVYMHQP-- 663
Q S V + L++AL ++ I QLD+ + +L+ +++E +Y+ P
Sbjct: 1308 IQHPDTYDTGMQSNTVHHYALMTSLSLALDNNYYITQLDISSAYLYADIKEELYIRPPPH 1367
Query: 664 MGFRDPIHPDYVCLLKKSLYGLKQAPRAWYQRFADFAF-TIGFSHSKSDHSLFIYRKGND 722
+G D + LKKS YGLKQ+ WY+ + G + +F N
Sbjct: 1368 LGMNDKL-----IRLKKSHYGLKQSGANWYETIKSYLIKQCGMEEVRGWSCVF----KNS 1418
Query: 723 MTYILLYVDDIILTASSDVLRRSIMSLLASEFAMK 757
I L+VDD+IL + + I++ L ++ K
Sbjct: 1419 QVTICLFVDDMILFSKDLNANKKIITTLKKQYDTK 1453
>YMT5_YEAST (Q04214) Transposon Ty1 protein B
Length = 1328
Score = 101 bits (251), Expect = 1e-20
Identities = 104/408 (25%), Positives = 170/408 (41%), Gaps = 40/408 (9%)
Query: 19 NIVVGSGQEIPIRGYGQTYISPPYPPLSLNNVLHAPKLIKNLIFVRRFTIDNNVTIEFDP 78
N+V + IPI G + VLH P + +L+ + +
Sbjct: 58 NVVDAQKRNIPINAIGDLQFHFQDNTKTSIKVLHTPNIAYDLLSLNELAAVDITAC---- 113
Query: 79 FSFSVEDIQTGIPLMRCDSTGDLYPLTT-----------TTSHQSTSPATFAALSPTLWH 127
F+ +V + G L GD Y ++ T ++ TS +T P + H
Sbjct: 114 FTKNVLERSDGTVLAPIVKYGDFYWVSKKYLLPSNISVPTINNVHTSESTRKYPYPFI-H 172
Query: 128 NRLGHPGANVLSFLNKNK----FIECKQISSPSI---CQSCIYGK-----HVKLPFSISN 175
L H A + + KN F E S +I C C+ GK H+K +
Sbjct: 173 RMLAHANAQTIRYSLKNNTITYFNESDVDWSSAIDYQCPDCLIGKSTKHRHIK-GSRLKY 231
Query: 176 STTSKPFDIIHSDLWTSPI--LSSAGHKYYLFFLDDYTNFVWTFPIG--RKSQVPSIFSS 231
+ +PF +H+D++ P+ L + Y++ F D+ T F W +P+ R+ + +F++
Sbjct: 232 QNSYEPFQYLHTDIF-GPVHNLPKSAPSYFISFTDETTKFRWVYPLHDRREDSILDVFTT 290
Query: 232 FHAFIKTQFGTTIKCFQCDNGTEFNNEYFTQFCHKNGMVFRFSCPHTSPQNGKAERKIRA 291
AFIK QF ++ Q D G+E+ N +F KNG+ ++ S +G AER R
Sbjct: 291 ILAFIKNQFQASVLVIQMDRGSEYTNRTLHKFLEKNGITPCYTTTADSRAHGVAERLNRT 350
Query: 292 INNFIRTSLAHSSMPPSFWHHALQITTYLQNILPSKILSHHSPTQYLYHRDPSYTHLRVF 351
+ + RT L S +P W A++ +T ++N L S S S Q+ + L F
Sbjct: 351 LLDDCRTQLQCSGLPNHLWFSAIEFSTIVRNSLASP-KSKKSARQHAGLAGLDISTLLPF 409
Query: 352 GCLCYPLFPS--TAINKLQARSTPCAFLGYPQNHRGYKCYDLSSKKII 397
G P+ + +K+ R P L +N GY Y S KK +
Sbjct: 410 G---QPVIVNDHNPNSKIHPRGIPGYALHPSRNSYGYIIYLPSLKKTV 454
Score = 69.3 bits (168), Expect = 5e-11
Identities = 61/215 (28%), Positives = 99/215 (45%), Gaps = 30/215 (13%)
Query: 555 EFDALIKNKTW---------ELVPRPQNVNFIRSMWIFRHKKKSDGSFERYKARLVGDGR 605
E + L+K KTW E+ P+ I SM+IF K+ DG+ +KAR V G
Sbjct: 830 EVNQLLKMKTWDTDKYYDRKEIDPK----RVINSMFIFNRKR--DGT---HKARFVARGD 880
Query: 606 SQ*VGVDCDETFSPVVKPTTIRIVLTIALSKSWPIHQLDVKNVFLHGELQETVYMHQP-- 663
Q S V + L++AL ++ I QLD+ + +L+ +++E +Y+ P
