

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144563.2 + phase: 0
(576 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
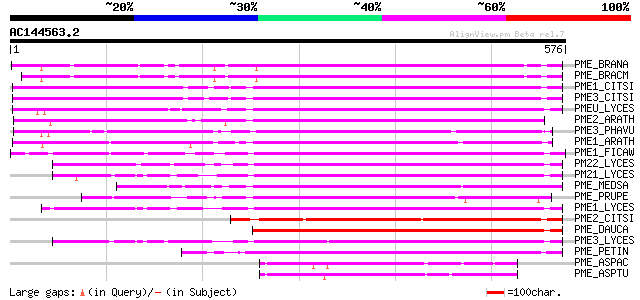
Score E
Sequences producing significant alignments: (bits) Value
PME_BRANA (P41510) Probable pectinesterase precursor (EC 3.1.1.1... 391 e-108
PME_BRACM (Q42608) Pectinesterase (EC 3.1.1.11) (Pectin methyles... 369 e-101
PME1_CITSI (O04886) Pectinesterase 1 precursor (EC 3.1.1.11) (Pe... 366 e-100
PME3_CITSI (P83948) Pectinesterase 3 precursor (EC 3.1.1.11) (Pe... 364 e-100
PMEU_LYCES (Q43143) Pectinesterase U1 precursor (EC 3.1.1.11) (P... 345 2e-94
PME2_ARATH (Q42534) Pectinesterase 2 precursor (EC 3.1.1.11) (Pe... 338 3e-92
PME3_PHAVU (Q43111) Pectinesterase 3 precursor (EC 3.1.1.11) (Pe... 328 2e-89
PME1_ARATH (Q43867) Pectinesterase 1 precursor (EC 3.1.1.11) (Pe... 327 4e-89
PME1_FICAW (P83947) Pectinesterase precursor (EC 3.1.1.11) (Pect... 313 1e-84
PM22_LYCES (Q96575) Pectinesterase 2 precursor (EC 3.1.1.11) (Pe... 307 4e-83
PM21_LYCES (P09607) Pectinesterase 2 precursor (EC 3.1.1.11) (Pe... 304 4e-82
PME_MEDSA (Q42920) Pectinesterase precursor (EC 3.1.1.11) (Pecti... 304 5e-82
PME_PRUPE (Q43062) Pectinesterase PPE8B precursor (EC 3.1.1.11) ... 301 2e-81
PME1_LYCES (P14280) Pectinesterase 1 precursor (EC 3.1.1.11) (Pe... 299 2e-80
PME2_CITSI (O04887) Pectinesterase 2 precursor (EC 3.1.1.11) (Pe... 296 1e-79
PME_DAUCA (P83218) Pectinesterase (EC 3.1.1.11) (Pectin methyles... 292 1e-78
PME3_LYCES (Q96576) Pectinesterase 3 precursor (EC 3.1.1.11) (Pe... 292 2e-78
PME_PETIN (Q43043) Pectinesterase precursor (EC 3.1.1.11) (Pecti... 288 3e-77
PME_ASPAC (Q12535) Pectinesterase precursor (EC 3.1.1.11) (Pecti... 122 3e-27
PME_ASPTU (P17872) Pectinesterase precursor (EC 3.1.1.11) (Pecti... 110 1e-23
>PME_BRANA (P41510) Probable pectinesterase precursor (EC 3.1.1.11)
(Pectin methylesterase) (PE)
Length = 584
Score = 391 bits (1005), Expect = e-108
Identities = 236/599 (39%), Positives = 337/599 (55%), Gaps = 47/599 (7%)
Query: 3 GKVIISAVSLILVVGVAIGVVVAVRKNGE---DPEVQTQQRNLRIMCQNSQDQKLCHETL 59
GK++IS S++LVVGVAIGVV V K G D + + Q+ + +C ++ D+ C +TL
Sbjct: 4 GKIVISVASMLLVVGVAIGVVTFVNKGGGAGGDKTLNSHQKAVESLCASATDKGSCAKTL 63
Query: 60 SSVHGADAADPKAYIAASVKAATDNVIKAFNMSERLTTEYGKENGA--KMALNDCKDLMQ 117
V + DP I A + A D V K+ N + GK A K L+ CK ++
Sbjct: 64 DPVK---SDDPSKLIKAFMLATKDAVTKSTNFTASTEEGMGKNINATSKAVLDYCKRVLM 120
Query: 118 FALDSLDLSTKCVHDNNIQAVHDQIADMRNWLSAVISYRQACMEGFDDANDGEKKIKEQF 177
+AL+ L+ + + ++ +Q ++ ++ WL+ V +Y+ C+ DD + E + +
Sbjct: 121 YALEDLETIVEEMGED-LQQSGSKMDQLKQWLTGVFNYQTDCI---DDIEESELR---KV 173
Query: 178 HVQSLDSVQKVTAVALDIVTGLSDILQQFNLKFD--------VKPASRRLLNSEVTVDDQ 229
+ + + +++ A+DI L+ + Q N+K D PA R L ++ D +
Sbjct: 174 MGEGIAHSKILSSNAIDIFHALTTAMSQMNVKVDDMKKGNLGETPAPDRDLLEDL--DQK 231
Query: 230 GYPSWISSSDRKLLAKMQRKNWRAN------------IMPNAVVAKDGSGQFKTIQAALA 277
G P W S DRKL+A+ R A+ I P VVAKDGSGQFKTI A+
Sbjct: 232 GLPKWHSDKDRKLMAQAGRPGAPADEGIGEGGGGGGKIKPTHVVAKDGSGQFKTISEAVK 291
Query: 278 SYPKGNKGRYVIYVKAGVYDEYITVPKDAVNILMYGDGPARTIVTGRKS--FAAGVKTMQ 335
+ P+ N GR +IY+KAGVY E +T+PK N+ M+GDG +TI+T +S + G T
Sbjct: 292 ACPEKNPGRCIIYIKAGVYKEQVTIPKKVNNVFMFGDGATQTIITFDRSVGLSPGTTTSL 351
Query: 336 TATFANTAMGFIGKAMTFENTAGPDGHQAVAFRNQGDMSALVGCHIVGYQDSLYVQSNRQ 395
+ T + GF+ K + F+NTAGP GHQAVAFR GD + + C GYQD+LYV + RQ
Sbjct: 352 SGTVQVESEGFMAKWIGFQNTAGPLGHQAVAFRVNGDRAVIFNCRFDGYQDTLYVNNGRQ 411
Query: 396 YYRNCLVSGTVDFIFGSSATLIQHSTIIVRKPGKGQFNTITADGSDT-MNLNTGIVIQDC 454
+YRN +VSGTVDFIFG SAT+IQ+S I+ RK GQ N +TADG++ + GIV+ +C
Sbjct: 412 FYRNIVVSGTVDFIFGKSATVIQNSLILCRKGSPGQTNHVTADGNEKGKAVKIGIVLHNC 471
Query: 455 NIIPEAALFPERFTIRSYLGRPWKYLAKTVVMESTIGDFIHPDGWTIWQGEQNHNTCYYA 514
I+ + L +R T++SYLGRPWK A T V+ + IGD I P GW WQGE+ H T Y
Sbjct: 472 RIMADKELEADRLTVKSYLGRPWKPFATTAVIGTEIGDLIQPTGWNEWQGEKFHLTATYV 531
Query: 515 EYANTGPGANVARRVKWKGYHGVISRAEANKFTAGIWLQAGPKSAAEWLNGLHVPHYLG 573
E+ N GPGAN A RV W S AE +FT WL + A W+ +VP LG
Sbjct: 532 EFNNRGPGANTAARVPWAKM--AKSAAEVERFTVANWL-----TPANWIQEANVPVQLG 583
>PME_BRACM (Q42608) Pectinesterase (EC 3.1.1.11) (Pectin
methylesterase) (PE) (Fragment)
Length = 571
Score = 369 bits (946), Expect = e-101
Identities = 228/589 (38%), Positives = 325/589 (54%), Gaps = 47/589 (7%)
Query: 13 ILVVGVAIGVVVAVRKNGE---DPEVQTQQRNLRIMCQNSQDQKLCHETLSSVHGADAAD 69
+LVVGVAIGVV V K G D + + Q+ + +C ++ D+ C +TL V + D
Sbjct: 1 LLVVGVAIGVVTFVNKGGGAGGDKTLNSHQKAVESLCASATDKGSCAKTLDPVK---SDD 57
Query: 70 PKAYIAASVKAATDNVIKAFNMSERLTTEYGKENGA--KMALNDCKDLMQFALDSLDLST 127
P I A + A D V K+ N + GK A K L+ CK ++ +AL+ L+
Sbjct: 58 PSKLIKAFMLATKDAVTKSTNFTASTEEGMGKNMNATSKAVLDYCKRVLMYALEDLETIV 117
Query: 128 KCVHDNNIQAVHDQIADMRNWLSAVISYRQACMEGFDDANDGEKKIKEQFHVQSLDSVQK 187
+ + ++ +Q ++ ++ WL+ V +Y+ C+ DD + E + + + + +
Sbjct: 118 EEMGED-LQQSGSKMDQLKQWLTGVFNYQTDCI---DDIEESELR---KVMGEGIRHSKI 170
Query: 188 VTAVALDIVTGLSDILQQFNLKFD--------VKPASRRLLNSEVTVDDQGYPSWISSSD 239
+++ A+DI L+ + Q N+K D PA R L ++ D +G P W S D
Sbjct: 171 LSSNAIDIFHALTTAMSQMNVKVDDMKKGNLGETPAPDRDLLEDL--DQKGLPKWHSDKD 228
