

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144541.15 - phase: 0
(593 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
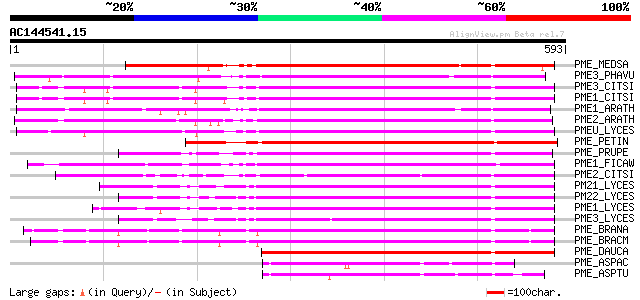
Score E
Sequences producing significant alignments: (bits) Value
PME_MEDSA (Q42920) Pectinesterase precursor (EC 3.1.1.11) (Pecti... 420 e-117
PME3_PHAVU (Q43111) Pectinesterase 3 precursor (EC 3.1.1.11) (Pe... 381 e-105
PME3_CITSI (P83948) Pectinesterase 3 precursor (EC 3.1.1.11) (Pe... 372 e-102
PME1_CITSI (O04886) Pectinesterase 1 precursor (EC 3.1.1.11) (Pe... 370 e-102
PME1_ARATH (Q43867) Pectinesterase 1 precursor (EC 3.1.1.11) (Pe... 369 e-101
PME2_ARATH (Q42534) Pectinesterase 2 precursor (EC 3.1.1.11) (Pe... 363 e-100
PMEU_LYCES (Q43143) Pectinesterase U1 precursor (EC 3.1.1.11) (P... 355 2e-97
PME_PETIN (Q43043) Pectinesterase precursor (EC 3.1.1.11) (Pecti... 347 4e-95
PME_PRUPE (Q43062) Pectinesterase PPE8B precursor (EC 3.1.1.11) ... 332 2e-90
PME1_FICAW (P83947) Pectinesterase precursor (EC 3.1.1.11) (Pect... 330 6e-90
PME2_CITSI (O04887) Pectinesterase 2 precursor (EC 3.1.1.11) (Pe... 323 1e-87
PM21_LYCES (P09607) Pectinesterase 2 precursor (EC 3.1.1.11) (Pe... 319 1e-86
PM22_LYCES (Q96575) Pectinesterase 2 precursor (EC 3.1.1.11) (Pe... 315 2e-85
PME1_LYCES (P14280) Pectinesterase 1 precursor (EC 3.1.1.11) (Pe... 308 2e-83
PME3_LYCES (Q96576) Pectinesterase 3 precursor (EC 3.1.1.11) (Pe... 303 8e-82
PME_BRANA (P41510) Probable pectinesterase precursor (EC 3.1.1.1... 296 8e-80
PME_BRACM (Q42608) Pectinesterase (EC 3.1.1.11) (Pectin methyles... 286 1e-76
PME_DAUCA (P83218) Pectinesterase (EC 3.1.1.11) (Pectin methyles... 283 9e-76
PME_ASPAC (Q12535) Pectinesterase precursor (EC 3.1.1.11) (Pecti... 112 2e-24
PME_ASPTU (P17872) Pectinesterase precursor (EC 3.1.1.11) (Pecti... 102 3e-21
>PME_MEDSA (Q42920) Pectinesterase precursor (EC 3.1.1.11) (Pectin
methylesterase) (PE) (P65)
Length = 447
Score = 420 bits (1079), Expect = e-117
Identities = 218/468 (46%), Positives = 293/468 (62%), Gaps = 32/468 (6%)
Query: 124 LDTCKQVMQLSIDEFQRSLERFSNFDLNSLDRVLTSLKVWLSGAITYQETCLDAFENTTT 183
++ C +V+ ++D +S+ FD + L LKVWL+G +++Q+TCLD F NTTT
Sbjct: 1 MEICNEVLDYAVDGIHKSVGTLDQFDFHKLSEYAFDLKVWLTGTLSHQQTCLDGFANTTT 60
Query: 184 DAGKKMKEVLQTSMHMSSNGLSIINQLS---KTFEEMKQPAGRRLLKESVDGEEDVLGHG 240
AG+ M +VL+TSM +SSN + +++ +S K F+ + RRLL + DG
Sbjct: 61 KAGETMTKVLKTSMELSSNAIDMMDAVSRILKGFDTSQYSVSRRLLSD--DG-------- 110
Query: 241 GDFELPEWVDDRAGVRKLLNKMTGRKLQAHVVVAKDGSGNFTTITEALKHVPKKNLKPFV 300
+P WV+D G R+LL G +Q + VVA+DGSG F T+T+ALK VP KN PFV
Sbjct: 111 ----IPSWVND--GHRRLL---AGGNVQPNAVVAQDGSGQFKTLTDALKTVPPKNAVPFV 161
Query: 301 IYIKEGVYKEYVEVTKTMTHVVFIGDGGRKTRITGNKNFIDGVGTFKTASVAITGDFFVG 360
I++K GVYKE V V K M +V IGDG KT+ TG+ N+ DG+ T+ TA+ + G F+
Sbjct: 162 IHVKAGVYKETVNVAKEMNYVTVIGDGPTKTKFTGSLNYADGINTYNTATFGVNGANFMA 221
Query: 361 IGIGFENSAGPEKHQAVALRVQSDRSIFYKCRMDGYQDTLYAHTMRQFYRDCIISGTIDF 420
IGFEN+AG KHQAVALRV +D++IFY C+MDG+QDTLY + RQFYRDC ISGTIDF
Sbjct: 222 KDIGFENTAGTGKHQAVALRVTADQAIFYNCQMDGFQDTLYVQSQRQFYRDCSISGTIDF 281
Query: 421 VFGDSIAVLQNCTFVVRKPLENQQCIVTAQGRKEKNQPTGLIIQGGSIVADPKYYPVRLK 480
VFG+ V QNC V R P + QQC+VTA GR+++N + L+ Q +P V K
Sbjct: 282 VFGERFGVFQNCKLVCRLPAKGQQCLVTAGGREKQNSASALVFQSSHFTGEPALTSVTPK 341
Query: 481 NKAYLARPWKDFSRTIFLDTYIGDMITPEGYMPWQTPAGITGTDTCYYGEYNNRGPGSDV 540
+YL RPWK +S+ + +D+ I + PEGYMPW G +TC Y EYNN+GPG+D
Sbjct: 342 -VSYLGRPWKQYSKVVIMDSTIDAIFVPEGYMPWM---GSAFKETCTYYEYNNKGPGADT 397
Query: 541 KQRVKWQGVKTITSEGAASFVPIRFFH------GDDWIRVTRVPYSPG 582
RVKW GVK +TS AA + P +FF D WI + VPYS G
Sbjct: 398 NLRVKWHGVKVLTSNVAAEYYPGKFFEIVNATARDTWIVKSGVPYSLG 445
>PME3_PHAVU (Q43111) Pectinesterase 3 precursor (EC 3.1.1.11)
(Pectin methylesterase 3) (PE 3)
Length = 581
Score = 381 bits (978), Expect = e-105
Identities = 222/577 (38%), Positives = 324/577 (55%), Gaps = 38/577 (6%)
Query: 6 DAKRNKRLAIIGVSTFLLVAMVVAVTVNVGFNNKKE---PGEETSKESHVSQSVKAVKTL 62
+ K KRL II VS+ +L+A+++A V +N+ P ++ ++ +S + ++K +
Sbjct: 22 EKKTRKRLIIIAVSSIVLIAVIIAAVAGVVIHNRNSESSPSSDSVPQTELSPAA-SLKAV 80
Query: 63 CAPTDYKKECEDSLIA-HAGNITEPKELIKIAFNITIAKISEGLKKTHLLQEAEKDERTK 121
C T Y C S+ + N T+P+ L K++ + I ++S K L AE+D R +
Sbjct: 81 CDTTRYPSSCFSSISSLPESNTTDPELLFKLSLRVAIDELSSFPSK--LRANAEQDARLQ 138
Query: 122 QALDTCKQVMQLSIDEFQRSLERFSNFDLNSLDRV-LTSLKVWLSGAITYQETCLDAFEN 180
+A+D C V ++D S+ +++++ WLS A+T Q+TCLDA
Sbjct: 139 KAIDVCSSVFGDALDRLNDSISALGTVAGRIASSASVSNVETWLSAALTDQDTCLDAVGE 198
Query: 181 TTTDAGKKMKEVLQTSMHMS----SNGLSIINQLSKTFEEMKQPAGRRLLKESVDGEEDV 236
+ A + + ++T+M S SN L+I+ ++ + P R L
Sbjct: 199 LNSTAARGALQEIETAMRNSTEFASNSLAIVTKILGLLSRFETPIHHRRL---------- 248
Query: 237 LGHGGDFELPEWVDDRAGVRKLLNKMTGRKLQAHVVVAKDGSGNFTTITEALKHVPKKNL 296
LG PEW+ A R+LL + VVAKDGSG F TI EALK V KK+
Sbjct: 249 LG------FPEWLG--AAERRLLEEKNNDSTP-DAVVAKDGSGQFKTIGEALKLVKKKSE 299
Query: 297 KPFVIYIKEGVYKEYVEVTKTMTHVVFIGDGGRKTRITGNKNFIDGVGTFKTASVAITGD 356
+ F +Y+KEG Y E +++ K +V+ GDG KT + G++NF+DG TF+TA+ A+ G
Sbjct: 300 ERFSVYVKEGRYVENIDLDKNTWNVMIYGDGKDKTFVVGSRNFMDGTPTFETATFAVKGK 359