Sbjct: 881 IQHPDTYDSGMQSNTVHHYALMTSLSLALDNNYYITQLDISSAYLYADIKEELYIRPPPH 940
Query: 664 MGFRDPIHPDYVCLLKKSLYGLKQAPRAWYQRFADFAF-TIGFSHSKSDHSLFIYRKGND 722
+G D + LKKSLYGLKQ+ WY+ + G + +F N
Sbjct: 941 LGMNDKL-----IRLKKSLYGLKQSGANWYETIKSYLIKQCGMEEVRGWSCVF----ENS 991
Query: 723 MTYILLYVDDIILTASSDVLRRSIMSLLASEFAMK 757
I L+VDD++L + + + I+ L ++ K
Sbjct: 992 QVTICLFVDDMVLFSKNLNSNKRIIDKLKMQYDTK 1026
>YCH4_YEAST (P25600) Transposon Ty5-1 34.5 kDa hypothetical protein
Length = 308
Score = 85.1 bits (209), Expect = 8e-16
Identities = 43/123 (34%), Positives = 66/123 (52%)
Query: 643 LDVKNVFLHGELQETVYMHQPMGFRDPIHPDYVCLLKKSLYGLKQAPRAWYQRFADFAFT 702
+DV FL+ + E +Y+ QP GF + +PDYV L +YGLKQAP W + +
Sbjct: 1 MDVDTAFLNSTMDEPIYVKQPPGFVNERNPDYVWELYGGMYGLKQAPLLWNEHINNTLKK 60
Query: 703 IGFSHSKSDHSLFIYRKGNDMTYILLYVDDIILTASSDVLRRSIMSLLASEFAMKDLGTL 762
IGF + +H L+ + YI +YVDD+++ A S + + L ++MKDLG +
Sbjct: 61 IGFCRHEGEHGLYFRSTSDGPIYIGVYVDDLLVAAPSPKIYDRVKQELTKLYSMKDLGKV 120
Query: 763 SYF 765
F
Sbjct: 121 DKF 123
>YJL3_YEAST (P47024) Transposon Ty4 207.7 kDa hypothetical protein
Length = 1803
Score = 73.2 bits (178), Expect = 3e-12
Identities = 53/171 (30%), Positives = 85/171 (48%), Gaps = 17/171 (9%)
Query: 596 YKARLVGDGRSQ*VGVDCDETFSPVVKPTT----IRIVLTIALSKSWPIHQLDVKNVFLH 651
YKAR+V G +Q +T+S + + I+I L IA +++ + LD+ + FL+
Sbjct: 1337 YKARIVCRGDTQ-----SPDTYSVITTESLNHNHIKIFLMIANNRNMFMKTLDINHAFLY 1391
Query: 652 GELQETVYMHQPMGFRDPIHPDYVCLLKKSLYGLKQAPRAWYQRFADFAFTIGFSHSKSD 711
+L+E +Y+ P R V L K+LYGLKQ+P+ W + IG +
Sbjct: 1392 AKLEEEIYIPHPHDRR------CVVKLNKALYGLKQSPKEWNDHLRQYLNGIGLKDNSYT 1445
Query: 712 HSLFIYRKGNDMTYILLYVDDIILTASSDVLRRSIMSLLASEFAMKDLGTL 762
L+ N M I +YVDD ++ AS++ ++ L S F +K GTL
Sbjct: 1446 PGLYQTEDKNLM--IAVYVDDCVIAASNEQRLDEFINKLKSNFELKITGTL 1494
Score = 66.6 bits (161), Expect = 3e-10
Identities = 85/339 (25%), Positives = 133/339 (39%), Gaps = 36/339 (10%)
Query: 127 HNRLGHPGANVLSFLNKNKFIE--CKQISSPSI--CQSCIYGKHVKLPF---SISN-STT 178
H R+GH G + K+ E I P+ CQ+C K K S++N ST
Sbjct: 562 HKRMGHTGIQQIENSIKHNHYEESLDLIKEPNEFWCQTCKISKATKRNHYTGSMNNHSTD 621
Query: 179 SKPFDIIHSDLWTSPILSSAGHK-YYLFFLDDYTNFVWTFPIGRKSQVPSIFSSFHA--- 234
+P D++ S+A K Y L +D+ T + T K+ +I +
Sbjct: 622 HEPGSSWCMDIFGPVSSSNADTKRYMLIMVDNNTRYCMTSTHFNKN-AETILAQVRKNIQ 680
Query: 235 FIKTQFGTTIKCFQCDNGTEFNNEYFTQFCHKNGMVFRFSCPHTSPQNGKAERKIRAINN 294
