

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144644.6 + phase: 0
(235 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
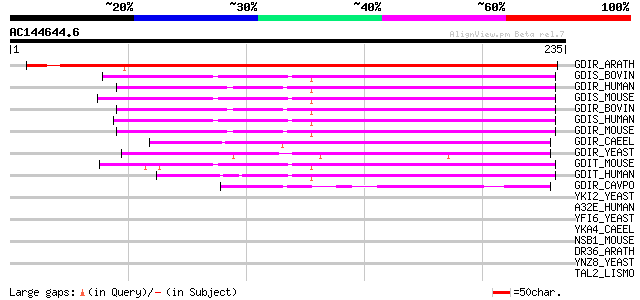
Score E
Sequences producing significant alignments: (bits) Value
GDIR_ARATH (Q9SFC6) Rho GDP-dissociation inhibitor 1 (Rho GDI-1)... 265 7e-71
GDIS_BOVIN (Q9TU03) Rho GDP-dissociation inhibitor 2 (Rho GDI 2)... 144 2e-34
GDIR_HUMAN (P52565) Rho GDP-dissociation inhibitor 1 (Rho GDI 1)... 144 2e-34
GDIS_MOUSE (Q61599) Rho GDP-dissociation inhibitor 2 (Rho GDI 2)... 142 5e-34
GDIR_BOVIN (P19803) Rho GDP-dissociation inhibitor 1 (Rho GDI 1)... 142 7e-34
GDIS_HUMAN (P52566) Rho GDP-dissociation inhibitor 2 (Rho GDI 2)... 142 9e-34
GDIR_MOUSE (Q99PT1) Rho GDP-dissociation inhibitor 1 (Rho GDI 1)... 140 2e-33
GDIR_CAEEL (Q20496) Probable rho GDP-dissociation inhibitor (Rho... 112 8e-25
GDIR_YEAST (Q12434) Rho GDP-dissociation inhibitor (Rho GDI) 101 2e-21
GDIT_MOUSE (Q62160) Rho GDP-dissociation inhibitor 3 (Rho GDI 3)... 100 3e-21
GDIT_HUMAN (Q99819) Rho GDP-dissociation inhibitor 3 (Rho GDI 3)... 98 2e-20
GDIR_CAVPO (P80237) Rho GDP-dissociation inhibitor 1 (Rho GDI 1)... 53 5e-07
YKI2_YEAST (P36080) Hypothetical 50.5 kDa protein in MDH1-VMA5 i... 36 0.090
A32E_HUMAN (Q9BTT0) Acidic leucine-rich nuclear phosphoprotein 3... 35 0.12
YFI6_YEAST (P43597) Hypothetical 137.7 kDa protein in UGS1-FAB1 ... 35 0.20
YKA4_CAEEL (P34256) Hypothetical protein B0303.4 in chromosome III 34 0.26
NSB1_MOUSE (Q9JL35) Nucleosome binding protein 1 (Nucleosome bin... 34 0.26
DR36_ARATH (Q8VZC7) Probable disease resistance protein At5g45510 34 0.34
YNZ8_YEAST (P53847) Hypothetical 88.1 kDa protein in ATX1-SIP3 i... 33 0.45
TAL2_LISMO (P66957) Probable transaldolase 2 (EC 2.2.1.2) 33 0.45
>GDIR_ARATH (Q9SFC6) Rho GDP-dissociation inhibitor 1 (Rho GDI-1)
(AtRhoGDI1)
Length = 240
Score = 265 bits (677), Expect = 7e-71
Identities = 131/240 (54%), Positives = 171/240 (70%), Gaps = 20/240 (8%)
Query: 8 VSTTKDVSFNAHLEEEHKNKGTNKVGVNYDEAEEIVPDED---------------EEEEP 52
VS +D+ F +++ N NK G + + + D+D EEE+
Sbjct: 4 VSGARDMGF-----DDNNNNKNNKDGDDENSSSRTRADDDALSRQMSESSLCATEEEEDD 58
