

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144564.8 - phase: 0
(655 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
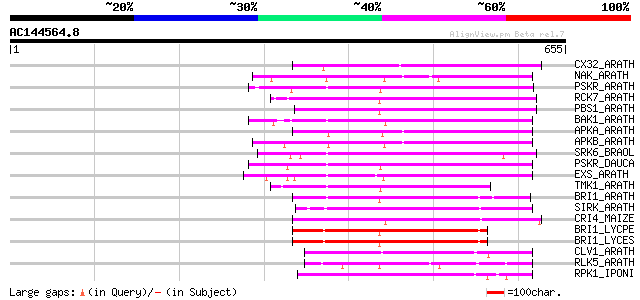
Score E
Sequences producing significant alignments: (bits) Value
CX32_ARATH (P27450) Probable serine/threonine-protein kinase Cx3... 222 2e-57
NAK_ARATH (P43293) Probable serine/threonine-protein kinase NAK ... 217 9e-56
PSKR_ARATH (Q9ZVR7) Putative phytosulfokine receptor precursor (... 214 6e-55
RCK7_ARATH (Q9LQQ8) Putative serine/threonine-protein kinase RLC... 211 6e-54
PBS1_ARATH (Q9FE20) Serine/threonine-protein kinase PBS1 (EC 2.7... 209 2e-53
BAK1_ARATH (Q94F62) BRASSINOSTEROID INSENSITIVE 1-associated rec... 209 2e-53
APKA_ARATH (Q06548) Protein kinase APK1A, chloroplast precursor ... 207 9e-53
APKB_ARATH (P46573) Protein kinase APK1B, chloroplast precursor ... 205 3e-52
SRK6_BRAOL (Q09092) Putative serine/threonine-protein kinase rec... 197 7e-50
PSKR_DAUCA (Q8LPB4) Phytosulfokine receptor precursor (EC 2.7.1.... 193 1e-48
EXS_ARATH (Q9LYN8) Leucine-rich repeat receptor protein kinase E... 192 2e-48
TMK1_ARATH (P43298) Putative receptor protein kinase TMK1 precur... 189 2e-47
BRI1_ARATH (O22476) BRASSINOSTEROID INSENSITIVE 1 precursor (EC ... 182 3e-45
SIRK_ARATH (O64483) Senescence-induced receptor-like serine/thre... 178 5e-44
CRI4_MAIZE (O24585) Putative receptor protein kinase CRINKLY4 pr... 176 2e-43
BRI1_LYCPE (Q8L899) Systemin receptor SR160 precursor (EC 2.7.1.... 176 2e-43
BRI1_LYCES (Q8GUQ5) Brassinosteroid LRR receptor kinase precurso... 175 3e-43
CLV1_ARATH (Q9SYQ8) Receptor protein kinase CLAVATA1 precursor (... 172 2e-42
RLK5_ARATH (P47735) Receptor-like protein kinase 5 precursor (EC... 169 3e-41
RPK1_IPONI (P93194) Receptor-like protein kinase precursor (EC 2... 168 4e-41
>CX32_ARATH (P27450) Probable serine/threonine-protein kinase Cx32,
chloroplast precursor (EC 2.7.1.37)
Length = 419
Score = 222 bits (566), Expect = 2e-57
Identities = 126/305 (41%), Positives = 181/305 (59%), Gaps = 12/305 (3%)
Query: 334 KKISYYELLNATNNFEETQKLGQGGFGGVYKGYFK---------DSNSVAAIKRISADSR 384
K ++ +L AT NF+ LGQGGFG VY+G+ S + AIKR++++S
Sbjct: 72 KVYNFLDLKTATKNFKPDSMLGQGGFGKVYRGWVDATTLAPSRVGSGMIVAIKRLNSESV 131
Query: 385 QGVKQYSAEVKIISQLRHRNLVKLTGWCHKKNELILIYEYMPNGSLDFHLFRGGSILPWN 444
QG ++ +EV + L HRNLVKL G+C + EL+L+YE+MP GSL+ HLFR PW+
Sbjct: 132 QGFAEWRSEVNFLGMLSHRNLVKLLGYCREDKELLLVYEFMPKGSLESHLFRRNDPFPWD 191
Query: 445 LRYNIALGLASALLYLQEEWEKCVIHRDIKSSNIMLDSDFNTKLGDFGLARL-MDHEKGS 503
LR I +G A L +L ++ VI+RD K+SNI+LDS+++ KL DFGLA+L EK
Sbjct: 192 LRIKIVIGAARGLAFL-HSLQREVIYRDFKASNILLDSNYDAKLSDFGLAKLGPADEKSH 250
Query: 504 ETTVVAGTRGYLAPEYIDTSKARKESDIFSFGVVLLEIACGKKAIHHQELEGEVSLVEWV 563
TT + GT GY APEY+ T +SD+F+FGVVLLEI G A + + G+ SLV+W+
Sbjct: 251 VTTRIMGTYGYAAPEYMATGHLYVKSDVFAFGVVLLEIMTGLTAHNTKRPRGQESLVDWL 310
Query: 564 W-ELYGLRNLIVAADPKLCGIFDVKQLECLLVVGLWCANPDITSRPSIKKVIKVLNFEAP 622
EL + D + G + K + + L C PD +RP +K+V++VL
Sbjct: 311 RPELSNKHRVKQIMDKGIKGQYTTKVATEMARITLSCIEPDPKNRPHMKEVVEVLEHIQG 370
Query: 623 LPILP 627
L ++P
Sbjct: 371 LNVVP 375
>NAK_ARATH (P43293) Probable serine/threonine-protein kinase NAK (EC
2.7.1.37)
Length = 389
Score = 217 bits (552), Expect = 9e-56
Identities = 132/352 (37%), Positives = 197/352 (55%), Gaps = 24/352 (6%)
Query: 287 GVCLSWSLVAILIIFLWKKNK-----GKEDEPTSETTSDQDMDDEFQMGAGPKKISYYEL 341
G C S + + W +K G + T+ + + E A K S EL
Sbjct: 2 GGCFSNRIKTDIASSTWLSSKFLSRDGSKGSSTASFSYMPRTEGEILQNANLKNFSLSEL 61