+++TQF ++ D GTEF N+ ++ G+ + NG+AER IR I
Sbjct: 681 YVETQFDRKVREINSDRGTEFTNDQIEEYFISKGIHHILTSTQDHAANGRAERYIRTIIT 740
Query: 295 FIRTSLAHSSMPPSFWHHALQITTYLQNILPSKILSHHSPTQYLYHRDPSYTHLRVFGCL 354
T L S++ FW +A+ T ++N L K S R P L F
Sbjct: 741 DATTLLRQSNLRVKFWEYAVTSATNIRNYLEHK--STGKLPLKAISRQPVTVRLMSF--- 795
Query: 355 CYPLFPSTAI-----NKLQARSTPCAFLGYPQNHRGYKCYDLSSKKIIIS-RHVIFDETQ 408
P I KL+ P L N GYK + S KI+ S + I + T
Sbjct: 796 -LPFGEKGIIWNHNHKKLKPSGLPSIILCKDPNSYGYKFFIPSKNKIVTSDNYTIPNYTM 854
Query: 409 FPFAKTHTSSTHTYEFLNDSLHPLLHYHLQNDPKQDEPE 447
+ + +++F +D ND ++D+ E
Sbjct: 855 DGRVRNTQNINKSHQFSSD-----------NDDEEDQIE 882
>M300_ARATH (P93293) Hypothetical mitochondrial protein AtMg00300
(ORF145a) (ORF1451)
Length = 145
Score = 70.9 bits (172), Expect = 2e-11
Identities = 34/93 (36%), Positives = 48/93 (51%)
Query: 101 LYPLTTTTSHQSTSPATFAALSPTLWHNRLGHPGANVLSFLNKNKFIECKQISSPSICQS 160
LY L + ++ A A LWH+RL H + L K F++ ++SS C+
Sbjct: 47 LYILQGSVETGESNLAETAKDETRLWHSRLAHMSQRGMELLVKKGFLDSSKVSSLKFCED 106
Query: 161 CIYGKHVKLPFSISNSTTSKPFDIIHSDLWTSP 193
CIYGK ++ FS TT P D +HSDLW +P
Sbjct: 107 CIYGKTHRVNFSTGQHTTKNPLDYVHSDLWGAP 139
>POL4_DROME (P10394) Retrovirus-related Pol polyprotein from
transposon 412 [Contains: Protease (EC 3.4.23.-); Reverse
transcriptase (EC 2.7.7.49); Endonuclease]
Length = 1237
Score = 59.3 bits (142), Expect = 5e-08
Identities = 54/200 (27%), Positives = 87/200 (43%), Gaps = 8/200 (4%)
Query: 142 NKNKFIECKQISSPSICQSCIYGKHVKLPFSISNSTTSKPFDIIHSDLWTSPILSSAGHK 201
N +K+I+ + + CQ KH K P +I+ T FD + D S G++
Sbjct: 927 NMSKYIK-EYVRKCQKCQKAKTTKHTKTPMTITE-TPEHAFDRVVVDTIGPLPKSENGNE 984
Query: 202 YYLFFLDDYTNFVWTFPIGRKSQVPSIFSSFHAFIKTQFGTTIKCFQCDNGTEFNNEYFT 261
Y + + D T ++ PI KS + F +FI ++G +K F D GTE+ N T
Sbjct: 985 YAVTLICDLTKYLVAIPIANKSAKTVAKAIFESFI-LKYG-PMKTFITDMGTEYKNSIIT 1042
Query: 262 QFCHKNGMVFRFSCPHTSPQNGKAERKIRAINNFIRTSLAHSSMPPSFWHHALQITTYLQ 321
C + S H G ER R +N +IR ++ S + W LQ Y
Sbjct: 1043 DLCKYLKIKNITSTAHHHQTVGVVERSHRTLNEYIR---SYISTDKTDWDVWLQYFVYCF 1099
Query: 322 NILPSKILSHHSPTQYLYHR 341
N S ++ ++ P + ++ R
Sbjct: 1100 NTTQS-MVHNYCPYELVFGR 1118
>POL_SFV1 (P23074) Pol polyprotein [Contains: Protease (EC 3.4.23.-);
Reverse transcriptase/ribonuclease H (EC 2.7.7.49) (EC
3.1.26.4) (RT); Integrase (IN)]
Length = 1161
Score = 57.8 bits (138), Expect = 1e-07
Identities = 48/187 (25%), Positives = 75/187 (39%), Gaps = 11/187 (5%)