Query: 53 KLELDLGPQFSLKEQLEKDKDDESLRKWKEQLLGNVDVSAVGEKIEPEVKIVSLTIICQG 112
+L LGPQ+++KE LEKDKDDESLRKWKEQLLG+VDV+ +GE ++PEV+I SL II G
Sbjct: 59 DSKLQLGPQYTIKEHLEKDKDDESLRKWKEQLLGSVDVTNIGETLDPEVRIDSLAIISPG 118
Query: 113 RPDLILPIPFTADSKKSMFTLKEGSQYRLKFSFTVSNNIVSGLRYTNVVWKTGVRVENTK 172
RPD++L +P + K FTLKEGS+Y LKF+F V+NNIVSGLRYTN VWKTGV+V+ K
Sbjct: 119 RPDIVLLVPENGNPKGMWFTLKEGSKYNLKFTFHVNNNIVSGLRYTNTVWKTGVKVDRAK 178
Query: 173 KMLGTYSPQQEPYTYELEEEITPSGLFARGTYSARTKFVDDDRKCYLDASYRFEIQKNWP 232
+MLGT+SPQ EPY + + EE TPSG+FARG+YSARTKF+DDD KCYL+ +Y F+I+K WP
Sbjct: 179 EMLGTFSPQLEPYNHVMPEETTPSGMFARGSYSARTKFLDDDNKCYLEINYSFDIRKEWP 238
>GDIS_BOVIN (Q9TU03) Rho GDP-dissociation inhibitor 2 (Rho GDI 2)
(Rho-GDI beta) (Ly-GDI) (D4-GDP-dissociation inhibitor)
(D4-GDI)
Length = 200
Score = 144 bits (362), Expect = 2e-34
Identities = 80/195 (41%), Positives = 114/195 (58%), Gaps = 6/195 (3%)
Query: 40 EEIVPDEDEEEEPKLELDLGPQFSLKEQLEKDKDDESLRKWKEQLLGNVDVSAVGEKIEP 99
E V ++D+E + KL PQ SLKE E DKDDESL K+K+ LLG D V + P
Sbjct: 7 EPHVEEDDDELDGKLNYKPPPQKSLKELQEMDKDDESLTKYKKTLLG--DGPVVADPTAP 64
Query: 100 EVKIVSLTIICQGRPDLILPIPFTADS---KKSMFTLKEGSQYRLKFSFTVSNNIVSGLR 156
V + LT++C+ P I + T D KK F LKEG +YR+K +F V+ +IVSGL+
Sbjct: 65 NVTVTRLTLVCESAPGPIT-MDLTGDLEALKKETFVLKEGVEYRVKINFKVNKDIVSGLK 123
Query: 157 YTNVVWKTGVRVENTKKMLGTYSPQQEPYTYELEEEITPSGLFARGTYSARTKFVDDDRK 216
Y ++TGV+V+ M+G+Y P+ E Y + E P G+ ARGTY ++ F DDD+
Sbjct: 124 YVQHTYRTGVKVDKATFMVGSYGPRPEEYEFLTPIEEAPKGMLARGTYHNKSFFTDDDKH 183
Query: 217 CYLDASYRFEIQKNW 231
+L + I+K+W
Sbjct: 184 DHLTWEWNLSIKKDW 198
>GDIR_HUMAN (P52565) Rho GDP-dissociation inhibitor 1 (Rho GDI 1)
(Rho-GDI alpha)
Length = 204
Score = 144 bits (362), Expect = 2e-34
Identities = 78/189 (41%), Positives = 113/189 (59%), Gaps = 6/189 (3%)
Query: 46 EDEEEEPKLELDLGPQFSLKEQLEKDKDDESLRKWKEQLLGNVDVSAVGEKIEPEVKIVS 105
E+EE+E + Q S++E E DKDDESLRK+KE LLG V VSA + P V +
Sbjct: 17 ENEEDEHSVNYKPPAQKSIQEIQELDKDDESLRKYKEALLGRVAVSA--DPNVPNVVVTG 74
Query: 106 LTIICQGRPDLILPIPFTADS---KKSMFTLKEGSQYRLKFSFTVSNNIVSGLRYTNVVW 162
LT++C P L + T D KK F LKEG +YR+K SF V+ IVSG++Y +
Sbjct: 75 LTLVCSSAPGP-LELDLTGDLESFKKQSFVLKEGVEYRIKISFRVNREIVSGMKYIQHTY 133
