

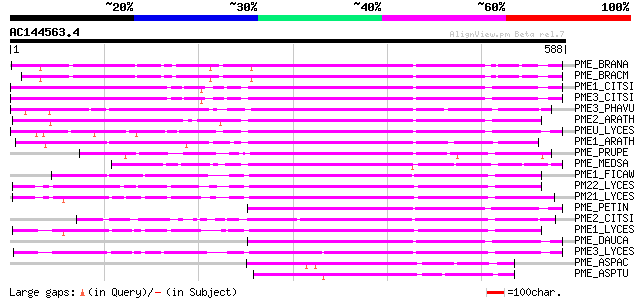
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144563.4 + phase: 0
(588 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
PME_BRANA (P41510) Probable pectinesterase precursor (EC 3.1.1.1... 351 3e-96
PME_BRACM (Q42608) Pectinesterase (EC 3.1.1.11) (Pectin methyles... 330 5e-90
PME1_CITSI (O04886) Pectinesterase 1 precursor (EC 3.1.1.11) (Pe... 326 9e-89
PME3_CITSI (P83948) Pectinesterase 3 precursor (EC 3.1.1.11) (Pe... 325 2e-88
PME3_PHAVU (Q43111) Pectinesterase 3 precursor (EC 3.1.1.11) (Pe... 310 9e-84
PME2_ARATH (Q42534) Pectinesterase 2 precursor (EC 3.1.1.11) (Pe... 309 2e-83
PMEU_LYCES (Q43143) Pectinesterase U1 precursor (EC 3.1.1.11) (P... 305 2e-82
PME1_ARATH (Q43867) Pectinesterase 1 precursor (EC 3.1.1.11) (Pe... 300 7e-81
PME_PRUPE (Q43062) Pectinesterase PPE8B precursor (EC 3.1.1.11) ... 278 4e-74
PME_MEDSA (Q42920) Pectinesterase precursor (EC 3.1.1.11) (Pecti... 278 4e-74
PME1_FICAW (P83947) Pectinesterase precursor (EC 3.1.1.11) (Pect... 271 3e-72
PM22_LYCES (Q96575) Pectinesterase 2 precursor (EC 3.1.1.11) (Pe... 270 6e-72
PM21_LYCES (P09607) Pectinesterase 2 precursor (EC 3.1.1.11) (Pe... 266 1e-70
PME_PETIN (Q43043) Pectinesterase precursor (EC 3.1.1.11) (Pecti... 265 2e-70
PME2_CITSI (O04887) Pectinesterase 2 precursor (EC 3.1.1.11) (Pe... 261 3e-69
PME1_LYCES (P14280) Pectinesterase 1 precursor (EC 3.1.1.11) (Pe... 259 1e-68
PME_DAUCA (P83218) Pectinesterase (EC 3.1.1.11) (Pectin methyles... 258 4e-68
PME3_LYCES (Q96576) Pectinesterase 3 precursor (EC 3.1.1.11) (Pe... 253 1e-66
PME_ASPAC (Q12535) Pectinesterase precursor (EC 3.1.1.11) (Pecti... 123 2e-27
PME_ASPTU (P17872) Pectinesterase precursor (EC 3.1.1.11) (Pecti... 107 7e-23
>PME_BRANA (P41510) Probable pectinesterase precursor (EC 3.1.1.11)
(Pectin methylesterase) (PE)
Length = 584
Score = 351 bits (901), Expect = 3e-96
Identities = 222/612 (36%), Positives = 329/612 (53%), Gaps = 61/612 (9%)
Query: 3 GKVIISVVSLILVVGVIIGVVVDIRKNGE---DPKVQTQQRNLRIMCQNAQDQKLCHDTL 59
GK++ISV S++LVVGV IGVV + K G D + + Q+ + +C +A D+ C TL
Sbjct: 4 GKIVISVASMLLVVGVAIGVVTFVNKGGGAGGDKTLNSHQKAVESLCASATDKGSCAKTL 63
Query: 60 SSVRGADAADPKAYIAAAVKAATDNVIKAFNMSERLTTEYGKKNGA--KMALDDCKDLMQ 117
V+ + DP I A + A D V K+ N + GK A K LD CK ++
Sbjct: 64 DPVK---SDDPSKLIKAFMLATKDAVTKSTNFTASTEEGMGKNINATSKAVLDYCKRVLM 120
Query: 118 FALDSLDLSNNCVRDNNIEAVHDQTADMRNWLSAVISYKQGCMEGFDDANDGEKKIKEQF 177
+AL+ L+ + ++ ++ + ++ WL+ V +Y+ C+ DD + E + +
Sbjct: 121 YALEDLETIVEEMGED-LQQSGSKMDQLKQWLTGVFNYQTDCI---DDIEESELR---KV 173
Query: 178 HVQSLYSVQKVTAVALDIVTGLSDILQQFNLNFD---------IKPPSRRLLNSEVTVDD 228
+ + + +++ A+DI L+ + Q N+ D P R LL +D
Sbjct: 174 MGEGIAHSKILSSNAIDIFHALTTAMSQMNVKVDDMKKGNLGETPAPDRDLLED---LDQ 230
Query: 229 QGYPSWISSSGRKLLAKMQRKGWRAN------------IRPNAVVANDGSGQFKTIQAAL 276
+G P W S RKL+A+ R G A+ I+P VVA DGSGQFKTI A+
Sbjct: 231 KGLPKWHSDKDRKLMAQAGRPGAPADEGIGEGGGGGGKIKPTHVVAKDGSGQFKTISEAV 290
Query: 277 ASYPKGNKDRYVIYVKAGVYDEYITVPKEAVNILMYGDGPAKTIVTGRKNQ--MAGTNTQ 334
+ P+ N R +IY+KAGVY E +T+PK+ N+ M+GDG +TI+T ++ GT T
Sbjct: 291 KACPEKNPGRCIIYIKAGVYKEQVTIPKKVNNVFMFGDGATQTIITFDRSVGLSPGTTTS 350
Query: 335 NTATFSNTAMGFIGKAMTFENTAGPAGMQAVAFRNIGDMSALVGCHIVGYQDTLYVQTNR 394
+ T + GF+ K + F+NTAGP G QAVAFR GD + + C GYQDTLYV R
Sbjct: 351 LSGTVQVESEGFMAKWIGFQNTAGPLGHQAVAFRVNGDRAVIFNCRFDGYQDTLYVNNGR 410
Query: 395 QFYRNCVISGTIDFIFGTSATLIQSSTIIVRKGNYDHNEYNVIVADGSPLVN-MNTGIVI 453
QFYRN V+SGT+DFIFG SAT+IQ+S I+ RKG+ + N + ADG+ + GIV+
Sbjct: 411 QFYRNIVVSGTVDFIFGKSATVIQNSLILCRKGS--PGQTNHVTADGNEKGKAVKIGIVL 468
Query: 454 QDCNIIPEAALVPEKFTVRSYLGRPWQNESKAVIMESTIGDFIHQDGWTTWPEEQNKPEK 513
+C I+ + L ++ TV+SYLGRPW+ + ++ + IGD I GW W + EK
Sbjct: 469 HNCRIMADKELEADRLTVKSYLGRPWKPFATTAVIGTEIGDLIQPTGWNEW-----QGEK 523
Query: 514 HHENTCYFAEYANTGPGANVARRVKWKGYKGVISRSEATKYTASIWLDAGPKTAPKSAVE 573
H T + E+ N GPGAN A RV W K S +E ++T + WL +
Sbjct: 524 FH-LTATYVEFNNRGPGANTAARVPWA--KMAKSAAEVERFTVANWL---------TPAN 571
Query: 574 WLHDLHVPHYLG 585
W+ + +VP LG
Sbjct: 572 WIQEANVPVQLG 583
>PME_BRACM (Q42608) Pectinesterase (EC 3.1.1.11) (Pectin
methylesterase) (PE) (Fragment)
Length = 571
Score = 330 bits (847), Expect = 5e-90
Identities = 214/602 (35%), Positives = 317/602 (52%), Gaps = 61/602 (10%)
Query: 13 ILVVGVIIGVVVDIRKNGE---DPKVQTQQRNLRIMCQNAQDQKLCHDTLSSVRGADAAD 69
+LVVGV IGVV + K G D + + Q+ + +C +A D+ C TL V+ + D
Sbjct: 1 LLVVGVAIGVVTFVNKGGGAGGDKTLNSHQKAVESLCASATDKGSCAKTLDPVK---SDD 57
Query: 70 PKAYIAAAVKAATDNVIKAFNMSERLTTEYGKKNGA--KMALDDCKDLMQFALDSLDLSN 127
P I A + A D V K+ N + GK A K LD CK ++ +AL+ L+
Sbjct: 58 PSKLIKAFMLATKDAVTKSTNFTASTEEGMGKNMNATSKAVLDYCKRVLMYALEDLETIV 117
Query: 128 NCVRDNNIEAVHDQTADMRNWLSAVISYKQGCMEGFDDANDGEKKIKEQFHVQSLYSVQK 187
+ ++ ++ + ++ WL+ V +Y+ C+ DD + E + + + + +
Sbjct: 118 EEMGED-LQQSGSKMDQLKQWLTGVFNYQTDCI---DDIEESELR---KVMGEGIRHSKI 170
Query: 188 VTAVALDIVTGLSDILQQFNLNFD---------IKPPSRRLLNSEVTVDDQGYPSWISSS 238
+++ A+DI L+ + Q N+ D P R LL +D +G P W S
Sbjct: 171 LSSNAIDIFHALTTAMSQMNVKVDDMKKGNLGETPAPDRDLLED---LDQKGLPKWHSDK 227
Query: 239 GRKLLAKMQRKGWRAN------------IRPNAVVANDGSGQFKTIQAALASYPKGNKDR 286
RKL+A+ R G A+ I+P VVA DGSGQFKTI A+ + P+ N R