Query: 342 LNATNNFEETQKLGQGGFGGVYKGYFKDSNS---------VAAIKRISADSRQGVKQYSA 392
+AT NF +G+GGFG V+KG+ +S+ V A+KR++ + QG +++ A
Sbjct: 62 KSATRNFRPDSVVGEGGFGCVFKGWIDESSLAPSKPGTGIVIAVKRLNQEGFQGHREWLA 121
Query: 393 EVKIISQLRHRNLVKLTGWCHKKNELILIYEYMPNGSLDFHLFRGGSI---LPWNLRYNI 449
E+ + QL H NLVKL G+C ++ +L+YE+M GSL+ HLFR G+ L WN R +
Sbjct: 122 EINYLGQLDHPNLVKLIGYCLEEEHRLLVYEFMTRGSLENHLFRRGTFYQPLSWNTRVRM 181
Query: 450 ALGLASALLYLQEEWEKCVIHRDIKSSNIMLDSDFNTKLGDFGLARLMDHEKGSE---TT 506
ALG A L +L + VI+RD K+SNI+LDS++N KL DFGLAR D G +T
Sbjct: 182 ALGAARGLAFLHNA-QPQVIYRDFKASNILLDSNYNAKLSDFGLAR--DGPMGDNSHVST 238
Query: 507 VVAGTRGYLAPEYIDTSKARKESDIFSFGVVLLEIACGKKAIHHQELEGEVSLVEWVWE- 565
V GT+GY APEY+ T +SD++SFGVVLLE+ G++AI + GE +LV+W
Sbjct: 239 RVMGTQGYAAPEYLATGHLSVKSDVYSFGVVLLELLSGRRAIDKNQPVGEHNLVDWARPY 298
Query: 566 LYGLRNLIVAADPKLCGIFDVKQLECLLVVGLWCANPDITSRPSIKKVIKVL 617
L R L+ DP+L G + + + + V+ L C + D SRP++ +++K +
Sbjct: 299 LTNKRRLLRVMDPRLQGQYSLTRALKIAVLALDCISIDAKSRPTMNEIVKTM 350
>PSKR_ARATH (Q9ZVR7) Putative phytosulfokine receptor precursor (EC
2.7.1.37) (Phytosulfokine LRR receptor kinase)
Length = 1008
Score = 214 bits (545), Expect = 6e-55
Identities = 127/348 (36%), Positives = 198/348 (56%), Gaps = 15/348 (4%)
Query: 282 GLAVGGVCLSWSLVAILIIFLWKKNKGKEDEPTSETTSDQDMDDEFQMGA--------GP 333
G+A G V L L + +I L + + E +P E + + + ++G+
Sbjct: 663 GIAFGSVFL---LTLLSLIVLRARRRSGEVDPEIEESESMNRKELGEIGSKLVVLFQSND 719
Query: 334 KKISYYELLNATNNFEETQKLGQGGFGGVYKGYFKDSNSVAAIKRISADSRQGVKQYSAE 393
K++SY +LL++TN+F++ +G GGFG VYK D VA IK++S D Q +++ AE
Sbjct: 720 KELSYDDLLDSTNSFDQANIIGCGGFGMVYKATLPDGKKVA-IKKLSGDCGQIEREFEAE 778
Query: 394 VKIISQLRHRNLVKLTGWCHKKNELILIYEYMPNGSLDFHLFR---GGSILPWNLRYNIA 450
V+ +S+ +H NLV L G+C KN+ +LIY YM NGSLD+ L G ++L W R IA
Sbjct: 779 VETLSRAQHPNLVLLRGFCFYKNDRLLIYSYMENGSLDYWLHERNDGPALLKWKTRLRIA 838
Query: 451 LGLASALLYLQEEWEKCVIHRDIKSSNIMLDSDFNTKLGDFGLARLMDHEKGSETTVVAG 510
G A LLYL E + ++HRDIKSSNI+LD +FN+ L DFGLARLM + +T + G
Sbjct: 839 QGAAKGLLYLHEGCDPHILHRDIKSSNILLDENFNSHLADFGLARLMSPYETHVSTDLVG 898
Query: 511 TRGYLAPEYIDTSKARKESDIFSFGVVLLEIACGKKAIHHQELEGEVSLVEWVWELYGLR 570
T GY+ PEY S A + D++SFGVVLLE+ K+ + + +G L+ WV ++
Sbjct: 899 TLGYIPPEYGQASVATYKGDVYSFGVVLLELLTDKRPVDMCKPKGCRDLISWVVKMKHES 958
Query: 571 NLIVAADPKLCGIFDVKQLECLLVVGLWCANPDITSRPSIKKVIKVLN 618
DP + + K++ +L + C + + RP+ ++++ L+
Sbjct: 959 RASEVFDPLIYSKENDKEMFRVLEIACLCLSENPKQRPTTQQLVSWLD 1006
>RCK7_ARATH (Q9LQQ8) Putative serine/threonine-protein kinase
RLCKVII (EC 2.7.1.37)
Length = 423
Score = 211 bits (536), Expect = 6e-54
Identities = 127/319 (39%), Positives = 179/319 (55%), Gaps = 7/319 (2%)
Query: 308 GKEDEPTSETTSDQDMDDEFQMGAGPKKISYYELLNATNNFEETQKLGQGGFGGVYKGYF 367
GKED+ S +++D+ G + ++ EL AT NF LG+GGFG V+KG
Sbjct: 65 GKEDQ-LSLDVKGLNLNDQVT-GKKAQTFTFQELAEATGNFRSDCFLGEGGFGKVFKGTI 122
Query: 368 KDSNSVAAIKRISADSRQGVKQYSAEVKIISQLRHRNLVKLTGWCHKKNELILIYEYMPN 427
+ + V AIK++ + QG++++ EV +S H NLVKL G+C + ++ +L+YEYMP
Sbjct: 123 EKLDQVVAIKQLDRNGVQGIREFVVEVLTLSLADHPNLVKLIGFCAEGDQRLLVYEYMPQ 182
Query: 428 GSLDFHLF---RGGSILPWNLRYNIALGLASALLYLQEEWEKCVIHRDIKSSNIMLDSDF 484
GSL+ HL G L WN R IA G A L YL + VI+RD+K SNI+L D+
Sbjct: 183 GSLEDHLHVLPSGKKPLDWNTRMKIAAGAARGLEYLHDRMTPPVIYRDLKCSNILLGEDY 242
Query: 485 NTKLGDFGLARL-MDHEKGSETTVVAGTRGYLAPEYIDTSKARKESDIFSFGVVLLEIAC 543
KL DFGLA++ +K +T V GT GY AP+Y T + +SDI+SFGVVLLE+
Sbjct: 243 QPKLSDFGLAKVGPSGDKTHVSTRVMGTYGYCAPDYAMTGQLTFKSDIYSFGVVLLELIT 302
Query: 544 GKKAIHHQELEGEVSLVEWVWELY-GLRNLIVAADPKLCGIFDVKQLECLLVVGLWCANP 602