Query: 240 RKLLAKMQRKNWRAN------------IMPNAVVAKDGSGQFKTIQAALASYPKGNKGRY 287
RKL+A+ R A+ I P VVAKDGSGQFKTI A+ + P+ N GR
Sbjct: 229 RKLMAQAGRPGAPADEGIGEGGGGGGKIKPTHVVAKDGSGQFKTISEAVKACPEKNPGRC 288
Query: 288 VIYVKAGVYDEYITVPKDAVNILMYGDGPARTIVTGRKS--FAAGVKTMQTATFANTAMG 345
+IY+KAGVY E +T+PK N+ M+GDG +TI+T +S + G T + T + G
Sbjct: 289 IIYIKAGVYKEQVTIPKKVNNVFMFGDGATQTIITFDRSVGLSPGTTTSLSGTVQVESEG 348
Query: 346 FIGKAMTFENTAGPDGHQAVAFRNQGDMSALVGCHIVGYQDSLYVQSNRQYYRNCLVSGT 405
F+ K + F+NTAGP G+QAVAFR GD + + C GYQD+LYV + RQ+YRN +VSGT
Sbjct: 349 FMAKWIGFQNTAGPLGNQAVAFRVNGDRAVIFNCRFDGYQDTLYVNNGRQFYRNIVVSGT 408
Query: 406 VDFIFGSSATLIQHSTIIVRKPGKGQFNTITADGSDT-MNLNTGIVIQDCNIIPEAALFP 464
VDFI G SAT+IQ+S I+ RK GQ N +TADG + GIV+ +C I+ + L
Sbjct: 409 VDFINGKSATVIQNSLILCRKGSPGQTNHVTADGKQKGKAVKIGIVLHNCRIMADKELEA 468
Query: 465 ERFTIRSYLGRPWKYLAKTVVMESTIGDFIHPDGWTIWQGEQNHNTCYYAEYANTGPGAN 524
+R T++SYLGRPWK A T V+ + IGD I P GW WQGE+ H T Y E+ N GPGAN
Sbjct: 469 DRLTVKSYLGRPWKPFATTAVIGTEIGDLIQPTGWNEWQGEKFHLTATYVEFNNRGPGAN 528
Query: 525 VARRVKWKGYHGVISRAEANKFTAGIWLQAGPKSAAEWLNGLHVPHYLG 573
A RV W S AE +FT WL + A W+ +V LG
Sbjct: 529 PAARVPWAKM--AKSAAEVERFTVANWL-----TPANWIQEANVTVQLG 570
>PME1_CITSI (O04886) Pectinesterase 1 precursor (EC 3.1.1.11)
(Pectin methylesterase) (PE)
Length = 584
Score = 366 bits (939), Expect = e-100
Identities = 213/577 (36%), Positives = 320/577 (54%), Gaps = 29/577 (5%)
Query: 4 KVIISAVSLILVVGVAIGVVVAV--RKNGEDPEVQTQQRNLRIMCQNSQDQKLCHETLSS 61
K+ ++ + +LVV IG+V V RKN D + L+ C +++ LC +++
Sbjct: 29 KLFLALFATLLVVAAVIGIVAGVNSRKNSGDNGNEPHHAILKSSCSSTRYPDLCFSAIAA 88
Query: 62 VHGAD--AADPKAYIAASVKAATDNVIKAFNMSERLTTEYGKENGAKMALNDCKDLMQFA 119
V A K I S+ T V + ++L K+AL+DC + +
Sbjct: 89 VPEASKKVTSQKDVIEMSLNITTTAVEHNYFGIQKLLKRTNLTKREKVALHDCLETIDET 148
Query: 120 LDSLDLSTKCVHDN-NIQAVHDQIADMRNWLSAVISYRQACMEGF--DDANDGEKKIKEQ 176
LD L + + + + N +++ D++ +SA ++ + C++GF DDAN +
Sbjct: 149 LDELHKAVEDLEEYPNKKSLSQHADDLKTLMSAAMTNQGTCLDGFSHDDANKHVRDALSD 208
Query: 177 FHVQSLDSVQKVTAVALDIVTGLSDILQQFNLKFDVKPASRRLLNSEVTVDDQGYPSWIS 236
V V+K+ + AL ++ ++D ++ ++ R L E + D G+P+W+S
Sbjct: 209 GQVH----VEKMCSNALAMIKNMTDTDMMI-----MRTSNNRKLTEETSTVD-GWPAWLS 258
Query: 237 SSDRKLLAKMQRKNWRANIMPNAVVAKDGSGQFKTIQAALASYPKGNKGRYVIYVKAGVY 296
DR+LL +++ PNAVVA DGSG FKT+ AA+A+ P+G RY+I +KAGVY
Sbjct: 259 PGDRRLLQS-------SSVTPNAVVAADGSGNFKTVAAAVAAAPQGGTKRYIIRIKAGVY 311
Query: 297 DEYITVPKDAVNILMYGDGPARTIVTGRKSFAAGVKTMQTATFANTAMGFIGKAMTFENT 356
E + V K NI+ GDG RTI+TG ++ G T ++AT A GF+ + +TF+NT
Sbjct: 312 RENVEVTKKHKNIMFIGDGRTRTIITGSRNVVDGSTTFKSATAAVVGEGFLARDITFQNT 371
Query: 357 AGPDGHQAVAFRNQGDMSALVGCHIVGYQDSLYVQSNRQYYRNCLVSGTVDFIFGSSATL 416
AGP HQAVA R D+SA C ++ YQD+LYV SNRQ++ NCL++GTVDFIFG++A +
Sbjct: 372 AGPSKHQAVALRVGADLSAFYNCDMLAYQDTLYVHSNRQFFVNCLIAGTVDFIFGNAAAV 431
Query: 417 IQHSTIIVRKPGKGQFNTITADGSDTMNLNTGIVIQDCNIIPEAALFPERFTIRSYLGRP 476
+Q+ I RKP GQ N +TA G N NTGIVIQ I + L P + + +YLGRP
Sbjct: 432 LQNCDIHARKPNSGQKNMVTAQGRTDPNQNTGIVIQKSRIGATSDLKPVQGSFPTYLGRP 491
Query: 477 WKYLAKTVVMESTIGDFIHPDGWTIWQGEQNHNTCYYAEYANTGPGANVARRVKWKGYHG 536
WK ++TV+M+S+I D IHP GW W G NT +Y E+ N+G GA + RVKWKG+
Sbjct: 492 WKEYSRTVIMQSSITDLIHPAGWHEWDGNFALNTLFYGEHQNSGAGAGTSGRVKWKGFRV 551
Query: 537 VISRAEANKFTAGIWLQAGPKSAAEWLNGLHVPHYLG 573
+ S EA FT G ++ + + WL P LG
Sbjct: 552 ITSATEAQAFTPGSFI-----AGSSWLGSTGFPFSLG 583
>PME3_CITSI (P83948) Pectinesterase 3 precursor (EC 3.1.1.11)
(Pectin methylesterase 3) (PE 3)
Length = 584
Score = 364 bits (934), Expect = e-100
Identities = 212/577 (36%), Positives = 320/577 (54%), Gaps = 29/577 (5%)
Query: 4 KVIISAVSLILVVGVAIGVVVAV--RKNGEDPEVQTQQRNLRIMCQNSQDQKLCHETLSS 61
K+ ++ + +LVV IG+V V RKN D + L+ C +++ LC +++
Sbjct: 29 KLFLALFATLLVVAAVIGIVAGVNSRKNSGDNGNEPHHAILKSSCSSTRYPDLCFSAIAA 88
Query: 62 VHGAD--AADPKAYIAASVKAATDNVIKAFNMSERLTTEYGKENGAKMALNDCKDLMQFA 119
V A K I S+ T V + ++L K+AL+DC + +
Sbjct: 89 VPEASKKVTSQKDVIEMSLNITTTAVEHNYFGIQKLLKRTNLTKREKVALHDCLETIDET 148
Query: 120 LDSLDLSTKCVHDN-NIQAVHDQIADMRNWLSAVISYRQACMEGF--DDANDGEKKIKEQ 176
LD L + + + + N +++ D++ +SA ++ + C++GF DDAN +
Sbjct: 149 LDELHKAVEDLEEYPNKKSLSQHADDLKTLMSAAMTNQGTCLDGFSHDDANKHVRDALSD 208
Query: 177 FHVQSLDSVQKVTAVALDIVTGLSDILQQFNLKFDVKPASRRLLNSEVTVDDQGYPSWIS 236
V V+K+ + AL ++ ++D ++ +R+L+ TVD G+P+W+S
Sbjct: 209 GQVH----VEKMCSNALAMIKNMTDT----DMMIMRTSNNRKLIEETSTVD--GWPAWLS 258
Query: 237 SSDRKLLAKMQRKNWRANIMPNAVVAKDGSGQFKTIQAALASYPKGNKGRYVIYVKAGVY 296
+ DR+LL +++ PN VVA DGSG FKT+ A++A+ P+G RY+I +KAGVY
Sbjct: 259 TGDRRLLQS-------SSVTPNVVVAADGSGNFKTVAASVAAAPQGGTKRYIIRIKAGVY 311
Query: 297 DEYITVPKDAVNILMYGDGPARTIVTGRKSFAAGVKTMQTATFANTAMGFIGKAMTFENT 356
E + V K NI+ GDG RTI+TG ++ G T ++AT A GF+ + +TF+NT
Sbjct: 312 RENVEVTKKHKNIMFIGDGRTRTIITGSRNVVDGSTTFKSATVAVVGEGFLARDITFQNT 371
Query: 357 AGPDGHQAVAFRNQGDMSALVGCHIVGYQDSLYVQSNRQYYRNCLVSGTVDFIFGSSATL 416
AGP HQAVA R D+SA C ++ YQD+LYV SNRQ++ NCL++GTVDFIFG++A +
Sbjct: 372 AGPSKHQAVALRVGADLSAFYNCDMLAYQDTLYVHSNRQFFVNCLIAGTVDFIFGNAAAV 431
Query: 417 IQHSTIIVRKPGKGQFNTITADGSDTMNLNTGIVIQDCNIIPEAALFPERFTIRSYLGRP 476