Sbjct: 228 DRKLMAQAGRPGAPADEGIGEGGGGGGKIKPTHVVAKDGSGQFKTISEAVKACPEKNPGR 287
Query: 287 YVIYVKAGVYDEYITVPKEAVNILMYGDGPAKTIVTGRKNQ--MAGTNTQNTATFSNTAM 344
+IY+KAGVY E +T+PK+ N+ M+GDG +TI+T ++ GT T + T +
Sbjct: 288 CIIYIKAGVYKEQVTIPKKVNNVFMFGDGATQTIITFDRSVGLSPGTTTSLSGTVQVESE 347
Query: 345 GFIGKAMTFENTAGPAGMQAVAFRNIGDMSALVGCHIVGYQDTLYVQTNRQFYRNCVISG 404
GF+ K + F+NTAGP G QAVAFR GD + + C GYQDTLYV RQFYRN V+SG
Sbjct: 348 GFMAKWIGFQNTAGPLGNQAVAFRVNGDRAVIFNCRFDGYQDTLYVNNGRQFYRNIVVSG 407
Query: 405 TIDFIFGTSATLIQSSTIIVRKGNYDHNEYNVIVADGSPLVN-MNTGIVIQDCNIIPEAA 463
T+DFI G SAT+IQ+S I+ RKG+ + N + ADG + GIV+ +C I+ +
Sbjct: 408 TVDFINGKSATVIQNSLILCRKGS--PGQTNHVTADGKQKGKAVKIGIVLHNCRIMADKE 465
Query: 464 LVPEKFTVRSYLGRPWQNESKAVIMESTIGDFIHQDGWTTWPEEQNKPEKHHENTCYFAE 523
L ++ TV+SYLGRPW+ + ++ + IGD I GW W + EK H T + E
Sbjct: 466 LEADRLTVKSYLGRPWKPFATTAVIGTEIGDLIQPTGWNEW-----QGEKFH-LTATYVE 519
Query: 524 YANTGPGANVARRVKWKGYKGVISRSEATKYTASIWLDAGPKTAPKSAVEWLHDLHVPHY 583
+ N GPGAN A RV W K S +E ++T + WL + W+ + +V
Sbjct: 520 FNNRGPGANPAARVPWA--KMAKSAAEVERFTVANWL---------TPANWIQEANVTVQ 568
Query: 584 LG 585
LG
Sbjct: 569 LG 570
>PME1_CITSI (O04886) Pectinesterase 1 precursor (EC 3.1.1.11)
(Pectin methylesterase) (PE)
Length = 584
Score = 326 bits (836), Expect = 9e-89
Identities = 205/595 (34%), Positives = 323/595 (53%), Gaps = 49/595 (8%)
Query: 2 KGKVIISVVSLILVVGVIIGVV--VDIRKNGEDPKVQTQQRNLRIMCQNAQDQKLCHDTL 59
K K+ +++ + +LVV +IG+V V+ RKN D + L+ C + + LC +
Sbjct: 27 KKKLFLALFATLLVVAAVIGIVAGVNSRKNSGDNGNEPHHAILKSSCSSTRYPDLCFSAI 86
Query: 60 SSVRGAD--AADPKAYIAAAVKAATDNVIKAFNMSERLTTEYGKKNGAKMALDDCKDLMQ 117
++V A K I ++ T V + ++L K+AL DC + +
Sbjct: 87 AAVPEASKKVTSQKDVIEMSLNITTTAVEHNYFGIQKLLKRTNLTKREKVALHDCLETID 146
Query: 118 FALDSLDLSNNCVRDN-NIEAVHDQTADMRNWLSAVISYKQGCMEGF--DDANDGEKKIK 174
LD L + + + N +++ D++ +SA ++ + C++GF DDAN K ++
Sbjct: 147 ETLDELHKAVEDLEEYPNKKSLSQHADDLKTLMSAAMTNQGTCLDGFSHDDAN---KHVR 203
Query: 175 EQFHVQSLYSVQKVTAVALDIVTGLSD----ILQQFNLNFDIKPPSRRLLNSEVTVDDQG 230
+ ++ V+K+ + AL ++ ++D I++ N +R+L TVD G
Sbjct: 204 DALSDGQVH-VEKMCSNALAMIKNMTDTDMMIMRTSN--------NRKLTEETSTVD--G 252
Query: 231 YPSWISSSGRKLLAKMQRKGWRANIRPNAVVANDGSGQFKTIQAALASYPKGNKDRYVIY 290
+P+W+S R+LL +++ PNAVVA DGSG FKT+ AA+A+ P+G RY+I
Sbjct: 253 WPAWLSPGDRRLLQS-------SSVTPNAVVAADGSGNFKTVAAAVAAAPQGGTKRYIIR 305
Query: 291 VKAGVYDEYITVPKEAVNILMYGDGPAKTIVTGRKNQMAGTNTQNTATFSNTAMGFIGKA 350
+KAGVY E + V K+ NI+ GDG +TI+TG +N + G+ T +AT + GF+ +
Sbjct: 306 IKAGVYRENVEVTKKHKNIMFIGDGRTRTIITGSRNVVDGSTTFKSATAAVVGEGFLARD 365
Query: 351 MTFENTAGPAGMQAVAFRNIGDMSALVGCHIVGYQDTLYVQTNRQFYRNCVISGTIDFIF 410
+TF+NTAGP+ QAVA R D+SA C ++ YQDTLYV +NRQF+ NC+I+GT+DFIF
Sbjct: 366 ITFQNTAGPSKHQAVALRVGADLSAFYNCDMLAYQDTLYVHSNRQFFVNCLIAGTVDFIF 425
Query: 411 GTSATLIQSSTIIVRKGNYDHNEYNVIVADGSPLVNMNTGIVIQDCNIIPEAALVPEKFT 470
G +A ++Q+ I RK N + N++ A G N NTGIVIQ I + L P + +
Sbjct: 426 GNAAAVLQNCDIHARKPN--SGQKNMVTAQGRTDPNQNTGIVIQKSRIGATSDLKPVQGS 483
Query: 471 VRSYLGRPWQNESKAVIMESTIGDFIHQDGWTTWPEEQNKPEKHHENTCYFAEYANTGPG 530
+YLGRPW+ S+ VIM+S+I D IH GW W NT ++ E+ N+G G
Sbjct: 484 FPTYLGRPWKEYSRTVIMQSSITDLIHPAGWHEW------DGNFALNTLFYGEHQNSGAG 537
Query: 531 ANVARRVKWKGYKGVISRSEATKYTASIWLDAGPKTAPKSAVEWLHDLHVPHYLG 585
A + RVKWKG++ + S +EA +T ++ + WL P LG
Sbjct: 538 AGTSGRVKWKGFRVITSATEAQAFTPGSFI---------AGSSWLGSTGFPFSLG 583
>PME3_CITSI (P83948) Pectinesterase 3 precursor (EC 3.1.1.11)
(Pectin methylesterase 3) (PE 3)
Length = 584
Score = 325 bits (833), Expect = 2e-88
Identities = 203/595 (34%), Positives = 323/595 (54%), Gaps = 49/595 (8%)
Query: 2 KGKVIISVVSLILVVGVIIGVV--VDIRKNGEDPKVQTQQRNLRIMCQNAQDQKLCHDTL 59
K K+ +++ + +LVV +IG+V V+ RKN D + L+ C + + LC +
Sbjct: 27 KKKLFLALFATLLVVAAVIGIVAGVNSRKNSGDNGNEPHHAILKSSCSSTRYPDLCFSAI 86
Query: 60 SSVRGAD--AADPKAYIAAAVKAATDNVIKAFNMSERLTTEYGKKNGAKMALDDCKDLMQ 117
++V A K I ++ T V + ++L K+AL DC + +
Sbjct: 87 AAVPEASKKVTSQKDVIEMSLNITTTAVEHNYFGIQKLLKRTNLTKREKVALHDCLETID 146
Query: 118 FALDSLDLSNNCVRDN-NIEAVHDQTADMRNWLSAVISYKQGCMEGF--DDANDGEKKIK 174
LD L + + + N +++ D++ +SA ++ + C++GF DDAN K ++
Sbjct: 147 ETLDELHKAVEDLEEYPNKKSLSQHADDLKTLMSAAMTNQGTCLDGFSHDDAN---KHVR 203
Query: 175 EQFHVQSLYSVQKVTAVALDIVTGLSD----ILQQFNLNFDIKPPSRRLLNSEVTVDDQG 230
+ ++ V+K+ + AL ++ ++D I++ N +R+L+ TVD G
Sbjct: 204 DALSDGQVH-VEKMCSNALAMIKNMTDTDMMIMRTSN--------NRKLIEETSTVD--G 252
Query: 231 YPSWISSSGRKLLAKMQRKGWRANIRPNAVVANDGSGQFKTIQAALASYPKGNKDRYVIY 290
+P+W+S+ R+LL +++ PN VVA DGSG FKT+ A++A+ P+G RY+I
Sbjct: 253 WPAWLSTGDRRLLQS-------SSVTPNVVVAADGSGNFKTVAASVAAAPQGGTKRYIIR 305
Query: 291 VKAGVYDEYITVPKEAVNILMYGDGPAKTIVTGRKNQMAGTNTQNTATFSNTAMGFIGKA 350
+KAGVY E + V K+ NI+ GDG +TI+TG +N + G+ T +AT + GF+ +
Sbjct: 306 IKAGVYRENVEVTKKHKNIMFIGDGRTRTIITGSRNVVDGSTTFKSATVAVVGEGFLARD 365
Query: 351 MTFENTAGPAGMQAVAFRNIGDMSALVGCHIVGYQDTLYVQTNRQFYRNCVISGTIDFIF 410
+TF+NTAGP+ QAVA R D+SA C ++ YQDTLYV +NRQF+ NC+I+GT+DFIF
Sbjct: 366 ITFQNTAGPSKHQAVALRVGADLSAFYNCDMLAYQDTLYVHSNRQFFVNCLIAGTVDFIF 425
Query: 411 GTSATLIQSSTIIVRKGNYDHNEYNVIVADGSPLVNMNTGIVIQDCNIIPEAALVPEKFT 470
G +A ++Q+ I RK N + N++ A G N NTGIVIQ I + L P + +
Sbjct: 426 GNAAAVLQNCDIHARKPN--SGQKNMVTAQGRADPNQNTGIVIQKSRIGATSDLKPVQGS 483