Query: 158 CQSCIYGKHVKL--PFSISNSTTSKPFDIIHSDLWTSPILSSAGHKYYLFFLDDYTNFVW 215
C+ C+ L P + KPFD + D + P+ S G+ + L +D T FVW
Sbjct: 858 CKQCLVTNATNLTSPPILRPVKPLKPFDKFYID-YIGPLPPSNGYLHVLVVVDSMTGFVW 916
Query: 216 TFPIGRKSQVPSIFSSFHAFIKTQFGTTIKCFQCDNGTEFNNEYFTQFCHKNGMVFRFSC 275
+P ++ PS ++ A K D G F + F + + G+ FS
Sbjct: 917 LYP----TKAPSTSATVKALNMLTSIAIPKVLHSDQGAAFTSSTFADWAKEKGIQLEFST 972
Query: 276 PHTSPQNGKAERKIRAINNFIRTSLAHSSMPPSFWHHALQITTYLQNILPSKILSHHSPT 335
P+ +GK ERK I + L P+ W+ L + N S S ++P
Sbjct: 973 PYHPQSSGKVERKNSDIKRLLTKLLIGR---PAKWYDLLPVVQLALNNSYSP-SSKYTPH 1028
Query: 336 QYLYHRD 342
Q L+ D
Sbjct: 1029 QLLFGVD 1035
>POL_SFV3L (P27401) Pol polyprotein [Contains: Protease (EC 3.4.23.-);
Reverse transcriptase/ribonuclease H (EC 2.7.7.49) (EC
3.1.26.4) (RT); Integrase (IN)]
Length = 1157
Score = 57.0 bits (136), Expect = 2e-07
Identities = 48/187 (25%), Positives = 73/187 (38%), Gaps = 11/187 (5%)
Query: 158 CQSCIYGKHVKL--PFSISNSTTSKPFDIIHSDLWTSPILSSAGHKYYLFFLDDYTNFVW 215
C+ C+ L P + KPFD D + P+ S G+ + L +D T FVW
Sbjct: 860 CKQCLVTNAATLAAPPILRPERPVKPFDKFFID-YIGPLPPSNGYLHVLVVVDSMTGFVW 918
Query: 216 TFPIGRKSQVPSIFSSFHAFIKTQFGTTIKCFQCDNGTEFNNEYFTQFCHKNGMVFRFSC 275
+P ++ PS ++ A K D G F + F + G+ FS
Sbjct: 919 LYP----TKAPSTSATVKALNMLTSIAVPKVIHSDQGAAFTSATFADWAKNKGIQLEFST 974
Query: 276 PHTSPQNGKAERKIRAINNFIRTSLAHSSMPPSFWHHALQITTYLQNILPSKILSHHSPT 335
P+ +GK ERK I + L P+ W+ L + N S S ++P
Sbjct: 975 PYHPQSSGKVERKNSDIKRLLTKLLVGR---PAKWYDLLPVVQLALNNSYSP-SSKYTPH 1030
Query: 336 QYLYHRD 342
Q L+ D
Sbjct: 1031 QLLFGID 1037
>POL_FOAMV (P14350) Pol polyprotein [Contains: Reverse
transcriptase/ribonuclease H (EC 2.7.7.49) (EC 3.1.26.4)
(RT); Integrase (IN)]
Length = 886
Score = 55.8 bits (133), Expect = 5e-07
Identities = 42/163 (25%), Positives = 66/163 (39%), Gaps = 9/163 (5%)
Query: 180 KPFDIIHSDLWTSPILSSAGHKYYLFFLDDYTNFVWTFPIGRKSQVPSIFSSFHAFIKTQ 239
KPFD D + P+ S G+ Y L +D T F W +P ++ PS ++ +
Sbjct: 674 KPFDKFFID-YIGPLPPSQGYLYVLVVVDGMTGFTWLYP----TKAPSTSATVKSLNVLT 728
Query: 240 FGTTIKCFQCDNGTEFNNEYFTQFCHKNGMVFRFSCPHTSPQNGKAERKIRAINNFIRTS 299
K D G F + F ++ + G+ FS P+ K ERK I +
Sbjct: 729 SIAIPKVIHSDQGAAFTSSTFAEWAKERGIHLEFSTPYHPQSGSKVERKNSDIKRLLTKL 788
Query: 300 LAHSSMPPSFWHHALQITTYLQNILPSKILSHHSPTQYLYHRD 342
L P+ W+ L + N S +L ++P Q L+ D
Sbjct: 789 LVGR---PTKWYDLLPVVQLALNNTYSPVLK-YTPHQLLFGID 827