G+KAI + + + +LV W L+ RN DP L G + V+ L L + C
Sbjct: 303 GRKAIDNTKTRKDQNLVGWARPLFKDRRNFPKMVDPLLQGQYPVRGLYQALAISAMCVQE 362
Query: 603 DITSRPSIKKVIKVLNFEA 621
T RP + V+ LNF A
Sbjct: 363 QPTMRPVVSDVVLALNFLA 381
>PBS1_ARATH (Q9FE20) Serine/threonine-protein kinase PBS1 (EC
2.7.1.37) (AvrPphB susceptible protein 1)
Length = 456
Score = 209 bits (532), Expect = 2e-53
Identities = 121/290 (41%), Positives = 166/290 (56%), Gaps = 5/290 (1%)
Query: 337 SYYELLNATNNFEETQKLGQGGFGGVYKGYFKDSNSVAAIKRISADSRQGVKQYSAEVKI 396
++ EL AT NF LG+GGFG VYKG + V A+K++ + QG +++ EV +
Sbjct: 75 AFRELAAATMNFHPDTFLGEGGFGRVYKGRLDSTGQVVAVKQLDRNGLQGNREFLVEVLM 134
Query: 397 ISQLRHRNLVKLTGWCHKKNELILIYEYMPNGSLDFHLF---RGGSILPWNLRYNIALGL 453
+S L H NLV L G+C ++ +L+YE+MP GSL+ HL L WN+R IA G
Sbjct: 135 LSLLHHPNLVNLIGYCADGDQRLLVYEFMPLGSLEDHLHDLPPDKEALDWNMRMKIAAGA 194
Query: 454 ASALLYLQEEWEKCVIHRDIKSSNIMLDSDFNTKLGDFGLARL-MDHEKGSETTVVAGTR 512
A L +L ++ VI+RD KSSNI+LD F+ KL DFGLA+L +K +T V GT
Sbjct: 195 AKGLEFLHDKANPPVIYRDFKSSNILLDEGFHPKLSDFGLAKLGPTGDKSHVSTRVMGTY 254
Query: 513 GYLAPEYIDTSKARKESDIFSFGVVLLEIACGKKAIHHQELEGEVSLVEWVWELYG-LRN 571
GY APEY T + +SD++SFGVV LE+ G+KAI + GE +LV W L+ R
Sbjct: 255 GYCAPEYAMTGQLTVKSDVYSFGVVFLELITGRKAIDSEMPHGEQNLVAWARPLFNDRRK 314
Query: 572 LIVAADPKLCGIFDVKQLECLLVVGLWCANPDITSRPSIKKVIKVLNFEA 621
I ADP+L G F + L L V C +RP I V+ L++ A
Sbjct: 315 FIKLADPRLKGRFPTRALYQALAVASMCIQEQAATRPLIADVVTALSYLA 364
>BAK1_ARATH (Q94F62) BRASSINOSTEROID INSENSITIVE 1-associated
receptor kinase 1 precursor (EC 2.7.1.37)
(BRI1-associated receptor kinase 1) (Somatic
embryogenesis receptor-like kinase 3)
Length = 615
Score = 209 bits (532), Expect = 2e-53
Identities = 128/345 (37%), Positives = 195/345 (56%), Gaps = 19/345 (5%)
Query: 282 GLAVGGVCLSWSLVAILIIFLWKKNKGKE---DEPTSETTSDQDMDDEFQMGAGPKKISY 338
G G L +++ AI + + W++ K ++ D P E D E +G K+ S
Sbjct: 229 GGVAAGAALLFAVPAIALAW-WRRKKPQDHFFDVPAEE-------DPEVHLGQ-LKRFSL 279
Query: 339 YELLNATNNFEETQKLGQGGFGGVYKGYFKDSNSVAAIKRISADSRQGVK-QYSAEVKII 397
EL A++NF LG+GGFG VYKG D VA +KR+ + QG + Q+ EV++I
Sbjct: 280 RELQVASDNFSNKNILGRGGFGKVYKGRLADGTLVA-VKRLKEERTQGGELQFQTEVEMI 338
Query: 398 SQLRHRNLVKLTGWCHKKNELILIYEYMPNGSLDFHLFRGGSILP---WNLRYNIALGLA 454
S HRNL++L G+C E +L+Y YM NGS+ L P W R IALG A
Sbjct: 339 SMAVHRNLLRLRGFCMTPTERLLVYPYMANGSVASCLRERPESQPPLDWPKRQRIALGSA 398
Query: 455 SALLYLQEEWEKCVIHRDIKSSNIMLDSDFNTKLGDFGLARLMDHEKGSETTVVAGTRGY 514
L YL + + +IHRD+K++NI+LD +F +GDFGLA+LMD++ TT V GT G+
Sbjct: 399 RGLAYLHDHCDPKIIHRDVKAANILLDEEFEAVVGDFGLAKLMDYKDTHVTTAVRGTIGH 458
Query: 515 LAPEYIDTSKARKESDIFSFGVVLLEIACGKKAIHHQEL--EGEVSLVEWVWELYGLRNL 572
+APEY+ T K+ +++D+F +GV+LLE+ G++A L + +V L++WV L + L
Sbjct: 459 IAPEYLSTGKSSEKTDVFGYGVMLLELITGQRAFDLARLANDDDVMLLDWVKGLLKEKKL 518
Query: 573 IVAADPKLCGIFDVKQLECLLVVGLWCANPDITSRPSIKKVIKVL 617
D L G + +++E L+ V L C RP + +V+++L
Sbjct: 519 EALVDVDLQGNYKDEEVEQLIQVALLCTQSSPMERPKMSEVVRML 563
>APKA_ARATH (Q06548) Protein kinase APK1A, chloroplast precursor (EC
2.7.1.-)
Length = 410
Score = 207 bits (526), Expect = 9e-53
Identities = 119/298 (39%), Positives = 173/298 (57%), Gaps = 15/298 (5%)
Query: 334 KKISYYELLNATNNFEETQKLGQGGFGGVYKGYFKDSNSVA---------AIKRISADSR 384
K S+ EL +AT NF LG+GGFG V+KG+ + + A A+K+++ D
Sbjct: 54 KSFSFAELKSATRNFRPDSVLGEGGFGCVFKGWIDEKSLTASRPGTGLVIAVKKLNQDGW 113
Query: 385 QGVKQYSAEVKIISQLRHRNLVKLTGWCHKKNELILIYEYMPNGSLDFHLFRGG---SIL 441
QG +++ AEV + Q HR+LVKL G+C + +L+YE+MP GSL+ HLFR G L
Sbjct: 114 QGHQEWLAEVNYLGQFSHRHLVKLIGYCLEDEHRLLVYEFMPRGSLENHLFRRGLYFQPL 173