Query: 471 VRSYLGRPWQNESKAVIMESTIGDFIHQDGWTTWPEEQNKPEKHHENTCYFAEYANTGPG 530
+YLGRPW+ S+ VIM+S+I D IH GW W NT ++ E+ N G G
Sbjct: 484 FPTYLGRPWKEYSRTVIMQSSITDVIHPAGWHEW------DGNFALNTLFYGEHQNAGAG 537
Query: 531 ANVARRVKWKGYKGVISRSEATKYTASIWLDAGPKTAPKSAVEWLHDLHVPHYLG 585
A + RVKWKG++ + S +EA +T ++ + WL P LG
Sbjct: 538 AGTSGRVKWKGFRVITSATEAQAFTPGSFI---------AGSSWLGSTGFPFSLG 583
>PME3_PHAVU (Q43111) Pectinesterase 3 precursor (EC 3.1.1.11)
(Pectin methylesterase 3) (PE 3)
Length = 581
Score = 310 bits (793), Expect = 9e-84
Identities = 199/584 (34%), Positives = 305/584 (52%), Gaps = 44/584 (7%)
Query: 2 KGKVIISVVSLILV---VGVIIGVVVDIRKNGEDPKVQTQQR-------NLRIMCQNAQD 51
K +II+V S++L+ + + GVV+ R + P + + +L+ +C +
Sbjct: 27 KRLIIIAVSSIVLIAVIIAAVAGVVIHNRNSESSPSSDSVPQTELSPAASLKAVCDTTRY 86
Query: 52 QKLCHDTLSSVRGADAADPKAYIAAAVKAATDNVIKAFNMSERLTTEYGKKNGAKMALDD 111
C ++SS+ ++ DP+ +++ A D + +F R E + + A+D
Sbjct: 87 PSSCFSSISSLPESNTTDPELLFKLSLRVAIDE-LSSFPSKLRANAEQDAR--LQKAIDV 143
Query: 112 CKDLMQFALDSLDLSNNCVRD-NNIEAVHDQTADMRNWLSAVISYKQGCMEGFDDANDGE 170
C + ALD L+ S + + A +++ WLSA ++ + C++ + N
Sbjct: 144 CSSVFGDALDRLNDSISALGTVAGRIASSASVSNVETWLSAALTDQDTCLDAVGELNSTA 203
Query: 171 KKIKEQFHVQSLYSVQKVTAVALDIVTGLSDILQQFNLNFDIKPPSRRLLNSEVTVDDQG 230
+ Q ++ + + + +L IVT + +L +F RRLL G
Sbjct: 204 ARGALQEIETAMRNSTEFASNSLAIVTKILGLLSRFETPIH----HRRLL---------G 250
Query: 231 YPSWISSSGRKLLAKMQRKGWRANIRPNAVVANDGSGQFKTIQAALASYPKGNKDRYVIY 290
+P W+ ++ R+LL + P+AVVA DGSGQFKTI AL K +++R+ +Y
Sbjct: 251 FPEWLGAAERRLLEEKNNDS-----TPDAVVAKDGSGQFKTIGEALKLVKKKSEERFSVY 305
Query: 291 VKAGVYDEYITVPKEAVNILMYGDGPAKTIVTGRKNQMAGTNTQNTATFSNTAMGFIGKA 350
VK G Y E I + K N+++YGDG KT V G +N M GT T TATF+ GFI K
Sbjct: 306 VKEGRYVENIDLDKNTWNVMIYGDGKDKTFVVGSRNFMDGTPTFETATFAVKGKGFIAKD 365
Query: 351 MTFENTAGPAGMQAVAFRNIGDMSALVGCHIVGYQDTLYVQTNRQFYRNCVISGTIDFIF 410
+ F N AG + QAVA R+ D S C G+QDTLY +NRQFYR+C I+GTIDFIF
Sbjct: 366 IGFVNNAGASKHQAVALRSGSDRSVFFRCSFDGFQDTLYAHSNRQFYRDCDITGTIDFIF 425
Query: 411 GTSATLIQSSTIIVRKGNYDHNEYNVIVADGSPLVNMNTGIVIQDCNIIPEAALVPEKFT 470
G +A + QS I+ R+ N++N I A G N NTGI+IQ I P T
Sbjct: 426 GNAAVVFQSCKIMPRQPL--PNQFNTITAQGKKDPNQNTGIIIQKSTITP----FGNNLT 479
Query: 471 VRSYLGRPWQNESKAVIMESTIGDFIHQDGWTTWPEEQNKPEKHHENTCYFAEYANTGPG 530
+YLGRPW++ S VIM+S IG ++ GW +W P T ++AEY N+GPG
Sbjct: 480 APTYLGRPWKDFSTTVIMQSDIGALLNPVGWMSWVPNVEPP-----TTIFYAEYQNSGPG 534
Query: 531 ANVARRVKWKGYKGVISRSEATKYTASIWLDAGPKTAPKSAVEW 574
A+V++RVKW GYK I+ A ++T ++ GP+ P +AV++
Sbjct: 535 ADVSQRVKWAGYKPTITDRNAEEFTVQSFIQ-GPEWLPNAAVQF 577
>PME2_ARATH (Q42534) Pectinesterase 2 precursor (EC 3.1.1.11)
(Pectin methylesterase 2) (PE 2)
Length = 587
Score = 309 bits (791), Expect = 2e-83
Identities = 196/579 (33%), Positives = 310/579 (52%), Gaps = 43/579 (7%)
Query: 4 KVIISVVSL-ILVVGVIIGVVVDIRKNGEDPKVQTQQRN----LRIMCQNAQDQKLCHDT 58
K+I+S ++ +L++ I+G+ ++ K+ T L+ +C + +LC
Sbjct: 19 KLILSSAAIALLLLASIVGIAATTTNQNKNQKITTLSSTSHAILKSVCSSTLYPELCFSA 78
Query: 59 LSSVRGADAADPKAYIAAAVKAATDNVI-KAFNMSERLTTEYGKKNGAKMALDDCKDLMQ 117
+++ G + K I A++ T V F + + + G AL DC + +
Sbjct: 79 VAATGGKELTSQKEVIEASLNLTTKAVKHNYFAVKKLIAKRKGLTPREVTALHDCLETID 138
Query: 118 FALDSLDLSNNCVRDN-NIEAVHDQTADMRNWLSAVISYKQGCMEGF--DDANDGEKK-- 172
LD L ++ + +++ D++ +S+ I+ + C++GF DDA+ +K
Sbjct: 139 ETLDELHVAVEDLHQYPKQKSLRKHADDLKTLISSAITNQGTCLDGFSYDDADRKVRKAL 198
Query: 173 IKEQFHVQSLYSVQKVTAVALDIVTGLSDILQQFNLNFDIKPPSRRLLNS------EVT- 225
+K Q HV+ + S A+A+ + ++ + NF+++ S N+ EVT
Sbjct: 199 LKGQVHVEHMCS----NALAM-----IKNMTETDIANFELRDKSSTFTNNNNRKLKEVTG 249
Query: 226 -VDDQGYPSWISSSGRKLLAKMQRKGWRANIRPNAVVANDGSGQFKTIQAALASYPKGNK 284
+D G+P W+S R+LL + I+ +A VA+DGSG F T+ AA+A+ P+ +
Sbjct: 250 DLDSDGWPKWLSVGDRRLLQG-------STIKADATVADDGSGDFTTVAAAVAAAPEKSN 302
Query: 285 DRYVIYVKAGVYDEYITVPKEAVNILMYGDGPAKTIVTGRKNQMAGTNTQNTATFSNTAM 344
R+VI++KAGVY E + V K+ NI+ GDG KTI+TG +N + G+ T ++AT +
Sbjct: 303 KRFVIHIKAGVYRENVEVTKKKTNIMFLGDGRGKTIITGSRNVVDGSTTFHSATVAAVGE 362
Query: 345 GFIGKAMTFENTAGPAGMQAVAFRNIGDMSALVGCHIVGYQDTLYVQTNRQFYRNCVISG 404
F+ + +TF+NTAGP+ QAVA R D SA C + YQDTLYV +NRQF+ C I+G
Sbjct: 363 RFLARDITFQNTAGPSKHQAVALRVGSDFSAFYQCDMFAYQDTLYVHSNRQFFVKCHITG 422
Query: 405 TIDFIFGTSATLIQSSTIIVRKGNYDHNEYNVIVADGSPLVNMNTGIVIQDCNIIPEAAL 464
T+DFIFG +A ++Q I R+ N + N++ A G N NTGIVIQ+C I + L
Sbjct: 423 TVDFIFGNAAAVLQDCDINARRPN--SGQKNMVTAQGRSDPNQNTGIVIQNCRIGGTSDL 480
Query: 465 VPEKFTVRSYLGRPWQNESKAVIMESTIGDFIHQDGWTTWPEEQNKPEKHHENTCYFAEY 524
+ K T +YLGRPW+ S+ VIM+S I D I +GW W +T + EY
Sbjct: 481 LAVKGTFPTYLGRPWKEYSRTVIMQSDISDVIRPEGWHEW------SGSFALDTLTYREY 534
Query: 525 ANTGPGANVARRVKWKGYKGVISRSEATKYTASIWLDAG 563
N G GA A RVKWKGYK + S +EA +TA ++ G
Sbjct: 535 LNRGGGAGTANRVKWKGYKVITSDTEAQPFTAGQFIGGG 573
>PMEU_LYCES (Q43143) Pectinesterase U1 precursor (EC 3.1.1.11)
(Pectin methylesterase) (PE)
Length = 583
Score = 305 bits (781), Expect = 2e-82
Identities = 194/602 (32%), Positives = 315/602 (52%), Gaps = 55/602 (9%)
Query: 2 KGKVIISVVSLILVVGVIIGVVVDIR---KNGEDPK-----VQTQQRNLRIMCQNAQDQK 53
K K+ +++V+ +L+V +IGVV ++ KN +D + ++ C N +
Sbjct: 18 KKKIYLAIVASVLLVAAVIGVVAGVKSHSKNSDDHADIMAISSSAHAIVKSACSNTLHPE 77
Query: 54 LCHDTLSSVRGADAADPKAYIAAAVKAATDNVIKA-----FNMSERLTTEYGKKNGAKMA 108