Query: 163 KTGVRVENTKKMLGTYSPQQEPYTYELEEEITPSGLFARGTYSARTKFVDDDRKCYLDAS 222
+ GV+++ T M+G+Y P+ E Y + E P G+ ARG+YS +++F DDD+ +L
Sbjct: 134 RKGVKIDKTDYMVGSYGPRAEEYEFLTPVEEAPKGMLARGSYSIKSRFTDDDKTDHLSWE 193
Query: 223 YRFEIQKNW 231
+ I+K+W
Sbjct: 194 WNLTIKKDW 202
>GDIS_MOUSE (Q61599) Rho GDP-dissociation inhibitor 2 (Rho GDI 2)
(Rho-GDI beta) (D4)
Length = 200
Score = 142 bits (359), Expect = 5e-34
Identities = 78/197 (39%), Positives = 116/197 (58%), Gaps = 6/197 (3%)
Query: 38 EAEEIVPDEDEEEEPKLELDLGPQFSLKEQLEKDKDDESLRKWKEQLLGNVDVSAVGEKI 97
+A+ + + D++ + KL PQ SLKE E DKDDESL K+K+ LLG DV V +
Sbjct: 5 DAQPQLEEADDDLDSKLNYKPPPQKSLKELQEMDKDDESLTKYKKTLLG--DVPVVADPT 62
Query: 98 EPEVKIVSLTIICQGRPDLILPIPFTADS---KKSMFTLKEGSQYRLKFSFTVSNNIVSG 154
P V + L+++C P I + T D KK F LKEG +YR+K +F V+ +IVSG
Sbjct: 63 VPNVTVTRLSLVCDSAPGPIT-MDLTGDLEALKKDTFVLKEGIEYRVKINFKVNKDIVSG 121
Query: 155 LRYTNVVWKTGVRVENTKKMLGTYSPQQEPYTYELEEEITPSGLFARGTYSARTKFVDDD 214
L+Y ++TG+RV+ M+G+Y P+ E Y + E P G+ ARGTY ++ F DDD
Sbjct: 122 LKYVQHTYRTGMRVDKATFMVGSYGPRPEEYEFLTPVEEAPKGMLARGTYHNKSFFTDDD 181
Query: 215 RKCYLDASYRFEIQKNW 231
++ +L + I+K+W
Sbjct: 182 KQDHLTWEWNLAIKKDW 198
>GDIR_BOVIN (P19803) Rho GDP-dissociation inhibitor 1 (Rho GDI 1)
(Rho-GDI alpha)
Length = 204
Score = 142 bits (358), Expect = 7e-34
Identities = 78/189 (41%), Positives = 112/189 (58%), Gaps = 6/189 (3%)
Query: 46 EDEEEEPKLELDLGPQFSLKEQLEKDKDDESLRKWKEQLLGNVDVSAVGEKIEPEVKIVS 105
E+EE+E + Q S++E E DKDDESLRK+KE LLG V VSA + P V +
Sbjct: 17 ENEEDEHSVNYKPPAQKSIQEIQELDKDDESLRKYKEALLGRVAVSA--DPNVPNVVVTR 74
Query: 106 LTIICQGRPDLILPIPFTADS---KKSMFTLKEGSQYRLKFSFTVSNNIVSGLRYTNVVW 162
LT++C P L + T D KK F LKEG +YR+K SF V+ IVSG++Y +
Sbjct: 75 LTLVCSTAPGP-LELDLTGDLESFKKQSFVLKEGVEYRIKISFRVNREIVSGMKYIQHTY 133
Query: 163 KTGVRVENTKKMLGTYSPQQEPYTYELEEEITPSGLFARGTYSARTKFVDDDRKCYLDAS 222
+ GV+++ T M+G+Y P+ E Y + E P G+ ARG+Y+ +++F DDDR +L
Sbjct: 134 RKGVKIDKTDYMVGSYGPRAEEYEFLTPMEEAPKGMLARGSYNIKSRFTDDDRTDHLSWE 193
Query: 223 YRFEIQKNW 231
+ I+K W
Sbjct: 194 WNLTIKKEW 202
>GDIS_HUMAN (P52566) Rho GDP-dissociation inhibitor 2 (Rho GDI 2)
(Rho-GDI beta) (Ly-GDI)
