

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144541.11 - phase: 0
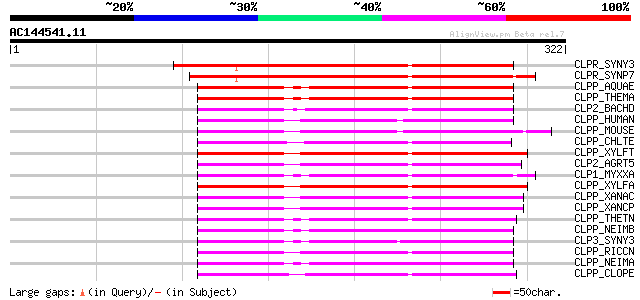
(322 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
CLPR_SYNY3 (P74466) Putative ATP-dependent Clp protease proteoly... 168 1e-41
CLPR_SYNP7 (Q9L4P4) Putative ATP-dependent Clp protease proteoly... 167 4e-41
CLPP_AQUAE (O67357) ATP-dependent Clp protease proteolytic subun... 137 5e-32
CLPP_THEMA (Q9WZF9) ATP-dependent Clp protease proteolytic subun... 133 5e-31
CLP2_BACHD (Q9K888) ATP-dependent Clp protease proteolytic subun... 127 3e-29
CLPP_HUMAN (Q16740) Putative ATP-dependent Clp protease proteoly... 125 2e-28
CLPP_MOUSE (O88696) Putative ATP-dependent Clp protease proteoly... 124 4e-28
CLPP_CHLTE (Q8KC73) ATP-dependent Clp protease proteolytic subun... 122 9e-28
CLPP_XYLFT (Q87E51) ATP-dependent Clp protease proteolytic subun... 121 2e-27
CLP2_AGRT5 (Q8UFY6) ATP-dependent Clp protease proteolytic subun... 121 3e-27
CLP1_MYXXA (O30612) ATP-dependent Clp protease proteolytic subun... 121 3e-27
CLPP_XYLFA (Q9PE41) ATP-dependent Clp protease proteolytic subun... 120 3e-27
CLPP_XANAC (Q8PNI5) ATP-dependent Clp protease proteolytic subun... 120 5e-27
CLPP_XANCP (Q8PBY6) ATP-dependent Clp protease proteolytic subun... 119 8e-27
CLPP_THETN (Q8RC25) ATP-dependent Clp protease proteolytic subun... 119 1e-26
CLPP_NEIMB (Q9JZ38) ATP-dependent Clp protease proteolytic subun... 119 1e-26
CLP3_SYNY3 (P74467) Probable ATP-dependent Clp protease proteoly... 119 1e-26
CLPP_RICCN (Q92HM5) ATP-dependent Clp protease proteolytic subun... 117 4e-26
CLPP_NEIMA (Q9JU33) ATP-dependent Clp protease proteolytic subun... 117 5e-26
CLPP_CLOPE (Q8XKK1) ATP-dependent Clp protease proteolytic subun... 117 5e-26
>CLPR_SYNY3 (P74466) Putative ATP-dependent Clp protease proteolytic
subunit-like (Endopeptidase Clp-like)
Length = 225
Score = 168 bits (426), Expect = 1e-41
Identities = 88/207 (42%), Positives = 135/207 (64%), Gaps = 12/207 (5%)
Query: 96 ASYSQQRRPKRPPPDLPSLLLNGRIVYIGMPLVPA----------VTELVIAELMFLQWM 145
+SY K PPPDL SLLL RIVY+GMPL + VT+L+IA+L++LQ+
Sbjct: 8 SSYYGDMAFKTPPPDLESLLLKERIVYLGMPLFSSDEVKQQVGIDVTQLIIAQLLYLQFD 67
Query: 146 APKEPIYIYINSTGTTRADGETVAMESEGFAIYDAMMQMKTEINTVALGAAVGQACLLLS 205
P +PIY YINSTGT+ G+ V E+E FAI D + +K ++T+ +G A+G A ++LS
Sbjct: 68 DPDKPIYFYINSTGTSWYTGDAVGFETEAFAICDTLNYIKPPVHTICIGQAMGTAAMILS 127
Query: 206 AGTKGRRYMTPHAKAMIQQPRVPSSGLRPASDVLIHAKEVMVNRDTLVKLLAKHTENSEE 265
+GTKG R PHA ++ Q R + G A+D+ I AKEV+ N+ T++++L+ +T ++E
Sbjct: 128 SGTKGYRASLPHATIVLNQNRTGAQG--QATDIQIRAKEVISNKQTMLEILSLNTGQTQE 185