LC+ + +V +D + ++ + + +KA + + E + T G K+A
Sbjct: 78 LCYSAIVNV--SDFSKKVTSQKDVIELSLNITVKAVRRNYYAVKELIKTRKGLTPREKVA 135
Query: 109 LDDCKDLMQFALDSLDLSNNCVRD----NNIEAVHDQTADMRNWLSAVISYKQGCMEGFD 164
L DC + M LD L + V D N +++ + D++ +S+ I+ ++ C++GF
Sbjct: 136 LHDCLETMDETLDEL---HTAVEDLELYPNKKSLKEHVEDLKTLISSAITNQETCLDGFS 192
Query: 165 DANDGEKKIKEQFHVQSLYSVQKVTAVALDIVTGLSDILQQFNLNFDIKPPSRRLLNSEV 224
++ +KK+++ ++ V+K+ + AL ++ ++D + +R+L
Sbjct: 193 H-DEADKKVRKVL-LKGQKHVEKMCSNALAMICNMTDTDIANEMKLSAPANNRKL----- 245
Query: 225 TVDDQG-YPSWISSSGRKLLAKMQRKGWRANIRPNAVVANDGSGQFKTIQAALASYPKGN 283
V+D G +P W+S+ R+LL + + P+ VVA DGSG +KT+ A+ P+ +
Sbjct: 246 -VEDNGEWPEWLSAGDRRLLQS-------STVTPDVVVAADGSGDYKTVSEAVRKAPEKS 297
Query: 284 KDRYVIYVKAGVYDEYITVPKEAVNILMYGDGPAKTIVTGRKNQMAGTNTQNTATFSNTA 343
RYVI +KAGVY E + VPK+ NI+ GDG + TI+T +N G+ T ++AT A
Sbjct: 298 SKRYVIRIKAGVYRENVDVPKKKTNIMFMGDGKSNTIITASRNVQDGSTTFHSATVVRVA 357
Query: 344 MGFIGKAMTFENTAGPAGMQAVAFRNIGDMSALVGCHIVGYQDTLYVQTNRQFYRNCVIS 403
+ + +TF+NTAG + QAVA D+SA C ++ YQDTLYV +NRQF+ C+++
Sbjct: 358 GKVLARDITFQNTAGASKHQAVALCVGSDLSAFYRCDMLAYQDTLYVHSNRQFFVQCLVA 417
Query: 404 GTIDFIFGTSATLIQSSTIIVRKGNYDHNEYNVIVADGSPLVNMNTGIVIQDCNIIPEAA 463
GT+DFIFG A + Q I R+ + N++ A G N NTGIVIQ C I +
Sbjct: 418 GTVDFIFGNGAAVFQDCDIHARRPG--SGQKNMVTAQGRTDPNQNTGIVIQKCRIGATSD 475
Query: 464 LVPEKFTVRSYLGRPWQNESKAVIMESTIGDFIHQDGWTTWPEEQNKPEKHHENTCYFAE 523
L P + + +YLGRPW+ S+ VIM+S+I D I GW W +T ++ E
Sbjct: 476 LRPVQKSFPTYLGRPWKEYSRTVIMQSSITDVIQPAGWHEW------NGNFALDTLFYGE 529
Query: 524 YANTGPGANVARRVKWKGYKGVISRSEATKYTASIWLDAGPKTAPKSAVEWLHDLHVPHY 583
YANTG GA + RVKWKG+K + S +EA YT ++ G WL P
Sbjct: 530 YANTGAGAPTSGRVKWKGHKVITSSTEAQAYTPGRFIAGG---------SWLSSTGFPFS 580
Query: 584 LG 585
LG
Sbjct: 581 LG 582
>PME1_ARATH (Q43867) Pectinesterase 1 precursor (EC 3.1.1.11)
(Pectin methylesterase 1) (PE 1)
Length = 586
Score = 300 bits (768), Expect = 7e-81
Identities = 189/564 (33%), Positives = 292/564 (51%), Gaps = 37/564 (6%)
Query: 7 ISVVSLI-LVVGVIIGVVVDIRKNGEDPKVQ---TQQRNLRIMCQNAQDQKLCHDTLSSV 62
ISVV LI +++ ++ VV KN P T +L+ +C + + C ++S +
Sbjct: 34 ISVVVLIAVIIAAVVATVVHKNKNESTPSPPPELTPSTSLKAICSVTRFPESCISSISKL 93
Query: 63 RGADAADPKAYIAAAVKAATDNVIKAFNMSERLTTEYGKKNGAKMALDDCKDLMQFALDS 122
++ DP+ ++K D + ++ E+L+ E + K AL C DL++ ALD
Sbjct: 94 PSSNTTDPETLFKLSLKVIIDELDSISDLPEKLSKETEDER-IKSALRVCGDLIEDALDR 152
Query: 123 LDLSNNCVRDNNIEAV--HDQTADMRNWLSAVISYKQGCMEGFDDANDGEKKIKEQFHVQ 180
L+ + + + D + + D++ WLSA ++ + C + D+ + + Q
Sbjct: 153 LNDTVSAIDDEEKKKTLSSSKIEDLKTWLSATVTDHETCFDSLDELKQNKTEYANSTITQ 212
Query: 181 SLYSVQ----KVTAVALDIVTGLSDILQQFNLNFDIKPPSRRLLNSEVTVDDQGYPSWIS 236
+L S + T+ +L IV+ + L + RR L S + W
Sbjct: 213 NLKSAMSRSTEFTSNSLAIVSKILSALSDLGIPIH----RRRRLMSHHHQQSVDFEKW-- 266
Query: 237 SSGRKLLAKMQRKGWRANIRPNAVVANDGSGQFKTIQAALASYPKGNKDRYVIYVKAGVY 296
+ R+LL A ++P+ VA DG+G T+ A+A PK + +VIYVK+G Y
Sbjct: 267 -ARRRLLQT-------AGLKPDVTVAGDGTGDVLTVNEAVAKVPKKSLKMFVIYVKSGTY 318
Query: 297 DEYITVPKEAVNILMYGDGPAKTIVTGRKNQMAGTNTQNTATFSNTAMGFIGKAMTFENT 356
E + + K N+++YGDG KTI++G KN + GT T TATF+ GFI K + NT
Sbjct: 319 VENVVMDKSKWNVMIYGDGKGKTIISGSKNFVDGTPTYETATFAIQGKGFIMKDIGIINT 378
Query: 357 AGPAGMQAVAFRNIGDMSALVGCHIVGYQDTLYVQTNRQFYRNCVISGTIDFIFGTSATL 416
AG A QAVAFR+ D S C G+QDTLY +NRQFYR+C ++GTIDFIFG++A +
Sbjct: 379 AGAAKHQAVAFRSGSDFSVYYQCSFDGFQDTLYPHSNRQFYRDCDVTGTIDFIFGSAAVV 438
Query: 417 IQSSTIIVRKGNYDHNEYNVIVADGSPLVNMNTGIVIQDCNIIPEAALVPEKFTVRSYLG 476
Q I+ R+ N++N I A G N ++G+ IQ C I ++ +YLG
Sbjct: 439 FQGCKIMPRQPL--SNQFNTITAQGKKDPNQSSGMSIQRCTISANGNVI-----APTYLG 491
Query: 477 RPWQNESKAVIMESTIGDFIHQDGWTTWPEEQNKPEKHHENTCYFAEYANTGPGANVARR 536
RPW+ S VIME+ IG + GW +W + P + + EY NTGPG++V +R
Sbjct: 492 RPWKEFSTTVIMETVIGAVVRPSGWMSWVSGVDPPA-----SIVYGEYKNTGPGSDVTQR 546
Query: 537 VKWKGYKGVISRSEATKYTASIWL 560
VKW GYK V+S +EA K+T + L
Sbjct: 547 VKWAGYKPVMSDAEAAKFTVATLL 570
>PME_PRUPE (Q43062) Pectinesterase PPE8B precursor (EC 3.1.1.11)
(Pectin methylesterase) (PE)
Length = 522
Score = 278 bits (710), Expect = 4e-74
Identities = 181/511 (35%), Positives = 267/511 (51%), Gaps = 58/511 (11%)
Query: 75 AAAVKAATDNVIKAFNMSERLTTEYGKKNGAKMALDDCKDLMQFALD----SLDLSNNCV 130
A ++K D V + ++ + +G A A+ DC DL+ F+ D SL S N
Sbjct: 57 AGSLKDTIDAVQQVASILSQFANAFGDFRLAN-AISDCLDLLDFSADELNWSLSASQNQK 115
Query: 131 RDNNIEAVHDQTADMRNWLSAVISYKQGCMEGFDDANDGEKKIKEQFHVQSLYSVQKVTA 190
NN + ++D+R WLSA + + C GF+ N VQ + +
Sbjct: 116 GKNN--STGKLSSDLRTWLSAALVNQDTCSNGFEGTNS---------------IVQGLIS 158
Query: 191 VALDIVTGL-SDILQQFNLNFDIKPPSRRLLNSEVTVDDQGYPSWISSSGRKLLAKMQRK 249
L VT L ++L Q + N + + P+ ++ PSW+ + RKLL Q
Sbjct: 159 AGLGQVTSLVQELLTQVHPNSNQQGPNGQI------------PSWVKTKDRKLL---QAD 203
Query: 250 GWRANIRPNAVVANDGSGQFKTIQAALASYPKGNKDRYVIYVKAGVYDEYITVPKEAVNI 309
G + +A+VA DG+G F + A+ + P + RYVIY+K G Y E + + K+ N+
Sbjct: 204 G----VSVDAIVAQDGTGNFTNVTDAVLAAPDYSMRRYVIYIKRGTYKENVEIKKKKWNL 259
Query: 310 LMYGDGPAKTIVTGRKNQMAGTNTQNTATFSNTAMGFIGKAMTFENTAGPAGMQAVAFRN 369
+M GDG TI++G ++ + G T +ATF+ + GFI + +TFENTAGP QAVA R+
Sbjct: 260 MMIGDGMDATIISGNRSFVDGWTTFRSATFAVSGRGFIARDITFENTAGPEKHQAVALRS 319
Query: 370 IGDMSALVGCHIVGYQDTLYVQTNRQFYRNCVISGTIDFIFGTSATLIQSSTIIVRKGNY 429