+Q+ I RKP GQ N +TA G N NTGIVIQ I + L P + + +YLGRP
Sbjct: 432 LQNCDIHARKPNSGQKNMVTAQGRADPNQNTGIVIQKSRIGATSDLKPVQGSFPTYLGRP 491
Query: 477 WKYLAKTVVMESTIGDFIHPDGWTIWQGEQNHNTCYYAEYANTGPGANVARRVKWKGYHG 536
WK ++TV+M+S+I D IHP GW W G NT +Y E+ N G GA + RVKWKG+
Sbjct: 492 WKEYSRTVIMQSSITDVIHPAGWHEWDGNFALNTLFYGEHQNAGAGAGTSGRVKWKGFRV 551
Query: 537 VISRAEANKFTAGIWLQAGPKSAAEWLNGLHVPHYLG 573
+ S EA FT G ++ + + WL P LG
Sbjct: 552 ITSATEAQAFTPGSFI-----AGSSWLGSTGFPFSLG 583
>PMEU_LYCES (Q43143) Pectinesterase U1 precursor (EC 3.1.1.11)
(Pectin methylesterase) (PE)
Length = 583
Score = 345 bits (884), Expect = 2e-94
Identities = 204/583 (34%), Positives = 317/583 (53%), Gaps = 33/583 (5%)
Query: 4 KVIISAVSLILVVGVAIGVVVAVR---KNGEDPE-----VQTQQRNLRIMCQNSQDQKLC 55
K+ ++ V+ +L+V IGVV V+ KN +D + ++ C N+ +LC
Sbjct: 20 KIYLAIVASVLLVAAVIGVVAGVKSHSKNSDDHADIMAISSSAHAIVKSACSNTLHPELC 79
Query: 56 HETLSSVH--GADAADPKAYIAASVKAATDNVIKAFN-MSERLTTEYGKENGAKMALNDC 112
+ + +V K I S+ V + + + E + T G K+AL+DC
Sbjct: 80 YSAIVNVSDFSKKVTSQKDVIELSLNITVKAVRRNYYAVKELIKTRKGLTPREKVALHDC 139
Query: 113 KDLMQFALDSLDLSTKCVH-DNNIQAVHDQIADMRNWLSAVISYRQACMEGFDDANDGEK 171
+ M LD L + + + N +++ + + D++ +S+ I+ ++ C++GF ++ +K
Sbjct: 140 LETMDETLDELHTAVEDLELYPNKKSLKEHVEDLKTLISSAITNQETCLDGFSH-DEADK 198
Query: 172 KIKEQFHVQSLDSVQKVTAVALDIVTGLSDILQQFNLKFDVKPASRRLLNSEVTVDDQG- 230
K+++ ++ V+K+ + AL ++ ++D +K +R+L V+D G
Sbjct: 199 KVRKVL-LKGQKHVEKMCSNALAMICNMTDTDIANEMKLSAPANNRKL------VEDNGE 251
Query: 231 YPSWISSSDRKLLAKMQRKNWRANIMPNAVVAKDGSGQFKTIQAALASYPKGNKGRYVIY 290
+P W+S+ DR+LL + + P+ VVA DGSG +KT+ A+ P+ + RYVI
Sbjct: 252 WPEWLSAGDRRLLQS-------STVTPDVVVAADGSGDYKTVSEAVRKAPEKSSKRYVIR 304
Query: 291 VKAGVYDEYITVPKDAVNILMYGDGPARTIVTGRKSFAAGVKTMQTATFANTAMGFIGKA 350
+KAGVY E + VPK NI+ GDG + TI+T ++ G T +AT A + +
Sbjct: 305 IKAGVYRENVDVPKKKTNIMFMGDGKSNTIITASRNVQDGSTTFHSATVVRVAGKVLARD 364
Query: 351 MTFENTAGPDGHQAVAFRNQGDMSALVGCHIVGYQDSLYVQSNRQYYRNCLVSGTVDFIF 410
+TF+NTAG HQAVA D+SA C ++ YQD+LYV SNRQ++ CLV+GTVDFIF
Sbjct: 365 ITFQNTAGASKHQAVALCVGSDLSAFYRCDMLAYQDTLYVHSNRQFFVQCLVAGTVDFIF 424
Query: 411 GSSATLIQHSTIIVRKPGKGQFNTITADGSDTMNLNTGIVIQDCNIIPEAALFPERFTIR 470
G+ A + Q I R+PG GQ N +TA G N NTGIVIQ C I + L P + +
Sbjct: 425 GNGAAVFQDCDIHARRPGSGQKNMVTAQGRTDPNQNTGIVIQKCRIGATSDLRPVQKSFP 484
Query: 471 SYLGRPWKYLAKTVVMESTIGDFIHPDGWTIWQGEQNHNTCYYAEYANTGPGANVARRVK 530
+YLGRPWK ++TV+M+S+I D I P GW W G +T +Y EYANTG GA + RVK
Sbjct: 485 TYLGRPWKEYSRTVIMQSSITDVIQPAGWHEWNGNFALDTLFYGEYANTGAGAPTSGRVK 544
Query: 531 WKGYHGVISRAEANKFTAGIWLQAGPKSAAEWLNGLHVPHYLG 573
WKG+ + S EA +T G ++ G WL+ P LG
Sbjct: 545 WKGHKVITSSTEAQAYTPGRFIAGG-----SWLSSTGFPFSLG 582
>PME2_ARATH (Q42534) Pectinesterase 2 precursor (EC 3.1.1.11)
(Pectin methylesterase 2) (PE 2)
Length = 587
Score = 338 bits (866), Expect = 3e-92
Identities = 201/569 (35%), Positives = 307/569 (53%), Gaps = 34/569 (5%)
Query: 5 VIISAVSLILVVGVAIGVVVAVRKNGEDPEVQTQQRN----LRIMCQNSQDQKLCHETLS 60
++ SA +L++ +G+ ++ ++ T L+ +C ++ +LC ++
Sbjct: 21 ILSSAAIALLLLASIVGIAATTTNQNKNQKITTLSSTSHAILKSVCSSTLYPELCFSAVA 80
Query: 61 SVHGADAADPKAYIAASVKAATDNVI-KAFNMSERLTTEYGKENGAKMALNDCKDLMQFA 119
+ G + K I AS+ T V F + + + G AL+DC + +
Sbjct: 81 ATGGKELTSQKEVIEASLNLTTKAVKHNYFAVKKLIAKRKGLTPREVTALHDCLETIDET 140
Query: 120 LDSLDLSTKCVHDNNIQ-AVHDQIADMRNWLSAVISYRQACMEGF--DDANDGEKK--IK 174
LD L ++ + +H Q ++ D++ +S+ I+ + C++GF DDA+ +K +K
Sbjct: 141 LDELHVAVEDLHQYPKQKSLRKHADDLKTLISSAITNQGTCLDGFSYDDADRKVRKALLK 200
Query: 175 EQFHVQSLDSVQKVTAVALDIVTGLSDILQQFNLKFDVKPASRRLLNS------EVT--V 226
Q HV+ + S A+A+ + ++ + F+++ S N+ EVT +
Sbjct: 201 GQVHVEHMCS----NALAM-----IKNMTETDIANFELRDKSSTFTNNNNRKLKEVTGDL 251
Query: 227 DDQGYPSWISSSDRKLLAKMQRKNWRANIMPNAVVAKDGSGQFKTIQAALASYPKGNKGR 286
D G+P W+S DR+LL + I +A VA DGSG F T+ AA+A+ P+ + R
Sbjct: 252 DSDGWPKWLSVGDRRLLQG-------STIKADATVADDGSGDFTTVAAAVAAAPEKSNKR 304
Query: 287 YVIYVKAGVYDEYITVPKDAVNILMYGDGPARTIVTGRKSFAAGVKTMQTATFANTAMGF 346
+VI++KAGVY E + V K NI+ GDG +TI+TG ++ G T +AT A F
Sbjct: 305 FVIHIKAGVYRENVEVTKKKTNIMFLGDGRGKTIITGSRNVVDGSTTFHSATVAAVGERF 364
Query: 347 IGKAMTFENTAGPDGHQAVAFRNQGDMSALVGCHIVGYQDSLYVQSNRQYYRNCLVSGTV 406
+ + +TF+NTAGP HQAVA R D SA C + YQD+LYV SNRQ++ C ++GTV
Sbjct: 365 LARDITFQNTAGPSKHQAVALRVGSDFSAFYQCDMFAYQDTLYVHSNRQFFVKCHITGTV 424
Query: 407 DFIFGSSATLIQHSTIIVRKPGKGQFNTITADGSDTMNLNTGIVIQDCNIIPEAALFPER 466
DFIFG++A ++Q I R+P GQ N +TA G N NTGIVIQ+C I + L +
Sbjct: 425 DFIFGNAAAVLQDCDINARRPNSGQKNMVTAQGRSDPNQNTGIVIQNCRIGGTSDLLAVK 484
Query: 467 FTIRSYLGRPWKYLAKTVVMESTIGDFIHPDGWTIWQGEQNHNTCYYAEYANTGPGANVA 526
T +YLGRPWK ++TV+M+S I D I P+GW W G +T Y EY N G GA A
Sbjct: 485 GTFPTYLGRPWKEYSRTVIMQSDISDVIRPEGWHEWSGSFALDTLTYREYLNRGGGAGTA 544
Query: 527 RRVKWKGYHGVISRAEANKFTAGIWLQAG 555
RVKWKGY + S EA FTAG ++ G
Sbjct: 545 NRVKWKGYKVITSDTEAQPFTAGQFIGGG 573
>PME3_PHAVU (Q43111) Pectinesterase 3 precursor (EC 3.1.1.11)
(Pectin methylesterase 3) (PE 3)
Length = 581
Score = 328 bits (842), Expect = 2e-89
Identities = 203/569 (35%), Positives = 308/569 (53%), Gaps = 40/569 (7%)