Query: 266 TVSNVMKRSYYMDALLAKEFGVIDKIL 292
++ M R++Y+ AKE+G+ID++L
Sbjct: 186 KLAKDMDRTFYLTPAQAKEYGLIDRVL 212
>CLPR_SYNP7 (Q9L4P4) Putative ATP-dependent Clp protease proteolytic
subunit-like (Endopeptidase Clp-like)
Length = 228
Score = 167 bits (422), Expect = 4e-41
Identities = 87/211 (41%), Positives = 134/211 (63%), Gaps = 13/211 (6%)
Query: 105 KRPPPDLPSLLLNGRIVYIGMPLVPA----------VTELVIAELMFLQWMAPKEPIYIY 154
+ PPPDLPSLLL RI+Y+GMPL + VTEL+IA+L++L++ P++PIY Y
Sbjct: 19 RTPPPDLPSLLLKERIIYLGMPLFSSDDVKRQVGFDVTELIIAQLLYLEFDNPEKPIYFY 78
Query: 155 INSTGTTRADGETVAMESEGFAIYDAMMQMKTEINTVALGAAVGQACLLLSAGTKGRRYM 214
INSTGT+ G+ + E+E FAI D M +K ++T+ +G A+G A ++LS GT G R
Sbjct: 79 INSTGTSWYTGDAIGYETEAFAICDTMRYIKPPVHTICIGQAMGTAAMILSGGTPGNRAS 138
Query: 215 TPHAKAMIQQPRVPSSGLRPASDVLIHAKEVMVNRDTLVKLLAKHTENSEETVSNVMKRS 274
PHA ++ QPR + G ASD+ I AKEV+ N+ T++++ A++T + ++ R
Sbjct: 139 LPHATIVLNQPRTGAQG--QASDIQIRAKEVLANKRTMLEIFARNTGQDPDRLARDTDRM 196
Query: 275 YYMDALLAKEFGVIDKILWRGQEKIMADVPS 305
YM A E+G+ID++L ++ + A +PS
Sbjct: 197 LYMTPAQAVEYGLIDRVL-DSRKDLPAPLPS 226
>CLPP_AQUAE (O67357) ATP-dependent Clp protease proteolytic subunit
(EC 3.4.21.92) (Endopeptidase Clp)
Length = 201
Score = 137 bits (344), Expect = 5e-32
Identities = 71/183 (38%), Positives = 112/183 (60%), Gaps = 11/183 (6%)
Query: 110 DLPSLLLNGRIVYIGMPLVPAVTELVIAELMFLQWMAPKEPIYIYINSTGTTRADGETVA 169
D+ S LL RIV +G P+ V L++A+L+FL+ P + IY+YINS G G A
Sbjct: 26 DIYSRLLQDRIVLLGSPIDDHVANLIVAQLLFLESQDPDKDIYLYINSPG-----GSVTA 80
Query: 170 MESEGFAIYDAMMQMKTEINTVALGAAVGQACLLLSAGTKGRRYMTPHAKAMIQQPRVPS 229
G AIYD M +K ++ T+ +G A +LL+AG G+RY PH++ MI QP
Sbjct: 81 ----GLAIYDTMQYIKPDVVTICMGQAASMGAILLAAGAPGKRYALPHSRIMIHQPLGGI 136
Query: 230 SGLRPASDVLIHAKEVMVNRDTLVKLLAKHTENSEETVSNVMKRSYYMDALLAKEFGVID 289
G A+D++IHA+E+ ++ L+ +LAKHT ++ ++N ++R Y+M AK++G+ID
Sbjct: 137 QG--QATDIIIHAEEIKRIKEMLIDILAKHTGQPKDKIANDIERDYFMSPYEAKDYGLID 194
Query: 290 KIL 292
K++
Sbjct: 195 KVI 197
>CLPP_THEMA (Q9WZF9) ATP-dependent Clp protease proteolytic subunit
(EC 3.4.21.92) (Endopeptidase Clp)
Length = 203
Score = 133 bits (335), Expect = 5e-31
Identities = 74/183 (40%), Positives = 111/183 (60%), Gaps = 11/183 (6%)
Query: 110 DLPSLLLNGRIVYIGMPLVPAVTELVIAELMFLQWMAPKEPIYIYINSTGTTRADGETVA 169
D+ S LL RIV++G P+ V LVIA+L+FL+ P + +Y+YINS G G A
Sbjct: 28 DIFSRLLKDRIVFLGSPIDDYVANLVIAQLLFLEAEDPDKDVYLYINSPG-----GSVTA 82
Query: 170 MESEGFAIYDAMMQMKTEINTVALGAAVGQACLLLSAGTKGRRYMTPHAKAMIQQPRVPS 229
G AIYD M +K +++T+ +G A A +LL+AG KG+RY P+A+ MI QP +