D+S C+I GYQDTLY T RQFYR+C ISGT+DFIFG + + Q+ I+ +KG
Sbjct: 320 DSDLSVFYRCNIRGYQDTLYTHTMRQFYRDCKISGTVDFIFGDATVVFQNCQILAKKGL- 378
Query: 430 DHNEYNVIVADGSPLVNMNTGIVIQDCNIIPEAALVPEKFTVRS---YLGRPWQNESKAV 486
N+ N I A G N TGI IQ CNI ++ L E +V S YLGRPW+ S+ V
Sbjct: 379 -PNQKNSITAQGRKDPNEPTGISIQFCNITADSDL--EAASVNSTPTYLGRPWKLYSRTV 435
Query: 487 IMESTIGDFIHQDGWTTWPEEQNKPEKHHENTCYFAEYANTGPGANVARRVKWKGYKGVI 546
IM+S + + I +GW W + N+ ++ EY N GPGA + RVKW GY+
Sbjct: 436 IMQSFLSNVIRPEGWLEWNGD------FALNSLFYGEYMNYGPGAGLGSRVKWPGYQVFN 489
Query: 547 SRSEATKYTASIWLDAG---PKTAPKSAVEW 574
++A YT + +++ P T K E+
Sbjct: 490 ESTQAKNYTVAQFIEGNLWLPSTGVKYTAEF 520
>PME_MEDSA (Q42920) Pectinesterase precursor (EC 3.1.1.11) (Pectin
methylesterase) (PE) (P65)
Length = 447
Score = 278 bits (710), Expect = 4e-74
Identities = 179/481 (37%), Positives = 260/481 (53%), Gaps = 40/481 (8%)
Query: 109 LDDCKDLMQFALDSLDLSNNCVRDNNIEAVHDQTADMRNWLSAVISYKQGCMEGFDDAND 168
++ C +++ +A+D + S + + + + D++ WL+ +S++Q C++GF AN
Sbjct: 1 MEICNEVLDYAVDGIHKSVGTLDQFDFHKLSEYAFDLKVWLTGTLSHQQTCLDGF--ANT 58
Query: 169 GEKKIKEQFHVQSLYSVQKVTAVALDIVTGLSDILQQFNLN-FDIKPPSRRLLNSEVTVD 227
K + V L + ++++ A+D++ +S IL+ F+ + + + SRRLL+
Sbjct: 59 TTKAGETMTKV--LKTSMELSSNAIDMMDAVSRILKGFDTSQYSV---SRRLLS------ 107
Query: 228 DQGYPSWISSSGRKLLAKMQRKGWRANIRPNAVVANDGSGQFKTIQAALASYPKGNKDRY 287
D G PSW++ R+LLA N++PNAVVA DGSGQFKT+ AL + P N +
Sbjct: 108 DDGIPSWVNDGHRRLLAG-------GNVQPNAVVAQDGSGQFKTLTDALKTVPPKNAVPF 160
Query: 288 VIYVKAGVYDEYITVPKEAVNILMYGDGPAKTIVTGRKNQMAGTNTQNTATFSNTAMGFI 347
VI+VKAGVY E + V KE + + GDGP KT TG N G NT NTATF F+
Sbjct: 161 VIHVKAGVYKETVNVAKEMNYVTVIGDGPTKTKFTGSLNYADGINTYNTATFGVNGANFM 220
Query: 348 GKAMTFENTAGPAGMQAVAFRNIGDMSALVGCHIVGYQDTLYVQTNRQFYRNCVISGTID 407
K + FENTAG QAVA R D + C + G+QDTLYVQ+ RQFYR+C ISGTID
Sbjct: 221 AKDIGFENTAGTGKHQAVALRVTADQAIFYNCQMDGFQDTLYVQSQRQFYRDCSISGTID 280
Query: 408 FIFGTSATLIQSSTIIVR---KGNYDHNEYNVIVADGSPLVNMNTGIVIQDCNIIPEAAL 464
F+FG + Q+ ++ R KG + ++ A G N + +V Q + E AL
Sbjct: 281 FVFGERFGVFQNCKLVCRLPAKG-----QQCLVTAGGREKQNSASALVFQSSHFTGEPAL 335
Query: 465 VPEKFTVRSYLGRPWQNESKAVIMESTIGDFIHQDGWTTWPEEQNKPEKHHENTCYFAEY 524
V SYLGRPW+ SK VIM+STI +G+ W K TC + EY
Sbjct: 336 TSVTPKV-SYLGRPWKQYSKVVIMDSTIDAIFVPEGYMPWMGSAFK------ETCTYYEY 388
Query: 525 ANTGPGANVARRVKWKGYKGVISRSEATKYTASIWLDAGPKTAPKSAVEWLHDLHVPHYL 584
N GPGA+ RVKW G K V++ + A +Y + + TA + W+ VP+ L
Sbjct: 389 NNKGPGADTNLRVKWHGVK-VLTSNVAAEYYPGKFFEIVNATARDT---WIVKSGVPYSL 444
Query: 585 G 585
G
Sbjct: 445 G 445
>PME1_FICAW (P83947) Pectinesterase precursor (EC 3.1.1.11) (Pectin
methylesterase) (PE)
Length = 545
Score = 271 bits (694), Expect = 3e-72
Identities = 161/520 (30%), Positives = 266/520 (50%), Gaps = 38/520 (7%)
Query: 45 MCQNAQDQKLCHDTLSSVRGADAADPKAYIAAAVKAATDNVIKAFNMSERLTTEYGKKNG 104
+C + +++ C +S V G + AD + + + ++ T + KAF + +
Sbjct: 47 ICDQSVNKESCLAMISEVTGLNMADHRNLLKSFLEKTTPRIQKAFETANDASRRINNPQ- 105
Query: 105 AKMALDDCKDLMQFALDSLDLSNNCVRDNNIEAVHDQTADMRNWLSAVISYKQGCMEGFD 164
+ AL DC +LM + + + S + + N+ + D+ WLS V++ C++G +
Sbjct: 106 ERTALLDCAELMDLSKERVVDSISILFHQNLTTRSHE--DLHVWLSGVLTNHVTCLDGLE 163
Query: 165 DANDGEKKIKEQFHVQSLYSVQKVT-AVALDIVTGLSDILQQFNLNFDIKPPSRRLLNSE 223
+ + K + H+ L + + A+ + + S++++ NF
Sbjct: 164 EGSTDYIKTLMESHLNELILRARTSLAIFVTLFPAKSNVIEPVTGNF------------- 210
Query: 224 VTVDDQGYPSWISSSGRKLLAKMQRKGWRANIRPNAVVANDGSGQFKTIQAALASYPKGN 283
P+W+++ R+LL + + +I P+ VVA DGSG ++T+ A+A+ P +
Sbjct: 211 --------PTWVTAGDRRLLQTLGK-----DIEPDIVVAKDGSGDYETLNEAVAAIPDNS 257
Query: 284 KDRYVIYVKAGVYDEYITVPKEAVNILMYGDGPAKTIVTGRKNQMAGTNTQNTATFSNTA 343
K R ++ V+ G+Y+E + + N+++ G+G TI+TG +N + G+ T ++AT +
Sbjct: 258 KKRVIVLVRTGIYEENVDFGYQKKNVMLVGEGMDYTIITGSRNVVDGSTTFDSATVAAVG 317
Query: 344 MGFIGKAMTFENTAGPAGMQAVAFRNIGDMSALVGCHIVGYQDTLYVQTNRQFYRNCVIS 403
GFI + + F+NTAGP QAVA R D + + C I YQDTLY RQFYR+ I+
Sbjct: 318 DGFIAQDICFQNTAGPEKYQAVALRIGADETVINRCRIDAYQDTLYPHNYRQFYRDRNIT 377
Query: 404 GTIDFIFGTSATLIQSSTIIVRKGNYDHNEYNVIVADGSPLVNMNTGIVIQDCNIIPEAA 463
GT+DFIFG +A + Q+ +I RK + N I A G N NTG IQ+C I A
Sbjct: 378 GTVDFIFGNAAVVFQNCNLIPRK--QMKGQENTITAQGRTDPNQNTGTSIQNCEIFASAD 435
Query: 464 LVPEKFTVRSYLGRPWQNESKAVIMESTIGDFIHQDGWTTWPEEQNKPEKHHENTCYFAE 523
L P + T +SYLGRPW+ S+ V+MES I D I GW W + T ++ E
Sbjct: 436 LEPVEDTFKSYLGRPWKEYSRTVVMESYISDVIDPAGWLEWDRD------FALKTLFYGE 489
Query: 524 YANTGPGANVARRVKWKGYKGVISRSEATKYTASIWLDAG 563
Y N GPG+ + RVKW GY + S A ++T + + G
Sbjct: 490 YRNGGPGSGTSERVKWPGYHVITSPEVAEQFTVAELIQGG 529
>PM22_LYCES (Q96575) Pectinesterase 2 precursor (EC 3.1.1.11)
(Pectin methylesterase 2) (PE 2)
Length = 550
Score = 270 bits (691), Expect = 6e-72
Identities = 180/564 (31%), Positives = 285/564 (49%), Gaps = 55/564 (9%)
Query: 4 KVIISVVSLILVVGVIIGVVVDIRKNGEDPKVQTQQRNLRIMCQNAQDQKLCHDTLSSVR 63
K++ VV+L + + +++ +V + + + NL C+ AQD +LC +S +
Sbjct: 22 KILTFVVTLFVALFLVVFLVAPYQ-------FEIKHSNL---CKTAQDSQLCLSYVSDLM 71
Query: 64 GADAADPKAYIAAAVKAATDNVIKAFNMSERLTTEYGKK-NGAKM--ALDDCKDLMQFAL 120
+ + + + N + N + + ++ + N + AL DC +L+
Sbjct: 72 SNEIVTTDSDGLSILMKFLVNYVHQMNNAIPVVSKMKNQINDIRQEGALTDCLELLD--- 128
Query: 121 DSLDLSNNCVRDNNIEAVHDQTADMRNWLSAVISYKQGCMEGFDDANDGEKKIKEQFHVQ 180
S+DL ++ + + + H + A+ ++WLS V++ C++ D K + ++