Length = 201
Score = 142 bits (357), Expect = 9e-34
Identities = 79/190 (41%), Positives = 111/190 (57%), Gaps = 6/190 (3%)
Query: 45 DEDEEEEPKLELDLGPQFSLKEQLEKDKDDESLRKWKEQLLGNVDVSAVGEKIEPEVKIV 104
D+D+E + KL PQ SLKE E DKDDESL K+K+ LLG D V + P V +
Sbjct: 13 DDDDELDSKLNYKPPPQKSLKELQEMDKDDESLIKYKKTLLG--DGPVVTDPKAPNVVVT 70
Query: 105 SLTIICQGRPDLILPIPFTADS---KKSMFTLKEGSQYRLKFSFTVSNNIVSGLRYTNVV 161
LT++C+ P I + T D KK LKEGS+YR+K F V+ +IVSGL+Y
Sbjct: 71 RLTLVCESAPGPIT-MDLTGDLEALKKETIVLKEGSEYRVKIHFKVNRDIVSGLKYVQHT 129
Query: 162 WKTGVRVENTKKMLGTYSPQQEPYTYELEEEITPSGLFARGTYSARTKFVDDDRKCYLDA 221
++TGV+V+ M+G+Y P+ E Y + E P G+ ARGTY ++ F DDD++ +L
Sbjct: 130 YRTGVKVDKATFMVGSYGPRPEEYEFLTPVEEAPKGMLARGTYHNKSFFTDDDKQDHLSW 189
Query: 222 SYRFEIQKNW 231
+ I+K W
Sbjct: 190 EWNLSIKKEW 199
>GDIR_MOUSE (Q99PT1) Rho GDP-dissociation inhibitor 1 (Rho GDI 1)
(Rho-GDI alpha) (GDI-1)
Length = 204
Score = 140 bits (354), Expect = 2e-33
Identities = 77/189 (40%), Positives = 112/189 (58%), Gaps = 6/189 (3%)
Query: 46 EDEEEEPKLELDLGPQFSLKEQLEKDKDDESLRKWKEQLLGNVDVSAVGEKIEPEVKIVS 105
E+EE+E + Q S++E E DKDDESLRK+KE LLG V VSA + P V +
Sbjct: 17 ENEEDEHSVNYKPPAQKSIQEIQELDKDDESLRKYKEALLGRVAVSA--DPNVPNVIVTR 74
Query: 106 LTIICQGRPDLILPIPFTADS---KKSMFTLKEGSQYRLKFSFTVSNNIVSGLRYTNVVW 162
LT++C P L + T D KK F LKEG +YR+K SF V+ IVSG++Y +
Sbjct: 75 LTLVCSTAPGP-LELDLTGDLESFKKQSFVLKEGVEYRIKISFRVNREIVSGMKYIQHTY 133
Query: 163 KTGVRVENTKKMLGTYSPQQEPYTYELEEEITPSGLFARGTYSARTKFVDDDRKCYLDAS 222
+ GV+++ T M+G+Y P+ E Y + E P G+ ARG+Y+ +++F DDD+ +L
Sbjct: 134 RKGVKIDKTDYMVGSYGPRAEEYEFLTPMEEAPKGMLARGSYNIKSRFTDDDKTDHLSWE 193
Query: 223 YRFEIQKNW 231
+ I+K W
Sbjct: 194 WNLTIKKEW 202
>GDIR_CAEEL (Q20496) Probable rho GDP-dissociation inhibitor (Rho
GDI)
Length = 191
Score = 112 bits (280), Expect = 8e-25
Identities = 67/172 (38%), Positives = 93/172 (53%), Gaps = 3/172 (1%)
Query: 60 PQFSLKEQLEKDKDDESLRKWKEQLLGNVDVSAVGEKIEPEVKIVSLTIICQGRP--DLI 117
PQ S+ E L DK+DESL+ +K +LLG V V EK V + S+ ++ G+
Sbjct: 20 PQKSIDELLNADKEDESLKVYKAKLLGQGTV-IVDEKNPLRVIVRSVELLINGKTAQSFD 78
Query: 118 LPIPFTADSKKSMFTLKEGSQYRLKFSFTVSNNIVSGLRYTNVVWKTGVRVENTKKMLGT 177