Sbjct: 83 ----GLAIYDTMQYIKCDVSTICVGQAASMAAVLLAAGAKGKRYALPNARIMIHQPLGGA 138
Query: 230 SGLRPASDVLIHAKEVMVNRDTLVKLLAKHTENSEETVSNVMKRSYYMDALLAKEFGVID 289
G PA DV I +E++ +D L ++L+KHT E + R ++M A AKE+G++D
Sbjct: 139 EG--PAKDVEIITRELLRIKDLLNRILSKHTGQPIEKIEKDTDRDFFMSAEEAKEYGIVD 196
Query: 290 KIL 292
K++
Sbjct: 197 KVV 199
>CLP2_BACHD (Q9K888) ATP-dependent Clp protease proteolytic subunit
2 (EC 3.4.21.92) (Endopeptidase Clp 2)
Length = 194
Score = 127 bits (320), Expect = 3e-29
Identities = 74/183 (40%), Positives = 110/183 (59%), Gaps = 11/183 (6%)
Query: 110 DLPSLLLNGRIVYIGMPLVPAVTELVIAELMFLQWMAPKEPIYIYINSTGTTRADGETVA 169
D+ S LL RIV++G + + ++A+L+FL+ PK+ IY+YINS G G T
Sbjct: 20 DIYSRLLKDRIVFLGAAIDDQIANSIVAQLLFLEAENPKKDIYLYINSPG-----GST-- 72
Query: 170 MESEGFAIYDAMMQMKTEINTVALGAAVGQACLLLSAGTKGRRYMTPHAKAMIQQPRVPS 229
S GFAIYD M +K I+T+ G A A +LL AGTKG+R+ P+++ MI QP +
Sbjct: 73 --SAGFAIYDTMQFVKPSIHTICTGMAASFAAILLLAGTKGKRFALPNSEIMIHQPSGGA 130
Query: 230 SGLRPASDVLIHAKEVMVNRDTLVKLLAKHTENSEETVSNVMKRSYYMDALLAKEFGVID 289
G ASD+ I AK ++ R+ LV + ++ T S E V+ M R Y+M A A E+G++D
Sbjct: 131 QG--QASDLAITAKRILGIREKLVTVTSERTGQSPEKVAKDMDRDYFMSAEEALEYGIVD 188
Query: 290 KIL 292
+I+
Sbjct: 189 QII 191
>CLPP_HUMAN (Q16740) Putative ATP-dependent Clp protease proteolytic
subunit, mitochondrial precursor (EC 3.4.21.92)
(Endopeptidase Clp)
Length = 277
Score = 125 bits (313), Expect = 2e-28
Identities = 74/184 (40%), Positives = 109/184 (59%), Gaps = 13/184 (7%)
Query: 110 DLPSLLLNGRIVYIGMPLVPAVTELVIAELMFLQWMAPKEPIYIYINSTGTTRADGETVA 169
D+ S LL RIV + P+ +V LVIA+L+FLQ + K+PI++YINS G
Sbjct: 74 DIYSRLLRERIVCVMGPIDDSVASLVIAQLLFLQSESNKKPIHMYINSPG---------G 124
Query: 170 MESEGFAIYDAMMQMKTEINTVALGAAVGQACLLLSAGTKGRRYMTPHAKAMIQQPRVPS 229
+ + G AIYD M + I T +G A LLL+AGT G R+ P+++ MI QP S
Sbjct: 125 VVTAGLAIYDTMQYILNPICTWCVGQAASMGSLLLAAGTPGMRHSLPNSRIMIHQP---S 181
Query: 230 SGLR-PASDVLIHAKEVMVNRDTLVKLLAKHTENSEETVSNVMKRSYYMDALLAKEFGVI 288
G R A+D+ I A+E+M + L + AKHT+ S + + + M+R YM + A+EFG++
Sbjct: 182 GGARGQATDIAIQAEEIMKLKKQLYNIYAKHTKQSLQVIESAMERDRYMSPMEAQEFGIL 241
Query: 289 DKIL 292
DK+L
Sbjct: 242 DKVL 245
>CLPP_MOUSE (O88696) Putative ATP-dependent Clp protease proteolytic
subunit, mitochondrial precursor (EC 3.4.21.92)
(Endopeptidase Clp)
Length = 272
Score = 124 bits (310), Expect = 4e-28
Identities = 78/206 (37%), Positives = 116/206 (55%), Gaps = 14/206 (6%)
Query: 110 DLPSLLLNGRIVYIGMPLVPAVTELVIAELMFLQWMAPKEPIYIYINSTGTTRADGETVA 169
D+ S LL RIV + P+ +V LVIA+L+FLQ + K+PI++YINS G
Sbjct: 70 DIYSRLLRERIVCVMGPIDDSVASLVIAQLLFLQSESNKKPIHMYINSPG---------G 120