Sbjct: 129 QSVDLVSDSIAAID-KRTHSEHANAQSWLSGVLTNHVTCLDELDSFT---KAMINGTNLD 184
Query: 181 SLYSVQKVTAVALDIVTGLSDILQQFNLNFDIKPPSRRLLNSEVTVDDQGYPSWISSSGR 240
L S KV L VT +D D+ P + PSW+SS R
Sbjct: 185 ELISRAKVALAMLASVTTPND---------DVLRPGLGKM-----------PSWVSSRDR 224
Query: 241 KLLAKMQRKGWRANIRPNAVVANDGSGQFKTIQAALASYPKGNKDRYVIYVKAGVYDEYI 300
KL+ + +I NAVVA DG+G+++T+ A+A+ P +K RYVIYVK G+Y E +
Sbjct: 225 KLMESSGK-----DIGANAVVAKDGTGKYRTLAEAVAAAPDKSKTRYVIYVKRGIYKENV 279
Query: 301 TVPKEAVNILMYGDGPAKTIVTGRKNQMAGTNTQNTATFSNTAMGFIGKAMTFENTAGPA 360
V + +++ GDG TI+TG N + G+ T ++AT + GFI + + +NTAGPA
Sbjct: 280 EVSSRKMKLMIVGDGMHATIITGNLNVVDGSTTFHSATLAAVGKGFILQDICIQNTAGPA 339
Query: 361 GMQAVAFRNIGDMSALVGCHIVGYQDTLYVQTNRQFYRNCVISGTIDFIFGTSATLIQSS 420
QAVA R D S + C I YQDTLY + RQFYR+ ++GTIDFIFG +A + Q
Sbjct: 340 KHQAVALRVGADKSVINRCRIDAYQDTLYAHSQRQFYRDSYVTGTIDFIFGNAAVVFQKC 399
Query: 421 TIIVRK-GNYDHNEYNVIVADGSPLVNMNTGIVIQDCNIIPEAALVPEKFTVRSYLGRPW 479
++ RK G Y + N++ A G N TG IQ CNII + L P +YLGRPW
Sbjct: 400 KLVARKPGKY---QQNMVTAQGRTDPNQATGTSIQFCNIIASSDLEPVLKEFPTYLGRPW 456
Query: 480 QNESKAVIMESTIGDFIHQDGWTTWPEEQNKPEKHHENTCYFAEYANTGPGANVARRVKW 539
+ S+ V+MES +G I+ GW W + T Y+ E+ N GPGA ++RVKW
Sbjct: 457 KKYSRTVVMESYLGGLINPAGWAEWDGD------FALKTLYYGEFMNNGPGAGTSKRVKW 510
Query: 540 KGYKGVISRSEATKYTASIWLDAG 563
GY + +EA +T + + G
Sbjct: 511 PGYHCITDPAEAMPFTVAKLIQGG 534
>PM21_LYCES (P09607) Pectinesterase 2 precursor (EC 3.1.1.11)
(Pectin methylesterase 2) (PE 2)
Length = 550
Score = 266 bits (679), Expect = 1e-70
Identities = 183/583 (31%), Positives = 292/583 (49%), Gaps = 63/583 (10%)
Query: 4 KVIISVVSLILVVGVIIGVVVDIRKNGEDPKVQTQQRNLRIMCQNAQDQKLC----HDTL 59
K++ VV+L + + +++ +V + + + NL C+ AQD +LC D +
Sbjct: 22 KILTFVVTLFVALFLVVFLVAPYQ-------FEIKHSNL---CKTAQDSQLCLSYVSDLI 71
Query: 60 SSVRGADAADPKAYIAAAVKAATDNVIKAFNMSERLTTEYG--KKNGAKMALDDCKDLMQ 117
S+ +D + + + + + A + ++ + ++ GA L DC +L+
Sbjct: 72 SNEIVTSDSDGLSILKKFLVYSVHQMNNAIPVVRKIKNQINDIREQGA---LTDCLELLD 128
Query: 118 FALDSLDLSNNCVRDNNIEAVHDQTADMRNWLSAVISYKQGCMEGFDDANDGEKKIKEQF 177
++D L + D + H A+ ++WLS V++ C++ D K +
Sbjct: 129 LSVD-LVCDSIAAIDKRSRSEH---ANAQSWLSGVLTNHVTCLDELDSFT---KAMINGT 181
Query: 178 HVQSLYSVQKVTAVALDIVTGLSDILQQFNLNFDIKPPSRRLLNSEVTVDDQGYPSWISS 237
++ L S KV L VT +D + ++P ++ PSW+SS
Sbjct: 182 NLDELISRAKVALAMLASVTTPNDEV--------LRPGLGKM------------PSWVSS 221
Query: 238 SGRKLLAKMQRKGWRANIRPNAVVANDGSGQFKTIQAALASYPKGNKDRYVIYVKAGVYD 297
RKL+ + +I NAVVA DG+G+++T+ A+A+ P +K RYVIYVK G Y
Sbjct: 222 RDRKLMESSGK-----DIGANAVVAKDGTGKYRTLAEAVAAAPDKSKTRYVIYVKRGTYK 276
Query: 298 EYITVPKEAVNILMYGDGPAKTIVTGRKNQMAGTNTQNTATFSNTAMGFIGKAMTFENTA 357
E + V +N+++ GDG TI+TG N + G+ T ++AT + GFI + + +NTA
Sbjct: 277 ENVEVSSRKMNLMIIGDGMYATIITGSLNVVDGSTTFHSATLAAVGKGFILQDICIQNTA 336
Query: 358 GPAGMQAVAFRNIGDMSALVGCHIVGYQDTLYVQTNRQFYRNCVISGTIDFIFGTSATLI 417
GPA QAVA R D S + C I YQDTLY + RQFYR+ ++GTIDFIFG +A +
Sbjct: 337 GPAKHQAVALRVGADKSVINRCRIDAYQDTLYAHSQRQFYRDSYVTGTIDFIFGNAAVVF 396
Query: 418 QSSTIIVRK-GNYDHNEYNVIVADGSPLVNMNTGIVIQDCNIIPEAALVPEKFTVRSYLG 476
Q ++ RK G Y + N++ A G N TG IQ C+II L P +YLG
Sbjct: 397 QKCQLVARKPGKY---QQNMVTAQGRTDPNQATGTSIQFCDIIASPDLKPVVKEFPTYLG 453
Query: 477 RPWQNESKAVIMESTIGDFIHQDGWTTWPEEQNKPEKHHENTCYFAEYANTGPGANVARR 536
RPW+ S+ V+MES +G I GW W + T Y+ E+ N GPGA ++R
Sbjct: 454 RPWKKYSRTVVMESYLGGLIDPSGWAEWHGD------FALKTLYYGEFMNNGPGAGTSKR 507
Query: 537 VKWKGYKGVISRSEATKYTASIWLDAGP--KTAPKSAVEWLHD 577
VKW GY + +EA +T + + G ++ + V+ L+D
Sbjct: 508 VKWPGYHVITDPAEAMSFTVAKLIQGGSWLRSTDVAYVDGLYD 550
>PME_PETIN (Q43043) Pectinesterase precursor (EC 3.1.1.11) (Pectin
methylesterase) (PE)
Length = 374
Score = 265 bits (678), Expect = 2e-70
Identities = 136/333 (40%), Positives = 195/333 (57%), Gaps = 15/333 (4%)
Query: 253 ANIRPNAVVANDGSGQFKTIQAALASYPKGNKDRYVIYVKAGVYDEYITVPKEAVNILMY 312
+N +PNA VA DGSGQ+KTI+ AL + PK N + ++I++KAGVY EYI +PK N+++
Sbjct: 53 SNAKPNATVALDGSGQYKTIKEALDAVPKKNTEPFIIFIKAGVYKEYIDIPKSMTNVVLI 112
Query: 313 GDGPAKTIVTGRKNQMAGTNTQNTATFSNTAMGFIGKAMTFENTAGPAGMQAVAFRNIGD 372
G+GP KT +TG K+ G +T +T T F+ K + FENTAGP QAVA R D
Sbjct: 113 GEGPTKTKITGNKSVKDGPSTFHTTTVGVNGANFVAKNIGFENTAGPEKEQAVALRVSAD 172
Query: 373 MSALVGCHIVGYQDTLYVQTNRQFYRNCVISGTIDFIFGTSATLIQSSTIIVRKGNYDHN 432
+ + C I GYQDTLYV T RQFYR+C I+GT+DFIFG ++Q+ +IVRK N
Sbjct: 173 KAIIYNCQIDGYQDTLYVHTYRQFYRDCTITGTVDFIFGNGEAVLQNCKVIVRKP--AQN 230
Query: 433 EYNVIVADGSPLVNMNTGIVIQDCNIIPEAALVPEKFTVRSYLGRPWQNESKAVIMESTI 492
+ ++ A G IV+Q+C I P+ ++YLGRPW+ S+ +IM+S I
Sbjct: 231 QSCMVTAQGRTEPIQKGAIVLQNCEIKPDTDYFSLSPPSKTYLGRPWKEYSRTIIMQSYI 290
Query: 493 GDFIHQDGWTTWPEEQNKPEKHHENTCYFAEYANTGPGANVARRVKWKGYKGVISRSEAT 552
FI +GW W +T Y+AEY N GPGA + +R+ WKG++ + A
Sbjct: 291 DKFIEPEGWAPW-----NITNFGRDTSYYAEYQNRGPGAALDKRITWKGFQKGFTGEAAQ 345
Query: 553 KYTASIWLDAGPKTAPKSAVEWLHDLHVPHYLG 585
K+TA ++++ + WL +VP+ G
Sbjct: 346 KFTAGVYIN--------NDENWLQKANVPYEAG 370
>PME2_CITSI (O04887) Pectinesterase 2 precursor (EC 3.1.1.11)
(Pectin methylesterase) (PE)
Length = 510
Score = 261 bits (668), Expect = 3e-69
Identities = 172/508 (33%), Positives = 257/508 (49%), Gaps = 59/508 (11%)
Query: 71 KAYIAAAVKAATDNVIKAFNMSERLTTEYGKKNGAKMALDDCKDLMQFALDSLDLSNNCV 130