Query: 5 VIISAVSLILVVGVAIGVVVAVRKNGE-----DPEVQTQ---QRNLRIMCQNSQDQKLCH 56
+ +S++ LI V+ A+ VV +N E D QT+ +L+ +C ++ C
Sbjct: 32 IAVSSIVLIAVIIAAVAGVVIHNRNSESSPSSDSVPQTELSPAASLKAVCDTTRYPSSCF 91
Query: 57 ETLSSVHGADAADPKAYIAASVKAATDNVIKAFNMSERLTTEYGKENGAKMALNDCKDLM 116
++SS+ ++ DP+ S++ A D + +F R E ++ + A++ C +
Sbjct: 92 SSISSLPESNTTDPELLFKLSLRVAIDE-LSSFPSKLRANAE--QDARLQKAIDVCSSVF 148
Query: 117 QFALDSLDLSTKCVHDNNIQ-AVHDQIADMRNWLSAVISYRQACMEGFDDANDGEKKIKE 175
ALD L+ S + + A ++++ WLSA ++ + C++ + N +
Sbjct: 149 GDALDRLNDSISALGTVAGRIASSASVSNVETWLSAALTDQDTCLDAVGELNSTAARGAL 208
Query: 176 QFHVQSLDSVQKVTAVALDIVTGLSDILQQFNLKFDVKPASRRLLNSEVTVDDQGYPSWI 235
Q ++ + + + +L IVT + +L +F RRLL G+P W+
Sbjct: 209 QEIETAMRNSTEFASNSLAIVTKILGLLSRFETPIH----HRRLL---------GFPEWL 255
Query: 236 SSSDRKLLAKMQRKNWRANIMPNAVVAKDGSGQFKTIQAALASYPKGNKGRYVIYVKAGV 295
+++R+LL + + P+AVVAKDGSGQFKTI AL K ++ R+ +YVK G
Sbjct: 256 GAAERRLLEEKNNDS-----TPDAVVAKDGSGQFKTIGEALKLVKKKSEERFSVYVKEGR 310
Query: 296 YDEYITVPKDAVNILMYGDGPARTIVTGRKSFAAGVKTMQTATFANTAMGFIGKAMTFEN 355
Y E I + K+ N+++YGDG +T V G ++F G T +TATFA GFI K + F N
Sbjct: 311 YVENIDLDKNTWNVMIYGDGKDKTFVVGSRNFMDGTPTFETATFAVKGKGFIAKDIGFVN 370
Query: 356 TAGPDGHQAVAFRNQGDMSALVGCHIVGYQDSLYVQSNRQYYRNCLVSGTVDFIFGSSAT 415
AG HQAVA R+ D S C G+QD+LY SNRQ+YR+C ++GT+DFIFG++A
Sbjct: 371 NAGASKHQAVALRSGSDRSVFFRCSFDGFQDTLYAHSNRQFYRDCDITGTIDFIFGNAAV 430
Query: 416 LIQHSTIIVRKPGKGQFNTITADGSDTMNLNTGIVIQDCNIIPEAALFPERFTIRSYLGR 475
+ Q I+ R+P QFNTITA G N NTGI+IQ I P F T +YLGR
Sbjct: 431 VFQSCKIMPRQPLPNQFNTITAQGKKDPNQNTGIIIQKSTITP----FGNNLTAPTYLGR 486
Query: 476 PWKYLAKTVVMESTIGDFIHPDGWTIW-QGEQNHNTCYYAEYANTGPGANVARRVKWKGY 534
PWK + TV+M+S IG ++P GW W + T +YAEY N+GPGA+V++RVKW GY
Sbjct: 487 PWKDFSTTVIMQSDIGALLNPVGWMSWVPNVEPPTTIFYAEYQNSGPGADVSQRVKWAGY 546
Query: 535 HGVISRAEANKFTAGIWLQAGPKSAAEWL 563
I+ A +FT ++Q GP EWL
Sbjct: 547 KPTITDRNAEEFTVQSFIQ-GP----EWL 570
>PME1_ARATH (Q43867) Pectinesterase 1 precursor (EC 3.1.1.11)
(Pectin methylesterase 1) (PE 1)
Length = 586
Score = 327 bits (839), Expect = 4e-89
Identities = 199/574 (34%), Positives = 301/574 (51%), Gaps = 39/574 (6%)
Query: 4 KVIISAVSLILVVGVAIGVVVA--VRKNGED-----PEVQTQQRNLRIMCQNSQDQKLCH 56
++++ ++S+++++ V I VVA V KN + P T +L+ +C ++ + C
Sbjct: 28 RLLLLSISVVVLIAVIIAAVVATVVHKNKNESTPSPPPELTPSTSLKAICSVTRFPESCI 87
Query: 57 ETLSSVHGADAADPKAYIAASVKAATDNVIKAFNMSERLTTEYGKENGAKMALNDCKDLM 116
++S + ++ DP+ S+K D + ++ E+L+ E E K AL C DL+
Sbjct: 88 SSISKLPSSNTTDPETLFKLSLKVIIDELDSISDLPEKLSKETEDER-IKSALRVCGDLI 146
Query: 117 QFALDSLDLSTKCVHDNNIQAV--HDQIADMRNWLSAVISYRQACMEGFDDANDGEKKIK 174
+ ALD L+ + + D + +I D++ WLSA ++ + C + D+ + +
Sbjct: 147 EDALDRLNDTVSAIDDEEKKKTLSSSKIEDLKTWLSATVTDHETCFDSLDELKQNKTEYA 206
Query: 175 EQFHVQSLDSVQ----KVTAVALDIVTGLSDILQQFNLKFDVKPASRRLLNSEVTVDDQG 230
Q+L S + T+ +L IV+ + L + RR L S
Sbjct: 207 NSTITQNLKSAMSRSTEFTSNSLAIVSKILSALSDLGIPIH----RRRRLMSHHHQQSVD 262
Query: 231 YPSWISSSDRKLLAKMQRKNWRANIMPNAVVAKDGSGQFKTIQAALASYPKGNKGRYVIY 290
+ W + R+LL A + P+ VA DG+G T+ A+A PK + +VIY
Sbjct: 263 FEKW---ARRRLLQT-------AGLKPDVTVAGDGTGDVLTVNEAVAKVPKKSLKMFVIY 312
Query: 291 VKAGVYDEYITVPKDAVNILMYGDGPARTIVTGRKSFAAGVKTMQTATFANTAMGFIGKA 350
VK+G Y E + + K N+++YGDG +TI++G K+F G T +TATFA GFI K
Sbjct: 313 VKSGTYVENVVMDKSKWNVMIYGDGKGKTIISGSKNFVDGTPTYETATFAIQGKGFIMKD 372
Query: 351 MTFENTAGPDGHQAVAFRNQGDMSALVGCHIVGYQDSLYVQSNRQYYRNCLVSGTVDFIF 410
+ NTAG HQAVAFR+ D S C G+QD+LY SNRQ+YR+C V+GT+DFIF
Sbjct: 373 IGIINTAGAAKHQAVAFRSGSDFSVYYQCSFDGFQDTLYPHSNRQFYRDCDVTGTIDFIF 432
Query: 411 GSSATLIQHSTIIVRKPGKGQFNTITADGSDTMNLNTGIVIQDCNIIPEAALFPERFTIR 470
GS+A + Q I+ R+P QFNTITA G N ++G+ IQ C I +
Sbjct: 433 GSAAVVFQGCKIMPRQPLSNQFNTITAQGKKDPNQSSGMSIQRCTISANGNVIAP----- 487
Query: 471 SYLGRPWKYLAKTVVMESTIGDFIHPDGWTIW-QGEQNHNTCYYAEYANTGPGANVARRV 529
+YLGRPWK + TV+ME+ IG + P GW W G + Y EY NTGPG++V +RV
Sbjct: 488 TYLGRPWKEFSTTVIMETVIGAVVRPSGWMSWVSGVDPPASIVYGEYKNTGPGSDVTQRV 547
Query: 530 KWKGYHGVISRAEANKFTAGIWLQAGPKSAAEWL 563
KW GY V+S AEA KFT L A+W+
Sbjct: 548 KWAGYKPVMSDAEAAKFTVATLLH-----GADWI 576
>PME1_FICAW (P83947) Pectinesterase precursor (EC 3.1.1.11) (Pectin
methylesterase) (PE)
Length = 545
Score = 313 bits (801), Expect = 1e-84
Identities = 191/576 (33%), Positives = 292/576 (50%), Gaps = 42/576 (7%)
Query: 2 SGKVIISAVSLILVVGVAIGVVVAVRKNGEDPEVQTQQRNLRIMCQNSQDQKLCHETLSS 61
+ K S ++ L+V I V PE+ + +C S +++ C +S
Sbjct: 11 ASKSCYSKITFFLLV---ISFAALVSTGFSSPELSLHHK----ICDQSVNKESCLAMISE 63
Query: 62 VHGADAADPKAYIAASVKAATDNVIKAFNMSERLTTEYGKENGAKMALNDCKDLMQFALD 121
V G + AD + + + ++ T + KAF + + + AL DC +LM + +
Sbjct: 64 VTGLNMADHRNLLKSFLEKTTPRIQKAFETANDASRRINNPQ-ERTALLDCAELMDLSKE 122
Query: 122 SL-DLSTKCVHDNNIQAVHDQIADMRNWLSAVISYRQACMEGFDDANDGEKKIKEQFHVQ 180
+ D + H N H+ D+ WLS V++ C++G ++ + K + H+
Sbjct: 123 RVVDSISILFHQNLTTRSHE---DLHVWLSGVLTNHVTCLDGLEEGSTDYIKTLMESHLN 179
Query: 181 SLDSVQKVTAVALDIVTGLSDILQQFNLKFDVKPASRRLLNSEVTVDDQGYPSWISSSDR 240
L L T L+ + F K +V + +P+W+++ DR
Sbjct: 180 EL---------ILRARTSLAIFVTLFPAKSNV-----------IEPVTGNFPTWVTAGDR 219