Query: 170 MESEGFAIYDAMMQMKTEINTVALGAAVGQACLLLSAGTKGRRYMTPHAKAMIQQPRVPS 229
+ + G AIYD M + I T +G A LLL+AG+ G R+ P+++ MI QP S
Sbjct: 121 VVTAGLAIYDTMQYILNPICTWCVGQAASMGSLLLAAGSPGMRHSLPNSRIMIHQP---S 177
Query: 230 SGLR-PASDVLIHAKEVMVNRDTLVKLLAKHTENSEETVSNVMKRSYYMDALLAKEFGVI 288
G R A+D+ I A+E+M + L + AKHT+ S + + + M+R YM + A+EFG++
Sbjct: 178 GGARGQATDIAIQAEEIMKLKKQLYNIYAKHTKQSLQVIESAMERDRYMSPMEAQEFGIL 237
Query: 289 DKILWRGQEKIMADVPSRDDRENGTA 314
DK+L + D P +E TA
Sbjct: 238 DKVLVHPPQD-GEDEPELVQKETATA 262
>CLPP_CHLTE (Q8KC73) ATP-dependent Clp protease proteolytic subunit
(EC 3.4.21.92) (Endopeptidase Clp)
Length = 225
Score = 122 bits (307), Expect = 9e-28
Identities = 71/182 (39%), Positives = 104/182 (57%), Gaps = 11/182 (6%)
Query: 110 DLPSLLLNGRIVYIGMPLVPAVTELVIAELMFLQWMAPKEPIYIYINSTGTTRADGETVA 169
D+ S LL RI+++G P+ V L+IA+L+FL+ P+ IYIYINS G +
Sbjct: 44 DIFSRLLRERIIFLGSPIDEHVAGLIIAQLIFLESEDPERDIYIYINSPGGS-------- 95
Query: 170 MESEGFAIYDAMMQMKTEINTVALGAAVGQACLLLSAGTKGRRYMTPHAKAMIQQPRVPS 229
S G IYD M ++ +I+TV +G A LL++GT G+R PH++ MI QP +
Sbjct: 96 -VSAGLGIYDTMQYIRPDISTVCVGMAASMGAFLLASGTSGKRASLPHSRIMIHQPSGGA 154
Query: 230 SGLRPASDVLIHAKEVMVNRDTLVKLLAKHTENSEETVSNVMKRSYYMDALLAKEFGVID 289
G +D+LI A+E+ R L LLAKHT + +R +M A+ AKE+G+ID
Sbjct: 155 QG--QETDILIQAREIEKIRHLLEDLLAKHTGQEVSRIREDSERDRWMSAVEAKEYGLID 212
Query: 290 KI 291
+I
Sbjct: 213 QI 214
>CLPP_XYLFT (Q87E51) ATP-dependent Clp protease proteolytic subunit
(EC 3.4.21.92) (Endopeptidase Clp)
Length = 208
Score = 121 bits (304), Expect = 2e-27
Identities = 65/191 (34%), Positives = 115/191 (60%), Gaps = 11/191 (5%)
Query: 110 DLPSLLLNGRIVYIGMPLVPAVTELVIAELMFLQWMAPKEPIYIYINSTGTTRADGETVA 169
D+ S LL R++++ P+ + L++A+L+FL+ P++ I IYINS G
Sbjct: 26 DIYSRLLKERLIFLVGPIDDYMANLIVAQLLFLEAENPEKDINIYINSPG---------G 76
Query: 170 MESEGFAIYDAMMQMKTEINTVALGAAVGQACLLLSAGTKGRRYMTPHAKAMIQQPRVPS 229
+ + G AIYD M +K ++T+ +G A LLL++G G+RY P+++ MI QP
Sbjct: 77 VVTAGMAIYDTMQYIKPAVSTICVGQAASMGALLLASGASGKRYALPNSRVMIHQPLGGF 136
Query: 230 SGLRPASDVLIHAKEVMVNRDTLVKLLAKHTENSEETVSNVMKRSYYMDALLAKEFGVID 289
G A+D+ IHA+E++ R L ++LAKHT S ET+++ +R + A+ A+ +G++D
Sbjct: 137 QG--QATDIDIHAREILALRARLNEILAKHTGQSLETIAHDTERDNFKSAVDAQAYGLVD 194
Query: 290 KILWRGQEKIM 300
++ + QE+++
Sbjct: 195 QVFGQRQEELI 205
>CLP2_AGRT5 (Q8UFY6) ATP-dependent Clp protease proteolytic subunit
2 (EC 3.4.21.92) (Endopeptidase Clp 2)
Length = 210
Score = 121 bits (303), Expect = 3e-27
Identities = 71/188 (37%), Positives = 109/188 (57%), Gaps = 11/188 (5%)
Query: 110 DLPSLLLNGRIVYIGMPLVPAVTELVIAELMFLQWMAPKEPIYIYINSTGTTRADGETVA 169