L P + ++KEGS YRL F+F V I SGL Y + V ++G+ VEN K M+G+
Sbjct: 79 LSDPAKLVNSDLSVSIKEGSNYRLSFAFHVQREITSGLHYKHKVKRSGITVENEKYMMGS 138
Query: 178 YSPQQEPYTYELEEEITPSGLFARGTYSARTKFVDDDRKCYLDASYRFEIQK 229
Y+P+ E Y+ E PSG+ RG Y +K DDD YLD + I K
Sbjct: 139 YAPKLEIQEYKSPNEEAPSGMMHRGKYKVYSKITDDDNNVYLDWQWTLHITK 190
>GDIR_YEAST (Q12434) Rho GDP-dissociation inhibitor (Rho GDI)
Length = 202
Score = 101 bits (251), Expect = 2e-21
Identities = 66/195 (33%), Positives = 102/195 (51%), Gaps = 18/195 (9%)
Query: 48 EEEEPKLELDLGPQFSLKEQLEKDKDDESLRKWKEQLLGNVDVSAV---GEKIEPEVKIV 104
EEE + + + ++ E D +DESL KWKE L + DV + G+K + V+ +
Sbjct: 12 EEERNNDQYKVSAKKTVDEYKNLDAEDESLAKWKESLGLSSDVLPLEFPGDKRKVVVQKI 71
Query: 105 SLTIICQGRPDLILPIPFTADSKKSM-------FTLKEGSQYRLKFSFTVSNNIVSGLRY 157
L + + P I F ++K++ + +KE S Y+LK F V + I++GLRY
Sbjct: 72 QLLVNTEPNP-----ITFDLTNEKTIKELASKRYKIKENSIYKLKIVFKVQHEIITGLRY 126
Query: 158 TNVVWKTGVRVENTKKMLGTYSPQQEP---YTYELEEEITPSGLFARGTYSARTKFVDDD 214
+ K G+ V+ LG+Y+P + Y EL E PSG ARG YSA +KF+DDD
Sbjct: 127 VQYIKKAGIAVDKIDDHLGSYAPNTKTKPFYEVELPESEAPSGFLARGNYSAVSKFIDDD 186
Query: 215 RKCYLDASYRFEIQK 229
+ +L ++ EI K
Sbjct: 187 KTNHLTLNWGVEIVK 201
>GDIT_MOUSE (Q62160) Rho GDP-dissociation inhibitor 3 (Rho GDI 3)
(Rho-GDI gamma) (Rho-GDI2)
Length = 225
Score = 100 bits (249), Expect = 3e-21
Identities = 63/204 (30%), Positives = 108/204 (52%), Gaps = 14/204 (6%)
Query: 39 AEEIVPDEDEEEEPKLEL--DLGPQF------SLKEQLEKDKDDESLRKWKEQLLGNVDV 90
A ++ D+D E P E+ ++ P++ S+ + D D SL K+K+ LLG +
Sbjct: 23 ARVLLADKDGESTPSDEVLDEIVPEYQAPGKKSMLAIWQLDPGDVSLVKYKQALLG--PL 80
Query: 91 SAVGEKIEPEVKIVSLTIICQGRPDLILPIPFTADS---KKSMFTLKEGSQYRLKFSFTV 147
+ + P V++ LT++ + P I+ + T D K +F LKEG +Y++K +F V
Sbjct: 81 PPIMDPSLPNVQVTRLTLLTEQAPGPII-MDLTGDLDALKNQVFVLKEGIEYKVKITFKV 139
Query: 148 SNNIVSGLRYTNVVWKTGVRVENTKKMLGTYSPQQEPYTYELEEEITPSGLFARGTYSAR 207
+ IVSGL+ + ++ G+RV+ M+G+Y P+ + Y + E P G ARG Y R
Sbjct: 140 NKEIVSGLKCLHHTYRRGLRVDKAIFMVGSYGPRAQEYEFVTSVEEAPRGALARGLYVVR 199
Query: 208 TKFVDDDRKCYLDASYRFEIQKNW 231
+ F DDDR +L + + ++W
Sbjct: 200 SLFTDDDRLNHLSWEWHLHVCQDW 223
>GDIT_HUMAN (Q99819) Rho GDP-dissociation inhibitor 3 (Rho GDI 3)
(Rho-GDI gamma)