K + A++ AT + + + + E K A +DC++L + + L+ ++N
Sbjct: 62 KISLQLALERATTAQSRTYTLGSKCRNER-----EKAAWEDCRELYELTVLKLNQTSN-- 114
Query: 131 RDNNIEAVHDQTADMRNWLSAVISYKQGCMEGFDDANDGEKKIKEQFHVQSLYSVQKVTA 190
+ D + WLS ++ + C +D E +V L S
Sbjct: 115 -----SSPGCTKVDKQTWLSTALTNLETCRASLEDLGVPE-------YVLPLLS------ 156
Query: 191 VALDIVTGLSDILQQFNLNFDIKPPSRRLLNSEVTVDDQGYPSWISSSGRKLLAKMQRKG 250
+ VT L I +LN ++ +E + D G+P+W+ RKLL
Sbjct: 157 ---NNVTKL--ISNTLSLN--------KVPYNEPSYKD-GFPTWVKPGDRKLL------- 195
Query: 251 WRANIRPNAVVANDGSGQFKTIQAALASYPKGNKDRYVIYVKAGVYDEYITVPKEAVNIL 310
+ R N VVA DGSG KTIQ A+A+ + RYVIY+KAG Y+E I V + NI+
Sbjct: 196 -QTTPRANIVVAQDGSGNVKTIQEAVAAASRAGGSRYVIYIKAGTYNENIEVKLK--NIM 252
Query: 311 MYGDGPAKTIVTGRKNQMAGTNTQNTATFSNTAMGFIGKAMTFENTAGPAGMQAVAFRNI 370
GDG KTI+TG K+ G T +AT + FI + +T NTAGP QAVA R+
Sbjct: 253 FVGDGIGKTIITGSKSVGGGATTFKSATVAVVGDNFIARDITIRNTAGPNNHQAVALRSG 312
Query: 371 GDMSALVGCHIVGYQDTLYVQTNRQFYRNCVISGTIDFIFGTSATLIQSSTIIVRKGNYD 430
D+S C GYQDTLYV + RQFYR C I GT+DFIFG +A ++Q+ I RK
Sbjct: 313 SDLSVFYRCSFEGYQDTLYVHSQRQFYRECDIYGTVDFIFGNAAVVLQNCNIFARK---P 369
Query: 431 HNEYNVIVADGSPLVNMNTGIVIQDCNIIPEAALVPEKFTVRSYLGRPWQNESKAVIMES 490
N N + A G N +TGI+I +C + + L P + +V+++LGRPW+ S+ V +++
Sbjct: 370 PNRTNTLTAQGRTDPNQSTGIIIHNCRVTAASDLKPVQSSVKTFLGRPWKQYSRTVYIKT 429
Query: 491 TIGDFIHQDGWTTWPEEQNKPEKHHENTCYFAEYANTGPGANVARRVKWKGYKGVISRSE 550
+ I+ GW W + NT Y+AEY NTGPG++ A RVKW+GY + S S+
Sbjct: 430 FLDSLINPAGWMEWSGD------FALNTLYYAEYMNTGPGSSTANRVKWRGYHVLTSPSQ 483
Query: 551 ATKYTASIWLDAGPKTAPKSAVEWLHDL 578
+++T ++ AG P + V + L
Sbjct: 484 VSQFTVGNFI-AGNSWLPATNVPFTSGL 510
>PME1_LYCES (P14280) Pectinesterase 1 precursor (EC 3.1.1.11)
(Pectin methylesterase 1) (PE 1)
Length = 546
Score = 259 bits (663), Expect = 1e-68
Identities = 177/568 (31%), Positives = 282/568 (49%), Gaps = 67/568 (11%)
Query: 4 KVIISVVSLILVVGVIIGVVVDIRKNGEDPKVQTQQRNLRIMCQNAQDQKLC----HDTL 59
K++ V++L + + ++ V+I+ + +C+ AQD +LC D +
Sbjct: 22 KILSFVITLFVALFLVAPYQVEIKHSN--------------LCKTAQDSQLCLSYVSDLI 67
Query: 60 SSVRGADAADPKAYIAAAVKAATDNVIKAFNMSERLTTEYG--KKNGAKMALDDCKDLMQ 117
S+ +D + + + + A + ++ + +++GA L DC +L+
Sbjct: 68 SNEIVTTESDGHSILMKFLVNYVHQMNNAIPVVRKMKNQINDIRQHGA---LTDCLELLD 124
Query: 118 FALDSLDLSNNCVRDNNIEAVHDQTADMRNWLSAVISYKQGCMEGFDDANDGEKKIKEQF 177
++D S + D + H A+ ++WLS V++ C++ D K +
Sbjct: 125 QSVDFASDSIAAI-DKRSRSEH---ANAQSWLSGVLTNHVTCLDELDSFT---KAMINGT 177
Query: 178 HVQSLYSVQKVT-AVALDIVTGLSDILQQFNLNFDIKPPSRRLLNSEVTVDDQGYPSWIS 236
+++ L S KV A+ + T D+ +TV + PSW+S
Sbjct: 178 NLEELISRAKVALAMLASLTTQDEDVF--------------------MTVLGK-MPSWVS 216
Query: 237 SSGRKLLAKMQRKGWRANIRPNAVVANDGSGQFKTIQAALASYPKGNKDRYVIYVKAGVY 296
S RKL+ + +I NAVVA DG+G ++T+ A+A+ P +K RYVIYVK G Y
Sbjct: 217 SMDRKLMESSGK-----DIIANAVVAQDGTGDYQTLAEAVAAAPDKSKTRYVIYVKRGTY 271
Query: 297 DEYITVPKEAVNILMYGDGPAKTIVTGRKNQMAGTNTQNTATFSNTAMGFIGKAMTFENT 356
E + V +N+++ GDG T +TG N + G+ T +AT + GFI + + +NT
Sbjct: 272 KENVEVASNKMNLMIVGDGMYATTITGSLNVVDGSTTFRSATLAAVGQGFILQDICIQNT 331
Query: 357 AGPAGMQAVAFRNIGDMSALVGCHIVGYQDTLYVQTNRQFYRNCVISGTIDFIFGTSATL 416
AGPA QAVA R DMS + C I YQDTLY + RQFYR+ ++GT+DFIFG +A +
Sbjct: 332 AGPAKDQAVALRVGADMSVINRCRIDAYQDTLYAHSQRQFYRDSYVTGTVDFIFGNAAVV 391
Query: 417 IQSSTIIVRK-GNYDHNEYNVIVADGSPLVNMNTGIVIQDCNIIPEAALVPEKFTVRSYL 475
Q ++ RK G Y + N++ A G N TG IQ CNII + L P +YL
Sbjct: 392 FQKCQLVARKPGKY---QQNMVTAQGRTDPNQATGTSIQFCNIIASSDLEPVLKEFPTYL 448
Query: 476 GRPWQNESKAVIMESTIGDFIHQDGWTTWPEEQNKPEKHHENTCYFAEYANTGPGANVAR 535
GRPW+ S+ V+MES +G I+ GW W + T Y+ E+ N GPGA ++
Sbjct: 449 GRPWKEYSRTVVMESYLGGLINPAGWAEWDGD------FALKTLYYGEFMNNGPGAGTSK 502
Query: 536 RVKWKGYKGVISRSEATKYTASIWLDAG 563
RVKW GY + ++A +T + + G
Sbjct: 503 RVKWPGYHVITDPAKAMPFTVAKLIQGG 530
>PME_DAUCA (P83218) Pectinesterase (EC 3.1.1.11) (Pectin
methylesterase) (PE)
Length = 319
Score = 258 bits (658), Expect = 4e-68
Identities = 140/333 (42%), Positives = 194/333 (58%), Gaps = 17/333 (5%)
Query: 253 ANIRPNAVVANDGSGQFKTIQAALASYPKGNKDRYVIYVKAGVYDEYITVPKEAVNILMY 312
+ + PN VVA DGSG +KT+ A+A+ P+ +K RYVI +KAGVY E + VPK+ NI+
Sbjct: 3 STVTPNVVVAADGSGDYKTVSEAVAAAPEDSKTRYVIRIKAGVYRENVDVPKKKKNIMFL 62
Query: 313 GDGPAKTIVTGRKNQMAGTNTQNTATFSNTAMGFIGKAMTFENTAGPAGMQAVAFRNIGD 372
GDG TI+T KN G+ T N+AT + GF+ + +TF+NTAG A QAVA R D
Sbjct: 63 GDGRTSTIITASKNVQDGSTTFNSATVAAVGAGFLARDITFQNTAGAAKHQAVALRVGSD 122
Query: 373 MSALVGCHIVGYQDTLYVQTNRQFYRNCVISGTIDFIFGTSATLIQSSTIIVRKGNYDHN 432
+SA C I+ YQD+LYV +NRQF+ NC I+GT+DFIFG +A ++Q I R+
Sbjct: 123 LSAFYRCDILAYQDSLYVHSNRQFFINCFIAGTVDFIFGNAAVVLQDCDIHARRPG--SG 180
Query: 433 EYNVIVADGSPLVNMNTGIVIQDCNIIPEAALVPEKFTVRSYLGRPWQNESKAVIMESTI 492
+ N++ A G N NTGIVIQ I + L P + + +YLGRPW+ S+ V+M+S+I
Sbjct: 181 QKNMVTAQGRTDPNQNTGIVIQKSRIGATSDLQPVQSSFPTYLGRPWKEYSRTVVMQSSI 240
Query: 493 GDFIHQDGWTTWPEEQNKPEKHHENTCYFAEYANTGPGANVARRVKWKGYKGVISRSEAT 552
+ I+ GW W +T Y+ EY NTG GA + RV WKG+K + S +EA
Sbjct: 241 TNVINPAGWFPW------DGNFALDTLYYGEYQNTGAGAATSGRVTWKGFKVITSSTEAQ 294
Query: 553 KYTASIWLDAGPKTAPKSAVEWLHDLHVPHYLG 585
+T ++ G WL P LG
Sbjct: 295 GFTPGSFIAGG---------SWLKATTFPFSLG 318
>PME3_LYCES (Q96576) Pectinesterase 3 precursor (EC 3.1.1.11)
(Pectin methylesterase 3) (PE 3)