D+ S LL RI+++ P+ + LV A+L+FL+ PK+ I +YINS G
Sbjct: 27 DIFSRLLKERIIFLTGPVEDQMASLVCAQLLFLEAENPKKEIALYINSPG---------G 77
Query: 170 MESEGFAIYDAMMQMKTEINTVALGAAVGQACLLLSAGTKGRRYMTPHAKAMIQQPRVPS 229
+ + G AIYD M ++ ++T+ +G A LLL+AG KG R+ TP+A+ M+ QP
Sbjct: 78 VVTAGMAIYDTMQFIRPAVSTLCVGQAASMGSLLLAAGEKGMRFATPNARIMVHQPSGGF 137
Query: 230 SGLRPASDVLIHAKEVMVNRDTLVKLLAKHTENSEETVSNVMKRSYYMDALLAKEFGVID 289
G ASD+ HA++++ + L ++ KHT + E V + R ++MDA AK++GVID
Sbjct: 138 QG--QASDIERHARDIIKMKRRLNEVYVKHTGRTLEEVEKTLDRDHFMDADEAKDWGVID 195
Query: 290 KILWRGQE 297
KIL QE
Sbjct: 196 KILTSRQE 203
>CLP1_MYXXA (O30612) ATP-dependent Clp protease proteolytic subunit
1 (EC 3.4.21.92) (Endopeptidase Clp 1)
Length = 203
Score = 121 bits (303), Expect = 3e-27
Identities = 70/196 (35%), Positives = 115/196 (57%), Gaps = 13/196 (6%)
Query: 110 DLPSLLLNGRIVYIGMPLVPAVTELVIAELMFLQWMAPKEPIYIYINSTGTTRADGETVA 169
DL S LL RI+ +G P+ V +++A+L+FL+ P + I +YINS G G A
Sbjct: 18 DLYSRLLKDRIIMLGTPVNDDVANIIVAQLLFLESEDPDKGINLYINSPG-----GSVTA 72
Query: 170 MESEGFAIYDAMMQMKTEINTVALGAAVGQACLLLSAGTKGRRYMTPHAKAMIQQPRVPS 229
G AIYD M +K ++T+ +G A LLL AG KG+RY P+++ MI QP +
Sbjct: 73 ----GLAIYDTMQYVKCPVSTICVGQAASMGALLLLAGAKGKRYALPNSRIMIHQPLGGA 128
Query: 230 SGLRPASDVLIHAKEVMVNRDTLVKLLAKHTENSEETVSNVMKRSYYMDALLAKEFGVID 289
G A+D+ I AKE++ R + L+ KHT ++ E + +R Y+M A A+++G+ID
Sbjct: 129 QG--QATDIDIQAKEILRLRSYINGLIVKHTGHTIERIEKDTERDYFMSAEDARQYGLID 186
Query: 290 KILWRGQEKIMADVPS 305
+++ +++++A P+
Sbjct: 187 EVV--EKQRVIAPTPA 200
>CLPP_XYLFA (Q9PE41) ATP-dependent Clp protease proteolytic subunit
(EC 3.4.21.92) (Endopeptidase Clp)
Length = 208
Score = 120 bits (302), Expect = 3e-27
Identities = 65/191 (34%), Positives = 115/191 (60%), Gaps = 11/191 (5%)
Query: 110 DLPSLLLNGRIVYIGMPLVPAVTELVIAELMFLQWMAPKEPIYIYINSTGTTRADGETVA 169
D+ S LL R++++ P+ + L++A+L+FL+ P++ I IYINS G
Sbjct: 26 DIYSRLLKERLIFLVGPIDDYMANLIVAQLLFLEAENPEKDINIYINSPG---------G 76
Query: 170 MESEGFAIYDAMMQMKTEINTVALGAAVGQACLLLSAGTKGRRYMTPHAKAMIQQPRVPS 229
+ + G AIYD M +K ++T+ +G A LLL++G G+RY P+++ MI QP
Sbjct: 77 VVTAGMAIYDTMQYIKPAVSTICVGQAASMGALLLASGASGKRYALPNSRVMIHQPLGGF 136
Query: 230 SGLRPASDVLIHAKEVMVNRDTLVKLLAKHTENSEETVSNVMKRSYYMDALLAKEFGVID 289
G A+D+ IHA+E++ R L ++LAKHT S ET+++ +R + A+ A+ +G++D
Sbjct: 137 QG--QATDIDIHAREILALRARLNEILAKHTGQSLETIAHDTERDNFKSAVDAQAYGLVD 194
Query: 290 KILWRGQEKIM 300
++L + E+++
Sbjct: 195 QVLGQRPEELI 205
>CLPP_XANAC (Q8PNI5) ATP-dependent Clp protease proteolytic subunit
(EC 3.4.21.92) (Endopeptidase Clp)
Length = 208
Score = 120 bits (301), Expect = 5e-27