Length = 225
Score = 97.8 bits (242), Expect = 2e-20
Identities = 62/172 (36%), Positives = 91/172 (52%), Gaps = 6/172 (3%)
Query: 63 SLKEQLEKDKDDESLRKWKEQLLGNVDVSAVGEKIEPEVKIVSLTIICQGRPDLILPIPF 122
SL E + D DD SL K+K LLG + AV + P V++ LT++ + P ++ +
Sbjct: 55 SLLEIRQLDPDDRSLAKYKRVLLGPLP-PAVDPSL-PNVQVTRLTLLSEQAPGPVV-MDL 111
Query: 123 TADS---KKSMFTLKEGSQYRLKFSFTVSNNIVSGLRYTNVVWKTGVRVENTKKMLGTYS 179
T D K +F LKEG YR+K SF V IVSGL+ + ++ G+RV+ T M+G+Y
Sbjct: 112 TGDLAVLKDQVFVLKEGVDYRVKISFKVHREIVSGLKCLHHTYRRGLRVDKTVYMVGSYG 171
Query: 180 PQQEPYTYELEEEITPSGLFARGTYSARTKFVDDDRKCYLDASYRFEIQKNW 231
P + Y + E P G RG Y + F DDDR +L + I ++W
Sbjct: 172 PSAQEYEFVTPVEEAPRGALVRGPYLVVSLFTDDDRTHHLSWEWGLCICQDW 223
>GDIR_CAVPO (P80237) Rho GDP-dissociation inhibitor 1 (Rho GDI 1)
(Rho-GDI alpha) (Fragments)
Length = 111
Score = 53.1 bits (126), Expect = 5e-07
Identities = 37/140 (26%), Positives = 60/140 (42%), Gaps = 29/140 (20%)
Query: 90 VSAVGEKIEPEVKIVSLTIICQGRPDLILPIPFTADSKKSMFTLKEGSQYRLKFSFTVSN 149
V+ + EP V + LT++C P L + T D + +++K SF
Sbjct: 1 VAVSADPNEPNVIVTRLTLVCSTAPGP-LELDLTGDLES----------FKIKISF---- 45
Query: 150 NIVSGLRYTNVVWKTGVRVENTKKMLGTYSPQQEPYTYELEEEITPSGLFARGTYSARTK 209
RY ++ GV+++ T M+G+Y P+ E Y + E P G+ AR
Sbjct: 46 ------RYIQHTYRKGVKIDKTDYMVGSYGPRAEEYEFLTPMEEAPKGMLAR-------- 91
Query: 210 FVDDDRKCYLDASYRFEIQK 229
F DDD+ +L I+K
Sbjct: 92 FTDDDKTDHLSGERNLTIKK 111
>YKI2_YEAST (P36080) Hypothetical 50.5 kDa protein in MDH1-VMA5
intergenic region
Length = 434
Score = 35.8 bits (81), Expect = 0.090
Identities = 24/97 (24%), Positives = 48/97 (48%), Gaps = 7/97 (7%)
Query: 12 KDVSFNAHLEEEHKNKGTNKVGVNYDEAEEIVPDEDEEEEPKLEL---DLGPQFSLKEQL 68
K + E K +G + + V ++ I PDEDEEEE +++ D G + L+ +
Sbjct: 89 KHMKMQKQKEATSKVEGDSDLNVEVNDPMIIAPDEDEEEEEDIKVIFDDEGNEIPLESKK 148
Query: 69 EKDKDDESLRK----WKEQLLGNVDVSAVGEKIEPEV 101
+ + D S+ K +E+L ++ A+ K++ ++
Sbjct: 149 DTTEPDRSVEKKSITEEEKLQRKKNLEALRSKLQAKI 185
>A32E_HUMAN (Q9BTT0) Acidic leucine-rich nuclear phosphoprotein 32
family member E (LANP-like protein) (LANP-L)
Length = 268
Score = 35.4 bits (80), Expect = 0.12
Identities = 19/63 (30%), Positives = 35/63 (55%), Gaps = 8/63 (12%)