Query: 357 FFVGIGIGFENSAGPEKHQAVALRVQSDRSIFYKCRMDGYQDTLYAHTMRQFYRDCIISG 416
F+ IGF N+AG KHQAVALR SDRS+F++C DG+QDTLYAH+ RQFYRDC I+G
Sbjct: 360 GFIAKDIGFVNNAGASKHQAVALRSGSDRSVFFRCSFDGFQDTLYAHSNRQFYRDCDITG 419
Query: 417 TIDFVFGDSIAVLQNCTFVVRKPLENQQCIVTAQGRKEKNQPTGLIIQGGSIVADPKYYP 476
TIDF+FG++ V Q+C + R+PL NQ +TAQG+K+ NQ TG+IIQ +I +
Sbjct: 420 TIDFIFGNAAVVFQSCKIMPRQPLPNQFNTITAQGKKDPNQNTGIIIQKSTITP----FG 475
Query: 477 VRLKNKAYLARPWKDFSRTIFLDTYIGDMITPEGYMPWQTPAGITGTDTCYYGEYNNRGP 536
L YL RPWKDFS T+ + + IG ++ P G+M W + T +Y EY N GP
Sbjct: 476 NNLTAPTYLGRPWKDFSTTVIMQSDIGALLNPVGWMSW--VPNVEPPTTIFYAEYQNSGP 533
Query: 537 GSDVKQRVKWQGVK-TITSEGAASFVPIRFFHGDDWI 572
G+DV QRVKW G K TIT A F F G +W+
Sbjct: 534 GADVSQRVKWAGYKPTITDRNAEEFTVQSFIQGPEWL 570
>PME3_CITSI (P83948) Pectinesterase 3 precursor (EC 3.1.1.11)
(Pectin methylesterase 3) (PE 3)
Length = 584
Score = 372 bits (955), Expect = e-102
Identities = 224/590 (37%), Positives = 336/590 (55%), Gaps = 46/590 (7%)
Query: 8 KRNKRLAIIGVSTFLLVAMVVAVTVNVGFNNKKEPGEETSKESHVSQSVKAVKTLCAPTD 67
K+ K+L + +T L+VA V+ + G N++K G+ ++ H +K+ C+ T
Sbjct: 25 KKKKKLFLALFATLLVVAAVIGIVA--GVNSRKNSGDNGNEPHHA-----ILKSSCSSTR 77
Query: 68 YKKECEDSLIA---HAGNITEPKELIKIAFNITIAKISE---GLKKTHLLQEAEKDERTK 121
Y C ++ A + +T K++I+++ NIT + G++K LL+ +R K
Sbjct: 78 YPDLCFSAIAAVPEASKKVTSQKDVIEMSLNITTTAVEHNYFGIQK--LLKRTNLTKREK 135
Query: 122 QALDTCKQVMQLSIDEFQRSLERFSNF-DLNSLDRVLTSLKVWLSGAITYQETCLDAFEN 180
AL C + + ++DE +++E + + SL + LK +S A+T Q TCLD F +
Sbjct: 136 VALHDCLETIDETLDELHKAVEDLEEYPNKKSLSQHADDLKTLMSAAMTNQGTCLDGFSH 195
Query: 181 TTTDAGKKMKEVLQTSM----HMSSNGLSIINQLSKT-FEEMKQPAGRRLLKES--VDGE 233
DA K +++ L M SN L++I ++ T M+ R+L++E+ VDG
Sbjct: 196 D--DANKHVRDALSDGQVHVEKMCSNALAMIKNMTDTDMMIMRTSNNRKLIEETSTVDG- 252
Query: 234 EDVLGHGGDFELPEWVDDRAGVRKLLNKMTGRKLQAHVVVAKDGSGNFTTITEALKHVPK 293
P W+ G R+LL + + +VVVA DGSGNF T+ ++ P+
Sbjct: 253 -----------WPAWLS--TGDRRLLQSSS---VTPNVVVAADGSGNFKTVAASVAAAPQ 296
Query: 294 KNLKPFVIYIKEGVYKEYVEVTKTMTHVVFIGDGGRKTRITGNKNFIDGVGTFKTASVAI 353
K ++I IK GVY+E VEVTK +++FIGDG +T ITG++N +DG TFK+A+VA+
Sbjct: 297 GGTKRYIIRIKAGVYRENVEVTKKHKNIMFIGDGRTRTIITGSRNVVDGSTTFKSATVAV 356
Query: 354 TGDFFVGIGIGFENSAGPEKHQAVALRVQSDRSIFYKCRMDGYQDTLYAHTMRQFYRDCI 413
G+ F+ I F+N+AGP KHQAVALRV +D S FY C M YQDTLY H+ RQF+ +C+
Sbjct: 357 VGEGFLARDITFQNTAGPSKHQAVALRVGADLSAFYNCDMLAYQDTLYVHSNRQFFVNCL 416
Query: 414 ISGTIDFVFGDSIAVLQNCTFVVRKPLENQQCIVTAQGRKEKNQPTGLIIQGGSIVADPK 473
I+GT+DF+FG++ AVLQNC RKP Q+ +VTAQGR + NQ TG++IQ I A
Sbjct: 417 IAGTVDFIFGNAAAVLQNCDIHARKPNSGQKNMVTAQGRADPNQNTGIVIQKSRIGATSD 476
Query: 474 YYPVRLKNKAYLARPWKDFSRTIFLDTYIGDMITPEGYMPWQTPAGITGTDTCYYGEYNN 533
PV+ YL RPWK++SRT+ + + I D+I P G+ W G +T +YGE+ N
Sbjct: 477 LKPVQGSFPTYLGRPWKEYSRTVIMQSSITDVIHPAGWHEWD---GNFALNTLFYGEHQN 533
Query: 534 RGPGSDVKQRVKWQGVKTITS-EGAASFVPIRFFHGDDWIRVTRVPYSPG 582
G G+ RVKW+G + ITS A +F P F G W+ T P+S G
Sbjct: 534 AGAGAGTSGRVKWKGFRVITSATEAQAFTPGSFIAGSSWLGSTGFPFSLG 583
>PME1_CITSI (O04886) Pectinesterase 1 precursor (EC 3.1.1.11)
(Pectin methylesterase) (PE)
Length = 584
Score = 370 bits (949), Expect = e-102
Identities = 222/590 (37%), Positives = 332/590 (55%), Gaps = 46/590 (7%)
Query: 8 KRNKRLAIIGVSTFLLVAMVVAVTVNVGFNNKKEPGEETSKESHVSQSVKAVKTLCAPTD 67
K+ K+L + +T L+VA V+ + G N++K G+ ++ H +K+ C+ T
Sbjct: 25 KKKKKLFLALFATLLVVAAVIGIVA--GVNSRKNSGDNGNEPHHA-----ILKSSCSSTR 77
Query: 68 YKKECEDSLIA---HAGNITEPKELIKIAFNITIAKISE---GLKKTHLLQEAEKDERTK 121
Y C ++ A + +T K++I+++ NIT + G++K LL+ +R K
Sbjct: 78 YPDLCFSAIAAVPEASKKVTSQKDVIEMSLNITTTAVEHNYFGIQK--LLKRTNLTKREK 135
Query: 122 QALDTCKQVMQLSIDEFQRSLERFSNF-DLNSLDRVLTSLKVWLSGAITYQETCLDAFEN 180
AL C + + ++DE +++E + + SL + LK +S A+T Q TCLD F +
Sbjct: 136 VALHDCLETIDETLDELHKAVEDLEEYPNKKSLSQHADDLKTLMSAAMTNQGTCLDGFSH 195
Query: 181 TTTDAGKKMKEVLQTSM----HMSSNGLSIINQLSKTFEEMKQPAGRRLLKE---SVDGE 233
DA K +++ L M SN L++I ++ T + + + R L E +VDG
Sbjct: 196 D--DANKHVRDALSDGQVHVEKMCSNALAMIKNMTDTDMMIMRTSNNRKLTEETSTVDG- 252
Query: 234 EDVLGHGGDFELPEWVDDRAGVRKLLNKMTGRKLQAHVVVAKDGSGNFTTITEALKHVPK 293
P W+ G R+LL + + + VVA DGSGNF T+ A+ P+
Sbjct: 253 -----------WPAWLSP--GDRRLLQSSS---VTPNAVVAADGSGNFKTVAAAVAAAPQ 296
Query: 294 KNLKPFVIYIKEGVYKEYVEVTKTMTHVVFIGDGGRKTRITGNKNFIDGVGTFKTASVAI 353
K ++I IK GVY+E VEVTK +++FIGDG +T ITG++N +DG TFK+A+ A+
Sbjct: 297 GGTKRYIIRIKAGVYRENVEVTKKHKNIMFIGDGRTRTIITGSRNVVDGSTTFKSATAAV 356
Query: 354 TGDFFVGIGIGFENSAGPEKHQAVALRVQSDRSIFYKCRMDGYQDTLYAHTMRQFYRDCI 413
G+ F+ I F+N+AGP KHQAVALRV +D S FY C M YQDTLY H+ RQF+ +C+
Sbjct: 357 VGEGFLARDITFQNTAGPSKHQAVALRVGADLSAFYNCDMLAYQDTLYVHSNRQFFVNCL 416
Query: 414 ISGTIDFVFGDSIAVLQNCTFVVRKPLENQQCIVTAQGRKEKNQPTGLIIQGGSIVADPK 473
I+GT+DF+FG++ AVLQNC RKP Q+ +VTAQGR + NQ TG++IQ I A
Sbjct: 417 IAGTVDFIFGNAAAVLQNCDIHARKPNSGQKNMVTAQGRTDPNQNTGIVIQKSRIGATSD 476
Query: 474 YYPVRLKNKAYLARPWKDFSRTIFLDTYIGDMITPEGYMPWQTPAGITGTDTCYYGEYNN 533
PV+ YL RPWK++SRT+ + + I D+I P G+ W G +T +YGE+ N
Sbjct: 477 LKPVQGSFPTYLGRPWKEYSRTVIMQSSITDLIHPAGWHEWD---GNFALNTLFYGEHQN 533
Query: 534 RGPGSDVKQRVKWQGVKTITS-EGAASFVPIRFFHGDDWIRVTRVPYSPG 582