Identities = 65/189 (34%), Positives = 113/189 (59%), Gaps = 11/189 (5%)
Query: 110 DLPSLLLNGRIVYIGMPLVPAVTELVIAELMFLQWMAPKEPIYIYINSTGTTRADGETVA 169
D+ S LL R++++ P+ + +++A+L+FL+ P++ I IYINS G
Sbjct: 26 DIYSRLLKERLIFLVGPIDDHMANVIVAQLLFLEADNPEKDISIYINSPG---------G 76
Query: 170 MESEGFAIYDAMMQMKTEINTVALGAAVGQACLLLSAGTKGRRYMTPHAKAMIQQPRVPS 229
+ + G AIYD M +K +++T+ +G A LLL++G G+RY P+++ MI QP
Sbjct: 77 VVTAGMAIYDTMQYIKPDVSTICVGQAASMGALLLASGAAGKRYALPNSRVMIHQPLGGF 136
Query: 230 SGLRPASDVLIHAKEVMVNRDTLVKLLAKHTENSEETVSNVMKRSYYMDALLAKEFGVID 289
G A+D+ IHA+E++ R L ++LAKHT S ET++ +R + A+ A+ +G++D
Sbjct: 137 QG--QATDIDIHAREILTLRSRLNEILAKHTGQSLETIARDTERDNFKSAVDAQAYGLVD 194
Query: 290 KILWRGQEK 298
++L R E+
Sbjct: 195 QVLERRPEE 203
>CLPP_XANCP (Q8PBY6) ATP-dependent Clp protease proteolytic subunit
(EC 3.4.21.92) (Endopeptidase Clp)
Length = 208
Score = 119 bits (299), Expect = 8e-27
Identities = 65/189 (34%), Positives = 112/189 (58%), Gaps = 11/189 (5%)
Query: 110 DLPSLLLNGRIVYIGMPLVPAVTELVIAELMFLQWMAPKEPIYIYINSTGTTRADGETVA 169
D+ S LL R++++ P+ + +++A+L+FL+ P++ I IYINS G
Sbjct: 26 DIYSRLLKERLIFLVGPIDDHMANVIVAQLLFLEADNPEKDISIYINSPG---------G 76
Query: 170 MESEGFAIYDAMMQMKTEINTVALGAAVGQACLLLSAGTKGRRYMTPHAKAMIQQPRVPS 229
+ + G AIYD M +K +++T+ +G A LLL++G G+RY P+++ MI QP
Sbjct: 77 VVTAGMAIYDTMQYIKPDVSTICVGQAASMGALLLASGAAGKRYALPNSRVMIHQPLGGF 136
Query: 230 SGLRPASDVLIHAKEVMVNRDTLVKLLAKHTENSEETVSNVMKRSYYMDALLAKEFGVID 289
G A+D+ IHA+E++ R L ++LAKHT S ET++ +R + A+ A+ +G++D
Sbjct: 137 QG--QATDIDIHAREILTLRSRLNEILAKHTGQSLETIARDTERDNFKSAVDAQAYGLVD 194
Query: 290 KILWRGQEK 298
+L R E+
Sbjct: 195 HVLERRPEE 203
>CLPP_THETN (Q8RC25) ATP-dependent Clp protease proteolytic subunit
(EC 3.4.21.92) (Endopeptidase Clp)
Length = 198
Score = 119 bits (297), Expect = 1e-26
Identities = 71/185 (38%), Positives = 108/185 (58%), Gaps = 11/185 (5%)
Query: 110 DLPSLLLNGRIVYIGMPLVPAVTELVIAELMFLQWMAPKEPIYIYINSTGTTRADGETVA 169
D+ S LL RIV++G + LVIA+++FL+ P + I++YINS G G A
Sbjct: 22 DIFSRLLKDRIVFLGDEINDTTASLVIAQMLFLEAEDPDKDIWLYINSPG-----GSITA 76
Query: 170 MESEGFAIYDAMMQMKTEINTVALGAAVGQACLLLSAGTKGRRYMTPHAKAMIQQPRVPS 229
G AIYD M +K ++ T+ +G A A LL+AG KG+R+ P+++ MI QP
Sbjct: 77 ----GLAIYDTMQYIKPDVVTLCVGMAASMAAFLLAAGAKGKRFALPNSEIMIHQPWGGM 132
Query: 230 SGLRPASDVLIHAKEVMVNRDTLVKLLAKHTENSEETVSNVMKRSYYMDALLAKEFGVID 289
G A+D+ IHA+ ++ RD L ++L+++T E + M+R Y+M A AK +G+ID
Sbjct: 133 QG--QATDIKIHAERLLRLRDKLERILSENTGQPLEKIKADMERDYFMTAEEAKTYGIID 190
Query: 290 KILWR 294
IL R
Sbjct: 191 DILVR 195
>CLPP_NEIMB (Q9JZ38) ATP-dependent Clp protease proteolytic subunit