Query: 442 PWNLRYNIALGLASALLYLQEEWEKCVIHRDIKSSNIMLDSDFNTKLGDFGLARLMD-HE 500
W LR +ALG A L +L E VI+RD K+SNI+LDS++N KL DFGLA+ +
Sbjct: 174 SWKLRLKVALGAAKGLAFLHSS-ETRVIYRDFKTSNILLDSEYNAKLSDFGLAKDGPIGD 232
Query: 501 KGSETTVVAGTRGYLAPEYIDTSKARKESDIFSFGVVLLEIACGKKAIHHQELEGEVSLV 560
K +T V GT GY APEY+ T +SD++SFGVVLLE+ G++A+ GE +LV
Sbjct: 233 KSHVSTRVMGTHGYAAPEYLATGHLTTKSDVYSFGVVLLELLSGRRAVDKNRPSGERNLV 292
Query: 561 EWVWE-LYGLRNLIVAADPKLCGIFDVKQLECLLVVGLWCANPDITSRPSIKKVIKVL 617
EW L R + D +L + +++ + + L C +I RP++ +V+ L
Sbjct: 293 EWAKPYLVNKRKIFRVIDNRLQDQYSMEEACKVATLSLRCLTTEIKLRPNMSEVVSHL 350
>APKB_ARATH (P46573) Protein kinase APK1B, chloroplast precursor (EC
2.7.1.-)
Length = 412
Score = 205 bits (522), Expect = 3e-52
Identities = 126/351 (35%), Positives = 187/351 (52%), Gaps = 21/351 (5%)
Query: 287 GVCLSWSLVAILIIFLWKKNKGKEDEPTSETTSDQDM------DDEFQMGAGPKKISYYE 340
G+CLS + A+ K + ++ +S + + E K ++ E
Sbjct: 2 GICLSAQIKAVSPGASPKYMSSEANDSLGSKSSSVSIRTNPRTEGEILQSPNLKSFTFAE 61
Query: 341 LLNATNNFEETQKLGQGGFGGVYKGYFKDSNSVA---------AIKRISADSRQGVKQYS 391
L AT NF LG+GGFG V+KG+ + A A+K+++ D QG +++
Sbjct: 62 LKAATRNFRPDSVLGEGGFGSVFKGWIDEQTLTASKPGTGVVIAVKKLNQDGWQGHQEWL 121
Query: 392 AEVKIISQLRHRNLVKLTGWCHKKNELILIYEYMPNGSLDFHLFRGGSI---LPWNLRYN 448
AEV + Q H NLVKL G+C + +L+YE+MP GSL+ HLFR GS L W LR
Sbjct: 122 AEVNYLGQFSHPNLVKLIGYCLEDEHRLLVYEFMPRGSLENHLFRRGSYFQPLSWTLRLK 181
Query: 449 IALGLASALLYLQEEWEKCVIHRDIKSSNIMLDSDFNTKLGDFGLAR-LMDHEKGSETTV 507
+ALG A L +L E VI+RD K+SNI+LDS++N KL DFGLA+ +K +T
Sbjct: 182 VALGAAKGLAFLHNA-ETSVIYRDFKTSNILLDSEYNAKLSDFGLAKDGPTGDKSHVSTR 240
Query: 508 VAGTRGYLAPEYIDTSKARKESDIFSFGVVLLEIACGKKAIHHQELEGEVSLVEWVWELY 567
+ GT GY APEY+ T +SD++S+GVVLLE+ G++A+ GE LVEW L
Sbjct: 241 IMGTYGYAAPEYLATGHLTTKSDVYSYGVVLLEVLSGRRAVDKNRPPGEQKLVEWARPLL 300
Query: 568 -GLRNLIVAADPKLCGIFDVKQLECLLVVGLWCANPDITSRPSIKKVIKVL 617
R L D +L + +++ + + L C +I RP++ +V+ L
Sbjct: 301 ANKRKLFRVIDNRLQDQYSMEEACKVATLALRCLTFEIKLRPNMNEVVSHL 351
>SRK6_BRAOL (Q09092) Putative serine/threonine-protein kinase
receptor precursor (EC 2.7.1.37) (S-receptor kinase)
(SRK)
Length = 849
Score = 197 bits (501), Expect = 7e-50
Identities = 125/357 (35%), Positives = 192/357 (53%), Gaps = 29/357 (8%)
Query: 293 SLVAILIIFLWKKNKGKEDEPTSETTSDQDMDDEFQMG----------AGPKKISYYEL- 341
S++ +LI+F K K K + ++ + ++ + M +G K EL
Sbjct: 455 SVLLLLIMFCLWKRKQKRAKASAISIANTQRNQNLPMNEMVLSSKREFSGEYKFEELELP 514
Query: 342 -------LNATNNFEETQKLGQGGFGGVYKGYFKDSNSVAAIKRISADSRQGVKQYSAEV 394
+ AT NF KLGQGGFG VYKG D +A +KR+S S QG ++ EV
Sbjct: 515 LIEMETVVKATENFSSCNKLGQGGFGIVYKGRLLDGKEIA-VKRLSKTSVQGTDEFMNEV 573
Query: 395 KIISQLRHRNLVKLTGWCHKKNELILIYEYMPNGSLDFHLF--RGGSILPWNLRYNIALG 452
+I++L+H NLV++ G C + +E +LIYEY+ N SLD +LF S L WN R++I G
Sbjct: 574 TLIARLQHINLVQVLGCCIEGDEKMLIYEYLENLSLDSYLFGKTRRSKLNWNERFDITNG 633
Query: 453 LASALLYLQEEWEKCVIHRDIKSSNIMLDSDFNTKLGDFGLARLMDHEKGSETTV-VAGT 511
+A LLYL ++ +IHRD+K SNI+LD + K+ DFG+AR+ + ++ T+ V GT
Sbjct: 634 VARGLLYLHQDSRFRIIHRDLKVSNILLDKNMIPKISDFGMARIFERDETEANTMKVVGT 693
Query: 512 RGYLAPEYIDTSKARKESDIFSFGVVLLEIACGKKAIHHQELEGEVSLVEWVWELYGLRN 571
GY++PEY ++SD+FSFGV++LEI GKK L+ E L+ +VW +
Sbjct: 694 YGYMSPEYAMYGIFSEKSDVFSFGVIVLEIVSGKKNRGFYNLDYENDLLSYVWSRWKEGR 753
Query: 572 LIVAADPKLC-------GIFDVKQLECLLVVGLWCANPDITSRPSIKKVIKVLNFEA 621
+ DP + IF +++ + +GL C RP++ V+ + EA
Sbjct: 754 ALEIVDPVIVDSLSSQPSIFQPQEVLKCIQIGLLCVQELAEHRPAMSSVVWMFGSEA 810
>PSKR_DAUCA (Q8LPB4) Phytosulfokine receptor precursor (EC 2.7.1.37)
(Phytosulfokine LRR receptor kinase)
Length = 1021