Query: 241 KLLAKMQRKNWRANIMPNAVVAKDGSGQFKTIQAALASYPKGNKGRYVIYVKAGVYDEYI 300
+LL + + +I P+ VVAKDGSG ++T+ A+A+ P +K R ++ V+ G+Y+E +
Sbjct: 220 RLLQTLGK-----DIEPDIVVAKDGSGDYETLNEAVAAIPDNSKKRVIVLVRTGIYEENV 274
Query: 301 TVPKDAVNILMYGDGPARTIVTGRKSFAAGVKTMQTATFANTAMGFIGKAMTFENTAGPD 360
N+++ G+G TI+TG ++ G T +AT A GFI + + F+NTAGP+
Sbjct: 275 DFGYQKKNVMLVGEGMDYTIITGSRNVVDGSTTFDSATVAAVGDGFIAQDICFQNTAGPE 334
Query: 361 GHQAVAFRNQGDMSALVGCHIVGYQDSLYVQSNRQYYRNCLVSGTVDFIFGSSATLIQHS 420
+QAVA R D + + C I YQD+LY + RQ+YR+ ++GTVDFIFG++A + Q+
Sbjct: 335 KYQAVALRIGADETVINRCRIDAYQDTLYPHNYRQFYRDRNITGTVDFIFGNAAVVFQNC 394
Query: 421 TIIVRKPGKGQFNTITADGSDTMNLNTGIVIQDCNIIPEAALFPERFTIRSYLGRPWKYL 480
+I RK KGQ NTITA G N NTG IQ+C I A L P T +SYLGRPWK
Sbjct: 395 NLIPRKQMKGQENTITAQGRTDPNQNTGTSIQNCEIFASADLEPVEDTFKSYLGRPWKEY 454
Query: 481 AKTVVMESTIGDFIHPDGWTIWQGEQNHNTCYYAEYANTGPGANVARRVKWKGYHGVISR 540
++TVVMES I D I P GW W + T +Y EY N GPG+ + RVKW GYH + S
Sbjct: 455 SRTVVMESYISDVIDPAGWLEWDRDFALKTLFYGEYRNGGPGSGTSERVKWPGYHVITSP 514
Query: 541 AEANKFTAGIWLQAGPKSAAEWLNGLHVPHYLGFKA 576
A +FT +Q G WL V + G A
Sbjct: 515 EVAEQFTVAELIQGG-----SWLGSTGVDYTAGLYA 545
>PM22_LYCES (Q96575) Pectinesterase 2 precursor (EC 3.1.1.11)
(Pectin methylesterase 2) (PE 2)
Length = 550
Score = 307 bits (787), Expect = 4e-83
Identities = 187/532 (35%), Positives = 277/532 (51%), Gaps = 40/532 (7%)
Query: 45 MCQNSQDQKLCHETLSSVHGADAADPKAYIAASVKAATDNVIKAFNMSERLTTEYGKE-N 103
+C+ +QD +LC +S + + + + + N + N + + ++ + N
Sbjct: 53 LCKTAQDSQLCLSYVSDLMSNEIVTTDSDGLSILMKFLVNYVHQMNNAIPVVSKMKNQIN 112
Query: 104 GAKM--ALNDCKDLMQFALDSLDLSTKCVHDNNIQAVHDQIADMRNWLSAVISYRQACME 161
+ AL DC +L+ ++D + S + + H + A+ ++WLS V++ C++
Sbjct: 113 DIRQEGALTDCLELLDQSVDLVSDSIAAID----KRTHSEHANAQSWLSGVLTNHVTCLD 168
Query: 162 GFDDANDGEKKIKEQFHVQSLDSVQKVTAVALDIVTGLSDILQQFNLKFDVKPASRRLLN 221
D K + ++ L S KV L VT +D + ++P ++
Sbjct: 169 ELDSFT---KAMINGTNLDELISRAKVALAMLASVTTPNDDV--------LRPGLGKM-- 215
Query: 222 SEVTVDDQGYPSWISSSDRKLLAKMQRKNWRANIMPNAVVAKDGSGQFKTIQAALASYPK 281
PSW+SS DRKL+ + +I NAVVAKDG+G+++T+ A+A+ P
Sbjct: 216 ----------PSWVSSRDRKLMESSGK-----DIGANAVVAKDGTGKYRTLAEAVAAAPD 260
Query: 282 GNKGRYVIYVKAGVYDEYITVPKDAVNILMYGDGPARTIVTGRKSFAAGVKTMQTATFAN 341
+K RYVIYVK G+Y E + V + +++ GDG TI+TG + G T +AT A
Sbjct: 261 KSKTRYVIYVKRGIYKENVEVSSRKMKLMIVGDGMHATIITGNLNVVDGSTTFHSATLAA 320
Query: 342 TAMGFIGKAMTFENTAGPDGHQAVAFRNQGDMSALVGCHIVGYQDSLYVQSNRQYYRNCL 401
GFI + + +NTAGP HQAVA R D S + C I YQD+LY S RQ+YR+
Sbjct: 321 VGKGFILQDICIQNTAGPAKHQAVALRVGADKSVINRCRIDAYQDTLYAHSQRQFYRDSY 380
Query: 402 VSGTVDFIFGSSATLIQHSTIIVRKPGKGQFNTITADGSDTMNLNTGIVIQDCNIIPEAA 461
V+GT+DFIFG++A + Q ++ RKPGK Q N +TA G N TG IQ CNII +
Sbjct: 381 VTGTIDFIFGNAAVVFQKCKLVARKPGKYQQNMVTAQGRTDPNQATGTSIQFCNIIASSD 440
Query: 462 LFPERFTIRSYLGRPWKYLAKTVVMESTIGDFIHPDGWTIWQGEQNHNTCYYAEYANTGP 521
L P +YLGRPWK ++TVVMES +G I+P GW W G+ T YY E+ N GP
Sbjct: 441 LEPVLKEFPTYLGRPWKKYSRTVVMESYLGGLINPAGWAEWDGDFALKTLYYGEFMNNGP 500
Query: 522 GANVARRVKWKGYHGVISRAEANKFTAGIWLQAGPKSAAEWLNGLHVPHYLG 573
GA ++RVKW GYH + AEA FT +Q G WL V + G
Sbjct: 501 GAGTSKRVKWPGYHCITDPAEAMPFTVAKLIQGG-----SWLRSTGVAYVDG 547
>PM21_LYCES (P09607) Pectinesterase 2 precursor (EC 3.1.1.11)
(Pectin methylesterase 2) (PE 2)
Length = 550
Score = 304 bits (779), Expect = 4e-82
Identities = 191/536 (35%), Positives = 276/536 (50%), Gaps = 48/536 (8%)
Query: 45 MCQNSQDQKLCHETLSSVHGADA----ADPKAYIAASVKAATDNVIKAFNMSERLTTEYG 100
+C+ +QD +LC +S + + +D + + + + + A + ++ +
Sbjct: 53 LCKTAQDSQLCLSYVSDLISNEIVTSDSDGLSILKKFLVYSVHQMNNAIPVVRKIKNQIN 112
Query: 101 --KENGAKMALNDCKDLMQFALDSLDLSTKCVHDNNIQAVHDQIADMRNWLSAVISYRQA 158
+E GA L DC +L+ ++D + S + D ++ H A+ ++WLS V++
Sbjct: 113 DIREQGA---LTDCLELLDLSVDLVCDSIAAI-DKRSRSEH---ANAQSWLSGVLTNHVT 165
Query: 159 CMEGFDDANDGEKKIKEQFHVQSLDSVQKVTAVALDIVTGLSDILQQFNLKFDVKPASRR 218
C++ D K + +LD + VAL ++ AS
Sbjct: 166 CLDELDSFT------KAMINGTNLDELISRAKVALAML------------------ASVT 201
Query: 219 LLNSEVTVDDQG-YPSWISSSDRKLLAKMQRKNWRANIMPNAVVAKDGSGQFKTIQAALA 277
N EV G PSW+SS DRKL+ + +I NAVVAKDG+G+++T+ A+A
Sbjct: 202 TPNDEVLRPGLGKMPSWVSSRDRKLMESSGK-----DIGANAVVAKDGTGKYRTLAEAVA 256
Query: 278 SYPKGNKGRYVIYVKAGVYDEYITVPKDAVNILMYGDGPARTIVTGRKSFAAGVKTMQTA 337
+ P +K RYVIYVK G Y E + V +N+++ GDG TI+TG + G T +A
Sbjct: 257 AAPDKSKTRYVIYVKRGTYKENVEVSSRKMNLMIIGDGMYATIITGSLNVVDGSTTFHSA 316
Query: 338 TFANTAMGFIGKAMTFENTAGPDGHQAVAFRNQGDMSALVGCHIVGYQDSLYVQSNRQYY 397
T A GFI + + +NTAGP HQAVA R D S + C I YQD+LY S RQ+Y
Sbjct: 317 TLAAVGKGFILQDICIQNTAGPAKHQAVALRVGADKSVINRCRIDAYQDTLYAHSQRQFY 376
Query: 398 RNCLVSGTVDFIFGSSATLIQHSTIIVRKPGKGQFNTITADGSDTMNLNTGIVIQDCNII 457
R+ V+GT+DFIFG++A + Q ++ RKPGK Q N +TA G N TG IQ C+II
Sbjct: 377 RDSYVTGTIDFIFGNAAVVFQKCQLVARKPGKYQQNMVTAQGRTDPNQATGTSIQFCDII 436
Query: 458 PEAALFPERFTIRSYLGRPWKYLAKTVVMESTIGDFIHPDGWTIWQGEQNHNTCYYAEYA 517
L P +YLGRPWK ++TVVMES +G I P GW W G+ T YY E+
Sbjct: 437 ASPDLKPVVKEFPTYLGRPWKKYSRTVVMESYLGGLIDPSGWAEWHGDFALKTLYYGEFM 496
Query: 518 NTGPGANVARRVKWKGYHGVISRAEANKFTAGIWLQAGPKSAAEWLNGLHVPHYLG 573
N GPGA ++RVKW GYH + AEA FT +Q G WL V + G