(EC 3.4.21.92) (Endopeptidase Clp)
Length = 204
Score = 119 bits (297), Expect = 1e-26
Identities = 69/183 (37%), Positives = 105/183 (56%), Gaps = 9/183 (4%)
Query: 110 DLPSLLLNGRIVYIGMPLVPAVTELVIAELMFLQWMAPKEPIYIYINSTGTTRADGETVA 169
D+ S LL RIV++ P+ LV+A+L+FL+ P + I+ YINS G G A
Sbjct: 23 DIYSRLLKERIVFLVGPVTDESANLVVAQLLFLESENPDKDIFFYINSPG-----GSVTA 77
Query: 170 MESEGFAIYDAMMQMKTEINTVALGAAVGQACLLLSAGTKGRRYMTPHAKAMIQQPRVPS 229
G +IYD M +K +++T+ LG A LLSAG KG+R+ P+++ MI QP +
Sbjct: 78 ----GMSIYDTMNFIKPDVSTLCLGQAASMGAFLLSAGEKGKRFALPNSRIMIHQPLISG 133
Query: 230 SGLRPASDVLIHAKEVMVNRDTLVKLLAKHTENSEETVSNVMKRSYYMDALLAKEFGVID 289
ASD+ IHA+E++ ++ L +L+AKH + + R +M A AKE+G+ID
Sbjct: 134 GLGGQASDIEIHARELLKIKEKLNRLMAKHCDRDLADLERDTDRDNFMSAEEAKEYGLID 193
Query: 290 KIL 292
+IL
Sbjct: 194 QIL 196
>CLP3_SYNY3 (P74467) Probable ATP-dependent Clp protease proteolytic
subunit 3 (EC 3.4.21.92) (Endopeptidase Clp 3)
Length = 202
Score = 119 bits (297), Expect = 1e-26
Identities = 64/183 (34%), Positives = 106/183 (56%), Gaps = 10/183 (5%)
Query: 110 DLPSLLLNGRIVYIGMPLVPAVTELVIAELMFLQWMAPKEPIYIYINSTGTTRADGETVA 169
D+ + L RI+++G + ++ ++A L++L P +PIY+YINS G G A
Sbjct: 22 DIYTRLSQERIIFLGQEVNDSIANRIVAFLLYLDSDDPSKPIYLYINSPG-----GSVTA 76
Query: 170 MESEGFAIYDAMMQMKTEINTVALGAAVGQACLLLSAGTKGRRYMTPHAKAMIQQPRVPS 229
G AIYD M +K E+ T+ +G A LL++G G+R PHA+ MI QP +
Sbjct: 77 ----GMAIYDTMQYIKAEVITICVGLAASMGAFLLASGAPGKRLALPHARIMIHQP-MGG 131
Query: 230 SGLRPASDVLIHAKEVMVNRDTLVKLLAKHTENSEETVSNVMKRSYYMDALLAKEFGVID 289
+G R A+D+ I A+E++ R L +++A+ T + E ++ R Y++ A AKE+G+ID
Sbjct: 132 TGRRQATDIDIEAREILRIRQQLNEIMAQRTGQTVEKIAKDTDRDYFLSAAEAKEYGLID 191
Query: 290 KIL 292
K++
Sbjct: 192 KVI 194
>CLPP_RICCN (Q92HM5) ATP-dependent Clp protease proteolytic subunit
(EC 3.4.21.92) (Endopeptidase Clp)
Length = 201
Score = 117 bits (293), Expect = 4e-26
Identities = 66/183 (36%), Positives = 103/183 (56%), Gaps = 11/183 (6%)
Query: 110 DLPSLLLNGRIVYIGMPLVPAVTELVIAELMFLQWMAPKEPIYIYINSTGTTRADGETVA 169
D+ S LL RI+++ + + L++A+L+FL+ PK+ IY+YINS G
Sbjct: 19 DIYSRLLKERIIFVCSTVEDHMANLIVAQLLFLEAENPKKDIYMYINSPG---------G 69
Query: 170 MESEGFAIYDAMMQMKTEINTVALGAAVGQACLLLSAGTKGRRYMTPHAKAMIQQPRVPS 229
+ + G AIYD M +K ++ T+ +G A LLL G KG RY PH++ MI QP
Sbjct: 70 VVTAGLAIYDTMQYIKPKVATLCIGQACSMGSLLLCGGEKGMRYSLPHSRIMIHQPSGGY 129
Query: 230 SGLRPASDVLIHAKEVMVNRDTLVKLLAKHTENSEETVSNVMKRSYYMDALLAKEFGVID 289
G A+D+ IHA+E + + L +L +KHTE + + M+R +M AK+FG++D
Sbjct: 130 KG--QATDIEIHAQETLKIKRLLNELYSKHTEQELKHIEKSMERDNFMSPEEAKKFGLVD 187
Query: 290 KIL 292
I+
Sbjct: 188 NIM 190