Score = 193 bits (490), Expect = 1e-48
Identities = 119/343 (34%), Positives = 184/343 (52%), Gaps = 9/343 (2%)
Query: 283 LAVG-GVCLSWSLVAILIIFLWKKNKGKEDEPTSETTSDQDMDDE----FQMGAGPKKIS 337
+AVG G+ + L L+I L ++G+ D + ++ F ++S
Sbjct: 673 VAVGTGLGTVFLLTVTLLIILRTTSRGEVDPEKKADADEIELGSRSVVLFHNKDSNNELS 732
Query: 338 YYELLNATNNFEETQKLGQGGFGGVYKGYFKDSNSVAAIKRISADSRQGVKQYSAEVKII 397
++L +T++F + +G GGFG VYK D VA IKR+S D+ Q +++ AEV+ +
Sbjct: 733 LDDILKSTSSFNQANIIGCGGFGLVYKATLPDGTKVA-IKRLSGDTGQMDREFQAEVETL 791
Query: 398 SQLRHRNLVKLTGWCHKKNELILIYEYMPNGSLDFHLFR---GGSILPWNLRYNIALGLA 454
S+ +H NLV L G+C+ KN+ +LIY YM NGSLD+ L G L W R IA G A
Sbjct: 792 SRAQHPNLVHLLGYCNYKNDKLLIYSYMDNGSLDYWLHEKVDGPPSLDWKTRLRIARGAA 851
Query: 455 SALLYLQEEWEKCVIHRDIKSSNIMLDSDFNTKLGDFGLARLMDHEKGSETTVVAGTRGY 514
L YL + E ++HRDIKSSNI+L F L DFGLARL+ TT + GT GY
Sbjct: 852 EGLAYLHQSCEPHILHRDIKSSNILLSDTFVAHLADFGLARLILPYDTHVTTDLVGTLGY 911
Query: 515 LAPEYIDTSKARKESDIFSFGVVLLEIACGKKAIHHQELEGEVSLVEWVWELYGLRNLIV 574
+ PEY S A + D++SFGVVLLE+ G++ + + G L+ WV ++ +
Sbjct: 912 IPPEYGQASVATYKGDVYSFGVVLLELLTGRRPMDVCKPRGSRDLISWVLQMKTEKRESE 971
Query: 575 AADPKLCGIFDVKQLECLLVVGLWCANPDITSRPSIKKVIKVL 617
DP + +++ +L + C + +RP+ ++++ L
Sbjct: 972 IFDPFIYDKDHAEEMLLVLEIACRCLGENPKTRPTTQQLVSWL 1014
>EXS_ARATH (Q9LYN8) Leucine-rich repeat receptor protein kinase EXS
precursor (EC 2.7.1.37) (Extra sporogenous cells protein)
(EXCESS MICROSPOROCYTES1 protein)
Length = 1192
Score = 192 bits (488), Expect = 2e-48
Identities = 120/370 (32%), Positives = 194/370 (52%), Gaps = 31/370 (8%)
Query: 277 TKLWEGLAVGGVCLSWSLVAILIIF------LWKKNKGKEDEPTSETTSDQDMDDE---F 327
TKL + G+ L ++++ + +F + K+ K ++D E + + D+ F
Sbjct: 822 TKLRSAWGIAGLMLGFTIIVFVFVFSLRRWAMTKRVKQRDDPERMEESRLKGFVDQNLYF 881
Query: 328 QMGAGPK---------------KISYYELLNATNNFEETQKLGQGGFGGVYKGYFKDSNS 372
G+ + K+ +++ AT++F + +G GGFG VYK +
Sbjct: 882 LSGSRSREPLSINIAMFEQPLLKVRLGDIVEATDHFSKKNIIGDGGFGTVYKACLPGEKT 941
Query: 373 VAAIKRISADSRQGVKQYSAEVKIISQLRHRNLVKLTGWCHKKNELILIYEYMPNGSLDF 432
VA +K++S QG +++ AE++ + +++H NLV L G+C E +L+YEYM NGSLD
Sbjct: 942 VA-VKKLSEAKTQGNREFMAEMETLGKVKHPNLVSLLGYCSFSEEKLLVYEYMVNGSLD- 999
Query: 433 HLFRGGS----ILPWNLRYNIALGLASALLYLQEEWEKCVIHRDIKSSNIMLDSDFNTKL 488
H R + +L W+ R IA+G A L +L + +IHRDIK+SNI+LD DF K+
Sbjct: 1000 HWLRNQTGMLEVLDWSKRLKIAVGAARGLAFLHHGFIPHIIHRDIKASNILLDGDFEPKV 1059
Query: 489 GDFGLARLMDHEKGSETTVVAGTRGYLAPEYIDTSKARKESDIFSFGVVLLEIACGKKAI 548
DFGLARL+ + +TV+AGT GY+ PEY +++A + D++SFGV+LLE+ GK+
Sbjct: 1060 ADFGLARLISACESHVSTVIAGTFGYIPPEYGQSARATTKGDVYSFGVILLELVTGKEPT 1119
Query: 549 HHQELEGE-VSLVEWVWELYGLRNLIVAADPKLCGIFDVKQLECLLVVGLWCANPDITSR 607
E E +LV W + + DP L + LL + + C R
Sbjct: 1120 GPDFKESEGGNLVGWAIQKINQGKAVDVIDPLLVSVALKNSQLRLLQIAMLCLAETPAKR 1179
Query: 608 PSIKKVIKVL 617
P++ V+K L
Sbjct: 1180 PNMLDVLKAL 1189
>TMK1_ARATH (P43298) Putative receptor protein kinase TMK1 precursor
(EC 2.7.1.-)
Length = 942
Score = 189 bits (480), Expect = 2e-47
Identities = 109/267 (40%), Positives = 152/267 (56%), Gaps = 10/267 (3%)
Query: 308 GKEDEPTSETTSDQDMDDEFQM-GAGPKKISYYELLNATNNFEETQKLGQGGFGGVYKGY 366
G D T TS+ + D QM AG IS L + TNNF LG GGFG VYKG
Sbjct: 549 GISDTYTLPGTSE--VGDNIQMVEAGNMLISIQVLRSVTNNFSSDNILGSGGFGVVYKGE 606
Query: 367 FKDSNSVAAIKRISAD--SRQGVKQYSAEVKIISQLRHRNLVKLTGWCHKKNELILIYEY 424
D +A +KR+ + +G ++ +E+ +++++RHR+LV L G+C NE +L+YEY
Sbjct: 607 LHDGTKIA-VKRMENGVIAGKGFAEFKSEIAVLTKVRHRHLVTLLGYCLDGNEKLLVYEY 665
Query: 425 MPNGSLDFHLFR----GGSILPWNLRYNIALGLASALLYLQEEWEKCVIHRDIKSSNIML 480
MP G+L HLF G L W R +AL +A + YL + IHRD+K SNI+L