G G+ RVKW+G + ITS A +F P F G W+ T P+S G
Sbjct: 534 SGAGAGTSGRVKWKGFRVITSATEAQAFTPGSFIAGSSWLGSTGFPFSLG 583
>PME1_ARATH (Q43867) Pectinesterase 1 precursor (EC 3.1.1.11)
(Pectin methylesterase 1) (PE 1)
Length = 586
Score = 369 bits (946), Expect = e-101
Identities = 223/586 (38%), Positives = 324/586 (55%), Gaps = 44/586 (7%)
Query: 8 KRNKRLAIIGVSTFLLVAMVVAVTV-NVGFNNKKEPGEETSKESHVSQSVKAVKTLCAPT 66
K KRL ++ +S +L+A+++A V V NK E E S S+KA+ C+ T
Sbjct: 24 KTRKRLLLLSISVVVLIAVIIAAVVATVVHKNKNESTPSPPPELTPSTSLKAI---CSVT 80
Query: 67 DYKKECEDSLIA-HAGNITEPKELIKIAFNITIAKISEGLKKTHLLQEAEKDERTKQALD 125
+ + C S+ + N T+P+ L K++ + I ++ L + +DER K AL
Sbjct: 81 RFPESCISSISKLPSSNTTDPETLFKLSLKVIIDELDSISDLPEKLSKETEDERIKSALR 140
Query: 126 TCKQVMQLSIDEFQRSLERFSNFDLNSLDRVLTS-----LKVWLSGAITYQETCLDAFE- 179
C +++ ++D ++ S D + L+S LK WLS +T ETC D+ +
Sbjct: 141 VCGDLIEDALDRLNDTV---SAIDDEEKKKTLSSSKIEDLKTWLSATVTDHETCFDSLDE 197
Query: 180 ---NTTTDAG----KKMKEVLQTSMHMSSNGLSIINQLSKTFEEMKQPAGRRLLKESVDG 232
N T A + +K + S +SN L+I++++ ++ P RR S
Sbjct: 198 LKQNKTEYANSTITQNLKSAMSRSTEFTSNSLAIVSKILSALSDLGIPIHRRRRLMSHHH 257
Query: 233 EEDVLGHGGDFELPEWVDDRAGVRKLLNKMTGRKLQAHVVVAKDGSGNFTTITEALKHVP 292
++ V DFE +W R+ L + G L+ V VA DG+G+ T+ EA+ VP
Sbjct: 258 QQSV-----DFE--KWA------RRRLLQTAG--LKPDVTVAGDGTGDVLTVNEAVAKVP 302
Query: 293 KKNLKPFVIYIKEGVYKEYVEVTKTMTHVVFIGDGGRKTRITGNKNFIDGVGTFKTASVA 352
KK+LK FVIY+K G Y E V + K+ +V+ GDG KT I+G+KNF+DG T++TA+ A
Sbjct: 303 KKSLKMFVIYVKSGTYVENVVMDKSKWNVMIYGDGKGKTIISGSKNFVDGTPTYETATFA 362
Query: 353 ITGDFFVGIGIGFENSAGPEKHQAVALRVQSDRSIFYKCRMDGYQDTLYAHTMRQFYRDC 412
I G F+ IG N+AG KHQAVA R SD S++Y+C DG+QDTLY H+ RQFYRDC
Sbjct: 363 IQGKGFIMKDIGIINTAGAAKHQAVAFRSGSDFSVYYQCSFDGFQDTLYPHSNRQFYRDC 422
Query: 413 IISGTIDFVFGDSIAVLQNCTFVVRKPLENQQCIVTAQGRKEKNQPTGLIIQGGSIVADP 472
++GTIDF+FG + V Q C + R+PL NQ +TAQG+K+ NQ +G+ IQ +I A+
Sbjct: 423 DVTGTIDFIFGSAAVVFQGCKIMPRQPLSNQFNTITAQGKKDPNQSSGMSIQRCTISANG 482
Query: 473 KYYPVRLKNKAYLARPWKDFSRTIFLDTYIGDMITPEGYMPWQTPAGITGTDTCYYGEYN 532
YL RPWK+FS T+ ++T IG ++ P G+M W +G+ + YGEY
Sbjct: 483 NVIA-----PTYLGRPWKEFSTTVIMETVIGAVVRPSGWMSW--VSGVDPPASIVYGEYK 535
Query: 533 NRGPGSDVKQRVKWQGVKTITSEG-AASFVPIRFFHGDDWIRVTRV 577
N GPGSDV QRVKW G K + S+ AA F HG DWI T V
Sbjct: 536 NTGPGSDVTQRVKWAGYKPVMSDAEAAKFTVATLLHGADWIPATGV 581
>PME2_ARATH (Q42534) Pectinesterase 2 precursor (EC 3.1.1.11)
(Pectin methylesterase 2) (PE 2)
Length = 587
Score = 363 bits (932), Expect = e-100
Identities = 231/594 (38%), Positives = 328/594 (54%), Gaps = 41/594 (6%)
Query: 6 DAKRNKRLAIIGVS-TFLLVAMVVAVTVNVGFNNKKEPGEETSKESHVSQSVKAVKTLCA 64
D K NK+L + + LL+A +V + NK + S SH +K++C+
Sbjct: 13 DFKNNKKLILSSAAIALLLLASIVGIAATTTNQNKNQKITTLSSTSHA-----ILKSVCS 67
Query: 65 PTDYKKECEDSLIAHAGN-ITEPKELIKIAFNITIAKISEGLKKTHLLQEAEKD--ERTK 121
T Y + C ++ A G +T KE+I+ + N+T + L K R
Sbjct: 68 STLYPELCFSAVAATGGKELTSQKEVIEASLNLTTKAVKHNYFAVKKLIAKRKGLTPREV 127
Query: 122 QALDTCKQVMQLSIDEFQRSLERFSNFDLN-SLDRVLTSLKVWLSGAITYQETCLDAFEN 180
AL C + + ++DE ++E + SL + LK +S AIT Q TCLD F
Sbjct: 128 TALHDCLETIDETLDELHVAVEDLHQYPKQKSLRKHADDLKTLISSAITNQGTCLDGF-- 185
Query: 181 TTTDAGKKMKEVLQTSM----HMSSNGLSIINQLSKT----FEEMKQPA-----GRRLLK 227
+ DA +K+++ L HM SN L++I +++T FE + + R LK
Sbjct: 186 SYDDADRKVRKALLKGQVHVEHMCSNALAMIKNMTETDIANFELRDKSSTFTNNNNRKLK 245
Query: 228 ESVDGEEDVLGHGGDFELPEWVDDRAGVRKLLNKMTGRKLQAHVVVAKDGSGNFTTITEA 287
E V G+ D G P+W+ G R+LL G ++A VA DGSG+FTT+ A
Sbjct: 246 E-VTGDLDSDGW------PKWLS--VGDRRLLQ---GSTIKADATVADDGSGDFTTVAAA 293
Query: 288 LKHVPKKNLKPFVIYIKEGVYKEYVEVTKTMTHVVFIGDGGRKTRITGNKNFIDGVGTFK 347
+ P+K+ K FVI+IK GVY+E VEVTK T+++F+GDG KT ITG++N +DG TF
Sbjct: 294 VAAAPEKSNKRFVIHIKAGVYRENVEVTKKKTNIMFLGDGRGKTIITGSRNVVDGSTTFH 353
Query: 348 TASVAITGDFFVGIGIGFENSAGPEKHQAVALRVQSDRSIFYKCRMDGYQDTLYAHTMRQ 407
+A+VA G+ F+ I F+N+AGP KHQAVALRV SD S FY+C M YQDTLY H+ RQ
Sbjct: 354 SATVAAVGERFLARDITFQNTAGPSKHQAVALRVGSDFSAFYQCDMFAYQDTLYVHSNRQ 413
Query: 408 FYRDCIISGTIDFVFGDSIAVLQNCTFVVRKPLENQQCIVTAQGRKEKNQPTGLIIQGGS 467
F+ C I+GT+DF+FG++ AVLQ+C R+P Q+ +VTAQGR + NQ TG++IQ
Sbjct: 414 FFVKCHITGTVDFIFGNAAAVLQDCDINARRPNSGQKNMVTAQGRSDPNQNTGIVIQNCR 473
Query: 468 IVADPKYYPVRLKNKAYLARPWKDFSRTIFLDTYIGDMITPEGYMPWQTPAGITGTDTCY 527
I V+ YL RPWK++SRT+ + + I D+I PEG+ W +G DT
Sbjct: 474 IGGTSDLLAVKGTFPTYLGRPWKEYSRTVIMQSDISDVIRPEGWHEW---SGSFALDTLT 530
Query: 528 YGEYNNRGPGSDVKQRVKWQGVKTITSEGAAS-FVPIRFFHGDDWIRVTRVPYS 580
Y EY NRG G+ RVKW+G K ITS+ A F +F G W+ T P+S
Sbjct: 531 YREYLNRGGGAGTANRVKWKGYKVITSDTEAQPFTAGQFIGGGGWLASTGFPFS 584
>PMEU_LYCES (Q43143) Pectinesterase U1 precursor (EC 3.1.1.11)
(Pectin methylesterase) (PE)
Length = 583
Score = 355 bits (910), Expect = 2e-97
Identities = 225/591 (38%), Positives = 328/591 (55%), Gaps = 40/591 (6%)
Query: 8 KRNKRLAIIGVSTFLLVAMVVAVTVNVGFNNKKEPGEETSKESHVSQSVKA-VKTLCAPT 66
KR K++ + V++ LLVA V+ V V ++K ++ + +S S A VK+ C+ T
Sbjct: 16 KRKKKIYLAIVASVLLVAAVIGVVAGVKSHSKNS--DDHADIMAISSSAHAIVKSACSNT 73
Query: 67 DYKKECEDSLIA---HAGNITEPKELIKIAFNITIAKISEGLKKTHLLQEAEKD--ERTK 121
+ + C +++ + +T K++I+++ NIT+ + L + K R K