Sbjct: 497 NNGPGAGTSKRVKWPGYHVITDPAEAMSFTVAKLIQGG-----SWLRSTDVAYVDG 547
>PME_MEDSA (Q42920) Pectinesterase precursor (EC 3.1.1.11) (Pectin
methylesterase) (PE) (P65)
Length = 447
Score = 304 bits (778), Expect = 5e-82
Identities = 172/462 (37%), Positives = 253/462 (54%), Gaps = 20/462 (4%)
Query: 112 CKDLMQFALDSLDLSTKCVHDNNIQAVHDQIADMRNWLSAVISYRQACMEGFDDANDGEK 171
C +++ +A+D + S + + + + D++ WL+ +S++Q C++GF AN K
Sbjct: 4 CNEVLDYAVDGIHKSVGTLDQFDFHKLSEYAFDLKVWLTGTLSHQQTCLDGF--ANTTTK 61
Query: 172 KIKEQFHVQSLDSVQKVTAVALDIVTGLSDILQQFNLKFDVKPASRRLLNSEVTVDDQGY 231
+ V L + ++++ A+D++ +S IL+ F+ SRRLL+ D G
Sbjct: 62 AGETMTKV--LKTSMELSSNAIDMMDAVSRILKGFDTS--QYSVSRRLLS------DDGI 111
Query: 232 PSWISSSDRKLLAKMQRKNWRANIMPNAVVAKDGSGQFKTIQAALASYPKGNKGRYVIYV 291
PSW++ R+LLA N+ PNAVVA+DGSGQFKT+ AL + P N +VI+V
Sbjct: 112 PSWVNDGHRRLLAG-------GNVQPNAVVAQDGSGQFKTLTDALKTVPPKNAVPFVIHV 164
Query: 292 KAGVYDEYITVPKDAVNILMYGDGPARTIVTGRKSFAAGVKTMQTATFANTAMGFIGKAM 351
KAGVY E + V K+ + + GDGP +T TG ++A G+ T TATF F+ K +
Sbjct: 165 KAGVYKETVNVAKEMNYVTVIGDGPTKTKFTGSLNYADGINTYNTATFGVNGANFMAKDI 224
Query: 352 TFENTAGPDGHQAVAFRNQGDMSALVGCHIVGYQDSLYVQSNRQYYRNCLVSGTVDFIFG 411
FENTAG HQAVA R D + C + G+QD+LYVQS RQ+YR+C +SGT+DF+FG
Sbjct: 225 GFENTAGTGKHQAVALRVTADQAIFYNCQMDGFQDTLYVQSQRQFYRDCSISGTIDFVFG 284
Query: 412 SSATLIQHSTIIVRKPGKGQFNTITADGSDTMNLNTGIVIQDCNIIPEAALFPERFTIRS 471
+ Q+ ++ R P KGQ +TA G + N + +V Q + E AL + S
Sbjct: 285 ERFGVFQNCKLVCRLPAKGQQCLVTAGGREKQNSASALVFQSSHFTGEPALTSVTPKV-S 343
Query: 472 YLGRPWKYLAKTVVMESTIGDFIHPDGWTIWQGEQNHNTCYYAEYANTGPGANVARRVKW 531
YLGRPWK +K V+M+STI P+G+ W G TC Y EY N GPGA+ RVKW
Sbjct: 344 YLGRPWKQYSKVVIMDSTIDAIFVPEGYMPWMGSAFKETCTYYEYNNKGPGADTNLRVKW 403
Query: 532 KGYHGVISRAEANKFTAGIWLQAGPKSAAEWLNGLHVPHYLG 573
G + S A + + + W+ VP+ LG
Sbjct: 404 HGVKVLTSNVAAEYYPGKFFEIVNATARDTWIVKSGVPYSLG 445
>PME_PRUPE (Q43062) Pectinesterase PPE8B precursor (EC 3.1.1.11)
(Pectin methylesterase) (PE)
Length = 522
Score = 301 bits (772), Expect = 2e-81
Identities = 184/501 (36%), Positives = 264/501 (51%), Gaps = 50/501 (9%)
Query: 75 AASVKAATDNVIKAFNMSERLTTEYGKENGAKMALNDCKDLMQFALDSLD--LSTKCVHD 132
A S+K D V + ++ + +G A A++DC DL+ F+ D L+ LS
Sbjct: 57 AGSLKDTIDAVQQVASILSQFANAFGDFRLAN-AISDCLDLLDFSADELNWSLSASQNQK 115
Query: 133 NNIQAVHDQIADMRNWLSAVISYRQACMEGFDDANDGEKKIKEQFHVQSLDSVQKVTAVA 192
+ +D+R WLSA + + C GF+ N VQ + +
Sbjct: 116 GKNNSTGKLSSDLRTWLSAALVNQDTCSNGFEGTNS---------------IVQGLISAG 160
Query: 193 LDIVTGL-SDILQQFNLKFDVKPASRRLLNSEVTVDDQGYPSWISSSDRKLLAKMQRKNW 251
L VT L ++L Q V P NS + PSW+ + DRKLL
Sbjct: 161 LGQVTSLVQELLTQ------VHP------NSNQQGPNGQIPSWVKTKDRKLLQA------ 202
Query: 252 RANIMPNAVVAKDGSGQFKTIQAALASYPKGNKGRYVIYVKAGVYDEYITVPKDAVNILM 311
+ +A+VA+DG+G F + A+ + P + RYVIY+K G Y E + + K N++M
Sbjct: 203 -DGVSVDAIVAQDGTGNFTNVTDAVLAAPDYSMRRYVIYIKRGTYKENVEIKKKKWNLMM 261
Query: 312 YGDGPARTIVTGRKSFAAGVKTMQTATFANTAMGFIGKAMTFENTAGPDGHQAVAFRNQG 371
GDG TI++G +SF G T ++ATFA + GFI + +TFENTAGP+ HQAVA R+
Sbjct: 262 IGDGMDATIISGNRSFVDGWTTFRSATFAVSGRGFIARDITFENTAGPEKHQAVALRSDS 321
Query: 372 DMSALVGCHIVGYQDSLYVQSNRQYYRNCLVSGTVDFIFGSSATLIQHSTIIVRKPGKGQ 431
D+S C+I GYQD+LY + RQ+YR+C +SGTVDFIFG + + Q+ I+ +K Q
Sbjct: 322 DLSVFYRCNIRGYQDTLYTHTMRQFYRDCKISGTVDFIFGDATVVFQNCQILAKKGLPNQ 381
Query: 432 FNTITADGSDTMNLNTGIVIQDCNIIPEAALFPERFTIRS---YLGRPWKYLAKTVVMES 488
N+ITA G N TGI IQ CNI ++ L E ++ S YLGRPWK ++TV+M+S
Sbjct: 382 KNSITAQGRKDPNEPTGISIQFCNITADSDL--EAASVNSTPTYLGRPWKLYSRTVIMQS 439
Query: 489 TIGDFIHPDGWTIWQGEQNHNTCYYAEYANTGPGANVARRVKWKGYHGVISRAEANKFT- 547
+ + I P+GW W G+ N+ +Y EY N GPGA + RVKW GY +A +T
Sbjct: 440 FLSNVIRPEGWLEWNGDFALNSLFYGEYMNYGPGAGLGSRVKWPGYQVFNESTQAKNYTV 499
Query: 548 -----AGIWL-QAGPKSAAEW 562
+WL G K AE+
Sbjct: 500 AQFIEGNLWLPSTGVKYTAEF 520
>PME1_LYCES (P14280) Pectinesterase 1 precursor (EC 3.1.1.11)
(Pectin methylesterase 1) (PE 1)
Length = 546
Score = 299 bits (765), Expect = 2e-80
Identities = 188/544 (34%), Positives = 282/544 (51%), Gaps = 45/544 (8%)
Query: 34 EVQTQQRNLRIMCQNSQDQKLCHETLSSVHGADAADPKAYIAASVKAATDNVIKAFNMSE 93
+V+ + NL C+ +QD +LC +S + + ++ + + N + N +
Sbjct: 41 QVEIKHSNL---CKTAQDSQLCLSYVSDLISNEIVTTESDGHSILMKFLVNYVHQMNNAI 97
Query: 94 RLTTEYGKE-NGAKM--ALNDCKDLMQFALDSLDLSTKCVHDNNIQAVHDQIADMRNWLS 150
+ + + N + AL DC +L+ ++D S + D ++ H A+ ++WLS
Sbjct: 98 PVVRKMKNQINDIRQHGALTDCLELLDQSVDFASDSIAAI-DKRSRSEH---ANAQSWLS 153
Query: 151 AVISYRQACMEGFDDANDGEKKIKEQFHVQSLDSVQKVTAVALDIVTGLSDILQQFNLKF 210
V++ C++ D K + +L+ + VAL ++ L+
Sbjct: 154 GVLTNHVTCLDELDSFT------KAMINGTNLEELISRAKVALAMLASLTT--------- 198
Query: 211 DVKPASRRLLNSEVTVDDQG-YPSWISSSDRKLLAKMQRKNWRANIMPNAVVAKDGSGQF 269
+ +V + G PSW+SS DRKL+ + +I+ NAVVA+DG+G +
Sbjct: 199 ---------QDEDVFMTVLGKMPSWVSSMDRKLMESSGK-----DIIANAVVAQDGTGDY 244
Query: 270 KTIQAALASYPKGNKGRYVIYVKAGVYDEYITVPKDAVNILMYGDGPARTIVTGRKSFAA 329
+T+ A+A+ P +K RYVIYVK G Y E + V + +N+++ GDG T +TG +
Sbjct: 245 QTLAEAVAAAPDKSKTRYVIYVKRGTYKENVEVASNKMNLMIVGDGMYATTITGSLNVVD 304
Query: 330 GVKTMQTATFANTAMGFIGKAMTFENTAGPDGHQAVAFRNQGDMSALVGCHIVGYQDSLY 389