Sbjct: 666 MPQGTLSRHLFEWSEEGLKPLLWKQRLTLALDVARGVEYLHGLAHQSFIHRDLKPSNILL 725
Query: 481 DSDFNTKLGDFGLARLMDHEKGSETTVVAGTRGYLAPEYIDTSKARKESDIFSFGVVLLE 540
D K+ DFGL RL KGS T +AGT GYLAPEY T + + D++SFGV+L+E
Sbjct: 726 GDDMRAKVADFGLVRLAPEGKGSIETRIAGTFGYLAPEYAVTGRVTTKVDVYSFGVILME 785
Query: 541 IACGKKAIHHQELEGEVSLVEWVWELY 567
+ G+K++ + E + LV W +Y
Sbjct: 786 LITGRKSLDESQPEESIHLVSWFKRMY 812
>BRI1_ARATH (O22476) BRASSINOSTEROID INSENSITIVE 1 precursor (EC
2.7.1.37) (AtBRI1) (Brassinosteroid LRR receptor kinase)
Length = 1196
Score = 182 bits (461), Expect = 3e-45
Identities = 107/287 (37%), Positives = 169/287 (58%), Gaps = 9/287 (3%)
Query: 334 KKISYYELLNATNNFEETQKLGQGGFGGVYKGYFKDSNSVAAIKRISADSRQGVKQYSAE 393
+K+++ +LL ATN F +G GGFG VYK KD ++VA IK++ S QG +++ AE
Sbjct: 869 RKLTFADLLQATNGFHNDSLIGSGGFGDVYKAILKDGSAVA-IKKLIHVSGQGDREFMAE 927
Query: 394 VKIISQLRHRNLVKLTGWCHKKNELILIYEYMPNGSLDFHLF---RGGSILPWNLRYNIA 450
++ I +++HRNLV L G+C +E +L+YE+M GSL+ L + G L W+ R IA
Sbjct: 928 METIGKIKHRNLVPLLGYCKVGDERLLVYEFMKYGSLEDVLHDPKKAGVKLNWSTRRKIA 987
Query: 451 LGLASALLYLQEEWEKCVIHRDIKSSNIMLDSDFNTKLGDFGLARLMD-HEKGSETTVVA 509
+G A L +L +IHRD+KSSN++LD + ++ DFG+ARLM + + +A
Sbjct: 988 IGSARGLAFLHHNCSPHIIHRDMKSSNVLLDENLEARVSDFGMARLMSAMDTHLSVSTLA 1047
Query: 510 GTRGYLAPEYIDTSKARKESDIFSFGVVLLEIACGKKAIHHQELEGEVSLVEWVWELYGL 569
GT GY+ PEY + + + D++S+GVVLLE+ GK+ + G+ +LV WV + L
Sbjct: 1048 GTPGYVPPEYYQSFRCSTKGDVYSYGVVLLELLTGKRPTDSPDF-GDNNLVGWVKQHAKL 1106
Query: 570 RNLIVAADPKLCGIFDVKQLECL--LVVGLWCANPDITSRPSIKKVI 614
R + DP+L ++E L L V + C + RP++ +V+
Sbjct: 1107 R-ISDVFDPELMKEDPALEIELLQHLKVAVACLDDRAWRRPTMVQVM 1152
>SIRK_ARATH (O64483) Senescence-induced receptor-like
serine/threonine kinase precursor (FLG22-induced
receptor-like kinase 1)
Length = 876
Score = 178 bits (451), Expect = 5e-44
Identities = 104/282 (36%), Positives = 159/282 (55%), Gaps = 7/282 (2%)
Query: 338 YYELLNATNNFEETQKLGQGGFGGVYKGYFKDSNSVAAIKRISADSRQGVKQYSAEVKII 397
Y E++N TNNFE +G+GGFG VY G A+K +S +S QG K++ AEV ++
Sbjct: 566 YSEVVNITNNFERV--IGKGGFGKVYHGVINGEQ--VAVKVLSEESAQGYKEFRAEVDLL 621
Query: 398 SQLRHRNLVKLTGWCHKKNELILIYEYMPNGSL-DFHLFRGGSILPWNLRYNIALGLASA 456
++ H NL L G+C++ N ++LIYEYM N +L D+ + IL W R I+L A
Sbjct: 622 MRVHHTNLTSLVGYCNEINHMVLIYEYMANENLGDYLAGKRSFILSWEERLKISLDAAQG 681
Query: 457 LLYLQEEWEKCVIHRDIKSSNIMLDSDFNTKLGDFGLARLMDHE-KGSETTVVAGTRGYL 515
L YL + ++HRD+K +NI+L+ K+ DFGL+R E G +TVVAG+ GYL
Sbjct: 682 LEYLHNGCKPPIVHRDVKPTNILLNEKLQAKMADFGLSRSFSVEGSGQISTVVAGSIGYL 741
Query: 516 APEYIDTSKARKESDIFSFGVVLLEIACGKKAIHHQELEGEVSLVEWVWELYGLRNLIVA 575
PEY T + ++SD++S GVVLLE+ G+ AI + E +V + + V + ++
Sbjct: 742 DPEYYSTRQMNEKSDVYSLGVVLLEVITGQPAIASSKTE-KVHISDHVRSILANGDIRGI 800
Query: 576 ADPKLCGIFDVKQLECLLVVGLWCANPDITSRPSIKKVIKVL 617
D +L +DV + + L C RP++ +V+ L
Sbjct: 801 VDQRLRERYDVGSAWKMSEIALACTEHTSAQRPTMSQVVMEL 842
>CRI4_MAIZE (O24585) Putative receptor protein kinase CRINKLY4
precursor (EC 2.7.1.-)
Length = 901
Score = 176 bits (446), Expect = 2e-43
Identities = 116/309 (37%), Positives = 164/309 (52%), Gaps = 17/309 (5%)
Query: 334 KKISYYELLNATNNFEETQKLGQGGFGGVYKGYFKDSNSVAAIKRISA-DSRQGVKQYSA 392
++ SY EL AT F E ++G+G F V+KG +D VA + I A D ++ K++
Sbjct: 491 QEFSYEELEQATGGFSEDSQVGKGSFSCVFKGILRDGTVVAVKRAIKASDVKKSSKEFHN 550
Query: 393 EVKIISQLRHRNLVKLTGWCHKKNELILIYEYMPNGSLDFHLFRGGSILP----WNLRYN 448
E+ ++S+L H +L+ L G+C +E +L+YE+M +GSL HL L W R
Sbjct: 551 ELDLLSRLNHAHLLNLLGYCEDGSERLLVYEFMAHGSLYQHLHGKDPNLKKRLNWARRVT 610
Query: 449 IALGLASALLYLQEEWEKCVIHRDIKSSNIMLDSDFNTKLGDFGLARLMDHEKGSETTVV 508