Sbjct: 74 LHPELCYSAIVNVSDFSKKVTSQKDVIELSLNITVKAVRRNYYAVKELIKTRKGLTPREK 133
Query: 122 QALDTCKQVMQLSIDEFQRSLERFSNF-DLNSLDRVLTSLKVWLSGAITYQETCLDAFEN 180
AL C + M ++DE ++E + + SL + LK +S AIT QETCLD F +
Sbjct: 134 VALHDCLETMDETLDELHTAVEDLELYPNKKSLKEHVEDLKTLISSAITNQETCLDGFSH 193
Query: 181 TTTDAGKKMKEVLQTSMH----MSSNGLSIINQLSKT--FEEMK--QPAGRRLLKESVDG 232
D KK+++VL M SN L++I ++ T EMK PA R L E
Sbjct: 194 DEAD--KKVRKVLLKGQKHVEKMCSNALAMICNMTDTDIANEMKLSAPANNRKLVED--- 248
Query: 233 EEDVLGHGGDFELPEWVDDRAGVRKLLNKMTGRKLQAHVVVAKDGSGNFTTITEALKHVP 292
+ E PEW+ AG R+LL T + VVVA DGSG++ T++EA++ P
Sbjct: 249 ---------NGEWPEWLS--AGDRRLLQSST---VTPDVVVAADGSGDYKTVSEAVRKAP 294
Query: 293 KKNLKPFVIYIKEGVYKEYVEVTKTMTHVVFIGDGGRKTRITGNKNFIDGVGTFKTASVA 352
+K+ K +VI IK GVY+E V+V K T+++F+GDG T IT ++N DG TF +A+V
Sbjct: 295 EKSSKRYVIRIKAGVYRENVDVPKKKTNIMFMGDGKSNTIITASRNVQDGSTTFHSATVV 354
Query: 353 ITGDFFVGIGIGFENSAGPEKHQAVALRVQSDRSIFYKCRMDGYQDTLYAHTMRQFYRDC 412
+ I F+N+AG KHQAVAL V SD S FY+C M YQDTLY H+ RQF+ C
Sbjct: 355 RVAGKVLARDITFQNTAGASKHQAVALCVGSDLSAFYRCDMLAYQDTLYVHSNRQFFVQC 414
Query: 413 IISGTIDFVFGDSIAVLQNCTFVVRKPLENQQCIVTAQGRKEKNQPTGLIIQGGSIVADP 472
+++GT+DF+FG+ AV Q+C R+P Q+ +VTAQGR + NQ TG++IQ I A
Sbjct: 415 LVAGTVDFIFGNGAAVFQDCDIHARRPGSGQKNMVTAQGRTDPNQNTGIVIQKCRIGATS 474
Query: 473 KYYPVRLKNKAYLARPWKDFSRTIFLDTYIGDMITPEGYMPWQTPAGITGTDTCYYGEYN 532
PV+ YL RPWK++SRT+ + + I D+I P G+ W G DT +YGEY
Sbjct: 475 DLRPVQKSFPTYLGRPWKEYSRTVIMQSSITDVIQPAGWHEWN---GNFALDTLFYGEYA 531
Query: 533 NRGPGSDVKQRVKWQGVKTITSE-GAASFVPIRFFHGDDWIRVTRVPYSPG 582
N G G+ RVKW+G K ITS A ++ P RF G W+ T P+S G
Sbjct: 532 NTGAGAPTSGRVKWKGHKVITSSTEAQAYTPGRFIAGGSWLSSTGFPFSLG 582
>PME_PETIN (Q43043) Pectinesterase precursor (EC 3.1.1.11) (Pectin
methylesterase) (PE)
Length = 374
Score = 347 bits (891), Expect = 4e-95
Identities = 181/399 (45%), Positives = 242/399 (60%), Gaps = 28/399 (7%)
Query: 189 MKEVLQTSMHMSSNGLSIINQLSKTFEEMKQPAGRRLLKESVDGEEDVLGHGGDFELPEW 248
M ++L ++ +S N LS++N + + L +S D
Sbjct: 1 MVKLLNSTRELSINALSMLNSFGDMVAQATGLNRKLLTTDSSD----------------- 43
Query: 249 VDDRAGVRKLLNKMTGRKLQAHVVVAKDGSGNFTTITEALKHVPKKNLKPFVIYIKEGVY 308
A R+LL + + VA DGSG + TI EAL VPKKN +PF+I+IK GVY
Sbjct: 44 ----ATARRLLQISNAKP---NATVALDGSGQYKTIKEALDAVPKKNTEPFIIFIKAGVY 96
Query: 309 KEYVEVTKTMTHVVFIGDGGRKTRITGNKNFIDGVGTFKTASVAITGDFFVGIGIGFENS 368
KEY+++ K+MT+VV IG+G KT+ITGNK+ DG TF T +V + G FV IGFEN+
Sbjct: 97 KEYIDIPKSMTNVVLIGEGPTKTKITGNKSVKDGPSTFHTTTVGVNGANFVAKNIGFENT 156
Query: 369 AGPEKHQAVALRVQSDRSIFYKCRMDGYQDTLYAHTMRQFYRDCIISGTIDFVFGDSIAV 428
AGPEK QAVALRV +D++I Y C++DGYQDTLY HT RQFYRDC I+GT+DF+FG+ AV
Sbjct: 157 AGPEKEQAVALRVSADKAIIYNCQIDGYQDTLYVHTYRQFYRDCTITGTVDFIFGNGEAV 216
Query: 429 LQNCTFVVRKPLENQQCIVTAQGRKEKNQPTGLIIQGGSIVADPKYYPVRLKNKAYLARP 488
LQNC +VRKP +NQ C+VTAQGR E Q +++Q I D Y+ + +K YL RP
Sbjct: 217 LQNCKVIVRKPAQNQSCMVTAQGRTEPIQKGAIVLQNCEIKPDTDYFSLSPPSKTYLGRP 276
Query: 489 WKDFSRTIFLDTYIGDMITPEGYMPWQTPAGITGTDTCYYGEYNNRGPGSDVKQRVKWQG 548
WK++SRTI + +YI I PEG+ PW G DT YY EY NRGPG+ + +R+ W+G
Sbjct: 277 WKEYSRTIIMQSYIDKFIEPEGWAPWNITN--FGRDTSYYAEYQNRGPGAALDKRITWKG 334
Query: 549 V-KTITSEGAASFVPIRFFHGDD-WIRVTRVPYSPGATK 585
K T E A F + + D+ W++ VPY G K
Sbjct: 335 FQKGFTGEAAQKFTAGVYINNDENWLQKANVPYEAGMMK 373
>PME_PRUPE (Q43062) Pectinesterase PPE8B precursor (EC 3.1.1.11)
(Pectin methylesterase) (PE)
Length = 522
Score = 332 bits (850), Expect = 2e-90
Identities = 184/468 (39%), Positives = 269/468 (57%), Gaps = 37/468 (7%)
Query: 117 DERTKQALDTCKQVMQLSIDEFQRSLERFSNFD--LNSLDRVLTSLKVWLSGAITYQETC 174
D R A+ C ++ S DE SL N NS ++ + L+ WLS A+ Q+TC
Sbjct: 83 DFRLANAISDCLDLLDFSADELNWSLSASQNQKGKNNSTGKLSSDLRTWLSAALVNQDTC 142
Query: 175 LDAFENTTTDAGKKMKEVLQTSMHMSSNGLSIINQLSKTFEEMKQPAGRRLLKESVDGEE 234
+ FE T + ++Q + S GL + L + P +
Sbjct: 143 SNGFEGTNS--------IVQG---LISAGLGQVTSLVQELLTQVHPNSNQ---------- 181
Query: 235 DVLGHGGDFELPEWVDDRAGVRKLLNKMTGRKLQAHVVVAKDGSGNFTTITEALKHVPKK 294
G + ++P WV + RKLL + +VA+DG+GNFT +T+A+ P
Sbjct: 182 ----QGPNGQIPSWVKTKD--RKLLQ---ADGVSVDAIVAQDGTGNFTNVTDAVLAAPDY 232
Query: 295 NLKPFVIYIKEGVYKEYVEVTKTMTHVVFIGDGGRKTRITGNKNFIDGVGTFKTASVAIT 354
+++ +VIYIK G YKE VE+ K +++ IGDG T I+GN++F+DG TF++A+ A++
Sbjct: 233 SMRRYVIYIKRGTYKENVEIKKKKWNLMMIGDGMDATIISGNRSFVDGWTTFRSATFAVS 292
Query: 355 GDFFVGIGIGFENSAGPEKHQAVALRVQSDRSIFYKCRMDGYQDTLYAHTMRQFYRDCII 414
G F+ I FEN+AGPEKHQAVALR SD S+FY+C + GYQDTLY HTMRQFYRDC I
Sbjct: 293 GRGFIARDITFENTAGPEKHQAVALRSDSDLSVFYRCNIRGYQDTLYTHTMRQFYRDCKI 352
Query: 415 SGTIDFVFGDSIAVLQNCTFVVRKPLENQQCIVTAQGRKEKNQPTGLIIQGGSIVADPKY 474
SGT+DF+FGD+ V QNC + +K L NQ+ +TAQGRK+ N+PTG+ IQ +I AD
Sbjct: 353 SGTVDFIFGDATVVFQNCQILAKKGLPNQKNSITAQGRKDPNEPTGISIQFCNITADSDL 412
Query: 475 YPVRLKN-KAYLARPWKDFSRTIFLDTYIGDMITPEGYMPWQTPAGITGTDTCYYGEYNN 533
+ + YL RPWK +SRT+ + +++ ++I PEG++ W G ++ +YGEY N
Sbjct: 413 EAASVNSTPTYLGRPWKLYSRTVIMQSFLSNVIRPEGWLEWN---GDFALNSLFYGEYMN 469
Query: 534 RGPGSDVKQRVKWQGVKTIT-SEGAASFVPIRFFHGDDWIRVTRVPYS 580
GPG+ + RVKW G + S A ++ +F G+ W+ T V Y+
Sbjct: 470 YGPGAGLGSRVKWPGYQVFNESTQAKNYTVAQFIEGNLWLPSTGVKYT 517
>PME1_FICAW (P83947) Pectinesterase precursor (EC 3.1.1.11) (Pectin