Query: 21 EEEHKNKGTNKVGVNYDEAEEIVPDEDEEEEP--------KLELDLGPQFSLKEQLEKDK 72
EEE +N+ G +E EE DEDE+E+ + E ++G + +KE+++ ++
Sbjct: 171 EEEEENEAGPPEGYEEEEEEEEEEDEDEDEDEDEAGSELGEGEEEVGLSYLMKEEIQDEE 230
Query: 73 DDE 75
DD+
Sbjct: 231 DDD 233
>YFI6_YEAST (P43597) Hypothetical 137.7 kDa protein in UGS1-FAB1
intergenic region
Length = 1233
Score = 34.7 bits (78), Expect = 0.20
Identities = 29/92 (31%), Positives = 45/92 (48%), Gaps = 13/92 (14%)
Query: 11 TKDVSFNAHLEEEHKNKGTNKVGVNYDEAEEIVPDEDEEEEP------KLELDLGPQFSL 64
TK +S N E++ GTN + V ++ EE +E+EEEE K E G Q ++
Sbjct: 444 TKRISDN----EKNLQHGTNDISVEVEKEEE---EEEEEEENSTFSKVKKENVTGEQEAV 496
Query: 65 KEQLEKDKDDESLRKWKEQLLGNVDVSAVGEK 96
+ ++ES K +E + G+ S GEK
Sbjct: 497 RNNEVSGTEEESTSKGEEIMGGDEKQSEAGEK 528
>YKA4_CAEEL (P34256) Hypothetical protein B0303.4 in chromosome III
Length = 616
Score = 34.3 bits (77), Expect = 0.26
Identities = 33/151 (21%), Positives = 70/151 (45%), Gaps = 13/151 (8%)
Query: 11 TKDVSFNAHLEEEHKNKGTNKVGVNYDEAEEIVPDEDEEEEPKLELDLGPQFSLKEQLEK 70
T+D+ + E++ ++ + E E +D++EE ++E+D G EQ+E
Sbjct: 244 TEDIVIDQQEEDDEEDDDDDSSDDAIIEDHEEEEGQDDDEETEIEIDRGGPIEEIEQVEV 303
Query: 71 DKDDESLRKWKEQLLGNVDVSAVGEKIEPEVKIVSLTII---CQGRPDLILPIPFTADSK 127
++++ + L + +V A E++ E K +I +G+ +++ + F D++
Sbjct: 304 EQNE-------DPPLDDQEVEAQPEQVLQETKESEFPVIEGPSEGK--ILIKLKFMNDTE 354
Query: 128 KSMFTLKEGSQYRLKFS-FTVSNNIVSGLRY 157
K+ + E + + K FT N V L Y
Sbjct: 355 KNTYASLEDTVAKFKVDHFTNLANQVIRLIY 385
>NSB1_MOUSE (Q9JL35) Nucleosome binding protein 1 (Nucleosome
binding protein 45) (NBP-45) (GARP45 protein)
Length = 406
Score = 34.3 bits (77), Expect = 0.26
Identities = 28/86 (32%), Positives = 47/86 (54%), Gaps = 11/86 (12%)
Query: 15 SFNAHLEEEHKNKGTNKVGVNYDEAEEIVPDEDEEEEPKLELDLGPQFSLKEQLEKDKDD 74
S +A + ++ + KG N+ G D EE DE EEE+ E D G + +KEQ ++ ++D
Sbjct: 167 SEDAEVSKDEEEKGDNEKGE--DGKEE--GDEKEEEKDDKEGDTGTEKEVKEQNKEAEED 222
Query: 75 ESLRKWKEQLLGNVDVSAVGEKIEPE 100
+ K KE+ + VG++ +PE
Sbjct: 223 DG--KCKEE-----ENKEVGKEGQPE 241
>DR36_ARATH (Q8VZC7) Probable disease resistance protein At5g45510
Length = 1202
Score = 33.9 bits (76), Expect = 0.34
Identities = 27/78 (34%), Positives = 38/78 (48%), Gaps = 18/78 (23%)