IA+ A + YL VIHRDIKSSNI++D D N ++ DFGL+ L + G+ + +
Sbjct: 611 IAVQAARGIEYLHGYACPPVIHRDIKSSNILIDEDHNARVADFGLSILGPADSGTPLSEL 670
Query: 509 -AGTRGYLAPEYIDTSKARKESDIFSFGVVLLEIACGKKAIHHQELEGEVSLVEWVWELY 567
AGT GYL PEY +SD++SFGVVLLEI G+KAI Q EG ++VEW L
Sbjct: 671 PAGTLGYLDPEYYRLHYLTTKSDVYSFGVVLLEILSGRKAIDMQFEEG--NIVEWAVPLI 728
Query: 568 GLRNLIVAADPKLCGIFDVKQLECLLVVGLWCANPDITSRPSIKKVIKVLNFEAPL---- 623
++ DP L D++ L+ + V C RPS+ KV L L
Sbjct: 729 KAGDIFAILDPVLSPPSDLEALKKIASVACKCVRMRGKDRPSMDKVTTALEHALALLMGS 788
Query: 624 -----PILP 627
PILP
Sbjct: 789 PCIEQPILP 797
>BRI1_LYCPE (Q8L899) Systemin receptor SR160 precursor (EC 2.7.1.37)
(Brassinosteroid LRR receptor kinase)
Length = 1207
Score = 176 bits (446), Expect = 2e-43
Identities = 96/234 (41%), Positives = 143/234 (61%), Gaps = 6/234 (2%)
Query: 334 KKISYYELLNATNNFEETQKLGQGGFGGVYKGYFKDSNSVAAIKRISADSRQGVKQYSAE 393
+K+++ +LL ATN F +G GGFG VYK KD SV AIK++ S QG ++++AE
Sbjct: 874 RKLTFADLLEATNGFHNDSLVGSGGFGDVYKAQLKDG-SVVAIKKLIHVSGQGDREFTAE 932
Query: 394 VKIISQLRHRNLVKLTGWCHKKNELILIYEYMPNGSLDFHLF---RGGSILPWNLRYNIA 450
++ I +++HRNLV L G+C E +L+YEYM GSL+ L + G L W R IA
Sbjct: 933 METIGKIKHRNLVPLLGYCKVGEERLLVYEYMKYGSLEDVLHDRKKTGIKLNWPARRKIA 992
Query: 451 LGLASALLYLQEEWEKCVIHRDIKSSNIMLDSDFNTKLGDFGLARLMD-HEKGSETTVVA 509
+G A L +L +IHRD+KSSN++LD + ++ DFG+ARLM + + +A
Sbjct: 993 IGAARGLAFLHHNCIPHIIHRDMKSSNVLLDENLEARVSDFGMARLMSAMDTHLSVSTLA 1052
Query: 510 GTRGYLAPEYIDTSKARKESDIFSFGVVLLEIACGKKAIHHQELEGEVSLVEWV 563
GT GY+ PEY + + + D++S+GVVLLE+ GK+ + G+ +LV WV
Sbjct: 1053 GTPGYVPPEYYQSFRCSTKGDVYSYGVVLLELLTGKQPTDSADF-GDNNLVGWV 1105
>BRI1_LYCES (Q8GUQ5) Brassinosteroid LRR receptor kinase precursor (EC
2.7.1.37) (tBRI1) (Altered brassinolide sensitivity 1)
(Systemin receptor SR160)
Length = 1207
Score = 175 bits (444), Expect = 3e-43
Identities = 96/234 (41%), Positives = 143/234 (61%), Gaps = 6/234 (2%)
Query: 334 KKISYYELLNATNNFEETQKLGQGGFGGVYKGYFKDSNSVAAIKRISADSRQGVKQYSAE 393
+K+++ +LL ATN F +G GGFG VYK KD SV AIK++ S QG ++++AE
Sbjct: 874 RKLTFADLLEATNGFHNDSLVGSGGFGDVYKAQLKDG-SVVAIKKLIHVSGQGDREFTAE 932
Query: 394 VKIISQLRHRNLVKLTGWCHKKNELILIYEYMPNGSLDFHLF---RGGSILPWNLRYNIA 450
++ I +++HRNLV L G+C E +L+YEYM GSL+ L + G L W R IA
Sbjct: 933 METIGKIKHRNLVPLLGYCKVGEERLLVYEYMKYGSLEDVLHDRKKIGIKLNWPARRKIA 992
Query: 451 LGLASALLYLQEEWEKCVIHRDIKSSNIMLDSDFNTKLGDFGLARLMD-HEKGSETTVVA 509
+G A L +L +IHRD+KSSN++LD + ++ DFG+ARLM + + +A
Sbjct: 993 IGAARGLAFLHHNCIPHIIHRDMKSSNVLLDENLEARVSDFGMARLMSAMDTHLSVSTLA 1052
Query: 510 GTRGYLAPEYIDTSKARKESDIFSFGVVLLEIACGKKAIHHQELEGEVSLVEWV 563
GT GY+ PEY + + + D++S+GVVLLE+ GK+ + G+ +LV WV
Sbjct: 1053 GTPGYVPPEYYQSFRCSTKGDVYSYGVVLLELLTGKQPTDSADF-GDNNLVGWV 1105
>CLV1_ARATH (Q9SYQ8) Receptor protein kinase CLAVATA1 precursor (EC
2.7.1.-)
Length = 980
Score = 172 bits (436), Expect = 2e-42
Identities = 96/279 (34%), Positives = 162/279 (57%), Gaps = 14/279 (5%)
Query: 349 EETQKLGQGGFGGVYKGYFKDSNSVAAIKRISADSRQGVKQYSAEVKIISQLRHRNLVKL 408
+E +G+GG G VY+G ++ VA + + + + ++AE++ + ++RHR++V+L
Sbjct: 693 KEENIIGKGGAGIVYRGSMPNNVDVAIKRLVGRGTGRSDHGFTAEIQTLGRIRHRHIVRL 752
Query: 409 TGWCHKKNELILIYEYMPNGSLD--FHLFRGGSILPWNLRYNIALGLASALLYLQEEWEK 466
G+ K+ +L+YEYMPNGSL H +GG L W R+ +A+ A L YL +
Sbjct: 753 LGYVANKDTNLLLYEYMPNGSLGELLHGSKGGH-LQWETRHRVAVEAAKGLCYLHHDCSP 811
Query: 467 CVIHRDIKSSNIMLDSDFNTKLGDFGLARLMDHEKGSE-TTVVAGTRGYLAPEYIDTSKA 525
++HRD+KS+NI+LDSDF + DFGLA+ + SE + +AG+ GY+APEY T K
Sbjct: 812 LILHRDVKSNNILLDSDFEAHVADFGLAKFLVDGAASECMSSIAGSYGYIAPEYAYTLKV 871