methylesterase) (PE)
Length = 545
Score = 330 bits (846), Expect = 6e-90
Identities = 199/565 (35%), Positives = 296/565 (52%), Gaps = 44/565 (7%)
Query: 20 TFLLVAMVVAVTVNVGFNNKKEPGEETSKESHVSQSVKAVKTLCAPTDYKKECEDSLIAH 79
TF L+ + A V+ GF+ S + +C + K+ C +
Sbjct: 20 TFFLLVISFAALVSTGFS---------------SPELSLHHKICDQSVNKESCLAMISEV 64
Query: 80 AG-NITEPKELIKIAFNITIAKISEGLKKTHLLQEAEKDERTKQALDTCKQVMQLSIDEF 138
G N+ + + L+K T +I + + + + + + AL C ++M LS +
Sbjct: 65 TGLNMADHRNLLKSFLEKTTPRIQKAFETANDASRRINNPQERTALLDCAELMDLSKERV 124
Query: 139 QRSLERFSNFDLNSLDRVLTSLKVWLSGAITYQETCLDAFENTTTDAGKKMKEVLQTSMH 198
S+ F N R L VWLSG +T TCLD E +TD ++T M
Sbjct: 125 VDSISIL--FHQNLTTRSHEDLHVWLSGVLTNHVTCLDGLEEGSTD-------YIKTLME 175
Query: 199 MSSNGLSIINQLSKTFEEMKQPAGRRLLKESVDGEEDVLGHGGDFELPEWVDDRAGVRKL 258
N L + + S PA ++ E V G+ P WV AG R+L
Sbjct: 176 SHLNELILRARTSLAIFVTLFPAKSNVI-------EPVTGN-----FPTWVT--AGDRRL 221
Query: 259 LNKMTGRKLQAHVVVAKDGSGNFTTITEALKHVPKKNLKPFVIYIKEGVYKEYVEVTKTM 318
L + G+ ++ +VVAKDGSG++ T+ EA+ +P + K ++ ++ G+Y+E V+
Sbjct: 222 LQTL-GKDIEPDIVVAKDGSGDYETLNEAVAAIPDNSKKRVIVLVRTGIYEENVDFGYQK 280
Query: 319 THVVFIGDGGRKTRITGNKNFIDGVGTFKTASVAITGDFFVGIGIGFENSAGPEKHQAVA 378
+V+ +G+G T ITG++N +DG TF +A+VA GD F+ I F+N+AGPEK+QAVA
Sbjct: 281 KNVMLVGEGMDYTIITGSRNVVDGSTTFDSATVAAVGDGFIAQDICFQNTAGPEKYQAVA 340
Query: 379 LRVQSDRSIFYKCRMDGYQDTLYAHTMRQFYRDCIISGTIDFVFGDSIAVLQNCTFVVRK 438
LR+ +D ++ +CR+D YQDTLY H RQFYRD I+GT+DF+FG++ V QNC + RK
Sbjct: 341 LRIGADETVINRCRIDAYQDTLYPHNYRQFYRDRNITGTVDFIFGNAAVVFQNCNLIPRK 400
Query: 439 PLENQQCIVTAQGRKEKNQPTGLIIQGGSIVADPKYYPVRLKNKAYLARPWKDFSRTIFL 498
++ Q+ +TAQGR + NQ TG IQ I A PV K+YL RPWK++SRT+ +
Sbjct: 401 QMKGQENTITAQGRTDPNQNTGTSIQNCEIFASADLEPVEDTFKSYLGRPWKEYSRTVVM 460
Query: 499 DTYIGDMITPEGYMPWQTPAGITGTDTCYYGEYNNRGPGSDVKQRVKWQGVKTITS-EGA 557
++YI D+I P G++ W + T +YGEY N GPGS +RVKW G ITS E A
Sbjct: 461 ESYISDVIDPAGWLEWDRDFAL---KTLFYGEYRNGGPGSGTSERVKWPGYHVITSPEVA 517
Query: 558 ASFVPIRFFHGDDWIRVTRVPYSPG 582
F G W+ T V Y+ G
Sbjct: 518 EQFTVAELIQGGSWLGSTGVDYTAG 542
>PME2_CITSI (O04887) Pectinesterase 2 precursor (EC 3.1.1.11)
(Pectin methylesterase) (PE)
Length = 510
Score = 323 bits (827), Expect = 1e-87
Identities = 198/538 (36%), Positives = 290/538 (53%), Gaps = 50/538 (9%)
Query: 50 SHVS--QSVKAVKTLCAPTDYKKECEDSLIAHAG--NITEPKELIKIAFNITIAKISEGL 105
SH S S + VK+ C T + CE L +I + + KI+ + + + +
Sbjct: 17 SHTSFGYSPEEVKSWCGKTPNPQPCEYFLTQKTDVTSIKQDTDFYKISLQLALERATTAQ 76
Query: 106 KKTHLLQEAEKDERTKQALDTCKQVMQLSIDEFQRSLERFSNFDLNSLDRVLTSLKVWLS 165
+T+ L ++ER K A + C+++ +L++ + ++ S+ +D+ + WLS
Sbjct: 77 SRTYTLGSKCRNEREKAAWEDCRELYELTVLKLNQTSN--SSPGCTKVDK-----QTWLS 129
Query: 166 GAITYQETCLDAFENTTTDAGKKMKEVLQTSMHMSSNGLSIINQLSKTFEEMKQPAGRRL 225
A+T ETC + E D G V + + + SN ++ + +S T K P
Sbjct: 130 TALTNLETCRASLE----DLG-----VPEYVLPLLSNNVTKL--ISNTLSLNKVPYNEPS 178
Query: 226 LKESVDGEEDVLGHGGDFELPEWVDDRAGVRKLLNKMTGRKLQAHVVVAKDGSGNFTTIT 285
K+ P WV + G RKLL +A++VVA+DGSGN TI
Sbjct: 179 YKDG---------------FPTWV--KPGDRKLLQTTP----RANIVVAQDGSGNVKTIQ 217
Query: 286 EALKHVPKKNLKPFVIYIKEGVYKEYVEVTKTMTHVVFIGDGGRKTRITGNKNFIDGVGT 345
EA+ + +VIYIK G Y E +EV + +++F+GDG KT ITG+K+ G T
Sbjct: 218 EAVAAASRAGGSRYVIYIKAGTYNENIEVK--LKNIMFVGDGIGKTIITGSKSVGGGATT 275
Query: 346 FKTASVAITGDFFVGIGIGFENSAGPEKHQAVALRVQSDRSIFYKCRMDGYQDTLYAHTM 405
FK+A+VA+ GD F+ I N+AGP HQAVALR SD S+FY+C +GYQDTLY H+
Sbjct: 276 FKSATVAVVGDNFIARDITIRNTAGPNNHQAVALRSGSDLSVFYRCSFEGYQDTLYVHSQ 335
Query: 406 RQFYRDCIISGTIDFVFGDSIAVLQNCTFVVRKPLENQQCIVTAQGRKEKNQPTGLIIQG 465
RQFYR+C I GT+DF+FG++ VLQNC RKP N+ +TAQGR + NQ TG+II
Sbjct: 336 RQFYRECDIYGTVDFIFGNAAVVLQNCNIFARKP-PNRTNTLTAQGRTDPNQSTGIIIHN 394
Query: 466 GSIVADPKYYPVRLKNKAYLARPWKDFSRTIFLDTYIGDMITPEGYMPWQTPAGITGTDT 525
+ A PV+ K +L RPWK +SRT+++ T++ +I P G+M W +G +T
Sbjct: 395 CRVTAASDLKPVQSSVKTFLGRPWKQYSRTVYIKTFLDSLINPAGWMEW---SGDFALNT 451
Query: 526 CYYGEYNNRGPGSDVKQRVKWQGVKTITSEGAAS-FVPIRFFHGDDWIRVTRVPYSPG 582
YY EY N GPGS RVKW+G +TS S F F G+ W+ T VP++ G
Sbjct: 452 LYYAEYMNTGPGSSTANRVKWRGYHVLTSPSQVSQFTVGNFIAGNSWLPATNVPFTSG 509
>PM21_LYCES (P09607) Pectinesterase 2 precursor (EC 3.1.1.11)
(Pectin methylesterase 2) (PE 2)
Length = 550
Score = 319 bits (818), Expect = 1e-86
Identities = 185/487 (37%), Positives = 268/487 (54%), Gaps = 33/487 (6%)
Query: 97 TIAKISEGLKKTHLLQEAEKDERTKQALDTCKQVMQLSIDEFQRSLERFSNFDLNSLDRV 156
++ +++ + ++ D R + AL C +++ LS+D S+ +
Sbjct: 93 SVHQMNNAIPVVRKIKNQINDIREQGALTDCLELLDLSVDLVCDSIAAIDKRSRSEH--- 149
Query: 157 LTSLKVWLSGAITYQETCLDAFENTTTDAGKKMKEVLQTSMHMSSNGLSIINQLSKTFEE 216
+ + WLSG +T TCLD ++ T K ++ NG ++ +S+
Sbjct: 150 -ANAQSWLSGVLTNHVTCLDELDSFT-------KAMI--------NGTNLDELISRAKVA 193
Query: 217 MKQPAGRRLLKESVDGEEDVLGHGGDFELPEWVDDRAGVRKLLNKMTGRKLQAHVVVAKD 276
+ A SV D + G ++P WV R RKL+ +G+ + A+ VVAKD
Sbjct: 194 LAMLA-------SVTTPNDEVLRPGLGKMPSWVSSRD--RKLMES-SGKDIGANAVVAKD 243
Query: 277 GSGNFTTITEALKHVPKKNLKPFVIYIKEGVYKEYVEVTKTMTHVVFIGDGGRKTRITGN 336
G+G + T+ EA+ P K+ +VIY+K G YKE VEV+ +++ IGDG T ITG+
Sbjct: 244 GTGKYRTLAEAVAAAPDKSKTRYVIYVKRGTYKENVEVSSRKMNLMIIGDGMYATIITGS 303