Length = 544
Score = 253 bits (646), Expect = 1e-66
Identities = 179/583 (30%), Positives = 274/583 (46%), Gaps = 70/583 (12%)
Query: 5 VIISVVSLILVVGVIIGVVVDIRKNGEDPKVQTQQRNLRIMCQNAQDQKLCHDTLSSVRG 64
V+ V+L LVV ++ +I+ + +C+ AQD +LC +S +
Sbjct: 27 VVTLFVALFLVVFLVAPYQFEIKHSN--------------LCKTAQDSQLCLSYVSEIVT 72
Query: 65 ADAADPKAYIAAAVKAA--TDNVIKAFNMSERLTTEYGKKNGAKMALDDCKDLMQFALDS 122
++ VK +N I + + ++ AL DC +L+ ++D
Sbjct: 73 TESDGVTVLKKFLVKYVHQMNNAIPVVRKIKNQINDIRQQG----ALTDCLELLDQSVDL 128
Query: 123 LDLSNNCVRDNNIEAVHDQTADMRNWLSAVISYKQGCMEGFDDANDGEKKIKEQFHVQSL 182
+ S + D + H A+ ++WLS V++ C+ D+ K + L
Sbjct: 129 VSDSIAAI-DKRSRSEH---ANAQSWLSGVLTNHVTCL---DELTSFSLSTKNGTVLDEL 181
Query: 183 YSVQKVTAVALDIVTGLSDILQQFNLNFDIKPPSRRLLNSEVTVDDQGYPSWISSSGRKL 242
+ KV L VT +D + + L P W+SS RKL
Sbjct: 182 ITRAKVALAMLASVTTPNDEVLRQGLG--------------------KMPYWVSSRDRKL 221
Query: 243 LAKMQRKGWRANIRPNAVVANDGSGQFKTIQAALASYPKGNKDRYVIYVKAGVYDEYITV 302
+ + +I N VVA DG+G ++T+ A+A+ P NK RYVIYVK G+Y E + V
Sbjct: 222 MESSGK-----DIIANRVVAQDGTGDYQTLAEAVAAAPDKNKTRYVIYVKMGIYKENVVV 276
Query: 303 PKEAVNILMYGDGPAKTIVTGRKNQMAGTNTQNTATFSNTAMGFIGKAMTFENTAGPAGM 362
K+ +N+++ GDG TI+TG N + G+ T + T + GFI + + +NTAGP
Sbjct: 277 TKKKMNLMIVGDGMNATIITGSLNVVDGS-TFPSNTLAAVGQGFILQDICIQNTAGPEKD 335
Query: 363 QAVAFRNIGDMSALVGCHIVGYQDTLYVQTNRQFYRNCVISGTIDFIFGTSATLIQSSTI 422
QAVA R DMS + C I YQDTLY + RQFYR+ ++GT+DFIFG +A + Q I
Sbjct: 336 QAVALRVGADMSVINRCRIDAYQDTLYAHSQRQFYRDSYVTGTVDFIFGNAAVVFQKCQI 395
Query: 423 IVRKGNYDHNEYNVIVADGSPLVNMNTGIVIQDCNIIPEAALVPEKFTVRSYLGRPWQNE 482
+ RK N + N++ A G N TG IQ C+II L P ++YLGRPW+
Sbjct: 396 VARKPN--KRQKNMVTAQGRTDPNQATGTSIQFCDIIASPDLEPVMNEYKTYLGRPWKKH 453
Query: 483 SKAVIMESTIGDFIHQDGWTTWPEEQNKPEKHHENTCYFAEYANTGPGANVARRVKWKGY 542
S+ V+M+S + I GW W + T Y+ E+ N GPGA ++RVKW GY
Sbjct: 454 SRTVVMQSYLDGHIDPSGWFEWRGD------FALKTLYYGEFMNNGPGAGTSKRVKWPGY 507
Query: 543 KGVISRSEATKYTASIWLDAGPKTAPKSAVEWLHDLHVPHYLG 585
+ +EA +T + + G WL+ V + G
Sbjct: 508 HVITDPNEAMPFTVAELIQGG---------SWLNSTSVAYVEG 541
>PME_ASPAC (Q12535) Pectinesterase precursor (EC 3.1.1.11) (Pectin
methylesterase) (PE)
Length = 331
Score = 123 bits (308), Expect = 2e-27
Identities = 92/295 (31%), Positives = 129/295 (43%), Gaps = 24/295 (8%)
Query: 252 RANIRPNAVVANDGSGQFKTIQAALASYPKGNKDRYVIYVKAGVYDEYITVPKEAVNILM 311
R A+V G + TI A+ + D I+++ G YDE + +P +++
Sbjct: 20 RTTAPSGAIVVAKSGGDYTTIGDAIDALSTSTTDTQTIFIEEGTYDEQVYLPAMTGKVII 79
Query: 312 YG-----DGPAKTIVT---GRKNQMAGTNTQNTATFSNTAMGFIGKAMTFENTAGPAGMQ 363
YG D A +VT + AG + TATF N A+G + NT G A Q
Sbjct: 80 YGQTENTDSYADNLVTITHAISYEDAGESDDLTATFRNKAVGSQVYNLNIANTCGQACHQ 139
Query: 364 AVAFRNIGDMSALVGCHIVGYQDTLYVQTNRQFYRNCVISGTIDFIFGTSA-TLIQSSTI 422
A+A D GC+ GYQDTL QT Q Y N I G +DFIFG A Q+ I
Sbjct: 140 ALALSAWADQQGYYGCNFTGYQDTLLAQTGNQLYINSYIEGAVDFIFGQHARAWFQNVDI 199
Query: 423 IVRKGNYDHNEYNVIVADGSPLVNMNTGIVIQDCNIIPEAALVPEKFTVRSYLGRPWQNE 482
V +G + I A+G + VI + + + T YLGRPW
Sbjct: 200 RVVEGPTSAS----ITANGRSSETDTSYYVINKSTVAAKEGDDVAEGTY--YLGRPWSEY 253
Query: 483 SKAVIMESTIGDFIHQDGWTTWPEEQNKPEKHHENTCY--FAEYANTGPGANVAR 535
++ V ++++ + I+ GWT W NT Y F EYANTG G+ R
Sbjct: 254 ARVVFQQTSMTNVINSLGWTEWSTST-------PNTEYVTFGEYANTGAGSEGTR 301
>PME_ASPTU (P17872) Pectinesterase precursor (EC 3.1.1.11) (Pectin
methylesterase) (PE)
Length = 331
Score = 107 bits (268), Expect = 7e-23
Identities = 81/285 (28%), Positives = 117/285 (40%), Gaps = 19/285 (6%)
Query: 259 AVVANDGSGQFKTIQAALASYPKGNKDRYVIYVKAGVYDEYITVPKEAVNILMYGDGPAK 318
A+V G + TI AA+ + + + I+++ G YDE + +P + +++YG
Sbjct: 27 AIVVAKSGGDYDTISAAVDALSTTSTETQTIFIEEGSYDEQVYIPALSGKLIVYGQTEDT 86
Query: 319 TIVTGRKNQMAGT-------NTQNTATFSNTAMGFIGKAMTFENTAGPAGMQAVAFRNIG 371
T T + N TAT N A G + NT G A QA+A
Sbjct: 87 TTYTSNLVNITHAIALADVDNDDETATLRNYAEGSAIYNLNIANTCGQACHQALAVSAYA 146
Query: 372 DMSALVGCHIVGYQDTLYVQTNRQFYRNCVISGTIDFIFGTSA-TLIQSSTIIVRKGNYD 430
C GYQDTL +T Q Y I G +DFIFG A I V +G
Sbjct: 147 SEQGYYACQFTGYQDTLLAETGYQVYAGTYIEGAVDFIFGQHARAWFHECDIRVLEGPSS 206
Query: 431 HNEYNVIVADGSPLVNMNTGIVIQDCNIIPEAALVPEKFTVRSYLGRPWQNESKAVIMES 490
+ I A+G + ++ VI + AA + + YLGRPW ++ ++
Sbjct: 207 AS----ITANGRSSESDDSYYVIHKSTV--AAADGNDVSSGTYYLGRPWSQYARVCFQKT 260
Query: 491 TIGDFIHQDGWTTWPEEQNKPEKHHENTCYFAEYANTGPGANVAR 535
++ D I+ GWT W E F EY NTG GA R
Sbjct: 261 SMTDVINHLGWTEWSTSTPNTE-----NVTFVEYGNTGTGAEGPR 300
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.317 0.133 0.392
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 68,583,161
Number of Sequences: 164201
Number of extensions: 2882301
Number of successful extensions: 6987
Number of sequences better than 10.0: 35
Number of HSP's better than 10.0 without gapping: 24
Number of HSP's successfully gapped in prelim test: 11
Number of HSP's that attempted gapping in prelim test: 6847
Number of HSP's gapped (non-prelim): 41
length of query: 588
length of database: 59,974,054
effective HSP length: 116
effective length of query: 472
effective length of database: 40,926,738
effective search space: 19317420336
effective search space used: 19317420336
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 69 (31.2 bits)
Medicago: description of AC144563.4