>CLPP_NEIMA (Q9JU33) ATP-dependent Clp protease proteolytic subunit
(EC 3.4.21.92) (Endopeptidase Clp)
Length = 204
Score = 117 bits (292), Expect = 5e-26
Identities = 68/183 (37%), Positives = 104/183 (56%), Gaps = 9/183 (4%)
Query: 110 DLPSLLLNGRIVYIGMPLVPAVTELVIAELMFLQWMAPKEPIYIYINSTGTTRADGETVA 169
D+ S LL RIV++ P+ LV+A+L+FL+ P + I+ YINS G G A
Sbjct: 23 DIYSRLLKERIVFLVGPVTDESANLVVAQLLFLESENPDKDIFFYINSPG-----GSVTA 77
Query: 170 MESEGFAIYDAMMQMKTEINTVALGAAVGQACLLLSAGTKGRRYMTPHAKAMIQQPRVPS 229
G +IYD M +K +++T+ LG A LLSAG KG+R+ P+++ MI QP +
Sbjct: 78 ----GMSIYDTMNFIKPDVSTLCLGQAASMGAFLLSAGEKGKRFALPNSRIMIHQPLISG 133
Query: 230 SGLRPASDVLIHAKEVMVNRDTLVKLLAKHTENSEETVSNVMKRSYYMDALLAKEFGVID 289
ASD+ IHA+E++ ++ L +L+AKH + R +M A AKE+G+ID
Sbjct: 134 GLGGQASDIEIHARELLKIKEKLNRLMAKHCGRDLADLERDTDRDNFMSAEEAKEYGLID 193
Query: 290 KIL 292
++L
Sbjct: 194 QVL 196
>CLPP_CLOPE (Q8XKK1) ATP-dependent Clp protease proteolytic subunit
(EC 3.4.21.92) (Endopeptidase Clp)
Length = 194
Score = 117 bits (292), Expect = 5e-26
Identities = 65/185 (35%), Positives = 108/185 (58%), Gaps = 11/185 (5%)
Query: 110 DLPSLLLNGRIVYIGMPLVPAVTELVIAELMFLQWMAPKEPIYIYINSTGTTRADGETVA 169
D+ S LL RI+ + + LV+A+L+FL+ P + I++YINS G +
Sbjct: 20 DIFSRLLKDRIIMLSGEVNDVTANLVVAQLLFLESEDPDKDIHLYINSPGGSI------- 72
Query: 170 MESEGFAIYDAMMQMKTEINTVALGAAVGQACLLLSAGTKGRRYMTPHAKAMIQQPRVPS 229
+ G AIYD M +K +++T+ +G A LLS+G KG+R+ P+A+ MI QP
Sbjct: 73 --TSGMAIYDTMQYIKPDVSTICIGMAASMGAFLLSSGAKGKRFALPNAEIMIHQPLGGF 130
Query: 230 SGLRPASDVLIHAKEVMVNRDTLVKLLAKHTENSEETVSNVMKRSYYMDALLAKEFGVID 289
G A+D+ IHAK ++ +D L ++L+++T E + ++R Y+M+A A E+G+ID
Sbjct: 131 QG--QATDIDIHAKRILKIKDKLNQILSENTNQPLEKIKVDVERDYFMEASEAVEYGLID 188
Query: 290 KILWR 294
K++ R
Sbjct: 189 KVIER 193
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.317 0.131 0.374
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 36,680,614
Number of Sequences: 164201
Number of extensions: 1520942
Number of successful extensions: 4891
Number of sequences better than 10.0: 161
Number of HSP's better than 10.0 without gapping: 139
Number of HSP's successfully gapped in prelim test: 22
Number of HSP's that attempted gapping in prelim test: 4507
Number of HSP's gapped (non-prelim): 173
length of query: 322
length of database: 59,974,054
effective HSP length: 110
effective length of query: 212
effective length of database: 41,911,944
effective search space: 8885332128
effective search space used: 8885332128
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 66 (30.0 bits)
Medicago: description of AC144541.11