Query: 337 KNFIDGVGTFKTASVAITGDFFVGIGIGFENSAGPEKHQAVALRVQSDRSIFYKCRMDGY 396
N +DG TF +A++A G F+ I +N+AGP KHQAVALRV +D+S+ +CR+D Y
Sbjct: 304 LNVVDGSTTFHSATLAAVGKGFILQDICIQNTAGPAKHQAVALRVGADKSVINRCRIDAY 363
Query: 397 QDTLYAHTMRQFYRDCIISGTIDFVFGDSIAVLQNCTFVVRKPLENQQCIVTAQGRKEKN 456
QDTLYAH+ RQFYRD ++GTIDF+FG++ V Q C V RKP + QQ +VTAQGR + N
Sbjct: 364 QDTLYAHSQRQFYRDSYVTGTIDFIFGNAAVVFQKCQLVARKPGKYQQNMVTAQGRTDPN 423
Query: 457 QPTGLIIQGGSIVADPKYYPVRLKNKAYLARPWKDFSRTIFLDTYIGDMITPEGYMPWQT 516
Q TG IQ I+A P PV + YL RPWK +SRT+ +++Y+G +I P G+ W
Sbjct: 424 QATGTSIQFCDIIASPDLKPVVKEFPTYLGRPWKKYSRTVVMESYLGGLIDPSGWAEWH- 482
Query: 517 PAGITGTDTCYYGEYNNRGPGSDVKQRVKWQGVKTITSEG-AASFVPIRFFHGDDWIRVT 575
G T YYGE+ N GPG+ +RVKW G IT A SF + G W+R T
Sbjct: 483 --GDFALKTLYYGEFMNNGPGAGTSKRVKWPGYHVITDPAEAMSFTVAKLIQGGSWLRST 540
Query: 576 RVPYSPG 582
V Y G
Sbjct: 541 DVAYVDG 547
>PM22_LYCES (Q96575) Pectinesterase 2 precursor (EC 3.1.1.11)
(Pectin methylesterase 2) (PE 2)
Length = 550
Score = 315 bits (807), Expect = 2e-85
Identities = 182/467 (38%), Positives = 258/467 (54%), Gaps = 33/467 (7%)
Query: 117 DERTKQALDTCKQVMQLSIDEFQRSLERFSNFDLNSLDRVLTSLKVWLSGAITYQETCLD 176
D R + AL C +++ S+D S+ + + + WLSG +T TCLD
Sbjct: 113 DIRQEGALTDCLELLDQSVDLVSDSIAAIDKRTHSEH----ANAQSWLSGVLTNHVTCLD 168
Query: 177 AFENTTTDAGKKMKEVLQTSMHMSSNGLSIINQLSKTFEEMKQPAGRRLLKESVDGEEDV 236
++ T K ++ NG ++ +S+ +L +DV
Sbjct: 169 ELDSFT-------KAMI--------NGTNLDELISRA------KVALAMLASVTTPNDDV 207
Query: 237 LGHGGDFELPEWVDDRAGVRKLLNKMTGRKLQAHVVVAKDGSGNFTTITEALKHVPKKNL 296
L G ++P WV R RKL+ +G+ + A+ VVAKDG+G + T+ EA+ P K+
Sbjct: 208 LRPGLG-KMPSWVSSRD--RKLMES-SGKDIGANAVVAKDGTGKYRTLAEAVAAAPDKSK 263
Query: 297 KPFVIYIKEGVYKEYVEVTKTMTHVVFIGDGGRKTRITGNKNFIDGVGTFKTASVAITGD 356
+VIY+K G+YKE VEV+ ++ +GDG T ITGN N +DG TF +A++A G
Sbjct: 264 TRYVIYVKRGIYKENVEVSSRKMKLMIVGDGMHATIITGNLNVVDGSTTFHSATLAAVGK 323
Query: 357 FFVGIGIGFENSAGPEKHQAVALRVQSDRSIFYKCRMDGYQDTLYAHTMRQFYRDCIISG 416
F+ I +N+AGP KHQAVALRV +D+S+ +CR+D YQDTLYAH+ RQFYRD ++G
Sbjct: 324 GFILQDICIQNTAGPAKHQAVALRVGADKSVINRCRIDAYQDTLYAHSQRQFYRDSYVTG 383
Query: 417 TIDFVFGDSIAVLQNCTFVVRKPLENQQCIVTAQGRKEKNQPTGLIIQGGSIVADPKYYP 476
TIDF+FG++ V Q C V RKP + QQ +VTAQGR + NQ TG IQ +I+A P
Sbjct: 384 TIDFIFGNAAVVFQKCKLVARKPGKYQQNMVTAQGRTDPNQATGTSIQFCNIIASSDLEP 443
Query: 477 VRLKNKAYLARPWKDFSRTIFLDTYIGDMITPEGYMPWQTPAGITGTDTCYYGEYNNRGP 536
V + YL RPWK +SRT+ +++Y+G +I P G+ W G T YYGE+ N GP
Sbjct: 444 VLKEFPTYLGRPWKKYSRTVVMESYLGGLINPAGWAEWD---GDFALKTLYYGEFMNNGP 500
Query: 537 GSDVKQRVKWQGVKTITSEG-AASFVPIRFFHGDDWIRVTRVPYSPG 582
G+ +RVKW G IT A F + G W+R T V Y G
Sbjct: 501 GAGTSKRVKWPGYHCITDPAEAMPFTVAKLIQGGSWLRSTGVAYVDG 547
>PME1_LYCES (P14280) Pectinesterase 1 precursor (EC 3.1.1.11)
(Pectin methylesterase 1) (PE 1)
Length = 546
Score = 308 bits (790), Expect = 2e-83
Identities = 183/499 (36%), Positives = 273/499 (54%), Gaps = 42/499 (8%)
Query: 89 LIKIAFNITIAKISEGLKKTHLLQEAEKDERTKQALDTCKQVMQLSIDEFQRSLERFSNF 148
L+K N + +++ + ++ D R AL C +++ S+D F++
Sbjct: 82 LMKFLVNY-VHQMNNAIPVVRKMKNQINDIRQHGALTDCLELLDQSVD--------FASD 132
Query: 149 DLNSLDRVLTS----LKVWLSGAITYQETCLDAFENTTTDAGKKMKEVLQTSMHMSSNGL 204
+ ++D+ S + WLSG +T TCLD ++ T K ++ NG
Sbjct: 133 SIAAIDKRSRSEHANAQSWLSGVLTNHVTCLDELDSFT-------KAMI--------NGT 177
Query: 205 SIINQLSKTFEEMKQPAGRRLLKESVDGEEDVLGHGGDFELPEWVDDRAGVRKLLNKMTG 264
++ +S+ + A L + D VLG ++P WV RKL+ +G
Sbjct: 178 NLEELISRAKVALAMLAS--LTTQDEDVFMTVLG-----KMPSWVSSMD--RKLMES-SG 227
Query: 265 RKLQAHVVVAKDGSGNFTTITEALKHVPKKNLKPFVIYIKEGVYKEYVEVTKTMTHVVFI 324
+ + A+ VVA+DG+G++ T+ EA+ P K+ +VIY+K G YKE VEV +++ +
Sbjct: 228 KDIIANAVVAQDGTGDYQTLAEAVAAAPDKSKTRYVIYVKRGTYKENVEVASNKMNLMIV 287
Query: 325 GDGGRKTRITGNKNFIDGVGTFKTASVAITGDFFVGIGIGFENSAGPEKHQAVALRVQSD 384
GDG T ITG+ N +DG TF++A++A G F+ I +N+AGP K QAVALRV +D
Sbjct: 288 GDGMYATTITGSLNVVDGSTTFRSATLAAVGQGFILQDICIQNTAGPAKDQAVALRVGAD 347
Query: 385 RSIFYKCRMDGYQDTLYAHTMRQFYRDCIISGTIDFVFGDSIAVLQNCTFVVRKPLENQQ 444
S+ +CR+D YQDTLYAH+ RQFYRD ++GT+DF+FG++ V Q C V RKP + QQ
Sbjct: 348 MSVINRCRIDAYQDTLYAHSQRQFYRDSYVTGTVDFIFGNAAVVFQKCQLVARKPGKYQQ 407
Query: 445 CIVTAQGRKEKNQPTGLIIQGGSIVADPKYYPVRLKNKAYLARPWKDFSRTIFLDTYIGD 504
+VTAQGR + NQ TG IQ +I+A PV + YL RPWK++SRT+ +++Y+G
Sbjct: 408 NMVTAQGRTDPNQATGTSIQFCNIIASSDLEPVLKEFPTYLGRPWKEYSRTVVMESYLGG 467
Query: 505 MITPEGYMPWQTPAGITGTDTCYYGEYNNRGPGSDVKQRVKWQGVKTITSEG-AASFVPI 563
+I P G+ W G T YYGE+ N GPG+ +RVKW G IT A F
Sbjct: 468 LINPAGWAEWD---GDFALKTLYYGEFMNNGPGAGTSKRVKWPGYHVITDPAKAMPFTVA 524
Query: 564 RFFHGDDWIRVTRVPYSPG 582
+ G W+R T V Y G
Sbjct: 525 KLIQGGSWLRSTGVAYVDG 543
>PME3_LYCES (Q96576) Pectinesterase 3 precursor (EC 3.1.1.11)
(Pectin methylesterase 3) (PE 3)
Length = 544
Score = 303 bits (776), Expect = 8e-82
Identities = 177/467 (37%), Positives = 253/467 (53%), Gaps = 34/467 (7%)
Query: 117 DERTKQALDTCKQVMQLSIDEFQRSLERFSNFDLNSLDRVLTSLKVWLSGAITYQETCLD 176
D R + AL C +++ S+D S+ + + + WLSG +T TCLD
Sbjct: 108 DIRQQGALTDCLELLDQSVDLVSDSIAAIDKRSRSEH----ANAQSWLSGVLTNHVTCLD 163
Query: 177 AFENTTTDAGKKMKEVLQTSMHMSSNGLSIINQLSKTFEEMKQPAGRRLLKESVDGEEDV 236