Query: 35 NYDEAEEIVPDEDEEEEPKLE---LDLGPQ-----FSLKEQLEKDKDDESLRKWKEQLLG 86
+++E E DEDEEEE K+E DL P+ KE + K +D+ +K KE
Sbjct: 95 DFEETEVGERDEDEEEEKKVEDLLKDLKPKIEKYLLEKKEAVVKKLEDDKKKKEKE---- 150
Query: 87 NVDVSAVGEKIEPEVKIV 104
EK+E E K+V
Sbjct: 151 ------AAEKLEAEKKLV 162
>YNZ8_YEAST (P53847) Hypothetical 88.1 kDa protein in ATX1-SIP3
intergenic region
Length = 754
Score = 33.5 bits (75), Expect = 0.45
Identities = 21/76 (27%), Positives = 38/76 (49%), Gaps = 4/76 (5%)
Query: 9 STTKDVSFNAHLEEEHKNKGTNKVGVNYDEAEEIVPDEDEEEEPKLELDLGPQFSLKEQL 68
+ +KD +N +E++ + +++ VN D+ EE E E+E + E +++ E +
Sbjct: 406 AVSKDDDWNWEVEDDDADAWGDEIDVNIDDEEEKTNQEKEKEPEEEENAWDEAWAIDENI 465
Query: 69 EKDKDDESLRKWKEQL 84
DD SL KE L
Sbjct: 466 ----DDASLENGKEHL 477
>TAL2_LISMO (P66957) Probable transaldolase 2 (EC 2.2.1.2)
Length = 218
Score = 33.5 bits (75), Expect = 0.45
Identities = 25/110 (22%), Positives = 50/110 (44%), Gaps = 14/110 (12%)
Query: 86 GNVDVSAVGEKIEPEVKIVSLTIICQGRPDLILPIPFTADSKKSMFTL-KEGSQYRLKFS 144
G V +G +E + + I+ + P++++ IP T + K++ TL EG + +
Sbjct: 52 GPVSAEVIG--LEADKMVEEARILAKWAPNVVVKIPMTEEGLKAVHTLTAEGIKTNVTLI 109
Query: 145 FTVSNNIVSGL-----------RYTNVVWKTGVRVENTKKMLGTYSPQQE 183
FTVS +++ R ++ + ++N KK+L Y + E
Sbjct: 110 FTVSQGLMAAKAGATYISPFLGRLDDIGTDGMILIKNLKKVLDNYGLKAE 159
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.311 0.132 0.374
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 29,172,862
Number of Sequences: 164201
Number of extensions: 1343857
Number of successful extensions: 5100
Number of sequences better than 10.0: 128
Number of HSP's better than 10.0 without gapping: 37
Number of HSP's successfully gapped in prelim test: 97
Number of HSP's that attempted gapping in prelim test: 4762
Number of HSP's gapped (non-prelim): 289
length of query: 235
length of database: 59,974,054
effective HSP length: 107
effective length of query: 128
effective length of database: 42,404,547
effective search space: 5427782016
effective search space used: 5427782016
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.8 bits)
S2: 64 (29.3 bits)
Medicago: description of AC144644.6