G T ++AT A GFI + + +NTAGP QAVA R DMS + C I YQD+LY
Sbjct: 305 GSTTFRSATLAAVGQGFILQDICIQNTAGPAKDQAVALRVGADMSVINRCRIDAYQDTLY 364
Query: 390 VQSNRQYYRNCLVSGTVDFIFGSSATLIQHSTIIVRKPGKGQFNTITADGSDTMNLNTGI 449
S RQ+YR+ V+GTVDFIFG++A + Q ++ RKPGK Q N +TA G N TG
Sbjct: 365 AHSQRQFYRDSYVTGTVDFIFGNAAVVFQKCQLVARKPGKYQQNMVTAQGRTDPNQATGT 424
Query: 450 VIQDCNIIPEAALFPERFTIRSYLGRPWKYLAKTVVMESTIGDFIHPDGWTIWQGEQNHN 509
IQ CNII + L P +YLGRPWK ++TVVMES +G I+P GW W G+
Sbjct: 425 SIQFCNIIASSDLEPVLKEFPTYLGRPWKEYSRTVVMESYLGGLINPAGWAEWDGDFALK 484
Query: 510 TCYYAEYANTGPGANVARRVKWKGYHGVISRAEANKFTAGIWLQAGPKSAAEWLNGLHVP 569
T YY E+ N GPGA ++RVKW GYH + A+A FT +Q G WL V
Sbjct: 485 TLYYGEFMNNGPGAGTSKRVKWPGYHVITDPAKAMPFTVAKLIQGG-----SWLRSTGVA 539
Query: 570 HYLG 573
+ G
Sbjct: 540 YVDG 543
>PME2_CITSI (O04887) Pectinesterase 2 precursor (EC 3.1.1.11)
(Pectin methylesterase) (PE)
Length = 510
Score = 296 bits (758), Expect = 1e-79
Identities = 151/344 (43%), Positives = 211/344 (60%), Gaps = 16/344 (4%)
Query: 230 GYPSWISSSDRKLLAKMQRKNWRANIMPNAVVAKDGSGQFKTIQAALASYPKGNKGRYVI 289
G+P+W+ DRKLL R N VVA+DGSG KTIQ A+A+ + RYVI
Sbjct: 182 GFPTWVKPGDRKLLQTTPRAN--------IVVAQDGSGNVKTIQEAVAAASRAGGSRYVI 233
Query: 290 YVKAGVYDEYITVPKDAVNILMYGDGPARTIVTGRKSFAAGVKTMQTATFANTAMGFIGK 349
Y+KAG Y+E I V NI+ GDG +TI+TG KS G T ++AT A FI +
Sbjct: 234 YIKAGTYNENIEVKLK--NIMFVGDGIGKTIITGSKSVGGGATTFKSATVAVVGDNFIAR 291
Query: 350 AMTFENTAGPDGHQAVAFRNQGDMSALVGCHIVGYQDSLYVQSNRQYYRNCLVSGTVDFI 409
+T NTAGP+ HQAVA R+ D+S C GYQD+LYV S RQ+YR C + GTVDFI
Sbjct: 292 DITIRNTAGPNNHQAVALRSGSDLSVFYRCSFEGYQDTLYVHSQRQFYRECDIYGTVDFI 351
Query: 410 FGSSATLIQHSTIIVRKPGKGQFNTITADGSDTMNLNTGIVIQDCNIIPEAALFPERFTI 469
FG++A ++Q+ I RKP + NT+TA G N +TGI+I +C + + L P + ++
Sbjct: 352 FGNAAVVLQNCNIFARKP-PNRTNTLTAQGRTDPNQSTGIIIHNCRVTAASDLKPVQSSV 410
Query: 470 RSYLGRPWKYLAKTVVMESTIGDFIHPDGWTIWQGEQNHNTCYYAEYANTGPGANVARRV 529
+++LGRPWK ++TV +++ + I+P GW W G+ NT YYAEY NTGPG++ A RV
Sbjct: 411 KTFLGRPWKQYSRTVYIKTFLDSLINPAGWMEWSGDFALNTLYYAEYMNTGPGSSTANRV 470
Query: 530 KWKGYHGVISRAEANKFTAGIWLQAGPKSAAEWLNGLHVPHYLG 573
KW+GYH + S ++ ++FT G ++ + WL +VP G
Sbjct: 471 KWRGYHVLTSPSQVSQFTVGNFI-----AGNSWLPATNVPFTSG 509
>PME_DAUCA (P83218) Pectinesterase (EC 3.1.1.11) (Pectin
methylesterase) (PE)
Length = 319
Score = 292 bits (748), Expect = 1e-78
Identities = 151/321 (47%), Positives = 197/321 (61%), Gaps = 5/321 (1%)
Query: 253 ANIMPNAVVAKDGSGQFKTIQAALASYPKGNKGRYVIYVKAGVYDEYITVPKDAVNILMY 312
+ + PN VVA DGSG +KT+ A+A+ P+ +K RYVI +KAGVY E + VPK NI+
Sbjct: 3 STVTPNVVVAADGSGDYKTVSEAVAAAPEDSKTRYVIRIKAGVYRENVDVPKKKKNIMFL 62
Query: 313 GDGPARTIVTGRKSFAAGVKTMQTATFANTAMGFIGKAMTFENTAGPDGHQAVAFRNQGD 372
GDG TI+T K+ G T +AT A GF+ + +TF+NTAG HQAVA R D
Sbjct: 63 GDGRTSTIITASKNVQDGSTTFNSATVAAVGAGFLARDITFQNTAGAAKHQAVALRVGSD 122
Query: 373 MSALVGCHIVGYQDSLYVQSNRQYYRNCLVSGTVDFIFGSSATLIQHSTIIVRKPGKGQF 432
+SA C I+ YQDSLYV SNRQ++ NC ++GTVDFIFG++A ++Q I R+PG GQ
Sbjct: 123 LSAFYRCDILAYQDSLYVHSNRQFFINCFIAGTVDFIFGNAAVVLQDCDIHARRPGSGQK 182
Query: 433 NTITADGSDTMNLNTGIVIQDCNIIPEAALFPERFTIRSYLGRPWKYLAKTVVMESTIGD 492
N +TA G N NTGIVIQ I + L P + + +YLGRPWK ++TVVM+S+I +
Sbjct: 183 NMVTAQGRTDPNQNTGIVIQKSRIGATSDLQPVQSSFPTYLGRPWKEYSRTVVMQSSITN 242
Query: 493 FIHPDGWTIWQGEQNHNTCYYAEYANTGPGANVARRVKWKGYHGVISRAEANKFTAGIWL 552
I+P GW W G +T YY EY NTG GA + RV WKG+ + S EA FT G ++
Sbjct: 243 VINPAGWFPWDGNFALDTLYYGEYQNTGAGAATSGRVTWKGFKVITSSTEAQGFTPGSFI 302
Query: 553 QAGPKSAAEWLNGLHVPHYLG 573
G WL P LG
Sbjct: 303 AGG-----SWLKATTFPFSLG 318
>PME3_LYCES (Q96576) Pectinesterase 3 precursor (EC 3.1.1.11)
(Pectin methylesterase 3) (PE 3)
Length = 544
Score = 292 bits (747), Expect = 2e-78
Identities = 187/532 (35%), Positives = 270/532 (50%), Gaps = 46/532 (8%)
Query: 45 MCQNSQDQKLCHETLSSVHGADAADPKAYIAASVKAA--TDNVIKAFNMSERLTTEYGKE 102
+C+ +QD +LC +S + ++ VK +N I + + ++
Sbjct: 53 LCKTAQDSQLCLSYVSEIVTTESDGVTVLKKFLVKYVHQMNNAIPVVRKIKNQINDIRQQ 112
Query: 103 NGAKMALNDCKDLMQFALDSLDLSTKCVHDNNIQAVHDQIADMRNWLSAVISYRQACMEG 162
AL DC +L+ ++D + S + D ++ H A+ ++WLS V++ C+
Sbjct: 113 G----ALTDCLELLDQSVDLVSDSIAAI-DKRSRSEH---ANAQSWLSGVLTNHVTCL-- 162
Query: 163 FDDANDGEKKIKEQFHVQSLDSVQKVTAVALDIVTGLSD-ILQQFNLKFDVKPASRRLLN 221
D+ K + L + KV L VT +D +L+Q K
Sbjct: 163 -DELTSFSLSTKNGTVLDELITRAKVALAMLASVTTPNDEVLRQGLGKM----------- 210
Query: 222 SEVTVDDQGYPSWISSSDRKLLAKMQRKNWRANIMPNAVVAKDGSGQFKTIQAALASYPK 281
P W+SS DRKL+ + +I+ N VVA+DG+G ++T+ A+A+ P
Sbjct: 211 ----------PYWVSSRDRKLMESSGK-----DIIANRVVAQDGTGDYQTLAEAVAAAPD 255
Query: 282 GNKGRYVIYVKAGVYDEYITVPKDAVNILMYGDGPARTIVTGRKSFAAGVKTMQTATFAN 341
NK RYVIYVK G+Y E + V K +N+++ GDG TI+TG + G T + T A
Sbjct: 256 KNKTRYVIYVKMGIYKENVVVTKKKMNLMIVGDGMNATIITGSLNVVDG-STFPSNTLAA 314
Query: 342 TAMGFIGKAMTFENTAGPDGHQAVAFRNQGDMSALVGCHIVGYQDSLYVQSNRQYYRNCL 401
GFI + + +NTAGP+ QAVA R DMS + C I YQD+LY S RQ+YR+
Sbjct: 315 VGQGFILQDICIQNTAGPEKDQAVALRVGADMSVINRCRIDAYQDTLYAHSQRQFYRDSY 374
Query: 402 VSGTVDFIFGSSATLIQHSTIIVRKPGKGQFNTITADGSDTMNLNTGIVIQDCNIIPEAA 461
V+GTVDFIFG++A + Q I+ RKP K Q N +TA G N TG IQ C+II