TS +S+ +++++L + +L ++V
Sbjct: 164 EL----------------TSFSLSTKNGTVLDELIT-----RAKVALAMLASVTTPNDEV 202
Query: 237 LGHGGDFELPEWVDDRAGVRKLLNKMTGRKLQAHVVVAKDGSGNFTTITEALKHVPKKNL 296
L G ++P WV R RKL+ +G+ + A+ VVA+DG+G++ T+ EA+ P KN
Sbjct: 203 LRQGLG-KMPYWVSSRD--RKLMES-SGKDIIANRVVAQDGTGDYQTLAEAVAAAPDKNK 258
Query: 297 KPFVIYIKEGVYKEYVEVTKTMTHVVFIGDGGRKTRITGNKNFIDGVGTFKTASVAITGD 356
+VIY+K G+YKE V VTK +++ +GDG T ITG+ N +DG TF + ++A G
Sbjct: 259 TRYVIYVKMGIYKENVVVTKKKMNLMIVGDGMNATIITGSLNVVDG-STFPSNTLAAVGQ 317
Query: 357 FFVGIGIGFENSAGPEKHQAVALRVQSDRSIFYKCRMDGYQDTLYAHTMRQFYRDCIISG 416
F+ I +N+AGPEK QAVALRV +D S+ +CR+D YQDTLYAH+ RQFYRD ++G
Sbjct: 318 GFILQDICIQNTAGPEKDQAVALRVGADMSVINRCRIDAYQDTLYAHSQRQFYRDSYVTG 377
Query: 417 TIDFVFGDSIAVLQNCTFVVRKPLENQQCIVTAQGRKEKNQPTGLIIQGGSIVADPKYYP 476
T+DF+FG++ V Q C V RKP + Q+ +VTAQGR + NQ TG IQ I+A P P
Sbjct: 378 TVDFIFGNAAVVFQKCQIVARKPNKRQKNMVTAQGRTDPNQATGTSIQFCDIIASPDLEP 437
Query: 477 VRLKNKAYLARPWKDFSRTIFLDTYIGDMITPEGYMPWQTPAGITGTDTCYYGEYNNRGP 536
V + K YL RPWK SRT+ + +Y+ I P G+ W+ G T YYGE+ N GP
Sbjct: 438 VMNEYKTYLGRPWKKHSRTVVMQSYLDGHIDPSGWFEWR---GDFALKTLYYGEFMNNGP 494
Query: 537 GSDVKQRVKWQGVKTITSEG-AASFVPIRFFHGDDWIRVTRVPYSPG 582
G+ +RVKW G IT A F G W+ T V Y G
Sbjct: 495 GAGTSKRVKWPGYHVITDPNEAMPFTVAELIQGGSWLNSTSVAYVEG 541
>PME_BRANA (P41510) Probable pectinesterase precursor (EC 3.1.1.11)
(Pectin methylesterase) (PE)
Length = 584
Score = 296 bits (759), Expect = 8e-80
Identities = 204/593 (34%), Positives = 299/593 (50%), Gaps = 41/593 (6%)
Query: 15 IIGVSTFLLVAMVVAVTVNVGFNNKKEPGEETSKESHVSQSVKAVKTLCAPTDYKKECED 74
+I V++ LLV + VA+ V N G + + SH KAV++LCA K C
Sbjct: 7 VISVASMLLV-VGVAIGVVTFVNKGGGAGGDKTLNSHQ----KAVESLCASATDKGSCAK 61
Query: 75 SLIAHAGNITEPKELIKIAFNITIAKISEGLKKTHLLQEA---EKDERTKQALDTCKQVM 131
+L + +P +LIK T +++ T +E + +K LD CK+V+
Sbjct: 62 TLDPVKSD--DPSKLIKAFMLATKDAVTKSTNFTASTEEGMGKNINATSKAVLDYCKRVL 119
Query: 132 QLSIDEFQRSLERFSNFDLNSLDRVLTSLKVWLSGAITYQETCLDAFENTTTDAGKKMKE 191
++++ + +E DL + LK WL+G YQ C+D E + + K M E
Sbjct: 120 MYALEDLETIVEEMGE-DLQQSGSKMDQLKQWLTGVFNYQTDCIDDIEES--ELRKVMGE 176
Query: 192 VLQTSMHMSSNGLSIINQLSKTFEEMKQPAG---RRLLKESVDGEEDVLGHGGDFELPEW 248
+ S +SSN + I + L+ +M + L E+ + D+L LP+W
Sbjct: 177 GIAHSKILSSNAIDIFHALTTAMSQMNVKVDDMKKGNLGETPAPDRDLLEDLDQKGLPKW 236
Query: 249 VDDRAGVRKLLNKMT----------------GRKLQAHVVVAKDGSGNFTTITEALKHVP 292
D+ RKL+ + G K++ VVAKDGSG F TI+EA+K P
Sbjct: 237 HSDKD--RKLMAQAGRPGAPADEGIGEGGGGGGKIKPTHVVAKDGSGQFKTISEAVKACP 294
Query: 293 KKNLKPFVIYIKEGVYKEYVEVTKTMTHVVFIGDGGRKTRITGNKN--FIDGVGTFKTAS 350
+KN +IYIK GVYKE V + K + +V GDG +T IT +++ G T + +
Sbjct: 295 EKNPGRCIIYIKAGVYKEQVTIPKKVNNVFMFGDGATQTIITFDRSVGLSPGTTTSLSGT 354
Query: 351 VAITGDFFVGIGIGFENSAGPEKHQAVALRVQSDRSIFYKCRMDGYQDTLYAHTMRQFYR 410
V + + F+ IGF+N+AGP HQAVA RV DR++ + CR DGYQDTLY + RQFYR
Sbjct: 355 VQVESEGFMAKWIGFQNTAGPLGHQAVAFRVNGDRAVIFNCRFDGYQDTLYVNNGRQFYR 414
Query: 411 DCIISGTIDFVFGDSIAVLQNCTFVVRKPLENQQCIVTAQG-RKEKNQPTGLIIQGGSIV 469
+ ++SGT+DF+FG S V+QN + RK Q VTA G K K G+++ I+
Sbjct: 415 NIVVSGTVDFIFGKSATVIQNSLILCRKGSPGQTNHVTADGNEKGKAVKIGIVLHNCRIM 474
Query: 470 ADPKYYPVRLKNKAYLARPWKDFSRTIFLDTYIGDMITPEGYMPWQTPAGITGTDTCYYG 529
AD + RL K+YL RPWK F+ T + T IGD+I P G+ WQ G T Y
Sbjct: 475 ADKELEADRLTVKSYLGRPWKPFATTAVIGTEIGDLIQPTGWNEWQ---GEKFHLTATYV 531
Query: 530 EYNNRGPGSDVKQRVKWQGVKTITSEGAASFVPIRFFHGDDWIRVTRVPYSPG 582
E+NNRGPG++ RV W + +E F + +WI+ VP G
Sbjct: 532 EFNNRGPGANTAARVPWAKMAKSAAE-VERFTVANWLTPANWIQEANVPVQLG 583
>PME_BRACM (Q42608) Pectinesterase (EC 3.1.1.11) (Pectin
methylesterase) (PE) (Fragment)
Length = 571
Score = 286 bits (731), Expect = 1e-76
Identities = 197/585 (33%), Positives = 293/585 (49%), Gaps = 40/585 (6%)
Query: 23 LVAMVVAVTVNVGFNNKKEPGEETSKESHVSQSVKAVKTLCAPTDYKKECEDSLIAHAGN 82
L+ + VA+ V N G + + SH KAV++LCA K C +L +
Sbjct: 1 LLVVGVAIGVVTFVNKGGGAGGDKTLNSHQ----KAVESLCASATDKGSCAKTLDPVKSD 56
Query: 83 ITEPKELIKIAFNITIAKISEGLKKTHLLQEA---EKDERTKQALDTCKQVMQLSIDEFQ 139
+P +LIK T +++ T +E + +K LD CK+V+ ++++ +
Sbjct: 57 --DPSKLIKAFMLATKDAVTKSTNFTASTEEGMGKNMNATSKAVLDYCKRVLMYALEDLE 114
Query: 140 RSLERFSNFDLNSLDRVLTSLKVWLSGAITYQETCLDAFENTTTDAGKKMKEVLQTSMHM 199
+E DL + LK WL+G YQ C+D E + + K M E ++ S +
Sbjct: 115 TIVEEMGE-DLQQSGSKMDQLKQWLTGVFNYQTDCIDDIEES--ELRKVMGEGIRHSKIL 171
Query: 200 SSNGLSIINQLSKTFEEMKQPAG---RRLLKESVDGEEDVLGHGGDFELPEWVDDRAGVR 256
SSN + I + L+ +M + L E+ + D+L LP+W D+ R
Sbjct: 172 SSNAIDIFHALTTAMSQMNVKVDDMKKGNLGETPAPDRDLLEDLDQKGLPKWHSDKD--R 229
Query: 257 KLLNKMT----------------GRKLQAHVVVAKDGSGNFTTITEALKHVPKKNLKPFV 300
KL+ + G K++ VVAKDGSG F TI+EA+K P+KN +
Sbjct: 230 KLMAQAGRPGAPADEGIGEGGGGGGKIKPTHVVAKDGSGQFKTISEAVKACPEKNPGRCI 289
Query: 301 IYIKEGVYKEYVEVTKTMTHVVFIGDGGRKTRITGNKN--FIDGVGTFKTASVAITGDFF 358
IYIK GVYKE V + K + +V GDG +T IT +++ G T + +V + + F
Sbjct: 290 IYIKAGVYKEQVTIPKKVNNVFMFGDGATQTIITFDRSVGLSPGTTTSLSGTVQVESEGF 349
Query: 359 VGIGIGFENSAGPEKHQAVALRVQSDRSIFYKCRMDGYQDTLYAHTMRQFYRDCIISGTI 418
+ IGF+N+AGP +QAVA RV DR++ + CR DGYQDTLY + RQFYR+ ++SGT+
Sbjct: 350 MAKWIGFQNTAGPLGNQAVAFRVNGDRAVIFNCRFDGYQDTLYVNNGRQFYRNIVVSGTV 409