Query: 526 RKESDIFSFGVVLLEIACGKKAIHHQELEGEVSLVEWV-------WELYGLRNLIVAADP 578
++SD++SFGVVLLE+ GKK + E V +V WV + ++ DP
Sbjct: 872 DEKSDVYSFGVVLLELIAGKKPV--GEFGEGVDIVRWVRNTEEEITQPSDAAIVVAIVDP 929
Query: 579 KLCGIFDVKQLECLLVVGLWCANPDITSRPSIKKVIKVL 617
+L G + + + + + + C + +RP++++V+ +L
Sbjct: 930 RLTG-YPLTSVIHVFKIAMMCVEEEAAARPTMREVVHML 967
>RLK5_ARATH (P47735) Receptor-like protein kinase 5 precursor (EC
2.7.1.37)
Length = 999
Score = 169 bits (427), Expect = 3e-41
Identities = 105/286 (36%), Positives = 164/286 (56%), Gaps = 22/286 (7%)
Query: 349 EETQKLGQGGFGGVYKGYFKDSNSVAAIKRISADSRQGVKQYS----------AEVKIIS 398
+E +G G G VYK + V A+K+++ + G +YS AEV+ +
Sbjct: 684 DEKNVIGFGSSGKVYKVELR-GGEVVAVKKLNKSVKGGDDEYSSDSLNRDVFAAEVETLG 742
Query: 399 QLRHRNLVKLTGWCHKKNELILIYEYMPNGSLDFHLF---RGGSILPWNLRYNIALGLAS 455
+RH+++V+L C + +L+YEYMPNGSL L +GG +L W R IAL A
Sbjct: 743 TIRHKSIVRLWCCCSSGDCKLLVYEYMPNGSLADVLHGDRKGGVVLGWPERLRIALDAAE 802
Query: 456 ALLYLQEEWEKCVIHRDIKSSNIMLDSDFNTKLGDFGLARLMDHEKGSET----TVVAGT 511
L YL + ++HRD+KSSNI+LDSD+ K+ DFG+A+ + GS+T + +AG+
Sbjct: 803 GLSYLHHDCVPPIVHRDVKSSNILLDSDYGAKVADFGIAK-VGQMSGSKTPEAMSGIAGS 861
Query: 512 RGYLAPEYIDTSKARKESDIFSFGVVLLEIACGKKAIHHQELEGEVSLVEWVWELYGLRN 571
GY+APEY+ T + ++SDI+SFGVVLLE+ GK+ + G+ + +WV
Sbjct: 862 CGYIAPEYVYTLRVNEKSDIYSFGVVLLELVTGKQPTDSE--LGDKDMAKWVCTALDKCG 919
Query: 572 LIVAADPKLCGIFDVKQLECLLVVGLWCANPDITSRPSIKKVIKVL 617
L DPKL F +++ ++ +GL C +P +RPS++KV+ +L
Sbjct: 920 LEPVIDPKLDLKFK-EEISKVIHIGLLCTSPLPLNRPSMRKVVIML 964
>RPK1_IPONI (P93194) Receptor-like protein kinase precursor (EC
2.7.1.37)
Length = 1109
Score = 168 bits (426), Expect = 4e-41
Identities = 104/288 (36%), Positives = 155/288 (53%), Gaps = 13/288 (4%)
Query: 340 ELLNATNNFEETQKLGQGGFGGVYKGYFKDSNSVAAIKRISADSRQGVKQYSAEVKIISQ 399
++L AT N + +G+G G +YK A K + + G E++ I +
Sbjct: 808 KVLEATENLNDKYVIGKGAHGTIYKATLSPDKVYAVKKLVFTGIKNGSVSMVREIETIGK 867
Query: 400 LRHRNLVKLTGWCHKKNELILIYEYMPNGSLD--FHLFRGGSILPWNLRYNIALGLASAL 457
+RHRNL+KL + +K +++Y YM NGSL H L W+ R+NIA+G A L
Sbjct: 868 VRHRNLIKLEEFWLRKEYGLILYTYMENGSLHDILHETNPPKPLDWSTRHNIAVGTAHGL 927
Query: 458 LYLQEEWEKCVIHRDIKSSNIMLDSDFNTKLGDFGLARLMDHEKGS-ETTVVAGTRGYLA 516
YL + + ++HRDIK NI+LDSD + DFG+A+L+D S + V GT GY+A
Sbjct: 928 AYLHFDCDPAIVHRDIKPMNILLDSDLEPHISDFGIAKLLDQSATSIPSNTVQGTIGYMA 987
Query: 517 PEYIDTSKARKESDIFSFGVVLLEIACGKKAIHHQELEGEVSLVEW---VWELYGLRNLI 573
PE T+ +ESD++S+GVVLLE+ KKA+ GE +V W VW G I
Sbjct: 988 PENAFTTVKSRESDVYSYGVVLLELITRKKAL-DPSFNGETDIVGWVRSVWTQTGEIQKI 1046
Query: 574 VAADPKLCG-IFD---VKQLECLLVVGLWCANPDITSRPSIKKVIKVL 617
V DP L + D ++Q+ L + L CA ++ RP+++ V+K L
Sbjct: 1047 V--DPSLLDELIDSSVMEQVTEALSLALRCAEKEVDKRPTMRDVVKQL 1092
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.319 0.137 0.416
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 80,725,452
Number of Sequences: 164201
Number of extensions: 3575942
Number of successful extensions: 12984
Number of sequences better than 10.0: 1687
Number of HSP's better than 10.0 without gapping: 1216
Number of HSP's successfully gapped in prelim test: 471
Number of HSP's that attempted gapping in prelim test: 8629
Number of HSP's gapped (non-prelim): 1975
length of query: 655
length of database: 59,974,054
effective HSP length: 117
effective length of query: 538
effective length of database: 40,762,537
effective search space: 21930244906
effective search space used: 21930244906
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 69 (31.2 bits)
Medicago: description of AC144564.8