Sbjct: 375 VTGTVDFIFGNAAVVFQKCQIVARKPNKRQKNMVTAQGRTDPNQATGTSIQFCDIIASPD 434
Query: 462 LFPERFTIRSYLGRPWKYLAKTVVMESTIGDFIHPDGWTIWQGEQNHNTCYYAEYANTGP 521
L P ++YLGRPWK ++TVVM+S + I P GW W+G+ T YY E+ N GP
Sbjct: 435 LEPVMNEYKTYLGRPWKKHSRTVVMQSYLDGHIDPSGWFEWRGDFALKTLYYGEFMNNGP 494
Query: 522 GANVARRVKWKGYHGVISRAEANKFTAGIWLQAGPKSAAEWLNGLHVPHYLG 573
GA ++RVKW GYH + EA FT +Q G WLN V + G
Sbjct: 495 GAGTSKRVKWPGYHVITDPNEAMPFTVAELIQGG-----SWLNSTSVAYVEG 541
>PME_PETIN (Q43043) Pectinesterase precursor (EC 3.1.1.11) (Pectin
methylesterase) (PE)
Length = 374
Score = 288 bits (736), Expect = 3e-77
Identities = 155/396 (39%), Positives = 226/396 (56%), Gaps = 28/396 (7%)
Query: 179 VQSLDSVQKVTAVALDIVTGLSDILQQFNLKFDVKPASRRLLNSEVTVDDQGYPSWISSS 238
V+ L+S ++++ AL ++ D++ Q +R+LL ++ SS
Sbjct: 2 VKLLNSTRELSINALSMLNSFGDMVAQ------ATGLNRKLLTTD-------------SS 42
Query: 239 DRKLLAKMQRKNWRANIMPNAVVAKDGSGQFKTIQAALASYPKGNKGRYVIYVKAGVYDE 298
D A +R +N PNA VA DGSGQ+KTI+ AL + PK N ++I++KAGVY E
Sbjct: 43 D----ATARRLLQISNAKPNATVALDGSGQYKTIKEALDAVPKKNTEPFIIFIKAGVYKE 98
Query: 299 YITVPKDAVNILMYGDGPARTIVTGRKSFAAGVKTMQTATFANTAMGFIGKAMTFENTAG 358
YI +PK N+++ G+GP +T +TG KS G T T T F+ K + FENTAG
Sbjct: 99 YIDIPKSMTNVVLIGEGPTKTKITGNKSVKDGPSTFHTTTVGVNGANFVAKNIGFENTAG 158
Query: 359 PDGHQAVAFRNQGDMSALVGCHIVGYQDSLYVQSNRQYYRNCLVSGTVDFIFGSSATLIQ 418
P+ QAVA R D + + C I GYQD+LYV + RQ+YR+C ++GTVDFIFG+ ++Q
Sbjct: 159 PEKEQAVALRVSADKAIIYNCQIDGYQDTLYVHTYRQFYRDCTITGTVDFIFGNGEAVLQ 218
Query: 419 HSTIIVRKPGKGQFNTITADGSDTMNLNTGIVIQDCNIIPEAALFPERFTIRSYLGRPWK 478
+ +IVRKP + Q +TA G IV+Q+C I P+ F ++YLGRPWK
Sbjct: 219 NCKVIVRKPAQNQSCMVTAQGRTEPIQKGAIVLQNCEIKPDTDYFSLSPPSKTYLGRPWK 278
Query: 479 YLAKTVVMESTIGDFIHPDGWTIWQ-GEQNHNTCYYAEYANTGPGANVARRVKWKGYHGV 537
++T++M+S I FI P+GW W +T YYAEY N GPGA + +R+ WKG+
Sbjct: 279 EYSRTIIMQSYIDKFIEPEGWAPWNITNFGRDTSYYAEYQNRGPGAALDKRITWKGFQKG 338
Query: 538 ISRAEANKFTAGIWLQAGPKSAAEWLNGLHVPHYLG 573
+ A KFTAG+++ + WL +VP+ G
Sbjct: 339 FTGEAAQKFTAGVYI----NNDENWLQKANVPYEAG 370
>PME_ASPAC (Q12535) Pectinesterase precursor (EC 3.1.1.11) (Pectin
methylesterase) (PE)
Length = 331
Score = 122 bits (305), Expect = 3e-27
Identities = 92/280 (32%), Positives = 130/280 (45%), Gaps = 19/280 (6%)
Query: 260 VVAKDGSGQFKTIQAALASYPKGNKGRYVIYVKAGVYDEYITVPKDAVNILMYG-----D 314
VVAK G G + TI A+ + I+++ G YDE + +P +++YG D
Sbjct: 29 VVAKSG-GDYTTIGDAIDALSTSTTDTQTIFIEEGTYDEQVYLPAMTGKVIIYGQTENTD 87
Query: 315 GPARTIVTGRKSFA---AGVKTMQTATFANTAMGFIGKAMTFENTAGPDGHQAVAFRNQG 371
A +VT + + AG TATF N A+G + NT G HQA+A
Sbjct: 88 SYADNLVTITHAISYEDAGESDDLTATFRNKAVGSQVYNLNIANTCGQACHQALALSAWA 147
Query: 372 DMSALVGCHIVGYQDSLYVQSNRQYYRNCLVSGTVDFIFGSSA-TLIQHSTI-IVRKPGK 429
D GC+ GYQD+L Q+ Q Y N + G VDFIFG A Q+ I +V P
Sbjct: 148 DQQGYYGCNFTGYQDTLLAQTGNQLYINSYIEGAVDFIFGQHARAWFQNVDIRVVEGPTS 207
Query: 430 GQFNTITADGSDTMNLNTGIVIQDCNIIPEAALFPERFTIRSYLGRPWKYLAKTVVMEST 489
+ITA+G + + VI + + T YLGRPW A+ V +++
Sbjct: 208 A---SITANGRSSETDTSYYVINKSTVAAKEGDDVAEGTY--YLGRPWSEYARVVFQQTS 262
Query: 490 IGDFIHPDGWTIWQGEQNHNTCY--YAEYANTGPGANVAR 527
+ + I+ GWT W NT Y + EYANTG G+ R
Sbjct: 263 MTNVINSLGWTEW-STSTPNTEYVTFGEYANTGAGSEGTR 301
>PME_ASPTU (P17872) Pectinesterase precursor (EC 3.1.1.11) (Pectin
methylesterase) (PE)
Length = 331
Score = 110 bits (274), Expect = 1e-23
Identities = 83/276 (30%), Positives = 120/276 (43%), Gaps = 12/276 (4%)
Query: 260 VVAKDGSGQFKTIQAALASYPKGNKGRYVIYVKAGVYDEYITVPKDAVNILMYGDGPART 319
VVAK G G + TI AA+ + + I+++ G YDE + +P + +++YG T
Sbjct: 29 VVAKSG-GDYDTISAAVDALSTTSTETQTIFIEEGSYDEQVYIPALSGKLIVYGQTEDTT 87
Query: 320 IVTGRK-------SFAAGVKTMQTATFANTAMGFIGKAMTFENTAGPDGHQAVAFRNQGD 372
T + A +TAT N A G + NT G HQA+A
Sbjct: 88 TYTSNLVNITHAIALADVDNDDETATLRNYAEGSAIYNLNIANTCGQACHQALAVSAYAS 147
Query: 373 MSALVGCHIVGYQDSLYVQSNRQYYRNCLVSGTVDFIFGSSATLIQHSTIIVRKPGKGQF 432
C GYQD+L ++ Q Y + G VDFIFG A H I G
Sbjct: 148 EQGYYACQFTGYQDTLLAETGYQVYAGTYIEGAVDFIFGQHARAWFHECDIRVLEGPSS- 206
Query: 433 NTITADGSDTMNLNTGIVIQDCNIIPEAALFPERFTIRSYLGRPWKYLAKTVVMESTIGD 492
+ITA+G + + ++ VI + AA + + YLGRPW A+ ++++ D
Sbjct: 207 ASITANGRSSESDDSYYVIHKSTV--AAADGNDVSSGTYYLGRPWSQYARVCFQKTSMTD 264
Query: 493 FIHPDGWTIWQ-GEQNHNTCYYAEYANTGPGANVAR 527
I+ GWT W N + EY NTG GA R
Sbjct: 265 VINHLGWTEWSTSTPNTENVTFVEYGNTGTGAEGPR 300
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.318 0.133 0.394
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 66,626,078
Number of Sequences: 164201
Number of extensions: 2752353
Number of successful extensions: 6465
Number of sequences better than 10.0: 32
Number of HSP's better than 10.0 without gapping: 23
Number of HSP's successfully gapped in prelim test: 9
Number of HSP's that attempted gapping in prelim test: 6358
Number of HSP's gapped (non-prelim): 39
length of query: 576
length of database: 59,974,054
effective HSP length: 116
effective length of query: 460
effective length of database: 40,926,738
effective search space: 18826299480
effective search space used: 18826299480
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 69 (31.2 bits)
Medicago: description of AC144563.2