Query: 419 DFVFGDSIAVLQNCTFVVRKPLENQQCIVTAQGR-KEKNQPTGLIIQGGSIVADPKYYPV 477
DF+ G S V+QN + RK Q VTA G+ K K G+++ I+AD +
Sbjct: 410 DFINGKSATVIQNSLILCRKGSPGQTNHVTADGKQKGKAVKIGIVLHNCRIMADKELEAD 469
Query: 478 RLKNKAYLARPWKDFSRTIFLDTYIGDMITPEGYMPWQTPAGITGTDTCYYGEYNNRGPG 537
RL K+YL RPWK F+ T + T IGD+I P G+ WQ G T Y E+NNRGPG
Sbjct: 470 RLTVKSYLGRPWKPFATTAVIGTEIGDLIQPTGWNEWQ---GEKFHLTATYVEFNNRGPG 526
Query: 538 SDVKQRVKWQGVKTITSEGAASFVPIRFFHGDDWIRVTRVPYSPG 582
++ RV W + +E F + +WI+ V G
Sbjct: 527 ANPAARVPWAKMAKSAAE-VERFTVANWLTPANWIQEANVTVQLG 570
>PME_DAUCA (P83218) Pectinesterase (EC 3.1.1.11) (Pectin
methylesterase) (PE)
Length = 319
Score = 283 bits (724), Expect = 9e-76
Identities = 145/314 (46%), Positives = 198/314 (62%), Gaps = 4/314 (1%)
Query: 270 HVVVAKDGSGNFTTITEALKHVPKKNLKPFVIYIKEGVYKEYVEVTKTMTHVVFIGDGGR 329
+VVVA DGSG++ T++EA+ P+ + +VI IK GVY+E V+V K +++F+GDG
Sbjct: 8 NVVVAADGSGDYKTVSEAVAAAPEDSKTRYVIRIKAGVYRENVDVPKKKKNIMFLGDGRT 67
Query: 330 KTRITGNKNFIDGVGTFKTASVAITGDFFVGIGIGFENSAGPEKHQAVALRVQSDRSIFY 389
T IT +KN DG TF +A+VA G F+ I F+N+AG KHQAVALRV SD S FY
Sbjct: 68 STIITASKNVQDGSTTFNSATVAAVGAGFLARDITFQNTAGAAKHQAVALRVGSDLSAFY 127
Query: 390 KCRMDGYQDTLYAHTMRQFYRDCIISGTIDFVFGDSIAVLQNCTFVVRKPLENQQCIVTA 449
+C + YQD+LY H+ RQF+ +C I+GT+DF+FG++ VLQ+C R+P Q+ +VTA
Sbjct: 128 RCDILAYQDSLYVHSNRQFFINCFIAGTVDFIFGNAAVVLQDCDIHARRPGSGQKNMVTA 187
Query: 450 QGRKEKNQPTGLIIQGGSIVADPKYYPVRLKNKAYLARPWKDFSRTIFLDTYIGDMITPE 509
QGR + NQ TG++IQ I A PV+ YL RPWK++SRT+ + + I ++I P
Sbjct: 188 QGRTDPNQNTGIVIQKSRIGATSDLQPVQSSFPTYLGRPWKEYSRTVVMQSSITNVINPA 247
Query: 510 GYMPWQTPAGITGTDTCYYGEYNNRGPGSDVKQRVKWQGVKTITSE-GAASFVPIRFFHG 568
G+ PW G DT YYGEY N G G+ RV W+G K ITS A F P F G
Sbjct: 248 GWFPWD---GNFALDTLYYGEYQNTGAGAATSGRVTWKGFKVITSSTEAQGFTPGSFIAG 304
Query: 569 DDWIRVTRVPYSPG 582
W++ T P+S G
Sbjct: 305 GSWLKATTFPFSLG 318
>PME_ASPAC (Q12535) Pectinesterase precursor (EC 3.1.1.11) (Pectin
methylesterase) (PE)
Length = 331
Score = 112 bits (281), Expect = 2e-24
Identities = 85/279 (30%), Positives = 128/279 (45%), Gaps = 18/279 (6%)
Query: 271 VVVAKDGSGNFTTITEALKHVPKKNLKPFVIYIKEGVYKEYVEVTKTMTHVVFIGDGGRK 330
+VVAK G G++TTI +A+ + I+I+EG Y E V + V+ G
Sbjct: 28 IVVAKSG-GDYTTIGDAIDALSTSTTDTQTIFIEEGTYDEQVYLPAMTGKVIIYGQTENT 86
Query: 331 TRITGNKNFIDGVGTFKTASVA--ITGDF---FVG---IGIGFENSAGPEKHQAVALRVQ 382
N I +++ A + +T F VG + N+ G HQA+AL
Sbjct: 87 DSYADNLVTITHAISYEDAGESDDLTATFRNKAVGSQVYNLNIANTCGQACHQALALSAW 146
Query: 383 SDRSIFYKCRMDGYQDTLYAHTMRQFYRDCIISGTIDFVFGD-SIAVLQNCTF-VVRKPL 440
+D+ +Y C GYQDTL A T Q Y + I G +DF+FG + A QN VV P
Sbjct: 147 ADQQGYYGCNFTGYQDTLLAQTGNQLYINSYIEGAVDFIFGQHARAWFQNVDIRVVEGP- 205
Query: 441 ENQQCIVTAQGRKEKNQPTGLIIQGGSIVADPKYYPVRLKNKAYLARPWKDFSRTIFLDT 500
+TA GR + + +I ++ A K + YL RPW +++R +F T
Sbjct: 206 --TSASITANGRSSETDTSYYVINKSTVAA--KEGDDVAEGTYYLGRPWSEYARVVFQQT 261
Query: 501 YIGDMITPEGYMPWQTPAGITGTDTCYYGEYNNRGPGSD 539
+ ++I G+ W T T+ +GEY N G GS+
Sbjct: 262 SMTNVINSLGWTEWST--STPNTEYVTFGEYANTGAGSE 298
>PME_ASPTU (P17872) Pectinesterase precursor (EC 3.1.1.11) (Pectin
methylesterase) (PE)
Length = 331
Score = 102 bits (254), Expect = 3e-21
Identities = 83/310 (26%), Positives = 130/310 (41%), Gaps = 25/310 (8%)
Query: 271 VVVAKDGSGNFTTITEALKHVPKKNLKPFVIYIKEGVYKEYVEVTKTMTHVVFIGDGGRK 330
+VVAK G G++ TI+ A+ + + + I+I+EG Y E V + ++ G
Sbjct: 28 IVVAKSG-GDYDTISAAVDALSTTSTETQTIFIEEGSYDEQVYIPALSGKLIVYGQTEDT 86
Query: 331 TRITGNKNFI-------DGVGTFKTASVAITGDFFVGIGIGFENSAGPEKHQAVALRVQS 383
T T N I D +TA++ + + N+ G HQA+A+ +
Sbjct: 87 TTYTSNLVNITHAIALADVDNDDETATLRNYAEGSAIYNLNIANTCGQACHQALAVSAYA 146
Query: 384 DRSIFYKCRMDGYQDTLYAHTMRQFYRDCIISGTIDFVFGD-SIAVLQNCTF-VVRKPLE 441
+Y C+ GYQDTL A T Q Y I G +DF+FG + A C V+ P
Sbjct: 147 SEQGYYACQFTGYQDTLLAETGYQVYAGTYIEGAVDFIFGQHARAWFHECDIRVLEGP-- 204
Query: 442 NQQCIVTAQGRKEKNQPTGLIIQGGSIVADPKYYPVRLKNKAYLARPWKDFSRTIFLDTY 501
+TA GR ++ + +I ++ A YL RPW ++R F T
Sbjct: 205 -SSASITANGRSSESDDSYYVIHKSTVAAADGNDV--SSGTYYLGRPWSQYARVCFQKTS 261
Query: 502 IGDMITPEGYMPWQTPAGITGTDTCYYGEYNNRGPGSDVKQRVKWQGVKTITSEGAASFV 561
+ D+I G+ W T T+ + EY N G G+ +G + S +
Sbjct: 262 MTDVINHLGWTEWST--STPNTENVTFVEYGNTGTGA--------EGPRANFSSELTEPI 311
Query: 562 PIRFFHGDDW 571
I + G DW
Sbjct: 312 TISWLLGSDW 321
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.318 0.135 0.396
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 71,080,304
Number of Sequences: 164201
Number of extensions: 3102721
Number of successful extensions: 7950
Number of sequences better than 10.0: 54
Number of HSP's better than 10.0 without gapping: 23
Number of HSP's successfully gapped in prelim test: 31
Number of HSP's that attempted gapping in prelim test: 7845
Number of HSP's gapped (non-prelim): 63
length of query: 593
length of database: 59,974,054
effective HSP length: 116
effective length of query: 477
effective length of database: 40,926,738
effective search space: 19522054026
effective search space used: 19522054026
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 69 (31.2 bits)
Medicago: description of AC144541.15