>M810_ARATH (P92519) Hypothetical mitochondrial protein AtMg00810
(ORF240b)
Length = 240
Score = 49.7 bits (117), Expect = 4e-05
Identities = 23/41 (56%), Positives = 31/41 (75%)
Query: 725 YILLYVDDIILTASSDVLRRSIMSLLASEFAMKDLGTLSYF 765
Y+LLYVDDI+LT SS+ L ++ L+S F+MKDLG + YF
Sbjct: 2 YLLLYVDDILLTGSSNTLLNMLIFQLSSTFSMKDLGPVHYF 42
>M710_ARATH (P92512) Hypothetical mitochondrial protein AtMg00710
(ORF120)
Length = 120
Score = 43.9 bits (102), Expect = 0.002
Identities = 20/67 (29%), Positives = 34/67 (49%)
Query: 290 RAINNFIRTSLAHSSMPPSFWHHALQITTYLQNILPSKILSHHSPTQYLYHRDPSYTHLR 349
R I +R+ L +P +F A ++ N PS ++ H P + + P+Y++LR
Sbjct: 3 RTIIEKVRSMLCECGLPKTFRADAANTAVHIINKYPSTAINFHVPDEVWFQSVPTYSYLR 62
Query: 350 VFGCLCY 356
FGC+ Y
Sbjct: 63 RFGCVAY 69
>YL52_CAEEL (P34431) Hypothetical protein F44E2.2 in chromosome III
Length = 2186
Score = 42.7 bits (99), Expect = 0.005
Identities = 40/166 (24%), Positives = 62/166 (37%), Gaps = 7/166 (4%)
Query: 158 CQSCIYGK-HVKLPFSISNSTTSKPFDIIHSDLWTSPILSSAGHKYYLFFLDDYTNFVWT 216
C C+ H KL S++ + P +I+ DL LS G++Y L +D +T +
Sbjct: 1510 CAKCLCANDHSKLTSSLTPYRMTFPLEIVACDLMDVG-LSVQGNRYILTIIDLFTKYGTA 1568
Query: 217 FPIGRKSQVPSIFSSFHAFIKTQFGTTIKCFQCDNGTEFNNEYFTQFCHKNGMVFRFSCP 276
PI K + ++ +F G D G EF N F QF H + +
Sbjct: 1569 VPIPDK-KAETVLKAFVERWAIGEGRIPLKLLTDQGKEFVNGLFAQFTH----MLKIEHI 1623
Query: 277 HTSPQNGKAERKIRAINNFIRTSLAHSSMPPSFWHHALQITTYLQN 322
T N +A + N I + + P W + Y N
Sbjct: 1624 TTKGYNSRANGAVERFNKTIMHIMKKKTAVPMEWDDQVVYAVYAYN 1669
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.332 0.142 0.470
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 118,967,173
Number of Sequences: 164201
Number of extensions: 5133682
Number of successful extensions: 16209
Number of sequences better than 10.0: 108
Number of HSP's better than 10.0 without gapping: 25
Number of HSP's successfully gapped in prelim test: 85
Number of HSP's that attempted gapping in prelim test: 15681
Number of HSP's gapped (non-prelim): 385
length of query: 993
length of database: 59,974,054
effective HSP length: 120
effective length of query: 873
effective length of database: 40,269,934
effective search space: 35155652382
effective search space used: 35155652382
T: 11
A: 40
X1: 15 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (22.0 bits)
S2: 71 (32.0 bits)
Medicago: description of AC144645.3


